Abstract
Premise of the study:
Here we report the complete chloroplast genome of the important medicinal species Huperzia serrata (Lycopodiaceae) and compare it to the chloroplast genome of the congeneric species H. lucidula.
Methods and Results:
The whole chloroplast genome of H. serrata was sequenced using an Illumina platform and assembled with Geneious version R9.0.5. The genome size of H. serrata was 154,176 bp, with 36.3% GC content. The complete chloroplast genome contained 120 unique genes, including 86 coding genes, four rRNA genes, and 30 tRNA genes. Comparison with the chloroplast genome of H. lucidula revealed three highly variable regions (rps16-chlB, ycf12-trnR, and ycf1) between these two species and 252 mutation events including 27 insertion/deletion polymorphisms and 225 single-nucleotide polymorphisms (SNPs). Ninety-two SNPs were identified in the gene-coding regions. In addition, 18 microsatellite sites were found, which can potentially be used in phylogeographic studies.
Conclusions:
The complete chloroplast genome of H. serrata is reported here, and will be a valuable genome resource for further phylogenetic, evolutionary, and medical studies of medicinal plants in the genus Huperzia.
Keywords: Huperzia serrata, lycophytes, Lycopodiaceae, mutation, next-generation sequencing
The structure of chloroplast genomes in land plants is generally highly conserved in terms of gene order, organization, and content, which makes them suitable for characterizing genetic relationships among species (Bock, 2007). Portions of these genomes have also been widely used by many plant taxonomists as effective DNA barcoding tools. Most of the chloroplast genomes of land plants have a pair of inverted repeats (IRs), separated by one large single copy region (LSC) and one small single copy region (SSC) (Jansen et al., 2005). However, variations occur in certain lineages, and these variations have been proven to be useful in identifying some critical events during the evolution of land plants (Dong et al., 2014; Song et al., 2015). One typical example is the 30-Kb inversion (from trnC to ycf2) detected in the LSC region from bryophytes and lycophytes to other land plants, supporting the hypothesis that lycophytes are a sister clade to all other extant vascular plants (Raubeson and Jansen, 1992).
Compared with those on seed plants, studies on chloroplast genomes of ferns and lycophytes have been relatively sparse (Lu et al., 2015). The North American firmoss Huperzia lucidula (Michx.) Trevis. (Lycopodiaceae) was the first lycophyte species with a complete chloroplast genome sequence (Wolf et al., 2005; GenBank accession no. NC_006861). Because H. lucidula belongs to a significant sister clade of all extant vascular plants, sequencing its complete chloroplast genome facilitates the exploration of the relationships between lycophytes and other vascular plants. Both the rearrangement structure of the chloroplast genome and the phylogenomic analyses of 73 protein-coding genes supported the hypothesis that lycophytes were a sister to both extant fern and seed plant lineages (Wolf et al., 2005).
However, the phylogenetic relationships within this family and particularly within the genus Huperzia Bernh. (ca. 55 species) are still unclear because of insufficient phylogenetic data (Zhang and Iwatsuki, 2013). Here we describe the complete chloroplast genome sequence of a valuable species (H. serrata (Thunb.) Trevis.) within this genus and compare it to existing chloroplast genome data of H. lucidula to better understand the mutation patterns in chloroplast genomes of Huperzia. Both H. lucidula and H. serrata belong to Huperzia sect. Serratae (Rothm.) Holub and form a clade based on matK sequences showing a close phylogenetic relationship (Zhang, 2004; Ji et al., 2007). Furthermore, H. serrata is an important medicinal plant containing huperzine A, which several studies have found to be effective in the treatment of Alzheimer’s disease (Tang, 1996; Wang et al., 1998; Guo et al., 2005). Thus, this draft genome may not only facilitate investigations into genetic variation but also elucidate relationships within the genus to guide further exploration of compounds in closely related species.
