Abstract
Brain glycogen and its metabolism are increasingly recognized as major players in brain functions. Moreover, alteration of glycogen metabolism in the brain contributes to neurodegenerative processes. In the brain, both muscle and brain glycogen phosphorylase isozymes regulate glycogen mobilization. However, given their distinct regulatory features, these two isozymes could confer distinct metabolic functions of glycogen in brain. Interestingly, recent proteomics studies have identified isozyme-specific reactive cysteine residues in brain glycogen phosphorylase (bGP). In this study, we show that the activity of human bGP is redox-regulated through the formation of a disulfide bond involving a highly reactive cysteine unique to the bGP isozyme. We found that this disulfide bond acts as a redox switch that precludes the allosteric activation of the enzyme by AMP without affecting its activation by phosphorylation. This unique regulatory feature of bGP sheds new light on the isoform-specific regulation of glycogen phosphorylase and glycogen metabolism.
Keywords: allosteric regulation, brain metabolism, carbohydrate metabolism, disulfide, glycogen, glycogen storage disease, redox regulation
Introduction
Glycogen is the main carbohydrate store in mammalian cells. In the human body, glycogen is found in the liver, the muscles, and the brain (1). Brain glycogen is primarily located in glial cells, especially in the astrocytes, but also to a lesser extent in the neurons (2, 3). For many decades, brain glycogen has been assigned to an emergency store of glucose because it is found at relatively low concentrations when compared with those found in the liver and the muscles. However, over the past 20 years, important progress has been made in revealing the critical role of brain glycogen in brain function. Indeed, astrocytic and neuronal activities are highly dependent on glycogenolysis (glycogen mobilization) and glycogen store during stress episodes, including hypoglycemia and ischemia (4–7). In addition, astrocytic glycogen sustains synaptic plasticity and is critical for high cognitive processes such as learning and memory consolidation (8, 9). Indeed, astrocytic glycogen has been shown to provide energy for K+ uptake and substrates for the synthesis of neurotransmitters (such as glutamate or GABA) and is used as a lactate supply to feed the astrocyte-neuron lactate shuttle (8). The alteration of glycogen metabolism and the subsequent glycogen accumulation in neurons have been assigned to neurodegeneration and the pathogenesis of Lafora disease (10). Such accumulation has also been observed in aging brain as well as in neurodegenerative diseases, including Alzheimer's disease or amyotrophic lateral sclerosis (11, 12). Glycogen phosphorylase (GP)2 is a key enzyme of glycogen mobilization. GP catalyzes the breakdown of glycogen in glucose 1-phosphate, also termed as glycogenolysis. This enzyme is found in humans as three isozymes, named after the tissue they predominate: the muscle (mGP), the liver (lGP), and the brain GP (bGP). In the brain, astrocytes express both the brain and the muscle isoform, whereas neurons only express the bGP (3, 13). GPs are allosteric enzymes that oscillate between an inactive T-state and an active R-state. The allosteric transition is mediated by phosphorylation of Ser14 in response to extracellular signals, such as norepinephrine or serotonin. Additionally, GP enzymes are also regulated by the binding of allosteric effectors, such as AMP (allosteric activator) and ATP and glucose 6-phosphate (allosteric inhibitors), allowing the enzyme to respond to the local energy requirements of the cell (1, 14).
Reactive oxygen species (ROS) are reactive compounds derived from molecular oxygen. In cells, most of the ROS are formed as a byproduct of the mitochondrial chain or are produced by enzymes including NADPH oxidase and xanthine oxidase (15–17). In normal conditions, ROS are mediators of intracellular signaling, through the reversible modification of proteins. The tight regulation of intracellular oxidant levels is allowed by antioxidant systems, including GSH and enzymatic systems such as superoxide dismutase (SOD) and catalase (15–17). However, the disruption of the oxidant/antioxidant balance and the resulting oxidative stress lead to DNA and protein damage, which contributes to cellular senescence, apoptosis, and tumorigenesis (16, 18). Hydrogen peroxide (H2O2) is a cellular oxidant involved in both the regulation of cell functions and the pathogenesis of several diseases, including cancer, neurodegenerative diseases, and aging (16). Proteins are primary targets of H2O2, particularly through the covalent modification of cysteine residues. In the presence of oxidants, cysteines can be oxidized to form sulfenic acid (−SO). Sulfenic acid is generally unstable and is converted to a disulfide in the presence of another thiol group. In the presence of high concentrations of oxidants, sulfenic acid may also undergo further oxidation and thus form a sulfinic (−SO2) or sulfonic (−SO3) acid (15, 19). Many studies aim to understand the impact of cysteine oxidation on protein functions to rapidly adjust their activity to the redox environment (20, 21). Furthermore, the development of new trapping techniques combined with high-throughput proteomics methods allows the identification of potential proteins, presenting reactive cysteine residues and thiol modifications in oxidative conditions. Energy metabolism enzymes are one of the main classes of proteins that contain highly reactive cysteines. Among them, several studies identified critical cysteine residues from the brain form of GP (22–26).
In the present study, we investigated the redox regulation of bGP. By using molecular, cellular, and in silico modeling approaches, we demonstrate that bGP is reversibly inhibited by H2O2. This inhibition occurs through the formation of an intramolecular disulfide bond involving Cys318 and Cys326, a reactive cysteine residue unique to the brain isoform of GPs. These two cysteines are located in the adenine loop, which belongs to the AMP-binding site (allosteric site). Interestingly, we found that the formation of the Cys318–Cys326 disulfide bond alters the AMP-dependent activation of the bGP without affecting the phosphorylation-dependent activation. Our results suggest that the brain isoform of GP is regulated by H2O2 and more broadly by redox conditions, through the formation of an isoform-specific disulfide bond in the AMP-binding site. This unique feature of bGP sheds new light on the isoform-selective regulation of glycogen phosphorylase activities and glycogen metabolism in the brain.
