From mouse pre-implantation and early post-implantation embryos, stem cells at different pluripotent states can be captured in in vitro culture. For example, naïve embryonic stem cells (ESC) are derived from inner cell mass (ICM) of the blastocyst, while primed epiblast stem cells (EpiSC) are captured from the post-implantation embryo epiblast. Though both ESCs and EpiSCs are pluripotent stem cells, they have distinct characters in terms of pluripotent gene expression profile, epigenetic status, metabolic pathways, growth factor requirement, and in female, the X chromosome inactivation status (1). On the other hand, these two pluripotent states are interchangeable, once switching the culture condition of ESCs to that for EpiSCs, naïve ESCs convert to EpiSCs. The efficiency of converting EpiSCs to ESCs, however, is much low, which usually requires over-expressing pluripotency-associated genes such as Nanog, Klf4, Esrrb, Tfcp2l1 and Nr5a2 (2-6). Similar to somatic cell reprogramming to induced pluripotent stem cells, converting EpiSCs to ESCs is an epigenome-resetting process. Characterizing these epigenetic barriers represents an important approach to further improve reprogramming efficiency (7).
In the April issue of Cell Stem Cell, Zhang et al. discovered that histone H3K4 methyltransferase MLL1 is one of the major barriers that hinders EpiSCs conversion to ESCs (8). First, they analysed the expression of MLL family members during ESC differentiation, and found that expression of MLL1 was particularly up-regulated and correlated with the expression of epiblast markers, Fgf5, Cer1, etc. To understand the biological role of MLL1 during ESC differentiation, they used an inhibitor of MLL1, MM-401 in the study. Treatment of ESCs with MM-401 delayed conversion to EpiSCs, implicating MLL1’s functional roles in this process.
Next, they examined the role of MLL1 in the conversion of EpiSCs to ESCs by MLL1 inhibition. Surprisingly, when EpiSCs were cultured in MM-401 with either LIF/KSR or bFGF/KSR for 72 h, the colonies acquired dome shaped morphology similar to ESCs, and stable ESC like lines were established with continued culture. Remarkably, after 72 h treatment of MM-401, 49.1% and 32.0% of cells became PECAM1+ in LIF/KSR and bFGF/KSR, respectively, demonstrating high conversion efficiencies. Genetically, deletion of Mll1 in EpiSCs or knocking down Mll1 robustly induced ESCs reversion.
In female pluripotent cells, X chromosome reactivation is one of the important criteria to differentiate naïve and primed pluripotent stem cells. Both X chromosomes are activated in ESCs, while in EpiSCs, one X chromosome is randomly inactivated (1). To assess MLL1 inhibition on X chromosome reactivation, the authors used two EpiSC lines, one is F1 hybrid XLabXJF1 EpiSC with XLab harbors GFP transgene and truncated Tsix and wild-type XJF1 (12F); the other line has two wild type X chromosomes (9F). Upon MLL1 inhibition, at day 3, about 45% and 30% of colonies are GFP+ in LIF/KSR and bFGF/KSR, respectively, similar to the numbers by PECAM1 staining. For 12F EpiSCs, RNA-FISH showed loss of Xist coating, bi-allelic expression of X-linked genes in the converted ESCs (MLL1-rESCs), indicating the successful conversion.
The converted MLL1-rESCs were pluripotent. First, they formed mature teratomas. And then when injected into blastocysts, these cells contributed to ICM and to the germline in the chimeras.
How did MLL1 inhibition lead to the high efficient conversion from EpiSCs to ESCs? To understand the mechanism, the authors performed transcriptomic analysis. MM-401 treatment induced rapid changes of the transcriptome, which eventually became similar to that of ESCs when the conversion was complete. ChIP-seq analysis identified that the majority of MLL1 binding sites in EpiSCs were in intergenic regions or introns, indicating that MLL1 functions by a mechanism via regulatory elements. Furthermore, the authors found that there were substantial differences between ESCs and EpiSCs for H3K4me1 sites (usually marking enhancers) in the genome. By combining RNA-Seq and CHIP-Seq data, they identified potential MLL1 target genes enriched with those involved in cell adhesion and development processes. Surprisingly, MLL1 did not appear to directly regulate the known pluripotency genes, demonstrating that repressing EpiSC features is a major mechanism for MLL1 inhibition induced EpiSC to ESC conversion.
