Abstract
Embryonic stem cells (ESCs) and the post-implantation epiblast stem cells (EpiSCs) portray two different states of pluripotency. They differ with respect to epigenetic signatures, dependency of growth factor signaling circuit and cell morphology. They are interconvertible, however, with poor reconversion efficiency. This is indicative of existence of other unknown regulatory pathways govern developmental stage transition. Zhang and colleagues have recently demonstrated that pharmacological inhibition of MLL1 histone methyltransferase is casually linked to efficient reprogramming of EpiSCs to developmentally competent ESCs. MLL1 controlled H3K4me1 serves as an epigenetic valve that ensures maintenance of EpiSCs. Removing this barrier leads to global redistribution of H3K4me1 at enhancers and target gene promoters, in turn represses EpiSC specific genes and reactivates ESC specific transcriptional network. This study underscores the critical role of MLL1 in establishing discrete chromatin states indispensible for early mammalian developmental events.
Keywords: Epiblast stem cells (EpiSC), H3K4me, MLL1, MM-401
Covalent modification of histone 3 by methylation on lysine 4 residues (H3K4me) is an evolutionary conserved epigenetic modification in eukaryotes (1). H3K4 can be mono-, di-, and tri-methylated and is generally found to be associated with transcriptionally permissive chromatin and developmentally regulated bivalent promoters (2-4). Depending on domain of chromatin, the type of methylation varies. For example, H3K4 trimethylation (H3K4me3) is associated primarily with the promoter region whereas H3K4 monomethylation (H3K4me1) usually marks the regulatory enhancers (4,5). While MLL1/2 catalyzes H3K4me3, bulk of H3K4me1 is catalyzed by MLL3/4. MLL methyltransferase belongs to the complex of proteins associated with Set1 (COMPASS) family of proteins and are highly conserved from yeast to mammals. While in budding and fission yeast only one member is found, mammals have evolved six distinct SET1/MLL complexes viz., SET1A and SET1B, MLL1-2 (homolog of Drosophila Trx) and MLL3-4 (homolog of Drosophila Trr) collectively known as MLL family proteins (6). Since the basic function of these enzymes is H3K4 methylation, the existence of multiple SET1/MLL variants raises the issue of functional overlap and redundancy among these enzymes. However, studies in mll knockouts mice have shown that deletion of one set of enzyme could not be compensated by others. Moreover, all individual set1/mll knockout mice show embryonic lethality (1,5,7). These studies established two important facts (I) MLL complexes have non-redundant functions and (II) Enzymatic activity (H3K4me) of MLL complexes is absolutely crucial for mammalian development.
The roles of MLL complexes—particularly MLL1—in metazoan development and diseases are well documented. Although Mll1 homozygous mice exhibit embryonic lethality, deletion of MLL1 in ESCs had no significant influence on their self-renewal (8). The exact role of MLL1 and H3K4me in pluripotent stem cells therefore remained incompletely understood. Recently, Zhang et al. (9) unraveled a new role of MLL1 in early developmental processes. By employing a MLL1 specific small molecule inhibitor MM-401, they found that MLL1 inhibition is sufficient to reprogram primed epiblast stem cells (EpiSC) to naïve pluripotent embryonic stem cells (ESC).
After several rounds of asymmetric cell division of blastomeres, mammalian zygote forms blastocyst. This structure consists of outer trophoblasts and inner cell mass (ICM) in the inside. The ICM further produces the hypoblast (differentiates into extraembryonic lineages) and the epiblast. ESCs derived from this stage of epiblast cells are known as naïve epiblast cells (henceforth as ESCs), which are considered as developmental ground state and can differentiate to all cell lineages (10). EpiSCs are derived from ESCs at post-implantation stage and are an in vitro equivalent of primed epiblast cells. EpiScs are also known as primed epiblast cells, although pluripotent, they differ from ESCs with respect to their potency to form germline, epigenetic state, cell morphology, etc. (10). Lack of inactivated X-chromosome is another hallmark of ESCs but not of EpiSCs. As naïve epiblast cells or ESCs represent a developmental ground state and rich source of true pluripotency, many successful efforts (such as overexpression of Klf2 and Nanog) have been made to revert the EpiSC into ESCs (11,12). The reprogramming efficiency of these methods however remained very poor even in the presence of leukemia inhibitory factor (LIF) and 2i (GSK3 and ERK inhibitor). The very fact of this poor reprogramming efficiency points to an alternative mechanism associated with this process.