METHODS AND RESULTS
Specimens of H. serrata were collected from Helong, Jilin Province, northeastern China. A voucher specimen (X. C. Zhang 6972) has been deposited in the Herbarium of the Institute of Botany, Chinese Academy of Sciences (PE). Total DNA was extracted with a modified cetyltrimethylammonium bromide (CTAB) method (Li et al., 2013). The DNAs were sheared into ∼350-bp fragments using the Covaris M220 focused-ultrasonicator (Covaris, Woburn, Massachusetts, USA). The NEBNext DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, Massachusetts, USA) was used for library construction. Paired-end reads of 2 × 150 bp then were generated using an Illumina HiSeq PE150 (Illumina, San Diego, California, USA). A total of 9,391,796 paired-end sequence reads of 150 bp were generated, of which 406,164 reads belong to the chloroplast genome. The chloroplast genome data were extracted using H. lucidula as a reference and assembled de novo with Geneious version R9.0.5 (Kearse et al., 2012). The first de novo assembly generated eight contigs, and the eight contigs were then extended by mapping raw reads to the contigs several times until all contigs were merged into one whole sequence of 138,841 bp. The four ends of IR regions were located through BLAST with the whole sequence itself to assemble into the complete chloroplast genome sequence. The annotation of all the genes encoding proteins, tRNAs, and rRNAs was constructed with Dual Organellar GenoMe Annotator (DOGMA; Wyman et al., 2004) and was uploaded to GenBank. The tRNAs were further verified using tRNAscan-SE version 1.21 (Lowe and Eddy, 1997; Schattner et al., 2005). The genome map was drawn with OGDraw version 1.2 (Lohse et al., 2007).
The chloroplast genome sequence of H. lucidula was downloaded from GenBank and aligned with H. serrata using MAFFT version 7 (Katoh and Standley, 2013). A sliding window analysis was conducted with DnaSP version 5.1 (Librado and Rozas, 2009) to evaluate the genetic diversity (π) across whole genomes within the genus Huperzia. The window length was set to 600 bp with a 200-bp step size based on the proposed length of DNA barcoding regions (Song et al., 2015). DnaSP was also used for identifying insertion/deletion polymorphisms (indels) with the chloroplast genome of H. lucidula as a reference. A custom Python script (https://www.biostars.org/p/119214/) based on single-nucleotide polymorphism (SNP) definition (a variation in a single nucleotide that occurs at a specific position in the genome) was employed to call SNPs. The SNPs in coding regions were classified in two ways: synonymous and nonsynonymous; transition and transversion. The simple sequence repeats (SSRs) in H. serrata were detected using NWISRL-Imperfect SSR Finder version 1.0 (Stieneke and Eujayl, 2007). The repeats unit length was set to two to nine base pairs with at least five copies for dinucleotide and four copies for other multinucleotide repeats.
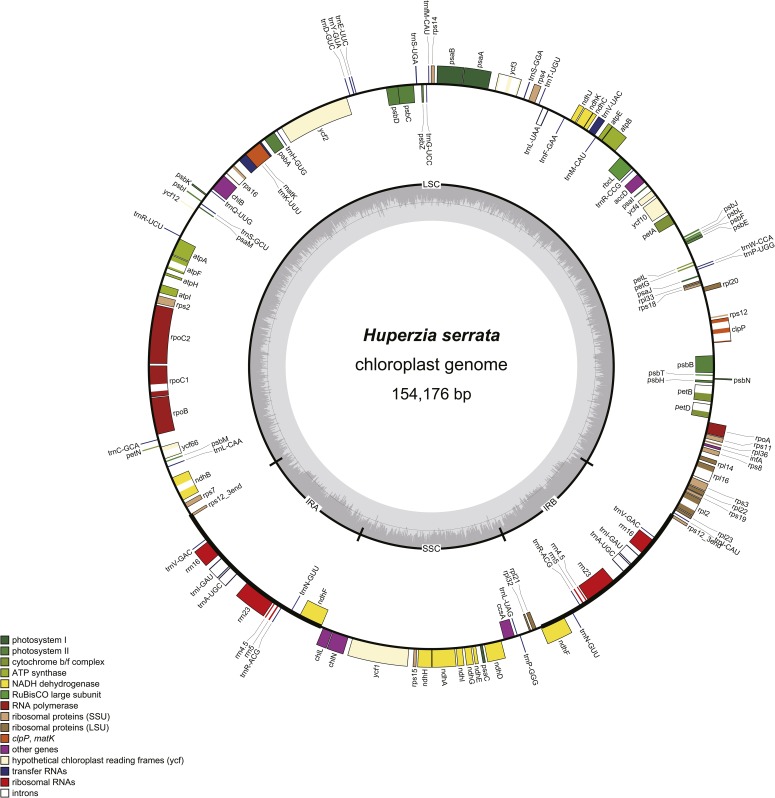
