Abstract
Genomic disorders and rare copy number abnormalities are identified in 15–25% of patients with syndromic conditions, but their prevalence in individuals with isolated birth defects is less clear. A spectrum of congenital heart defects (CHDs) is seen in heterotaxy, a highly heritable and genetically heterogeneous multiple congenital anomaly syndrome resulting from failure to properly establish left–right (L-R) organ asymmetry during early embryonic development. To identify novel genetic causes of heterotaxy, we analysed copy number variants (CNVs) in 225 patients with heterotaxy and heterotaxy-spectrum CHDs using array-based genotyping methods. Clinically relevant CNVs were identified in approximately 20% of patients and encompassed both known and putative heterotaxy genes. Patients were carefully phenotyped, revealing a significant association of abdominal situs inversus with pathogenic or likely pathogenic CNVs, while d-transposition of the great arteries was more frequently associated with common CNVs. Identified cytogenetic abnormalities ranged from large unbalanced translocations to smaller, kilobase-scale CNVs, including a rare, single exon deletion in ZIC3, a gene known to cause X-linked heterotaxy. Morpholino loss-of-function experiments in Xenopus support a role for one of these novel candidates, the platelet isoform of phosphofructokinase-1 (PFKP) in heterotaxy. Collectively, our results confirm a high CNV yield for array-based testing in patients with heterotaxy, and support use of CNV analysis for identification of novel biological processes relevant to human laterality.
This article is part of the themed issue ‘Provocative questions in left–right asymmetry’.
Keywords: copy number variation, heterotaxy, left–right patterning, congenital heart defects, phosphofructokinase
1. Introduction
Heterotaxy is a relatively infrequent (approx. 1 : 10 000) multiple congenital anomaly syndrome resulting from abnormal specification of the left–right (L–R) body axis during early embryonic development [1]. In its classic form, heterotaxy is characterized by combined occurrence of visceral situs abnormalities (gut malrotation, stomach and liver situs anomalies, abnormalities of spleen positioning or number) and congenital heart defects (CHDs) of varying complexity, which account for the majority of associated morbidity and mortality. Over 96% of patients with heterotaxy exhibit some form of CHD [2], often requiring surgical intervention. Clinical outcomes are disproportionately poorer than in patients without heterotaxy who have similar CHDs and are typified by prolonged courses and significantly greater likelihood for post-surgical complications [3–5]. This clinical picture firmly establishes heterotaxy as not only a disease of significant phenotypic heterogeneity, but also one of considerable medical and economic consequence.
Although heterotaxy typically occurs sporadically and with unknown cause, its relative risk is highest among all classes of CHDs, supporting existence of a strong genetic component [6]. Autosomal recessive, autosomal dominant and X-linked inheritance patterns have all been described [7]. Mutations in the zinc finger of the cerebellum 3 (ZIC3) gene are particularly well documented and are considered to be causative in the majority (approx. 75%) of familial X-linked pedigrees. Surprisingly, however, ZIC3 mutations underlie only a minority (3–5%) of sporadic heterotaxy cases [8,9]. Likewise, despite a conserved and central role for Nodal signalling in establishment of early molecular asymmetries, point mutations in Nodal pathway components are also not routinely identified and collectively explain only 5–10% of heterotaxy cases [10–12]. Mutations in other causative genes are detected at similar or even lower frequencies, indicating significant genetic heterogeneity. As a specific genetic etiology is currently identifiable in only a minority of patients, there remains enormous potential for novel gene and pathway discovery.
Copy number variants (CNVs) in the form of complex chromosomal rearrangements and submicroscopic duplications and deletions are increasingly recognized as important causes of birth defects and neurodevelopmental disease [13]. Depending on size and genomic position, CNVs can encompass complete or partial intronic or exonic regions, can include one or multiple genes, or can disrupt regulatory regions such as promoters and enhancers. If a CNV shifts the normal reading frame, this change may lead to premature truncation and loss-of-function through nonsense-mediated decay or to gain-of-function if the transcript escapes decay [14]. Gene interruptions and fusions, positional effects, recessive mutation unmasking and allelic transvection events are all additional potential molecular consequences. The pathogenic significance of a particular CNV is, therefore, highly dependent on the location of its breakpoints, the genomic content of the intervening deleted or duplicated segment, and the genomic landscape in which the CNV is situated.
Studies of patients with CHD indicate that CNVs are a major genetic cause of cardiovascular disease, occurring in 3–25% of patients with extra-cardiac abnormalities and in 3–10% with isolated heart defects [15]. To date, two studies have employed genome-wide approaches to explicitly examine the role of CNVs in heterotaxy [16,17]. Analysing a cohort of 262 patients with classic heterotaxy and/or isolated heterotaxy spectrum CHDs, Fakhro et al. [16] identified 45 rare CNVs in 39 patients, representing a 15% CNV yield in their phenotypically mixed population. A slightly higher rate of 26% (20 CNVs in 19/74 patients) was reported in a more recent study that restricted analysis solely to patients with classic heterotaxy [17]. Yields from both of these studies approximated those seen in other CHD cohorts [15] and together support CNVs as important contributors to human laterality defects.
While copy number variation is now regarded as a major cause of genetic disease, it is important to recognize that not all CNVs are pathogenic. Indeed, CNVs are estimated to encompass between 4.8 and 9.5% of the human genome [18]. In order to systematically catalogue this genetic variation, online and clinical databases have been established as repositories of commonly occurring CNVs. These databases serve as vital reference points for clinical and research efforts focused on identifying CNVs of pathogenic significance to genetic disease.
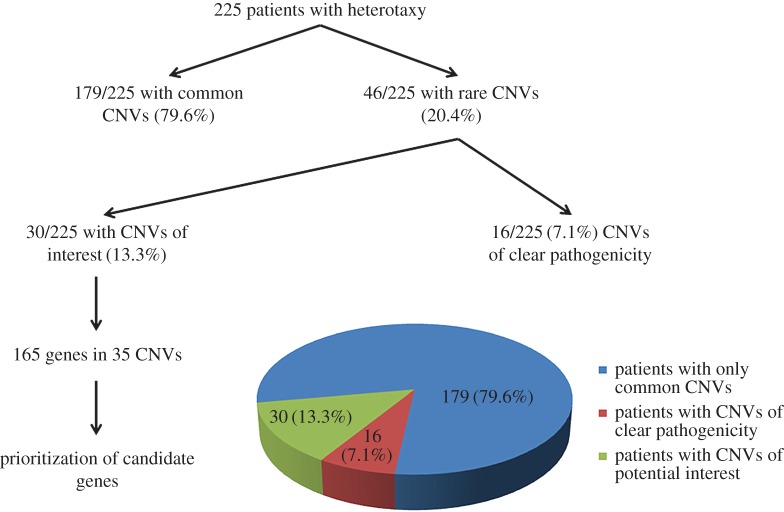
The flexibility and versatility of array comparative genomic hybridization (aCGH), and single-nucleotide polymorphism (SNP) microarrays have made them excellent tools for both gene discovery and clinical practice. In this study, we have screened a large cohort of patients with heterotaxy spectrum malformations using these complementary methods. In total, we identified rare CNVs in 46/225 (20.44%) patients ranging in size from large, megabase-scale translocations to smaller, kilobase-scale CNVs. Excluding CNVs that were considered to be clearly pathogenic owing to their overall size and complexity, or their genomic content (encompassing regions or genes previously associated with or identified as definitive causes of heterotaxy/CHD), we reduced the number of CNVs suitable for candidate gene analysis to 35 in 30/225 (13.33%) patients. Morpholino (MO)-based loss-of-function screens in Xenopus laevis were then used to confirm roles for gene candidates in L–R patterning. In this report, we describe overall findings from these CNV screens while highlighting functional testing results for platelet isoform of phosphofructokinase 1 (pfkp), a gene identified as a novel candidate. Collectively, our analyses support CNVs as a significant contributor to heterotaxy causation and reiterate the value of genome-wide CNV screening as a tool for identifying novel laterality genes and pathways.
2. Material and methods
(a). Patient recruitment and phenotypic classification
Detailed phenotypic information was collected from patient histories and chart review. Patients were classified as having situs inversus (SI) totalis, heterotaxy, or isolated heterotaxy spectrum CHD following previously defined criteria [9]. As some heterotaxy patients with ZIC3 mutations also exhibit VACTERL-like (vertebral anomalies, anal atresia, cardiovascular malformations, tracheo-esophageal fistula, renal anomalies, limb abnormalities) phenotypes, patients with these features are noted. Disease was considered to be ‘familial’ if (i) the pedigree demonstrated autosomal dominant, autosomal recessive or X-linked inheritance, (ii) there was more than one family member with heterotaxy or laterality disorder regardless of their degree of relationship to the proband, (iii) there was heterotaxy in the proband and a first degree relative with isolated CHD or (iv) there was heterotaxy in the proband and a first degree relative with situs-related defects. All other patients were considered to have sporadic disease. Patients with previously identified cytogenetic findings or with mutations in known heterotaxy genes were excluded from further analysis. No patient had a previously identified CNV from prior clinical screening. In total, the final cohort comprised 225 unrelated patients with assorted situs and/or cardiac abnormalities, including 139 males and 86 females. Full cohort demographics are summarized in table 1.
Table 1.