Recent studies by Zhang et al. (9) demonstrated that MLL1 expression is regulated during ESC differentiation to EpiSCs. Expression of MLL1 showed positive correlation with the expression of EpiSC markers such as Fgf5 and a reverse correlation with the expression of ESC markers such as Nanog. Pharmacological inhibition or Mll1 knock out was sufficient to reprogram the primed EpiSCs to naïve pluripotent ESCs within 6 passages of cell culture in presence of the inhibitor. The robust reprogramming event was true for mice across different genetic backgrounds and gender. Among many signatures of naïve pluripotent state, they observed reappearance of intense AKP (alkaline phosphatase) staining and dome shaped cell colonies with compact cells in the center of the colony. In an elegant approach by using female mouse EpiSC line containing polymorphic X chromosome, they could also demonstrate reactivation of X chromosome in EpiSCs upon MLL1 inhibition.
To further clarify how MLL1 inhibition resulted in reprogramming they performed gene expression analysis. Their finding showed MLL1i-rESCs (MLL1 inhibited EpiSCs reverted into ESCs) displayed molecular features reminiscent of ground state ESCs, that is higher expression of naïve specific markers such as klf2/4, tbx3, Sox2 and Nanog and lower expression of epiblast markers such as Fgf5, Wnt8a. Moreover, ChIP-seq revealed that MLL1 binding was reduced to its target lineage specific transcription factors such as Klf5. This was in line with the observation by Cao et al. (13) that MM-401 disrupts MLL1 complex. Consequently, they could also observe a strong reduction in H3K4me1—not H3K4me3—marks at MLL1 target genes and thus rapid down regulation of MLL1 target genes. This altered status of MLL1 function represses characteristic molecular features of EpiSCs and gained a transcriptional signature resembles more to ESCs. These observations directly point toward a scenario where MLL1-mediated epigenetic changes have decisive roles in gaining naïve pluripotent state.
One of the fascinating aspects of their study is the developmental competency of the MLL1i-rESCs as these reverted cells could successfully generate teratomas containing well-differentiated cells of all three germ layers. Besides, the ability of MLL1i-rESCs to give rise to chimeric mice underscores that MLL1 inhibition could efficiently reprogram them to authentic naïve pluripotent stem cell. Once the altered epigenetic event is initiated by MLL1 inhibition it can be inherited stably into future generations of ESCs. This is corroborated nicely by the fact that the reprogrammed status in MLL1i-rESCs could be maintained stably for at least 30 passages even after withdrawal of MM-401 from the culture medium. It indicates that MLL1 is an epigenetic gatekeeper supervising histone monomethylation in order to maintain primed pluripotent state (Figure 1). This would in turn drive further steps of embryogenesis. Interestingly, Mll1 knock out mice were able to develop till mid to late gestation (8). This argues that, although the loss of MLL1 is dispensable for early developmental events prior to formation of EpiSCs, it is however required to drive the later stages. In this context, it was proposed that MLL2 not MLL1 is the major methyltransferase during early embryonic development before the blastocyst stage, require to set up naïve epigenome (14,15).
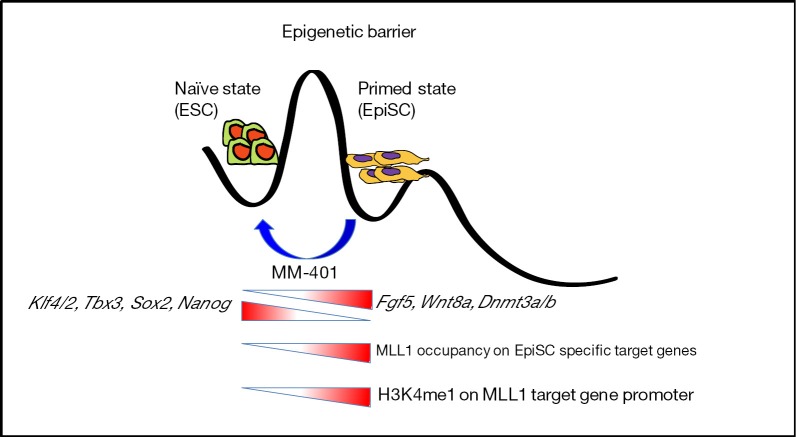
Figure 1.