The complete genome sequence of H. serrata (GenBank accession no. KX426071) was 154,176 bp long, 197 bp shorter than that of H. lucidula (154,373 bp; GenBank accession no. NC_006861). Both genomes had GC content of 36.3% (Table 1). A pair of IRs of 30,438 bp was separated by an LSC and a SSC of 104,080 bp and 19,658 bp, respectively, in H. serrata. The complete chloroplast genome contained 120 putative unique genes, including 86 coding genes, four rRNA genes, and 30 tRNA genes. The gene map of H. serrata is shown in Fig. 1. Based on our preliminary analysis, we found that 15 genes have one predicted intron (10 coding genes and five tRNA genes) and two coding genes have two introns (clpP and ycf3). Compared with H. lucidula, we found that the gene order and features are almost identical in genomes of H. serrata. Because comparisons between H. lucidula and other land plants have already been conducted in previous studies (Wolf et al., 2005), we did not repeat the work again. However, some unusual features also existed: an extra tRNA trnI-GAU between rrn16 and trnA-UGC in the IR region, and an intron within ycf66 in the LSC region were first annotated in H. serrata. These three genes were also annotated in the chloroplast genome sequence of another lycophyte plant, Isoetes flaccida A. Braun (GenBank accession no. NC_014675) (Karol et al., 2010). Similar to H. lucidula, nine predicted protein-coding genes (ndhJ, atpI, chlL, ndhH, ccsA, rpl36, ycf1, rps15, ndhD) lack their canonical start codons and/or stop codons at the expected positions. A triplet ACG, which is changed into a start codon by C to U RNA editing, and another triplet CAA, which is changed into a stop codon by C to U RNA editing, appear in the position of the expected start codon and stop codon, respectively (Tsuji et al., 2007). Furthermore, rps16 has two internal stop codons in the chloroplast genome and is therefore considered to be a pseudogene, but the chloroplast transcriptome evidence is needed to prove this hypothesis (Oldenkott et al., 2014).
Table 1.
Summary of Huperzia serrata and H. lucidula chloroplast features.
| Feature | H. lucidula | H. serrata |
| Total cpDNA size | 154,373 | 154,176 |
| LSC | 104,088 | 104,080 |
| SSC | 19,657 | 19,658 |
| IR | 30,628 | 30,438 |
| Total GC content (%) | 36.3 | 36.3 |
| LSC | 34.4 | 34.4 |
| SSC | 32.8 | 32.8 |
| IR | 44.9 | 45.0 |
| Total no. of genes | 119 | 120 |
| Protein encoding | 86 | 86 |
| tRNA | 29 | 30 |
| rRNA | 4 | 4 |
Note: IR = inverted repeat; LSC = large single copy; SSC = small single copy.
Fig. 1.
Gene map of the Huperzia serrata chloroplast reference genome. Genes outside of the outer circle are transcribed clockwise, whereas genes inside the outer circle are transcribed counterclockwise. The colored bars indicate different functional groups. The dashed darker gray area in the inner circle denotes GC content while the lighter gray area shows the AT content of the genome. IR = inverted repeat; LSC = large single copy; SSC = small single copy.