Heterotaxy cohort demographics. Percentages are in parentheses and relative to the 225 patient total.
| racial and ethnic categories | total (%) | gender |
inheritance |
|||
|---|---|---|---|---|---|---|
| females | males | familial | sporadic | unknown | ||
| Arabic | 2 (0.9) | 2 | 0 | 0 | 2 | 0 |
| Asian | 2 (0.9) | 0 | 2 | 0 | 2 | 0 |
| Black or African American | 13 (5.8) | 8 | 5 | 0 | 10 | 3 |
| Caucasian | 90 (40.0) | 33 | 57 | 9 | 64 | 18 |
| mixed | 8 (3.5) | 3 | 5 | 3 | 5 | 0 |
| unknown | 42 (18.7) | 19 | 23 | 9 | 11 | 21 |
| Hispanic/Latino | 68 (30.2) | 21 | 47 | 7 | 49 | 12 |
| total (%) | 225 (100.0) | 86 (38.2) | 139 (61.8) | 28 (12.4) | 143 (63.6) | 54 (24.0) |
(b). Chromosome microarray analysis
Genomic DNA was prepared from blood samples following standard protocols. To evaluate for CNVs, samples were analysed using genome-wide SNP analysis and aCGH, with platform choice being dictated by sample availability at time of testing. The majority of samples were analysed using both platforms, which allowed cross-validation of CNVs. SNP genotyping was performed using the Illumina Human370 DNA Analysis BeadChip (42 patients) or Illumina HumanOmni1-Quad Beadchip (106 patients) platforms (Illumina Inc., San Diego, CA, USA). These chips encompassed approximately 370 000 SNP markers (mean spacing 7.7 kb, median 5 kb) or 1 000 000 SNP markers (mean spacing 2.4 kb, median 1.2 kb), respectively. The Illumina Infinium Assay was performed as described by the manufacturer using 250 ng of patient DNA. B-allele frequency and log2 R ratio were analysed with Illumina GenomeStudio V2009.2 software, and DNA copy number changes were prioritized using output from cnvPartition Plug-in v. 2.3.4 software. Copy number changes called by each software were compared for samples run on both platforms, with manual re-inspection of the raw data when a CNV was identified on a single platform. This approach of cross-validation and manual data re-inspection identified CNVs that otherwise would have been missed on a single platform, proving the additive value of using the two methods.
Array-CGH experiments were completed using a custom array designed by Baylor Medical Genetics Laboratories (221 patients). This array has been previously described [19] and was designed to encompass approximately 180 000 oligonucleotides with whole genome coverage of approx. 30 kb resolution, and exon by exon coverage of over 1700 genes (mean spacing 4.2 probes/exon). Generated data were normalized using the Agilent Feature Extraction Software.
Genotyping data from both platforms were independently analysed and interpreted by two board-certified cytogeneticists. Spurious calls were eliminated from raw data, and CNVs meeting criteria for clinical relevancy were determined following accepted standards for clinical reporting at each institution, including publicly available online databases and clinical (Cincinnati Children's Hospital Medical Center (CCHMC), Baylor) databases of human genetic variation. Polymorphic CNVs, those located in introns or in non-genic regions, or those with fewer than three probes or 10 markers on autosomes were not reported. Common calls, observed more than 20 times in 20 000 cases analysed by Medical Genetics Laboratories at Baylor College of Medicine, were also not included.
Throughout this report, we provide results derived from each platform separately whenever possible. For CNVs identified by aCGH, size data are presented as minimum and maximum values, with gene content determined using maximum predicted breakpoints. Breakpoint predictions for all CNVs were completed using human reference sequence Build 36.1, hg18.
(c). In vitro fertilization and Xenopus laevis embryo staging
Sexually mature wild-type male and female Xenopus laevis were purchased from Nasco. Oocyte collection and in vitro fertilization protocols were performed as previously described [20]. Collected embryos were staged according to the Xenopus developmental table [21].
(d). Morpholino design
All morpholinos were obtained through GeneTools, LLC. Translation-blocking (TB) morpholinos targeting the translation start-site were designed for pfkp (5′- CTTCCCGAGCCCTCTCTCTACTCTC-3′; based on NM_001 097 850.1) and pitrm1 (5′-TCTGCCTCACAGCCGCACAACTATC-3′; NM_001086512) using NCBI reference sequences. A splice-blocking (SB) morpholino targeting the pfkp exon 8-intron eight splice junction (5′-AATGAACACAAGCCACTGTACCTCA-3′) was also generated using splice junction sequence obtained by PCR amplification of stage 47 Xenopus laevis genomic DNA (sense 5′-TGTTTATCCCTGAATACCC-3′; antisense 5′-CAAACTCCCCTAAACAAGTTA-3′). A fluorescein-conjugated zic3 morpholino (5′-ACAATAAAACTTACCTTCATGTGCT-3′) targeting the exon 2-intron two splice junction was used as a positive control. A standard negative control morpholino (5′-CCTCTTACCTCAGTTACAATTTATA-3′) was purchased to control for effects of microinjection.
(e). pfkp and pitrm1 knockdown
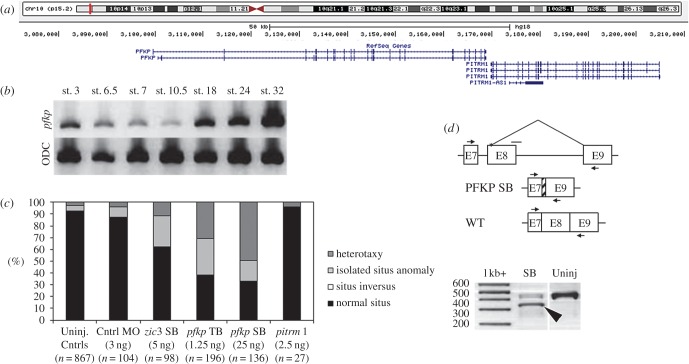
Expression of pfkp during L–R critical stages was confirmed by RT-PCR using the following cDNA primers: sense: 5′-AACCTGTTCCGGGAAGAATGGAGT-3′, antisense 5′-ACCCACAGTGTCTTCCCATGACTT-3′. ODC (ornithine decarboxylase) primers (sense 5′-GCCATTGTGAAGACTCTCTCCATTC-3′, antisense 5′-TTCGGGTGATTCCTTGCCAC-3′) were used as a PCR amplification control [20,22]. All morpholinos were injected into 2-cell stage Xenopus laevis embryos suspended in 4% Ficoll in 1/3 X Marc's modified Ringer's (MMR) buffer. Injections were targeted to the animal poles of both blastomeres. For organ situs assessments, embryos were cultured in 0.1X MMR until stage 47, at which time they were anaesthetized in 0.05% benzocaine and scored for heart, gut and gallbladder positioning. Embryos with abnormal situs of two or more organs were classified as having heterotaxy, while embryos with involvement of only a single organ system were considered to have an isolated situs anomaly. Embryos with mirror-image reversal of all three organs were given a classification of SI. Morpholinos were titrated to the lowest dosages required to yield organ situs defects (pfkp TB = 1.25 ng 3 nl−1 injection, pfkp SB = 25 ng 3 nl−1 injection, zic3 SB = 5.3 ng 3 nl−1 injection). pitrm1 morpholinos were tested across a range of 1–5 ng 3 nl−1 injection. Efficiency of pfkp splice blockade was determined at two stages by RT-PCR. Total RNA was extracted from stage 9 and stage 25 Xenopus laevis embryos by standard TRIzol methods (Life Technologies). Exon 8 deletion was confirmed by gel electrophoresis and bi-directional sequencing of the deletion product using primers targeting the 3′ end of exon 7 (5′-CCAGAGGACCTTTGTGCTGGAAGTTATG-3′) and the 5′ end of exon 9 (5′-ACACAACACAGGCTGGCGTTTC-3′). ODC primers were again used for control of RNA quality and PCR amplification.
(f). Synergy and rescue
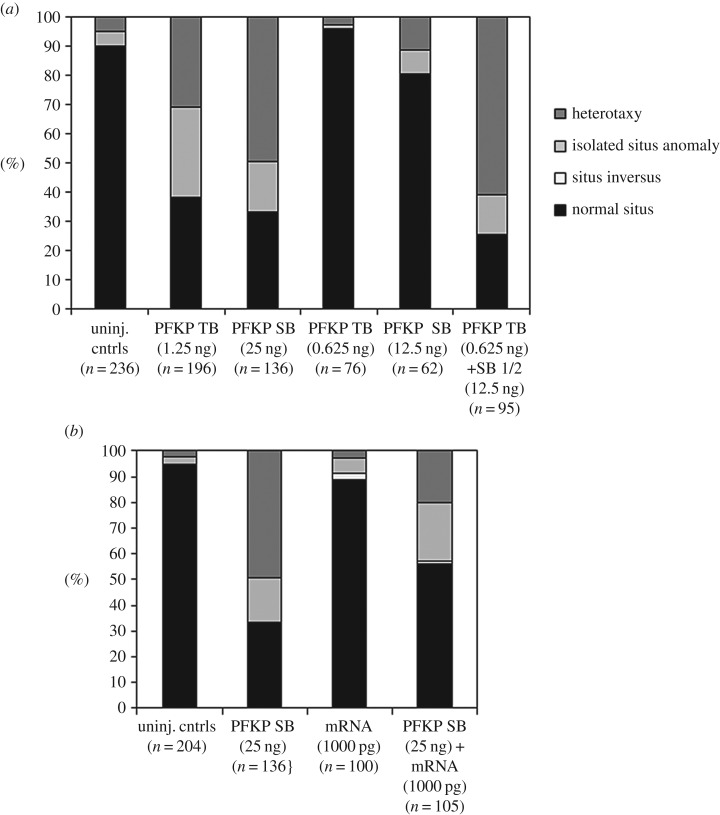
Morpholino synergy and mRNA rescue experiments were performed to confirm specificity of targeting. For synergy experiments, pfkp TB (0.83 ng) and SB (16.7 ng) morpholino doses that did not independently cause phenotypic abnormalities were co- or independently injected into both animal poles of 2-cell stage embryos. Morpholino synergy was demonstrated by a significant increase in organ situs defects in co-injected embryos relative to independently injected embryos at stage 47. For mRNA rescue experiments, in vitro transcribed full-length human PFKP mRNA was co-injected with pfkp SB morpholino into both animal poles of 2-cell stage embryos. Successful rescue was demonstrated by a significant decrease in organ situs defects in mRNA-treated embryos relative to untreated morphants. Full-length human PFKP cDNA for mRNA synthesis was generated from Origene cDNA clone SC118507 by PCR amplification (sense 5′-GCAGAGCTCGTTTAGTGAACCGC-3′, antisense 5′-GATGGGCACTCGCCGATTAG-3′). Identities of the cDNA restriction fragment and PCR product were both confirmed by Sanger sequencing. PFKP mRNA was subsequently in vitro transcribed by T7 polymerase using the mMessage mMachine kit (Ambion) following manufacturer's protocols and stored at −80°C. For rescue experiments, embryos were injected with either 500 or 1000 pg of transcribed mRNA, either alone or in combination with pfkp MO.