The standard human embryonic stem cells (hESCs) are more similar to mEpiSCs than mESCs (9). Recently, several human naïve ESC lines have been established and characterized (10-12). It’ll be interesting to examine MLL1 expression and its genome wide binding profiles in human pluripotent cells, and to test whether MLL1 inhibition in hESCs can also facilitate the conversion of hESCs to naïve hESCs. Besides its role in the conversion of different pluripotent cells, it’s important to investigate whether MLL1 is involved in reprogramming mouse and human somatic cells to induced pluripotent stem cells (iPSC) by Yamanaka factors, where epigenetic resetting has an essential role (13). What are the main targets of MLL1 in somatic cells, e.g., fibroblast? Are lineage specifiers regulated by MLL1? Two of the Yamanaka factors, Oct4 and Sox2 are known to down-regulate ectodermal (ECT) and mesendodermal (ME) genes, respectively (14), will MLL1 inhibition replace one or both of these factors? Also, lineage specifiers of ECT and ME can induce pluripotency without Oct4 and Sox2 (14,15), in this milieu, will MLL1 inhibition block the reprogramming?
Overall, to convert cells from one state to another, the epigenetic barriers in the cells must be overcome to turn off the genes of the parental cell identity and to turn on those ones for establishing and maintaining the new cell identity. Zhang et al. set a good example to prove that by simply inhibiting the activities of an epigenetic modifier, the pluripotent state can be efficiently changed as summarized in Figure 1. It is anticipated that more epigenetic modifiers will be identified for their functions in the switching of cell states.
Figure 1.
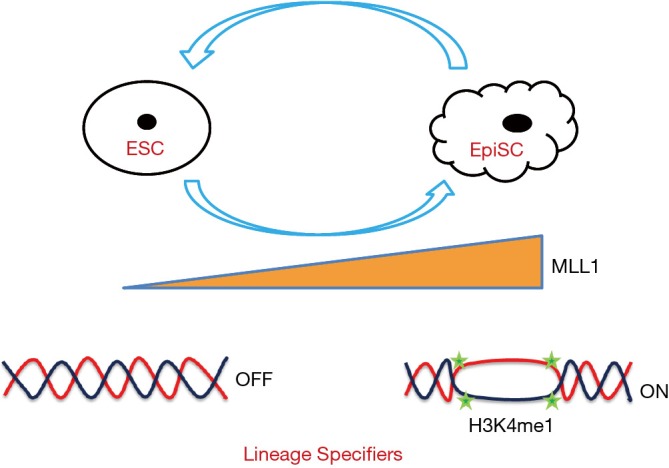
MLL1 in switching of pluripotent states. MLL1 is up-regulated when ESCs are differentiated. Inhibition of MLL1 in EpiSCs affects deposition of H3K4me1 on enhancers of lineage specifiers and facilitates the conversion to ESCs. ESC, embryonic stem cell; EpiSC, epiblast stem cell.
Acknowledgements
None.
Footnotes
Provenance: This is a Guest Commentary commissioned by Editor-in-Chief Zhizhuang Joe Zhao (Pathology Graduate Program, University of Oklahoma Health Sciences Center, Oklahoma City, USA).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell 2009;4:487-92. 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- 2.Silva J, Nichols J, Theunissen TW, et al. Nanog is the gateway to the pluripotent ground state. Cell 2009;138:722-37. 10.1016/j.cell.2009.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo G, Yang J, Nichols J, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 2009;136:1063-9. 10.1242/dev.030957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Festuccia N, Osorno R, Halbritter F, et al. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell 2012;11:477-90. 10.1016/j.stem.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye S, Li P, Tong C, et al. Embryonic stem cell self-renewal pathways converge on the transcription factor Tfcp2l1. EMBO J 2013;32:2548-60. 10.1038/emboj.2013.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo G, Smith A. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development 2010;137:3185-92. 10.1242/dev.052753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillich A, Bao S, Grabole N, et al. Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell 2012;10:425-39. 10.1016/j.stem.2012.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Gayen S, Xiong J, et al. MLL1 Inhibition Reprograms Epiblast Stem Cells to Naive Pluripotency. Cell Stem Cell 2016;18:481-94. 10.1016/j.stem.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007;448:196-9. 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- 10.Gafni O, Weinberger L, Mansour AA, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013;504:282-6. 10.1038/nature12745 [DOI] [PubMed] [Google Scholar]
- 11.Theunissen TW, Powell BE, Wang H, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014;15:471-87. 10.1016/j.stem.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo G, von Meyenn F, Santos F, et al. Naive Pluripotent Stem Cells Derived Directly from Isolated Cells of the Human Inner Cell Mass. Stem Cell Reports 2016;6:437-46. 10.1016/j.stemcr.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polo JM, Anderssen E, Walsh RM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012;151:1617-32. 10.1016/j.cell.2012.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu J, Wu C, Wu Y, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell 2013;153:963-75. 10.1016/j.cell.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montserrat N, Nivet E, Sancho-Martinez I, et al. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell 2013;13:341-50. 10.1016/j.stem.2013.06.019 [DOI] [PubMed] [Google Scholar]