The cartoon depicts two distinct states of pluripotency (naïve and primed) separated by epigenetic barrier resulted from distinct epigenetic events. This ensures the identity of each states characterized by lineage specific transcription factor expression and H3K4me1 on MLL1 target promoters. Pharmacological inhibition of MLL1 erases epigenetic script of EpiSCs and cross the barrier thereby reprogrammed into developmental ground state ESCs.
Since loss of H3K4me1 at promoter regions was casually linked with lower expression of EpiSCs specific genes, it would be intriguing to address the epigenetic alteration on ESC specific genes that were re-expressed in MLL1i-rESCs. Along this line, it would be interesting to check if the former event may be linked with the activation of another COMPASS family of histone methyltransferase targeting to those re-activated genes. This argument is based on the fact that MLL2 was assigned as a major methyltransferase in mouse ESCs (16). In a different context, enhancer and promoter H3K4me1 has been shown to be primarily catalyzed by MLL3/4 and promoter monomethylation maintains a repressive transcriptional output in progenitor myoblast cells (17). MLL3/4-mediated repression was found to act as an epigenetic checkpoint that prevents premature differentiation from myoblasts to myotubes. In this light, the role of MLL3/4 in the context of EpiSC reprogramming would be an interesting scenario that requires further investigations.
Finally, another crucial fact emerging from this study (9) concerns the homogeneity of reversion. Earlier manipulations generated only 1–5% conversion rate, compared to nearly 50% homogenous reprogrammed cells into ESCs after pharmacological inhibition of MLL1. Thus, this study offered developmental biologist a new tool to obtain invaluable source of highly homogenous ground state pluripotent stem cells.
Acknowledgements
Funding: This work was funded by the SFB 815/1177 and LOEWE Ub-Net.
Footnotes
Provenance: This is a Guest Perspective commissioned by Editor-in-Chief Zhizhuang Joe Zhao (Pathology Graduate Program, University of Oklahoma Health Sciences Center, Oklahoma City, USA).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 2006;75:243-69. 10.1146/annurev.biochem.75.103004.142422 [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006;125:315-26. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- 3.Azuara V, Perry P, Sauer S, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol 2006;8:532-8. 10.1038/ncb1403 [DOI] [PubMed] [Google Scholar]
- 4.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007;39:311-8. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- 5.Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016;352:aad9780. 10.1126/science.aad9780 [DOI] [PubMed] [Google Scholar]
- 6.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 2012;81:65-95. 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu BD, Hess JL, Horning SE, et al. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 1995;378:505-8. 10.1038/378505a0 [DOI] [PubMed] [Google Scholar]
- 8.Ernst P, Mabon M, Davidson AJ, et al. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr Biol 2004;14:2063-9. 10.1016/j.cub.2004.11.012 [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Gayen S, Xiong J, et al. MLL1 Inhibition Reprograms Epiblast Stem Cells to Naive Pluripotency. Cell Stem Cell 2016;18:481-94. 10.1016/j.stem.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell 2009;4:487-92. 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- 11.Bao S, Tang F, Li X, et al. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 2009;461:1292-5. 10.1038/nature08534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han DW, Tapia N, Joo JY, et al. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell 2010;143:617-27. 10.1016/j.cell.2010.10.015 [DOI] [PubMed] [Google Scholar]
- 13.Cao F, Townsend EC, Karatas H, et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell 2014;53:247-61. 10.1016/j.molcel.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser S, Lubitz S, Loveland KL, et al. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin 2009;2:5. 10.1186/1756-8935-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaser S, Schaft J, Lubitz S, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 2006;133:1423-32. 10.1242/dev.02302 [DOI] [PubMed] [Google Scholar]
- 16.Denissov S, Hofemeister H, Marks H, et al. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 2014;141:526-37. 10.1242/dev.102681 [DOI] [PubMed] [Google Scholar]
- 17.Cheng J, Blum R, Bowman C, et al. A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol Cell 2014;53:979-92. 10.1016/j.molcel.2014.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]