(g). Left–right marker analyses
Xenopus laevis embryos were fixed overnight at 4°C in MEMFA (4% paraformaldehyde in MEM salts). Fixed embryos were subsequently dehydrated through three consecutive 5 min 100% methanol washes and stored at −20°C. An antisense RNA probe for the L–R marker, coco was generated from a pCMV-SPORT6 plasmid (provided by Dr Mustafa Khokha) using a T7/SP6 DIG RNA Labeling Kit (Roche) following manufacturer's instructions. Whole-mount in situ hybridizations (WISH) were performed as previously described [23], with one modification: prior to hybridization, fixed stage 20–21 embryos were bisected along the transverse plane into separate anterior and posterior halves, briefly post-fixed (10 min in 4% MEMFA), and run through a graded methanol series. Hybridization was performed on both halves, which were then sorted and appropriately trimmed. Staining was accomplished using BM Purple alkaline phosphatase chromogenic substrate (Roche). Images were captured using a Nikon SMZ1500 stereomicroscope outfitted with a Nikon DXM1200F digital camera and processed using Nikon Act-1 (v. 2.62) imaging software. Coco expression was scored as left-sided biased, right-sided biased or right/left unbiased by visual inspection. Final embryo counts included only those embryos that could be unequivocally classified.
3. Results
(a). Genetic analyses of patients with heterotaxy identify rare copy number variants
Patients were carefully phenotyped with regard to CHD and extra-cardiac anomalies (table 2). In total, just under 20% of patients had isolated CHD, with the remainder having situs abnormalities of some kind. Of the 225 total patients tested, 148 (65.78%) were analysed by SNP array, 221 (98.22%) were analysed by aCGH and 144 (64.00%) were analysed using both platforms. To maximize our chances of identifying novel causes of heterotaxy, we focused attention on rare CNVs encompassing coding regions of one or more genes. Novel pathogenic or likely pathogenic CNVs were identified in 46/225 patients, representing an overall CNV yield of 20.4% (figure 1). We compared these 46 patients to the remainder of the heterotaxy cohort with common CNVs using two-tailed Fisher's exact tests (table 2). Notably, occurrence of abdominal situs abnormalities was significantly greater in this patient group compared with the remainder of the heterotaxy cohort. In addition, d-TGA was less likely to be associated with pathogenic or likely pathogenic CNVs (table 2). Thirteen patients were found to carry large/complex chromosomal abnormalities (table 3), including an approximate 3 Mb 22q11.2 critical region deletion consistent with a diagnosis of 22q11.2 deletion syndrome (DiGeorge syndrome) in a female patient with abdominal situs abnormalities and CHD (Patient 6). Another three CNVs affected genes previously associated with heterotaxy (table 4): a 553–686 kb 2p25.1 duplication encompassing ROCK2 in a male patient with sporadic and complex CHD (Patient 15), a 1.47–1.61 Mb 3p24.1 deletion encompassing TGFBR2 in a female patient with familial heterotaxy (Patient 16) and a 2.9–3.3 kb Xq26.2 deletion encompassing the third exon of ZIC3 in a male patient with sporadic heterotaxy (Patient 14).
Table 2.
Phenotypic summary of the 225 patient heterotaxy cohort. AS, aortic stenosis; ASD, atrial septal defect; AV, atrioventricular; BAV, bicuspid aortic valve; CHD, congenital heart disease; CNV, copy number variant; CoA, coarctation of the aorta; d-TGA, dextro-transposition of the great arteries; DILV, double inlet right ventricle; DORV, double outlet right ventricle; HJ bodies, Howell–Jolly bodies; HLHS, hypoplastic left heart syndrome; IVC, inferior vena cava; l-TGA, levo-transposition of the great arteries; PA, pulmonary atresia; PAPVR, partial anomalous pulmonary venous return; PS, pulmonic stenosis, including sub, valvar, and supravalvar but not branch PS; SI, situs inversus; SVC, superior vena cava; TAPVR, total anomalous pulmonary venous return; VACTERL, vertebral, anal, cardiac, tracheo-esophageal, renal, radial, limb; VSD, ventricular septal defect; n.s., non-significant; S, significant. Bold denotes findings that are statistically significant.
| heterotaxy cohort |
patients with pathogenic or likely pathogenic CNVs |
patients with common CNVs |
pathogenic/likely pathogenic versus common CNV |
||||
|---|---|---|---|---|---|---|---|
| n | % (n/225) | n | % (n/46) | n | % (n/179) | p-valuea | |
| overall phenotype | |||||||
| SI totalis | 18 | 8.00 | 4 | 8.70 | 14 | 7.82 | 0.7677 (n.s.) |
| heterotaxy | 153 | 68.00 | 36 | 78.26 | 117 | 65.36 | 0.1118 (n.s.) |
| isolated CHD | 43 | 19.11 | 5 | 10.87 | 38 | 21.23 | 0.1416 (n.s.) |
| VACTERL-like | 4 | 1.78 | 0 | 0.00 | 4 | 2.23 | 0.5841 (n.s.) |
| normal, familial disease | 1 | 0.44 | 0 | 0.00 | 1 | 0.56 | 1.0000 (n.s.) |
| no phenotype details | 6 | 2.67 | 1 | 2.17 | 5 | 2.79 | 1.0000 (n.s.) |
| abdomen/gastrointestinal | |||||||
| abdominal SI | 82 | 36.44 | 23 | 50.00 | 59 | 32.96 | 0.0394 (S) |
| bile duct hypoplasia/biliary atresia | 3 | 1.33 | 0 | 0.00 | 3 | 1.68 | 1.0000 (n.s.) |
| gallbladder abnormalities | 6 | 2.67 | 1 | 2.17 | 5 | 2.79 | 1.0000 (n.s.) |
| malrotation of the gut | 48 | 21.33 | 9 | 19.57 | 39 | 21.79 | 0.8418 (n.s.) |
| asplenia | 63 | 28.00 | 13 | 28.26 | 50 | 27.93 | 1.0000 (n.s.) |
| polysplenia | 22 | 9.78 | 4 | 8.70 | 18 | 10.06 | 1.0000 (n.s.) |
| multilobated spleen | 1 | 0.44 | 0 | 0.00 | 1 | 0.56 | 1.0000 (n.s.) |
| HJ bodies | 5 | 2.22 | 2 | 4.35 | 3 | 1.68 | 0.2710 (n.s.) |
| imperforate anus | 3 | 1.33 | 0 | 0.00 | 3 | 1.68 | 1.0000 (n.s.) |
| renal anomalies | 11 | 4.89 | 2 | 4.35 | 9 | 5.03 | 1.0000 (n.s.) |
| liver anomalies | 35 | 15.56 | 5 | 10.87 | 30 | 16.76 | 0.3725 (n.s.) |
| skeletal/limbs | |||||||
| vertebral anomalies | 10 | 4.44 | 2 | 4.35 | 8 | 4.47 | 1.0000 (n.s.) |
| rib anomalies | 4 | 1.78 | 1 | 2.17 | 3 | 1.68 | 1.0000 (n.s.) |
| scoliosis | 3 | 1.33 | 1 | 2.17 | 2 | 1.12 | 0.4982 (n.s.) |
| limb defects | 6 | 2.67 | 1 | 2.17 | 5 | 2.79 | 1.0000 (n.s.) |
| cardiac position | |||||||
| levocardia | 20 | 8.89 | 4 | 8.70 | 16 | 8.94 | 1.0000 (n.s.) |
| mesocardia | 5 | 2.22 | 1 | 2.17 | 4 | 2.23 | 1.0000 (n.s.) |
| dextrocardia | 13 | 5.78 | 2 | 4.35 | 11 | 6.15 | 1.0000 (n.s.) |
| atria/ventricles | |||||||
| common atrium | 16 | 7.11 | 1 | 2.17 | 15 | 8.38 | 0.2038 (n.s.) |
| atrial isomerism | 10 | 4.44 | 0 | 0.00 | 10 | 5.59 | 0.2203 (n.s.) |
| ASD | 76 | 33.78 | 20 | 43.48 | 56 | 31.28 | 0.1614 (n.s.) |
| AV canal | 55 | 24.44 | 11 | 23.91 | 44 | 24.58 | 1.0000 (n.s.) |
| single ventricle/HLHS | 37 | 16.44 | 10 | 21.74 | 27 | 15.08 | 0.2725 (n.s.) |
| VSD | 98 | 43.56 | 20 | 43.48 | 78 | 43.58 | 1.0000 (n.s.) |
| vessels | |||||||
| SVC abnormality | 49 | 21.78 | 14 | 30.43 | 35 | 19.55 | 0.1143 (n.s.) |
| IVC abnormality | 49 | 21.78 | 7 | 15.22 | 42 | 23.46 | 0.3163 (n.s.) |
| TAPVR | 32 | 14.22 | 7 | 15.22 | 25 | 13.97 | 0.8150 (n.s.) |
| PAPVR | 16 | 7.11 | 6 | 13.04 | 10 | 5.59 | 0.1038 (n.s.) |
| inflow/outflow | |||||||
| aortic arch abnormalities | 57 | 25.33 | 15 | 32.61 | 42 | 23.46 | 0.2535 (n.s.) |
| d-TGA | 81 | 36.00 | 9 | 19.57 | 72 | 40.22 | 0.0097 (S) |
| l-TGA | 31 | 13.78 | 8 | 17.39 | 31 | 17.32 | 1.0000 (n.s.) |
| DILV | 14 | 6.22 | 1 | 2.17 | 13 | 7.26 | 0.3107 (n.s.) |
| DORV | 65 | 28.89 | 18 | 39.13 | 47 | 26.26 | 0.1013 (n.s.) |
| PA | 49 | 21.78 | 12 | 26.09 | 37 | 20.67 | 0.4282 (n.s.) |
| PS | 58 | 25.78 | 11 | 23.91 | 47 | 26.26 | 0.8511 (n.s.) |
| AS | 4 | 1.78 | 2 | 4.35 | 2 | 1.12 | 0.1863 (n.s.) |
| CoA | 22 | 9.78 | 3 | 6.52 | 19 | 10.61 | 0.5796 (n.s.) |
| BAV | 2 | 0.89 | 1 | 2.17 | 1 | 0.56 | 0.3678 (n.s.) |
| other | |||||||
| arrhythmia | 16 | 7.11 | 2 | 4.35 | 14 | 7.82 | 0.7466 (n.s.) |
ap-values for statistical significance (p < 0.05) represent comparisons between patients with CNVs of clear or potential pathogenicity and patients with common CNVs by two-tailed Fischer's Exact tests using Graphpad statistical software (http://www.graphpad.com/quickcalcs/).