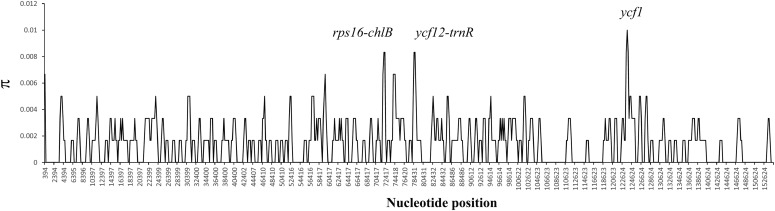
The nucleotide variability (π) of the aligned genome sequences of H. lucidula and H. serrata was calculated with DnaSP version 5.1 to explore the level of sequence divergence. The value varied from 0 to 0.1 with an average of 0.00143, showing that divergence between the genomes of these closely related species is small. However, three highly variable regions—rps16-chlB, ycf12-trnR, and ycf1—were located (Fig. 2). Only ycf1 is in the SSC region; the other two loci were in the LSC region. None of these highly variable regions have been employed in previous phylogenetic analyses of ferns and lycophytes (Kuo et al., 2011; Li et al., 2011). Based on these results, we infer that ycf1, ycf12-trnR, and rps16-chlB (π > 0.008) could be suitable for phylogenetic analyses at the species level.
Fig. 2.
Sliding window analysis of the whole chloroplast genomes of Huperzia serrata and H. lucidula. Window length = 600 bp; step size = 200 bp; x-axis = position of the midpoint of a window; y-axis = value of π of each window.
Eighteen potential SSR motifs were found, and most were located in the intergenic regions of the LSC region (Table 2). Only three types were identified, with the majority belonging to di- and trinucleotide motifs. ACT/TCT and AAT/TAA/TAG/TAT motifs were found among trinucleotide SSRs while only AT/TA motifs were identified for the dinucleotide motifs. Twenty-seven indels were revealed in the comparison between the chloroplast genome sequences of H. serrata and H. lucidula. Most indels ranged from one to nine base pairs in size and were located in noncoding regions, while three indels occurred within the coding region of the rpoC2 gene, with lengths of 24 bp, 30 bp, and 126 bp, respectively (Table 3). The three indels are all deletions in rpoC2 of H. serrata. Ninety-two SNPs, including 75 transitions and 17 transversions in gene-coding regions, and 133 SNPs, including 88 transitions and 45 transversions in noncoding regions, were detected (Table 4). Among gene-coding regions, 36 synonymous and 56 nonsynonymous substitution sites existed in the whole genomes. Thirty-seven out of 86 coding genes have nonsynonymous substitution sites. Among these genes, rpoC2, ycf1, and ycf2 have the most nonsynonymous substitution sites, showing that these three genes may have relatively fast rates of evolution and can be used in phylogenetic analyses.
Table 2.
Location of simple sequence repeats in Huperzia serrata.
| No. | Start | End | Location | Region | Motif | No. of repeats |
| 1 | 11,242 | 11,255 | petB | Intron | TA | 7 |
| 2 | 40,488 | 40,497 | trnF-trnL | Intergenic | TA | 5 |
| 3 | 43,483 | 43,492 | rps4-trnS | Intergenic | AT | 5 |
| 4 | 56,437 | 56,454 | psbD-trnE | Intergenic | AT | 9 |
| 5 | 70,509 | 70,522 | trnK-rps16 | Intergenic | AT | 7 |
| 6 | 74,093 | 74,104 | trnQ-psbK | Intergenic | TA | 6 |
| 7 | 84,161 | 84,178 | atpI-rps2 | Intergenic | TA | 9 |
| 8 | 85,147 | 85,161 | rps2-rpoC2 | Intergenic | TAG | 5 |
| 9 | 87,843 | 87,878 | rpoC2 | CDS | TGCTTCATC | 4 |
| 10 | 93,017 | 93,031 | rpoC1 | CDS | TCT | 5 |
| 11 | 97,444 | 97,457 | trnC-petN | Intergenic | TA | 7 |
| 12 | 99,074 | 99,097 | psbM-trnL | Intergenic | TAT | 8 |
| 13 | 99,938 | 99,947 | trnL-ndhB | Intergenic | TA | 5 |
| 14 | 100,289 | 100,298 | trnL-ndhB | Intergenic | AT | 5 |
| 15 | 121,906 | 121,917 | chlN-ycf1 | Intergenic | TAA | 4 |
| 16 | 127,339 | 127,350 | ycf1-rps15 | Intergenic | ACT | 4 |
| 17 | 130,250 | 130,261 | ndhA | Intron | AAT | 4 |
| 18 | 138,671 | 138,685 | rpl21-ndhF | Intergenic | TAA | 5 |
Note: CDS = coding DNA sequence.