Figure 1.
Overview of CNV findings. CNVs were designated as ‘common’ or ‘rare’ based on their presence in clinical and online databases of genomic variation (see §2). CNVs denoted as having ‘clear pathogenicity’ included a diverse array of large/complex chromosomal abnormalities (table 3), as well as CNVs containing genes previously associated with heterotaxy (table 4). CNVs ‘of interest’ represent CNVs containing potential candidate genes, including some identified at loci previously associated with CHD (electronic supplementary material, table S3) or with microduplication/deletion syndromes (electronic supplementary material, table S4).
Table 3.
Pathogenic chromosome abnormalities detected in the 225 patient heterotaxy cohort. abd., abdominal; abnl/abnls., abnormal/abnormalities; ASD, atrial septal defect; AVC, atrioventricular canal; bilat., bilateral; CAVC, complete atrioventricular canal; d-TGA, dextro-transposition of the great arteries; DD, developmental delay; dil., dilated; DILV, double inlet right ventricle; DORV, double outlet right ventricle; HLHS, hypoplastic left heart syndrome; IVC, inferior vena cava; l-TGA, levo-transposition of the great arteries; Lt., left; LV, left ventricle; PA, pulmonary atresia; PAPVR, partial anomalous pulmonary venous return; PDA, patent ductus arteriosus; PS, pulmonic stenosis, Rt., right; RA, right atrium; RV, right ventricle; SI, situs inversus; SVC, superior vena cava; TAPVR, total anomalous pulmonary venous return; UPD, uniparental disomy; VSD, ventricular septal defect.
| patient | gender | type | platforma | cytogenetic findings (breakpoints)b |
inheritance/phenotype | |||
|---|---|---|---|---|---|---|---|---|
| illumina SNP array | illumina size | agilent oligo array | agilent size | |||||
| 1 | M | gain | oligo | not analysed | not analysed | arr 9q33.2-q33.3(125204518-140138901)x3 | 14.9 Mb | sporadic/l-TGA, PA, ASD, VSD |
| 2 | M | unbalanced translocation/complex rearrangement | oligo | not analysed | not analysed | arr 5q15q35.3(94068223-176886171)x3 arr 9p24.1p23(8442856-12223883)x1 |
82 Mb, 3.8 Mb | familial/d-TGA, CAVC, VSD, ASD, asplenia |
| 3 | M | loss | 1 M, oligo | arr 22q11.1q11.21(15815765-16422571)x1 | 600 kb | arr 22q11.1q11.21(15815765-16422571)x1 | 600 kb | sporadic/situs ambiguus, abd. SI, VSD, DORV, PS, PA, SVC abnls., asplenia, renal abnls., pulmonary isomerism, heart malrotation, hypoplastic pulmonary artery, central gallbladder, pancreas rotated to Rt., stomach rotated to Lt., hypoplastic multicystic Rt. kidney, undescended Lt. testicle, absent Rt. testicle. |
| 4 | M | gain; UPD | 1 M | arr 2p12p11.2(82350453-84912738)x3, arr 11p14.2q13.3(26301682-70604313)x2 hmz |
2.6 Mb, 44.3 Mb | not analysed | not analysed | familial/oligohydramnios, hypoplastic lungs; ASD, PAPVR, DORV, CAVC, dextrocardia; UPD leads to homozygous BBS1 nonsense mutation |
| 5 | F | mosaic gain and loss | 1 M, oligo | arr 7p22.3p21.3(34332-12922987)x1-2 arr 7p21.3p15.1(12925517-28332937)x1-2 hmz arr 7p15.1p12.3(28333319-48346413)x1-2 arr 7p12.1(48346704-48898843)x2-3 arr 7p12.1p11.2(51513509-56362278)x2-3 |
12.9 Mb, 15.4 Mb, 20 Mb, 552.1 kb, 2.6 Mb | arr 7p22.3p12.3(136551-48338627)x1 arr 7p11.2(53925715-56351979)x3 |
48.2 Mb, 2.4 Mb | unknown/SI; PDA; Kartagener's: immotile cilia by nasal biopsy; consanguinity (parents are first cousins) |
| 6 | F | loss | oligo | not analysed | not analysed | arr 2p25.3(3274169-3431910)x1 arr 22q11.2(17020267-19761174)x1 |
157.7 kb, 2.7 Mb | unknown/Rt. aortic arch, VSD; abd. SI; gut malrotation |
| 7 | M | loss | 1 M, oligo | arr 8p23.3p21.2(146703-25298793)x1 | 25.2 Mb | arr 8p23.3p21.2(151470-25293991)x1 | 25.1 Mb | sporadic/DORV, ASD, VSD, PS, polysplenia, mitral atresia, duplicated IVC, midline pancreas, hypoplastic scrotum, microcephaly (FOC 45 cm), bilat. medial ulegyria of frontal hemispheres, organomegaly (liver, spleen, kidneys more than 95% for age) |
| 8 | M | mosaic monosomy 21 | 1 M, oligo | arr 21q11.2q22.3(13989691-46942324)x1-2 | 33 Mb | arr 21q11.2q22.3(13971999-46914745)x1 | 32.9 Mb | sporadic/dextrocardia, SVC abnls., hypoplastic Rt. thumb, chordee |
| 9 | M | complex rearrangement | 1 M, oligo | arr 2q35(216859560-217354800)x3 arr 18p11.32p11.31(99027-6681691)x1 arr 18p11.31p11.21(6683107-15400816)x3 |
494.2 kb, 6.6 Mb, 8.7 Mb | arr 2q35(216852776-217347523)x3 arr 18p11.32p11.31(121700-6596548)x1 arr 18p11.31p11.21(6684807-14969496)x3 |
494.7 kb, 6.5 Mb, 8.3 Mb | sporadic/d-TGA, HLHS, VSD, ASD, mitral atresia, hypoplastic LV, subvalvar and valvar PS |
| 10 | F | unbalanced translocation | 370 k | arr 2q37.3(237239911-242692820)x3, arr 10q26.13q26.3(125436928-135320785)x1 |
5.5 Mb, 9.9 Mb | not analysed | not analysed | familial/dextrocardia with Rt. aortic arch, atrial inversion, CAVC, SVC abnls., abd. SI, asplenia |
| 11 | M | mosaic unbalanced translocation | 1 M, oligo | arr (2)x2 hmz mos, 7q22.1q36.3(98613389-158815293)x2-3, arr 10q22.1q26.3(73126274-135374311)x1-2 |
62.2 Mb, 60.2 Mb | not analysed | not analysed | sporadic/unbalanced AVC, single outlet RV, TAPVR; abd. SI, asplenia |
| 12 | M | mosaic gain | 1 M, oligo | arr 5q11.2q35.3(51392113-180837635)x2-3 | 129.4 Mb | arr 5q11.2q35.3(51750061-180857866)x2-3 | 129.1 Mb | sporadic/CHD |
| 13 | F | loss | 1 M | arr 1p34.3p34.1(36507566-45013872)x2-3 | 8.5 Mb | not analysed | not analysed | sporadic/d-TGA, VSD, ASD, DORV, mod. dil. RV/RA, mild RV hypertrophy, large tortuous PDA, excessive aortopulmonary collaterals, DD (MRI: 2 small sites of leukomalacia in white matter of frontal lobes) |
aFor the purposes of this and all subsequent tables, array-CGH is listed as oligo array, while SNP-arrays are designated as 370 or 1 M depending on number of included markers (370 000 versus 1 000 000).
bLinear positions according to Build 36, hg18. Standard cytogenetic nomenclature is used.
Table 4.
Rare CNVs encompassing known heterotaxy genes. abd., abdominal; abnl., abnormal; ASD, atrial septal defect; AVVR, atrioventricular valve regurgitation CAVC, complete atrioventricular canal; CoA, coarctation of the aorta; d-TGA, dextro-transposition of the great arteries; DORV, double outlet right ventricle; IVC, inferior vena cava; mod., moderately; Lt., left; PA, pulmonary atresia; PAPVR, partial anomalous pulmonary venous return; PS, pulmonary stenosis, RV, right ventricle; SI, situs inversus; VSD, ventricular septal defect. Genes in bold have previously been described associated with heterotaxy.
| patient | gender | type | locus | platform | size illumina (bp) | size agilent (min–max) (bp) | # markers (illumina) | # probes (agilent) | affected genes | inheritance/phenotype | reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | M | loss | Xq26.2 | 1 M, oligo | 2876 | 3312 | 2 | 3 | ZIC3 | sporadic/dextrocardia, single ventricle, CAVC, AVVR, PA, PS, asplenia, mod. enlarged Lt. hepatic lobe (deceased) | [8] |
| 15 | M | gain | 2p25.1 | oligo | not analysed | 553 886–685 571 | not analysed | 13 | ROCK2, E2F6, GREB1, NTSR2, LPIN1 | sporadic/d-TGA, CoA, double chambered RV/DORV, VSD | [16] |
| 16 | F | loss | 3p24.1 | 1 M, oligo | 1 491 644 | 1 457 325–1 606 574 | 617 | 76 | RBMS3, TGFBR2, GADL1 | familial (sister with ASD)/abd SI, PAPVR, abnl. SVC, abnl. IVC, VSD, ASD, polysplenia, slightly upslanting palpebral fissures, long upper lip | [16] |
As it is important to rule out all potential heterotaxy gene candidates in a CNV of interest, we prioritized CNVs containing relatively fewer genes for further analysis. Twenty-two (62.86%) of these 35 ‘CNVs of interest’ involved gains of genetic material, while 13 (37.14%) involved genetic losses (electronic supplementary material, table S1). As an estimation of CNV gene content, we performed breakpoint predictions using available marker/probe positioning. These analyses were inherently limited by marker and probe spacing, but suggested 15/22 (68%) gains and 5/13 (38.46%) losses encompassed the entire coding region of at least one gene. Only one of the 22 identified gains (4.55%) and 4 of the 13 identified losses (30.77%) were predicted to lie entirely within a single gene. The remaining CNVs affected partial coding sequences or coding sequences of more than one gene. There was no obvious chromosome bias among detected CNVs, with CNVs of interest detected on 21/23 chromosomes, the exceptions being the X chromosome and chromosome 21 (data not shown).