Table 3.
Location of indels in the genomes of Huperzia serrata and H. lucidula.
| No. | Position | Location | Region | Motif | Size (bp) | Directiona |
| 1 | 5862 | rpl14-rps8 | Intergenic | A | 1 | Insertion |
| 2 | 7064 | rpl36-rps11 | Intergenic | G | 1 | Insertion |
| 3 | 9953 | petD | Intron | A | 1 | Insertion |
| 4 | 20,555–20,556 | rpl20-rps18 | Intergenic | GG | 2 | Insertion |
| 5 | 30,794 | psaI-accD | Intergenic | T | 1 | Deletion |
| 6 | 42,082–42,083 | trnL-UAA | Exon | CC | 2 | Insertion |
| 7 | 42,758–42,759 | trnT-rps4 | Intergenic | CC | 2 | Deletion |
| 8 | 44,361–44,363 | trnS-ycf3 | Intergenic | GGG | 3 | Deletion |
| 9 | 44,986–44,989 | ycf3 | Intron | TTC | 3 | Insertion |
| 10 | 51,669–51,673 | psaB-rps14 | Intergenic | GGGGG | 5 | Deletion |
| 11 | 51,749 | psaB-rps14 | Intergenic | G | 1 | Deletion |
| 12 | 58,365 | psbD-trnE | Intergenic | T | 1 | Insertion |
| 13 | 73,875 | chlB-trnQ | Intergenic | T | 1 | Insertion |
| 14 | 75,019 | psbK-psbI | Intergenic | A | 1 | Insertion |
| 15 | 76,144 | psaM-ycf12 | Intergenic | A | 1 | Deletion |
| 16 | 77,128 | ycf12-trnR | Intergenic | A | 1 | Insertion |
| 17 | 77,497–77,505 | ycf12-trnR | Intergenic | CGTAGTATT | 9 | Deletion |
| 18 | 78,117 | ycf12-trnR | Intergenic | G | 1 | Deletion |
| 19 | 81,577 | atpF | Intron | A | 1 | Deletion |
| 20 | 85,338–85,361 | rpoC2 | CDS | TCGGTTGCTTCACCAACAGTTTCC | 24 | Deletion |
| 21 | 85,557–85,586 | rpoC2 | CDS | TTATCACTAGTTTCTTCATCACTAGTTTCT | 30 | Deletion |
| 22 | 88,781–88,906 | rpoC2 | CDS | TTCAAATTCTGTCTGATCTTCTTCTAAAGAAGAATCAAATGATTCAAATTCTGTCTGATCTTCTTCTAAAGAAGAATCAAATGATTCAAATTCTGTCTGATCTTCTTCTAAAGAAGAATCAAATGA | 126 | Deletion |
| 23 | 92,802–92,803 | rpoC1 | Intron | TT | 2 | Insertion |
| 24 | 99,278–99,283 | psbM | CDS | TATTAT | 6 | Deletion |
| 25 | 100,234–100,235 | trnL-ndhB | Intergenic | CC | 2 | Deletion |
| 26 | 104,210 | rps7-rps12 | Intergenic | T | 1 | Deletion |
| 27 | 122,056 | chlN-ycf1 | Intergenic | A | 1 | Insertion |
Note: CDS = coding DNA sequence.
The plastome of Huperzia lucidula was used as a reference.
Table 4.
Comparisons of mutations, number of transitions (Ts) and transversions (Tv), and number of synonymous (S) and nonsynonymous (N) substitutions per gene of Huperzia serrata and H. lucidula.
| Gene type | Gene | Ts | Tv | S | N |
| Photosynthetic apparatus | petB | 1 | 0 | 0 | 1 |
| petD | 1 | 0 | 0 | 1 | |
| petN | 1 | 0 | 1 | 0 | |
| psaA | 0 | 1 | 1 | 0 | |
| psaB | 3 | 0 | 1 | 2 | |
| psbB | 3 | 0 | 1 | 2 | |
| psbD | 1 | 0 | 1 | 0 | |
| Photosynthetic metabolism | atpA | 1 | 0 | 0 | 1 |
| atpB | 3 | 0 | 1 | 2 | |
| atpE | 1 | 0 | 1 | 0 | |
| atpH | 2 | 0 | 2 | 0 | |
| atpI | 2 | 0 | 0 | 2 | |
| ndhA | 0 | 1 | 0 | 1 | |
| ndhB | 1 | 0 | 0 | 1 | |
| ndhC | 1 | 0 | 1 | 0 | |
| ndhF | 2 | 2 | 0 | 4 | |
| ndhG | 1 | 0 | 0 | 1 | |
| ndhH | 1 | 0 | 1 | 0 | |
| ndhK | 1 | 0 | 1 | 0 | |
| rbcL | 2 | 0 | 2 | 0 | |
| Gene expression | rpl21 | 1 | 0 | 1 | 0 |
| rpoB | 5 | 0 | 2 | 3 | |
| rpoC1 | 2 | 0 | 2 | 0 | |
| rpoC2 | 6 | 4 | 4 | 6 | |
| rps11 | 1 | 0 | 1 | 0 | |
| rps12 | 1 | 0 | 1 | 0 | |
| rps7 | 2 | 0 | 0 | 2 | |
| rps8 | 1 | 0 | 1 | 0 | |
| accD | 2 | 0 | 0 | 2 | |
| clpP | 1 | 0 | 0 | 1 | |
| matK | 1 | 1 | 0 | 2 | |
| Other genes | chlB | 2 | 0 | 2 | 0 |
| chlL | 2 | 0 | 0 | 2 | |
| chlN | 2 | 0 | 0 | 2 | |
| ycf1 | 13 | 3 | 6 | 10 | |
| ycf10 | 1 | 0 | 0 | 1 | |
| ycf2 | 4 | 5 | 2 | 7 | |
| Total | 75 | 17 | 36 | 56 |
CONCLUSIONS
Here we report the complete chloroplast genome sequence of H. serrata, an important and widely distributed medicinal plant. Availability of this chloroplast genome sequence and the existing H. lucidula chloroplast genome sequence enable us to evaluate the genome-wide mutational events within the genus Huperzia. The genome arrangement, gene order, gene size, and GC content of H. serrata and H. lucidula are almost identical. Three divergence hotspots (rps16-chlB, ycf12-trnR, and ycf1), 18 SSRs, 27 indels, and 225 SNPs across the whole genome were identified and could provide useful phylogenetic and phylogeographic information for closely related species. Moreover, conserved primers could be designed for the highly variable regions in Huperzia based on these two complete chloroplast genomes.