Of the 35 CNVs of interest, seven were more than 1 Mb in size (SNP size range 1.01–2.78 Mb, mean 1.96 Mb, median 2.06 Mb; aCGH size range 1.08–3.84 Mb, mean 2.02–2.30 Mb, median 1.86–2.58 Mb), and encompassed between 1 and 18 genes (mean 7; median 7). The remaining 28 CNVs individually affected less than 1 Mb of sequence (SNP size range 161.66–877.08 kb, mean 458.50 kb, median 472.45 kb; aCGH size range 4.90–960.81 kb, mean 322.04–387.01 kb, median 205.73–279.31 kb) impacting between 1 and 16 genes each (mean 3.96; median 2).
A total of 165 genes with diverse developmental and molecular functions were encompassed by the 35 CNVs of interest (electronic supplementary material, table S2 and figure S1). Among these genes were a subset with recognized roles in developmental processes and pathways important for L–R patterning, including ciliogenesis, transforming growth factor-β (TGFβ) signalling, and cell–cell communication. Eleven of the 35 CNVs of interest encompassed loci previously associated with CHD (electronic supplementary material, table S3), while one (a 1.70–2.59 Mb 2q13 deletion identified in a patient with sporadic heterotaxy) disrupted a block of 10 genes previously deleted in a similarly affected heterotaxy patient [24]. CNVs encompassing loci associated with known microduplication/deletion syndromes but not previously associated with heterotaxy or CHD are summarized in the electronic supplementary material, table S4. For candidate gene screens, we prioritized analysis of genes with known expression during critical L–R developmental stages in animal studies.
(b). Knockdown of pfkp results in left–right patterning defects in Xenopus laevis
Because L–R patterning processes are highly conserved across vertebrate species [25–29], significant knowledge regarding human laterality can be gleaned from animal studies. We undertook a Xenopus-based morpholino loss of function approach to screen identified CNVs for potential heterotaxy candidate genes. The merits of Xenopus as model for laterality are extensive and have been reviewed in detail elsewhere [30].
A single patient (Patient 34) from our heterotaxy cohort was found to carry a heterozygous 175 kb deletion encompassing the PFKP and PITRM1 genes. These genes encode the platelet isoform of phosphofructokinase-1 (PFK-1) and the protease pitrilysin metallopeptidase 1, respectively (figure 2a). This CNV was of interest as it was identified using both genotyping platforms and contained an easily testable number of genes, one of which (PFKP) is a recognized interaction partner of the H+-V-ATPase proton pump [31,32], a known regulator of left–right patterning [33]. Notably, the deletion was not previously described in either online or institutional cytogenetic laboratory databases (see §2). To assess the effect of pfkp and pitrm1 knockdown on L–R patterning, TB and SB morpholinos (GeneTools, LLC.) were designed and injected into both blastomeres of 2-cell stage Xenopus laevis embryos. Gut, heart and gallbladder were then scored for abnormal situs at stage 47 according to published criteria [34]. A significantly greater proportion of pfkp, but not pitrm1, morphants developed organ situs defects relative to uninjected or control morpholino-injected embryos (figure 2c). Organ situs defects in pfkp morphants were dose-dependent (data not shown). Consistent with previous reports demonstrating PFK-1 expression in early development [35–37], pfkp was expressed throughout stages known to be important for L–R patterning in Xenopus (figure 2b). These results indicated that pfkp but not pitrm1 was a suitable candidate for further study.
Figure 2.
pfkp but not pitrm1 Xenopus laevis morphants develop organ situs defects. (a) Genes in CNV interval. Source: USCS genome browser (https://genome.ucsc.edu/). (b) RT-PCR results demonstrate that pfkp is expressed throughout critical L–R patterning stages. ODC, ornithine decarboxylase. (c) pfkp knockdown in 2-cell embryos causes heterotaxy in Xenopus. Situs defects in st. 47 morphants are shown relative to zic3 control morphants and uninjected controls. (d) Confirmation of exon 8 deletion in pfkp SB morphants by RT-PCR targeting the exon 8–intron 8 splice junction. Upper schematic: small horizontal arrows denote primer annealing sites. The horizontal line without an arrowhead denotes the SB morpholino target site. The severely truncated exon 8 (see §3) is represented by cross-hatching. Lower panel (RT-PCR): large arrowhead denotes the pfkp deletion fragment. SB, pfkp splice block. (Online version in colour.)
In order to confirm pfkp mRNA knockdown in pfkp SB morphants, RT-PCR and gene-specific amplification of whole RNA from st. 9 (not shown) and st. 25 embryos (figure 2d, lane 2) was completed. Sequencing of the deletion product revealed activation of a cryptic splice site 13 bp downstream of the intron 7-exon 8 junction, but confirmed deletion of the majority of exon 8 (96/109 bp). The resulting pfkp transcript is predicted to encode a truncated, non-frameshifted 754aa (versus 786aa) protein lacking residues predicted to contribute to ATP binding. Synergy and rescue experiments using wild-type human PFKP mRNA- supported specificity of the SB morpholino effect (figure 3).
Figure 3.
Synergy and rescue experiments support MO specificity. (a) Sub-threshold dosages of pfkp TB and SB MOs yielded a higher proportion of embryos with situs defects when injected together than when injected alone. (b) Partial rescue of organ situs in SB morphants co-injected with WT human PFKP mRNA. SB, splice block; TB, translation block.
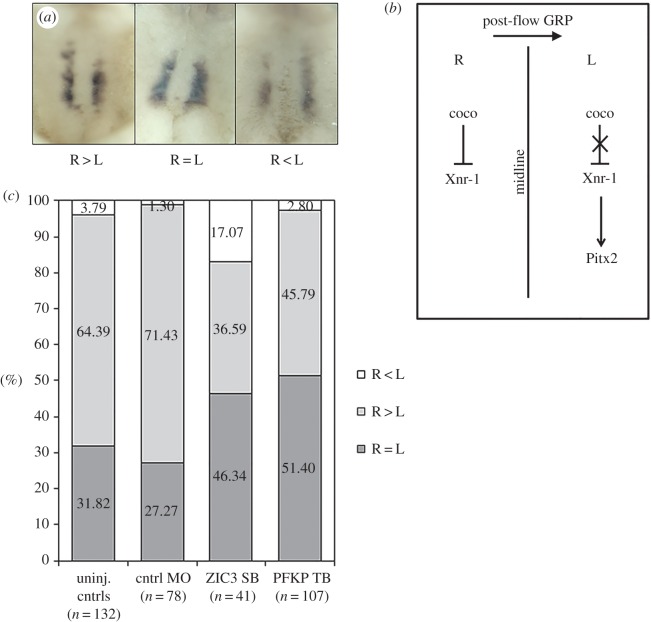
In order to confirm an effect for pfkp knockdown on L–R relevant signalling pathways, we examined expression of the conserved L–R patterning marker, coco, at the gastrocoel roof plate (GRP). Ciliated cells of the GRP generate a highly directional extracellular fluid flow that asymmetrically activates Nodal signalling pathways in the left lateral plate [27]. In this setting, coco serves as a potent inhibitor of the Nodal ligand, becoming increasingly restricted to the right side of the GRP and functioning to prevent right-sided Nodal expression during flow-relevant stages [38]. This right-sided bias is of critical importance for the laterality programme: without it, Nodal signalling asymmetries are improperly established and resulting embryos develop extensive laterality defects [39]. Consistent with previous reports [38], we observed a right-sided coco expression bias in approximately 60% of untreated late-flow stage embryos (stage 20–21; figure 4). This bias was significantly reduced in pfkp morphants (44.8%, p = 0.0058). Supporting functional impact. The magnitude of this reduction was comparable to that observed following knockdown of dnah9, which encodes an axonemal molecular motor essential for GRP ciliary motility and extracellular fluid flow [38]. On the basis of these morpholino studies, we identify pfkp as a novel gene associated with heterotaxy and conclude that the 175 kb CNV encompassing this gene is the most likely explanation for the laterality phenotype observed in our patient.
Figure 4.
pfkp knockdown disrupts right-sided coco bias. (a) Representative images of coco expression at the gastrocoel roof plate (GRP) in st. 20–21 embryos. (b) Schematic depicting right-sided coco bias during post-flow stages. Reduced coco expression on the left-side permits initiation of the Nodal signalling cascade by alleviating inhibition of Xenopus Nodal related-1 (Xnr-1). Downstream transcription factors (Pitx2) important for initiating asymmetric organ morphogenesis are activated. High levels of right-sided coco expression repress similar activation of Nodal signalling on the right side. (c) Relative to control embryos, significantly fewer pfkp morphants exhibited right-sided bias of coco expression (p = 0.0058, two-tailed Fisher's Exact test). (Online version in colour.)
4. Discussion
(a). The clinical importance of copy number variant analyses
We have performed comprehensive genome-wide CNV analyses on a large cohort of 225 patients with heterotaxy and CHD. These analyses identified clinically relevant CNVs in approximately 20% of patients, including 7.1% of clear pathogenicity, a diagnostic yield that indicates newborns with heterotaxy should have clinical chromosome microarray testing. Specific phenotypic abnormalities were not significant predictors of identifying a disease causing CNV, with the exception of abdominal SI (table 2). Future studies will be important to determine whether specific subclasses of CHD, such as d-TGA, are less likely to have CNV abnormalities. In addition, the burden of common CNVs present in individuals with heterotaxy should be further evaluated since this has been a noted correlation in other diseases [15].
(b). Copy number variant analyses identify novel heterotaxy gene candidates
As the majority of heterotaxy cases remain unexplained, the primary goal of study was to identify novel genetic variation of pathogenic significance to this patient population. Excluding CNVs of clear pathogenicity (tables 3 and 4), we detected 35 unique CNVs of potential interest for functional testing. Collectively, these variants encompassed 165 genes of diverse biological and molecular ontology (electronic supplementary material, table S2 and figure S1), including a subset with roles in pathways and processes relevant to laterality. These genes are individually described below.
The ‘nodal flow’ model of L–R patterning proposes that the vertebrate laterality programme is initiated by asymmetric activation of Nodal signalling in the left lateral plate shortly after gastrulation. Evidence from multiple species has solidified the importance of transient, ciliated organizers in establishing and propagating these signalling asymmetries [25–29]. Mutations affecting ciliary genes have subsequently been identified as a major cause of heterotaxy spectrum disorders ranging from ‘classic’ heterotaxy (CHD with visceral situs abnormalities) to primary ciliary dyskinesia (PCD) and other ciliopathies [40]. There is significant overlap between PCD and heterotaxy, with 6–12% of PCD patients having heterotaxy [41,42] and an unknown but significant percentage of heterotaxy patients having PCD [42]. Results from our CNV screens have identified rare variants encompassing a number of genes with cilia-related functions, including TTC21B, CEP290, TTBK2 and CFAP126. Two of these, CEP290 and TTC21B, are recognized causes of human ciliopathies and are known to play critical roles in cilia assembly and transport [43,44]. Mutations in TTC21B, which encodes the retrograde intraflagellar transport protein, THM1/IFT139, cause both nephronophthisis 12 (end-stage renal disease) and short-rib thoracic dysplasia 4 (skeletal anomalies with or without polydactyly) [45]. Similarly, TTC21B knockdown in mice is characterized by preaxial polydactyly, cleft palate, micro-ophthalmia and brain abnormalities. Retrograde ciliary transport is disrupted, leading to impaired cilia growth and downstream Sonic hedgehog (Shh) signalling [43]. The phenotypic spectrum resulting from CEP290 mutations is even more striking as pathogenic variants have been associated with a diverse array of ciliopathies without any obvious genotype–phenotype correlations [44]. Functionally, the CEP290 protein localizes to both the cilia transition zone [46] and to centriolar satellites, where it interacts with a number of other ciliary proteins required for cilia formation [47]. Knockdown of CEP290 in cultured mammalian cells disrupts satellite localization, preventing assembly of a functional cilium [47]. Similar disruption of centriolar satellites by morpholino-mediated knockdown of pericentrin (pcm1) in zebrafish yielded inverted heart looping in nearly 50% of morphant embryos [47]. These results suggest pathogenic potential for CEP290 mutations via loss of centrosomal protein targeting and ciliary assembly. While CEP290 mutations have not yet been reported as causes of human heterotaxy, at least one patient with SI has been identified to carry a CEP290 mutation [48].
Two other ciliary genes, TTBK2 and CFAP126, were identified in CNVs in the heterotaxy cohort. The first, TTBK2, encodes a serine/threonine kinase essential for intraflagellar transport (IFT) protein recruitment. Its localization to centrioles is an initiating step of ciliogenesis and is required to promote elongation of the ciliary axoneme [49]. Like TTC21B and CEP290, mutations in TTBK2 prevent formation of a functional cilium and are a recognized cause of disease in both humans (spinocerebellar ataxia 11) and bartleby (bby) mutant mice. TTBK2 mutations have yet to be reported among patients with heterotaxy; however, mice homozygous for bby mutations exhibit randomized heart looping indicative of impaired L–R patterning, in addition to other defects stemming from disrupted ciliogenesis (holoprosencephaly, limb defects, twisted body axis). Comparatively less is known about CFAP126, a newly characterized protein important for basal body docking and cilia positioning [50]. Nevertheless, expression has been noted at the mouse embryonic node as early as E7.75, placing CFAP126 at the correct location and time for impacting laterality [51].
In addition to these ciliary genes, two rare CNVs encompassing members of the TGFβ superfamily were identified. The first, a duplication involving the Nodal signalling modulator, NOMO3, was detected in a male patient with sporadic heterotaxy (Patient 19). NOMO3 exists as one of three highly similar genes on chromosome 16p12-p13 and is thought to act as a direct Nodal signalling antagonist, acting independently of other Nodal modulating pathways [52]. The second CNV, a duplication identified in a male patient with familial heterotaxy (Patient 40), encompassed LTBP1, a TGFβ binding protein with roles in TGFβ1 assembly, secretion, and targeting. Deletions involving LTBP1 have been previously associated with outflow tract (OFT) defects [53], but have not yet been reported among patients with heterotaxy. Intriguingly, experiments in Xenopus have identified LTBP1 as a potent activator of Activin and Nodal signalling, indicating potential for involvement in L–R relevant pathways [54].
Rare CNVs involving four distinct microRNAs (miR-148a, miR-455, miR-484 and miR-2117) were also identified in the heterotaxy cohort. Considering known functions in the context of L–R patterning, miR-418a is the most notable as it is a direct inhibitor of both ACVR1, an activin receptor [55] and ROCK1, a rho-associated kinase [56]. Although neither protein has been directly associated with laterality, family members of both (ACVR2B and ROCK2) are known genetic causes of heterotaxy in human patients [16,57].
A total of 10 rare CNVs were identified at loci previously associated with CHD (electronic supplementary material, table S3). To the best of our knowledge, only one of these, a large 1.7–2.6 Mb 2q13 duplication detected by oligoarray in a male patient with sporadic heterotaxy and complex CHD (Patient 29), affects a locus previously associated with heterotaxy [24]. The duplicated region encompassed 10 genes (electronic supplementary material, table S1) and was identical in gene content to a 1.62 Mb deletion detected in a patient with a similar laterality phenotype [24]. Genomic imbalances spanning approximately 1.7 Mb and encompassing the same 10 genes have been described in a number of other patients with CHD but who are otherwise lacking overt situs abnormalities [24,58–61]. No specific microduplication/deletion syndrome has been defined; however, brain abnormalities were suggestive of Joubert syndrome in one individual [60]. Recent knockdown studies in zebrafish have identified FBLN7 and TMEM87B as the genes most likely to be causative for CHD; however, their exact roles in cardiac development have yet to be determined [61]. In our patient, a second, larger CNV was also identified upstream of this region and included an additional 18 genes of uncertain pathogenic significance. Predicted breakpoints of the two intervals were non-overlapping but were in immediate proximity, suggesting the possibility of a single duplication event.
(c). PFKP as a novel cause of heterotaxy
PFKP, encoding the platelet isoform of the glycolytic enzyme, phosphofructokinase-1 (PFK-1), was among the top heterotaxy candidates genes identified in the cohort. Using established morphlino-based methods for candidate gene screening [16,20], we have identified pfkp as a novel regulator of laterality in Xenopus laevis.
PFK-1 is the rate-limiting enzyme of glycolysis, catalysing the irreversible conversion of fructose 6-phosphate to fructose 1,6-bisphosphate. In both human and frog, PFK-1 exists as three isoforms, each encoded by a unique locus and named in accordance with adult expression (muscle, M; liver, L and platelet/fibroblast, P). Bi-allelic loss of function mutations in PFKM result in diabetes as well as glycogen storage disease type VII (Tarui's disease) [62]. Our interest in PFKP was therefore additionally piqued by an unexplained, but long-recognized, increased risk for heterotaxy among the children of diabetic mothers [63].
Our study (figure 2b) and others [35–37] have shown that PFK-1 is expressed throughout early Xenopus development. Expression is most readily detected in the animal pole and marginal zone during gastrulation, and at high levels in the neural tube, neural crest and adjacent tissue during neurulation [64]. Overall levels of expression increase as development proceeds, particularly in dorsal tissues [64]. At least in humans, there does not appear to be a specific ‘fetal’ isoform as all three types are uniformly expressed in early embryonic stages: it is only as organ systems begin to develop that isoform-specific expression patterns begin to emerge [37,65]. Intriguingly, PFKP is the sole isoform identified at significant levels in fetal heart, which contrasts sharply with a preponderance of PFKM in adult cardiac tissue [65]. The significance of this finding, particularly with respect to CHD and heterotaxy, is currently uncertain. Because our morpholinos were designed to explicitly target the 5′ untranslated region of PFKP and not the M and L subtypes (which are encoded by different genes), it also remains to be seen whether embryonic knockdowns of these isoforms similarly disrupt L–R patterning.
In Xenopus, catalytic activity of PFK-1 isoforms [35] and overall rates of glycolysis [36] are very low until gastrulation [36]. As L–R patterning mechanisms may operate during early cleavage stages [66], roles for PFKP in L–R patterning could manifest in the pre-gastrula. Possible non-glycolytic functions for PFKP must therefore also be considered. It is potentially noteworthy that a non-glycolytic role in early dorsal patterning has recently been demonstrated for PFKFB4, one of five known isoforms of phosphofructokinase-2 (PFK-2) [67].
How might PFKP regulate L–R patterning independently of glycolysis? Isoforms of PFK-1 are known to bind the a-subunit of H+-V-ATPase [31,32], a plasma membrane and vacuolar proton pump whose inhibition in pre-gastrula stages results in significant defects in a number of L–R patterning processes including ion-flux (Xenopus/chick), LRO and cilia development (zebrafish) and Nodal signalling (chick/zebrafish) [33,68]. Together with observations from yeast that PFK-1 is required to maintain activity of the H+V-ATPase proton pump and that even a catalytically inactive PFK-1 is capable of completing this function [31], a reasonable hypothesis is that PFKP knockdown may impact L–R patterning through loss of a functionally critical interaction with the H+-V-ATPase a-subunit. As recent zebrafish studies indicate a primary role for H+-V-ATPase in KV formation [68], it is possible that PFKP may impact L–R patterning in a similar manner. Interestingly, a rare duplication involving the gene ATP6V1G1, which encodes the G-subunit of H+-V-ATPAase, was also identified in a male heterotaxy patient in our cohort (Patient 41). Like the a-subunit, the G-subunit contributes to the peripheral stator stalk, the non-rotating connection between the V1 and Vo halves of the ATPase [69]. Two of three dimers composed of E- and G-subunits attach to the a-subunit at its amino terminal domain, a site thought to be of importance for a-subunit function [70].
(d). Study limitations
Sample collection from trios (proband and parents) was not feasible in many cases. As a result, determinations of de novo inheritance were not possible. Similarly, while a few instances of copy number mosaicism were identified, direct comparisons between mosaic and non-mosaic CNV carriers were prevented by the rarity of individual CNVs. Cut-offs for CNV calling were also set at a minimum coverage of three markers/probes per CNV, restricting downstream genetic and functional analyses to variants exceeding these minimum size thresholds. While more difficult to detect reliably by microarray approaches, small exonic CNVs of 1–30 kb have been suggested to contribute to susceptibility of some genetic diseases [71]. Finally, analyses were restricted to CNVs involving coding regions of at least one gene, ignoring variants solely impacting intronic or regulatory sequences. Genetic variation in non-coding regions, while typically of less obvious functional significance, has been linked to genetic disease [72–74] but would not have been considered in our study.