LITERATURE CITED
- Bock R. 2007. Structure, function, and inheritance of plastid genomes. In R. Bock [ed.], Cell and molecular biology of plastids, 29–63. Springer, Berlin, Germany. [Google Scholar]
- Dong W., Liu H., Xu C., Zuo Y., Chen Z., Zhou S. 2014. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: A case study on ginsengs. BMC Genetics 15: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S.-L., Li P.-L., Fang F., Huang H., Cheng C.-G. 2005. FTIR spectra-principal component analysis of phenetic relationships of Huperzia serrata and its closely related species. Guangpuxue Yu Guangpu Fenxi 25: 693–697. [PubMed] [Google Scholar]
- Jansen R. K., Raubeson L. A., Boore J. L., Chumley T. W., Haberle R. C., Wyman S. K., Alverson A. J., et al. 2005. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods in Enzymology 395: 348–384. [DOI] [PubMed] [Google Scholar]
- Ji S., Pan S., Wang J., Huo K. 2007. Phylogeny relationship and molecular identification of ten Huperzia species (Huperziaceae) based on matK sequences. China Journal of Chinese Materia Medica 32: 1971–1975. [PubMed] [Google Scholar]
- Karol K. G., Arumuganathan K., Boore J. L., Duffy A. M., Everett K. D., Hall J. D., Hansen S. K., et al. 2010. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: Implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evolutionary Biology 10: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., et al. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L.-Y., Li F.-W., Chiou W.-L., Wang C.-N. 2011. First insights into fern matK phylogeny. Molecular Phylogenetics and Evolution 59: 556–566. [DOI] [PubMed] [Google Scholar]
- Li F.-W., Kuo L.-Y., Rothfels C. J., Ebihara A., Chiou W.-L., Windham M. D., Pryer K. M. 2011. rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS ONE 6: e26597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang S., Yu J., Wang L., Zhou S. 2013. A modified CTAB protocol for plant DNA extraction. Chinese Bulletin of Botany 48: 72–78. [Google Scholar]
- Librado P., Rozas J. 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics (Oxford, England) 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- Lohse M., Drechsel O., Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Current Genetics 52: 267–274. [DOI] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. M., Zhang N., Du X. Y., Wen J., Li D. Z. 2015. Chloroplast phylogenomics resolves key relationships in ferns. Journal of Systematics and Evolution 53: 448–457. [Google Scholar]
- Oldenkott B., Yamaguchi K., Tsuji-Tsukinoki S., Knie K., Knoop V. 2014. Chloroplast RNA editing going extreme: More than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 20: 1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raubeson L. A., Jansen R. K. 1992. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science 255: 1697–1699. [DOI] [PubMed] [Google Scholar]
- Schattner P., Brooks A. N., Lowe T. M. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Research 33: W686–W689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Dong W., Liu B., Xu C., Yao X., Gao J., Corlett R. T. 2015. Comparative analysis of complete chloroplast genome sequences of two tropical trees Machilus yunnanensis and Machilus balansae in the family Lauraceae. Frontiers in Plant Science 6: 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieneke D., Eujayl I. 2007. Imperfect SSR Finder, Version 1.0. USDA-ARS, Northwest Irrigation and Soils Research Lab, Kimberly, Idaho, USA. Website http://ssr.nwisrl.ars.usda.gov/ [accessed 14 October 2016].
- Tang X.-C. 1996. Huperzine A (shuangyiping): A promising drug for Alzheimer’s disease. Acta Pharmacologica Sinica 17: 481–484. [PubMed] [Google Scholar]
- Tsuji S., Ueda K., Nishiyama T., Hasebe M., Yoshikawa S., Konagaya A., Nishiuchi T., Yamaguchi K. 2007. The chloroplast genome from a lycophyte (microphyllophyte), Selaginella uncinata, has a unique inversion, transpositions and many gene losses. Journal of Plant Research 120: 281–290. [DOI] [PubMed] [Google Scholar]
- Wang L., Ji X., Wong Q., Yang J. 1998. Observations of 36 cases on Alzheimer’s disease treatment using Huperzine A. Journal of Nantong Medical College 18: 486–488. [Google Scholar]
- Wolf P. G., Karol K. G., Mandoli D. F., Kuehl J., Arumuganathan K., Ellis M. W., Mishler B. D., et al. 2005. The first complete chloroplast genome sequence of a lycophyte, Huperzia lucidula (Lycopodiaceae). Gene 350: 117–128. [DOI] [PubMed] [Google Scholar]
- Wyman S. K., Jansen R. K., Boore J. L. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics (Oxford, England) 20: 3252–3255. [DOI] [PubMed] [Google Scholar]
- Zhang L. B., Iwatsuki K. 2013. Lycopodiaceae. In Z. Y. Wu, P. H. Raven, and D. Y. Hong [eds.], Flora of China, vol. 2–3, 37–66. Science Press, Beijing, China, and Missouri Botanical Garden Press, St. Louis, Missouri, USA. [Google Scholar]
- Zhang L. B. 2004. Huperziaceae. In C. Y. Wu [ed.], Flora Reipublicae Popularis Sinicae, vol. 6, 86–219. Science Press, Beijing, China. [Google Scholar]