Our analyses of PFKP as an L–R patterning candidate were limited by an inability to definitively verify isoform-specific knockdown by Western blotting as the M and L isoforms are of similar size and commercially available antibodies demonstrate high likelihood for subtype cross-reactivity (data not shown). Nevertheless, confidence in the specificity of TB knockdown was provided by replication of the TB morphant phenotype in SB morphants and demonstration that sub-threshold dosages of both morpholinos synergized to produce organ situs defects when used in combination, but not in isolation.
5. Summary
In summary, we have performed CNV analyses on a cohort of 225 patients with heterotaxy and heterotaxy-spectrum CHDs and identified CNVs of potential pathogenic significance in a large proportion (20.4%). Detected CNVs ranged in size and complexity and collectively encompassed a number of genes with known or suspected functions in L–R patterning developmental programmes. Using rigorous morpholino-based studies, a role for the platelet isoform of phosphofructokinase-1, pfkp, in heterotaxy pathogenesis was confirmed in Xenopus laevis. Future work using both animal and in vitro culture systems will help to tease out temporal requirements as well as precise molecular and developmental functions for pfkp in L–R patterning.
Supplementary Material
Acknowledgements
We thank the patients and families for their participation and John Wells for independently scoring coco hybridized embryos.
Ethics
All studies were approved by the Institutional Review Board at Cincinnati Children's Hospital Medical Center (CCHMC) and informed consent was obtained from all study participants. Animal studies were carried out in compliance with local and federal animal use guidelines.
Authors' contributions
J.R.C. participated in study design, carried out the molecular and functional laboratory work, completed data and statistical analyses, and drafted the manuscript. M.T. carried out sample genotyping and participated in CNV analyses; C.S. and M.R. assisted with CNV calling; J.W.B. assisted in patient recruitment and phenotyping and provided DNA samples for genotyping, T.A.S. and S.R.L. reviewed and provided interpretation for all CNV data. S.M.W. conceived, designed and supervised the study. All authors gave final approval for publication.
Competing interests
The authors have no competing interests.
Funding
This work was supported by funding from the Burroughs Welcome Fund Clinical Scientist Award in Translational Research (grant no. 1008496) and March of Dimes Research Foundation (grant nos. 1FY10-401 and 6-FY13-167) (to S.M.W.).
References
- 1.Lin AE, Ticho BS, Houde K, Westgate MN, Holmes LB. 2000. Heterotaxy: associated conditions and hospital-based prevalence in newborns. Genet. Med. 2, 157–172. (doi:10.109700125817-200005000-00002) [DOI] [PubMed] [Google Scholar]
- 2.Lin AE, et al. 2014. Laterality defects in the national birth defects prevention study (1998–2007): birth prevalence and descriptive epidemiology. Am. J. Med. Genet. Part A 164A, 2581–2591. ( 10.1002/ajmg.a.36695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SJ, Kim WH, Lim HG, Lee JY. 2008. Outcome of 200 patients after an extracardiac Fontan procedure. J. Thorac. Cardiovasc. Surg. 136, 108–116. ( 10.1016/j.jtcvs.2007.12.032) [DOI] [PubMed] [Google Scholar]
- 4.Swisher M, Jonas R, Tian X, Lee ES, Lo CW, Leatherbury L. 2011. Increased postoperative and respiratory complications in patients with congenital heart disease associated with heterotaxy. J. Thorac. Cardiovasc. Surg. 141, 637–644. ( 10.1016/j.jtcvs.2010.07.082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartz PJ, et al. 2006. Early and late results of the modified fontan operation for heterotaxy syndrome 30 years of experience in 142 patients. J. Am. Coll. Cardiol. 48, 2301–2305. ( 10.1016/j.jacc.2006.07.053) [DOI] [PubMed] [Google Scholar]
- 6.Oyen N, Poulsen G, Boyd HA, Wohlfahrt J, Jensen PK, Melbye M. 2009. Recurrence of congenital heart defects in families. Circulation 120, 295–301. ( 10.1161/CIRCULATIONAHA.109.857987) [DOI] [PubMed] [Google Scholar]
- 7.Zhu L, Belmont JW, Ware SM. 2006. Genetics of human heterotaxias. Eur. J. Hum. Genet. 14, 17–25. ( 10.1038/sj.ejhg.5201506) [DOI] [PubMed] [Google Scholar]
- 8.Cowan J, Tariq M, Ware SM. 2014. Genetic and functional analyses of ZIC3 variants in congenital heart disease. Hum. Mutat. 35, 66–75. ( 10.1002/humu.22457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ware SM, Peng J, Zhu L, Fernbach S, Colicos S, Casey B, Towbin J, Belmont JW. 2004. Identification and functional analysis of ZIC3 mutations in heterotaxy and related congenital heart defects. Am. J. Hum. Genet. 74, 93–105. ( 10.1086/380998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sutherland MJ, Wang S, Quinn ME, Haaning A, Ware SM. 2013. Zic3 is required in the migrating primitive streak for node morphogenesis and left–right patterning. Hum. Mol. Genet. 22, 1913–1923. ( 10.1093/hmg/ddt001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohapatra B, et al. 2009. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum. Mol. Genet. 18, 861–871. ( 10.1093/hmg/ddn411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roessler E, et al. 2008. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am. J. Hum. Genet. 83, 18–29. ( 10.1016/j.ajhg.2008.05.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girirajan S, Campbell CD, Eichler EE. 2011. Human copy number variation and complex genetic disease. Annu. Rev. Genet. 45, 203–226. ( 10.1146/annurev-genet-102209-163544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang F, Gu W, Hurles ME, Lupski JR. 2009. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10, 451–481. ( 10.1146/annurev.genom.9.081307.164217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lander J, Ware SM. 2014. Copy number variation in congenital heart defects. Curr. Genet. Med. Rep. 2, 168–178. ( 10.1007/s40142-014-0049-3) [DOI] [Google Scholar]
- 16.Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. 2011. Rare copy number variations in congenital heart disease patients identify unique genes in left–right patterning. Proc. Natl Acad. Sci. USA 108, 2915–2920. ( 10.1073/pnas.1019645108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigler SL, et al. 2015. Novel copy-number variants in a population-based investigation of classic heterotaxy. Genet. Med. 17, 348–357. ( 10.1038/gim.2014.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zarrei M, MacDonald JR, Merico D, Scherer SW. 2015. A copy number variation map of the human genome. Nat. Rev. Genet. 16, 172–183. ( 10.1038/nrg3871) [DOI] [PubMed] [Google Scholar]
- 19.Boone PM, et al. 2010. Detection of clinically relevant exonic copy-number changes by array CGH. Hum. Mutat. 31, 1326–1342. ( 10.1002/humu.21360) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cast AE, Gao C, Amack JD, Ware SM. 2012. An essential and highly conserved role for Zic3 in left–right patterning, gastrulation and convergent extension morphogenesis. Dev. Biol. 364, 22–31. ( 10.1016/j.ydbio.2012.01.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nieuwkoop P, Faber J. 1994. Normal table of Xenopus laevis. New York, NY: Garland Publishing. [Google Scholar]
- 22.Heasman J, Kofron M, Wylie C. 2000. β-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev. Biol. 222, 124–134. ( 10.1006/dbio.2000.9720) [DOI] [PubMed] [Google Scholar]
- 23.Sive HL, Grainger RM, Harland RM. 2000. Early development of Xenopus laevis: a laboratory manual. 2000 ed. Cold Spring Harbor, NY: Cold Spring Harbor Press. [Google Scholar]
- 24.Rudd MK, Keene J, Bunke B, Kaminsky EB, Adam MP, Mulle JG, Ledbetter DH, Martin CL. 2009. Segmental duplications mediate novel, clinically relevant chromosome rearrangements. Hum. Mol. Genet. 18, 2957–2962. ( 10.1093/hmg/ddp233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. 2005. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left–right development of the brain, heart and gut. Development 132, 1247–1260. ( 10.1242/dev.01663) [DOI] [PubMed] [Google Scholar]
- 26.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. 1998. Randomization of left–right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837. ( 10.1016/S0092-8674(00)81705-5) [DOI] [PubMed] [Google Scholar]
- 27.Schweickert A, Weber T, Beyer T, Vick P, Bogusch S, Feistel K, Blum M. 2007. Cilia-driven leftward flow determines laterality in Xenopus. Curr. Biol. 17, 60–66. ( 10.1016/j.cub.2006.10.067) [DOI] [PubMed] [Google Scholar]
- 28.Sulik K, Dehart DB, Iangaki T, Carson JL, Vrablic T, Gesteland K, Schoenwolf GC. 1994. Morphogenesis of the murine node and notochordal plate. Dev. Dyn. 201, 260–278. ( 10.1002/aja.1002010309) [DOI] [PubMed] [Google Scholar]
- 29.Vick P, Schweickert A, Weber T, Eberhardt M, Mencl S, Shcherbakov D, Beyer T, Blum M. 2009. Flow on the right side of the gastrocoel roof plate is dispensable for symmetry breakage in the frog Xenopus laevis. Dev. Biol. 331, 281–291. ( 10.1016/j.ydbio.2009.05.547) [DOI] [PubMed] [Google Scholar]
- 30.Blum M, Beyer T, Weber T, Vick P, Andre P, Bitzer E, Schweickert A. 2009. Xenopus, an ideal model system to study vertebrate left–right asymmetry. Dev. Dyn. 238, 1215–1225. ( 10.1002/dvdy.21855) [DOI] [PubMed] [Google Scholar]
- 31.Su Y, Blake-Palmer KG, Sorrell S, Javid B, Bowers K, Zhou A, Chang SH, Qamar S, Karet FE. 2008. Human H+ATPase a4 subunit mutations causing renal tubular acidosis reveal a role for interaction with phosphofructokinase-1. Am. J. Physiol. Renal Physiol. 295, F950–F958. ( 10.1152/ajprenal.90258.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Y, Zhou A, Al-Lamki RS, Karet FE. 2003. The a-subunit of the V-type H+-ATPase interacts with phosphofructokinase-1 in humans. J. Biol. Chem. 278, 20 013–20 018. ( 10.1074/jbc.M210077200) [DOI] [PubMed] [Google Scholar]
- 33.Adams DS, Robinson KR, Fukumoto T, Yuan S, Albertson RC, Yelick P, Kuo L, McSweeney M, Levin M. 2006. Early, H+-V-ATPase-dependent proton flux is necessary for consistent left–right patterning of non-mammalian vertebrates. Development 133, 1657–1671. ( 10.1242/dev.02341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Branford WW, Essner JJ, Yost HJ. 2000. Regulation of gut and heart left–right asymmetry by context-dependent interactions between Xenopus lefty and BMP4 signaling. Dev. Biol. 223, 291–306. ( 10.1006/dbio.2000.9739) [DOI] [PubMed] [Google Scholar]
- 35.Raddatz E, Lovtrup-Rein H. 1986. Changes in activity of the regulatory glycolytic enzymes and of the pyruvate-dehydrogenase complex during the development of Xenopus laevis. Exp. Cell Biol. 54, 53–60. ( 10.1159/000163344) [DOI] [PubMed] [Google Scholar]
- 36.Dworkin MB, Dworkin-Rastl E. 1991. Carbon metabolism in early amphibian embryos. Trends Biochem. Sci. 16, 229–234. ( 10.1016/0968-0004(91)90091-9) [DOI] [PubMed] [Google Scholar]
- 37.Davidson M, Collins M, Byrne J, Vora S. 1983. Alterations in phosphofructokinase isoenzymes during early human development. Establishment of adult organ-specific patterns. Biochem. J. 214, 703–710. ( 10.1042/bj2140703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweickert A, Vick P, Getwan M, Weber T, Schneider I, Eberhardt M, Beyer T, Pachur A, Blum M. 2010. The nodal inhibitor Coco is a critical target of leftward flow in Xenopus. Curr. Biol. 20, 738–743. ( 10.1016/j.cub.2010.02.061) [DOI] [PubMed] [Google Scholar]
- 39.Blum M, Schweickert A, Vick P, Wright CV, Danilchik MV. 2014. Symmetry breakage in the vertebrate embryo: when does it happen and how does it work? Dev. Biol. 393, 109–123. ( 10.1016/j.ydbio.2014.06.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutherland MJ, Ware SM. 2009. Disorders of left–right asymmetry: heterotaxy and situs inversus. Am. J. Med. Genet. 151C, 307–317. ( 10.1002/ajmg.c.30228) [DOI] [PubMed] [Google Scholar]
- 41.Kennedy MP, et al. 2007. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation 115, 2814–2821. ( 10.1161/CIRCULATIONAHA.106.649038) [DOI] [PubMed] [Google Scholar]
- 42.Shapiro AJ, et al. 2014. Laterality defects other than situs inversus totalis in primary ciliary dyskinesia: insights into situs ambiguus and heterotaxy. Chest 146, 1176–1186. ( 10.1378/chest.13-1704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran PV, et al. 2008. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 40, 403–410. ( 10.1038/ng.105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coppieters F, Lefever S, Leroy BP, De Baere E. 2010. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum. Mutat. 31, 1097–1108. ( 10.1002/humu.21337) [DOI] [PubMed] [Google Scholar]
- 45.Davis EE, et al. 2011. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat. Genet. 43, 189–196. ( 10.1038/ng.756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, Witman GB. 2010. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190, 927–940. ( 10.1083/jcb.201006105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stowe TR, Wilkinson CJ, Iqbal A, Stearns T. 2012. The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol. Biol. Cell. 23, 3322–3335. ( 10.1091/mbc.E12-02-0134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brancati F, et al. 2007. CEP290 mutations are frequently identified in the oculo-renal form of Joubert syndrome-related disorders. Am. J. Hum. Genet. 81, 104–113. ( 10.1086/519026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goetz SC, Liem KF Jr, Anderson KV. 2012. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell 151, 847–858. ( 10.1016/j.cell.2012.10.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gegg M, Bottcher A, Burtscher I, Hasenoeder S, Van Campenhout C, Aichler M, Walch A, Grant SG, Lickert H. 2014. Flattop regulates basal body docking and positioning in mono- and multiciliated cells. eLife 3, e03842 ( 10.7554/eLife.03842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lange A, Gegg M, Burtscher I, Bengel D, Kremmer E, Lickert H. 2012. Fltp(T2AiCre): a new knock-in mouse line for conditional gene targeting in distinct mono- and multiciliated tissues. Differentiation. 83, S105–S113. ( 10.1016/j.diff.2011.11.003) [DOI] [PubMed] [Google Scholar]
- 52.Haffner C, Frauli M, Topp S, Irmler M, Hofmann K, Regula JT, Bally-Cuif L, Haass C. 2004. Nicalin and its binding partner Nomo are novel Nodal signaling antagonists. EMBO J. 23, 3041–3050. ( 10.1038/sj.emboj.7600307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Todorovic V, et al. 2007. Long form of latent TGF-β binding protein 1 (Ltbp1 L) is essential for cardiac outflow tract septation and remodeling. Development 134, 3723–3732. ( 10.1242/dev.008599) [DOI] [PubMed] [Google Scholar]
- 54.Altmann CR, Chang C, Munoz-Sanjuan I, Bell E, Heke M, Rifkin DB, Brivanlou AH. 2002. The latent-TGFβ-binding-protein-1 (LTBP-1) is expressed in the organizer and regulates nodal and activin signaling. Dev. Biol. 248, 118–127. ( 10.1006/dbio.2002.0716) [DOI] [PubMed] [Google Scholar]
- 55.Song H, Wang Q, Wen J, Liu S, Gao X, Cheng J, Zhang D. 2012. ACVR1, a therapeutic target of fibrodysplasia ossificans progressiva, is negatively regulated by miR-148a. AAPG Bull. 13, 2063–2077. ( 10.3390/ijms13022063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Ying ZZ, Tang ZL, Long LQ, Li K. 2012. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 287, 21 093–21 101. ( 10.1074/jbc.M111.330381) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kosaki R, Gebbia M, Kosaki K, Lewin M, Bowers P, Towbin JA, Casey B. 1999. Left–right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am. J. Med. Genet. 82, 70–76. ( 10.1002/(SICI)1096-8628(19990101)82:1%3C70::AID-AJMG14%3E3.0.CO;2-Y) [DOI] [PubMed] [Google Scholar]
- 58.Cooper GM, et al. 2011. A copy number variation morbidity map of developmental delay. Nat. Genet. 43, 838–846. ( 10.1038/ng.909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soemedi R, et al. 2012. Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am. J. Hum. Genet. 91, 489–501. ( 10.1016/j.ajhg.2012.08.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu HE, Hawash K, Picker J, Stoler J, Urion D, Wu BL, Shen Y. 2012. A recurrent 1.71 Mb genomic imbalance at 2q13 increases the risk of developmental delay and dysmorphism. Clin. Genet. 81, 257–264. ( 10.1111/j.1399-0004.2011.01637.x) [DOI] [PubMed] [Google Scholar]
- 61.Russell MW, Raeker MO, Geisler SB, Thomas PE, Simmons TA, Bernat JA, Thorsson T, Innis JW. 2014. Functional analysis of candidate genes in 2q13 deletion syndrome implicates FBLN7 and TMEM87B deficiency in congenital heart defects and FBLN7 in craniofacial malformations. Hum. Mol. Genet. 23, 4272–4284. ( 10.1093/hmg/ddu144) [DOI] [PubMed] [Google Scholar]
- 62.Ristow M, Vorgerd M, Mohlig M, Schatz H, Pfeiffer A. 1997. Deficiency of phosphofructo-1-kinase/muscle subtype in humans impairs insulin secretion and causes insulin resistance. J. Clin. Invest. 100, 2833–2841. ( 10.1172/JCI119831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lisowski LA, Verheijen PM, Copel JA, Kleinman CS, Wassink S, Visser GH, Meijboom EJ. 2010. Congenital heart disease in pregnancies complicated by maternal diabetes mellitus. An international clinical collaboration, literature review, and meta-analysis. Herz 35, 19–26. ( 10.1007/s00059-010-3244-3) [DOI] [PubMed] [Google Scholar]
- 64.Pegoraro C, Maczkowiak F, Monsoro-Burq AH. 2013. Pfkfb (6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase) isoforms display a tissue-specific and dynamic expression during Xenopus laevis development. Gene Expr. Patterns 13, 203–211. ( 10.1016/j.gep.2013.04.002) [DOI] [PubMed] [Google Scholar]
- 65.Kahn A, Cottreau D, Dreyfus JC. 1980. Phosphofructokinase in human fetus. Pediatr. Res. 14, 1162–1167. ( 10.1203/00006450-198011000-00003) [DOI] [PubMed] [Google Scholar]
- 66.Vandenberg LN, Levin M. 2010. Far from solved: a perspective on what we know about early mechanisms of left–right asymmetry. Dev. Dyn. 239, 3131–3146. ( 10.1002/dvdy.22450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pegoraro C, Figueiredo AL, Maczkowiak F, Pouponnot C, Eychene A, Monsoro-Burq AH. 2015. PFKFB4 controls embryonic patterning via Akt signalling independently of glycolysis. Nat. Commun. 6, 5953 ( 10.1038/ncomms6953) [DOI] [PubMed] [Google Scholar]
- 68.Gokey JJ, Dasgupta A, Amack JD. 2015. The V-ATPase accessory protein Atp6ap1b mediates dorsal forerunner cell proliferation and left–right asymmetry in zebrafish. Dev. Biol. 407, 115–130. ( 10.1016/j.ydbio.2015.08.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beyenbach KW, Wieczorek H. 2006. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J. Exp. Biol. 209, 577–589. ( 10.1242/jeb.02014) [DOI] [PubMed] [Google Scholar]
- 70.Holliday LS. 2014. Vacuolar H+-ATPase: an essential multitasking enzyme in physiology and pathophysiology. New J. Sci. 2014, 1–21. ( 10.1155/2014/675430) [DOI] [Google Scholar]
- 71.Poultney CS, et al. 2013. Identification of small exonic CNV from whole-exome sequence data and application to autism spectrum disorder. Am. J. Hum. Genet. 93, 607–619. ( 10.1016/j.ajhg.2013.09.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khurana E, et al. 2013. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science 342, 1235587 ( 10.1126/science.1235587) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maurano MT, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195. ( 10.1126/science.1222794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ward LD, Kellis M. 2012. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science 337, 1675–1678. ( 10.1126/science.1225057) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.