Results
Human bGP Is Reversibly Inhibited by Bolus Addition and by Continuous Generation of H2O2
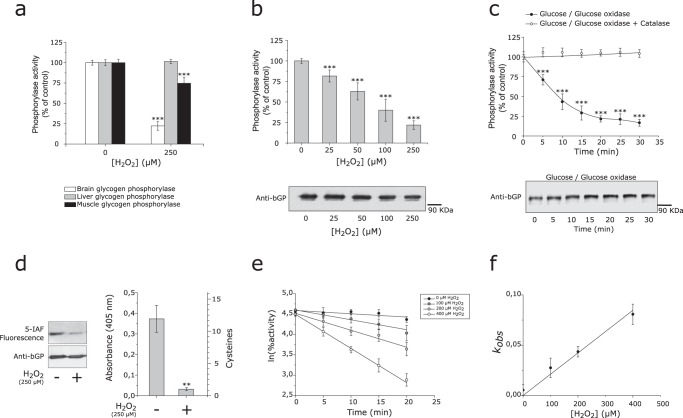
Recent proteomics studies identified putative reactive cysteine residues in human bGP isoenzyme, thus suggesting possible redox regulation of the activity of this key brain metabolic enzyme (22, 23, 26). Interestingly, it has been shown that oxidative stress and redox conditions in astrocytes could impact glycogen mobilization (3, 7, 27). As both mGP and bGP are expressed in the brain (in particular in astrocytes), we first tested whether bGP and mGP activity could be inhibited by H2O2. Although bGP was strongly inhibited by short exposure to 250 μm H2O2 (more than 80% inhibition), mGP activity was significantly less affected (Fig. 1a), which is in agreement with previous studies on mGP (40). In the same conditions, lGP isozyme was found not to be affected by exposure to H2O2 (Fig. 1a). These results are consistent with different proteomics studies showing reactive cysteine residues prone to oxidation and unique to the bGP isoform (22, 23, 26). We further investigated the inhibition of bGP by H2O2 by exposing the purified enzyme to a bolus of this oxidant at different concentrations (0–250 μm final concentration). As shown in Fig. 1b, bGP was significantly inhibited by H2O2 in a dose-dependent manner (Fig. 1b), with over 80% inhibition obtained with 250 μm H2O2. To make a more realistic and physiologic assessment of the effect of H2O2 on bGP activity, the purified enzyme was exposed to a continuous flux of H2O2 produced by the glucose/glucose oxidase system (27–29). A constant flux of 6 μm/min of H2O2 was used, which is considered as physiologically relevant (27, 28). bGP activity was found to be inhibited in a time-dependent manner with a residual activity of 20% after 20 min of exposure (Fig. 1c). The inhibition of bGP by the glucose-glucose oxidase system was completely avoided by the presence of catalase, thus demonstrating that bGP inhibition is due to the flux of H2O2 (Fig. 1c).
FIGURE 1.
Human bGP is inhibited by H2O2 through modification of cysteine residues. a, purified recombinant human bGP and lGP (white and gray bars) or purified muscle glycogen phosphorylase (mGP) (rabbit) (black bars) were incubated with or without 250 μm H2O2 for 30 min at 37 °C, prior to residual activity measurement. mGP and bGP activity was measured using AMP as an activator, whereas lGP, which only responds to phosphorylation, was phosphorylated by phosphorylase kinase for activation prior to activity measurement. Results are expressed as the percentage of the control without H2O2. Data represent mean values of three independent experiments ± S.D., ***, p < 0.001 when compared with control (no H2O2). b, reduced recombinant bGP was incubated with different concentrations of H2O2 (0–250 μm) for 30 min at 37 °C. Residual activity was assayed, and aliquots were also subjected to Western blotting analysis under non-reducing conditions. bGP was revealed using specific antibodies. Results are expressed as the percentage of the control. Data represent mean values of three independent experiments ± S.D., ***, p < 0.001 when compared with positive control. c, reduced recombinant bGP was incubated with glucose oxidase (1.2 units) and glucose (5 mm) for 30 min. In these conditions, H2O2 was continuously generated at a rate of 6 μm/min. An aliquot was removed every 5 min and assayed for bGP residual activity (filled circles). A control reaction was carried out in the presence of 300 units/ml catalase (open circles). Glucose, glucose oxidase, and catalase, independently, had no effect on bGP activity. Samples were analyzed by Western blotting, and bGP was revealed using specific antibodies. Results are expressed as the percentage of the control. Data represent mean values of three independent experiments ± S.D., ***, p < 0.001 when compared with t0. d, to confirm the oxidation of cysteine residues upon exposure to H2O2, recombinant bGP was inhibited by H2O2, and free cysteine residues were specifically labeled using the fluorescent probe 5-IAF. Samples were run on SDS-PAGE in the presence of 2-mercaptoethanol and blotted onto nitrocellulose membrane. 5-IAF fluorescence was measured (λex: 492 nm; λem: 520 nm), and bGP was revealed using specific antibodies. The untreated bGP was used as a positive control. To assess the number of oxidized cysteines after exposure to H2O2, thiol content was analyzed using the DTNB assay on both treated and untreated bGPs, as described under “Experimental Procedures.” The resulting TNB− formation was quantified by absorbance measurement at 405 nm. Absorbance of the non-treated protein was used as control. Data represent mean values of three independent experiments ± S.D., **, p < 0.01 when compared with reduced control. e, recombinant bGP was incubated with various concentrations of H2O2 (0–400 μm H2O2). Aliquots were removed every 5 min and assayed for residual activity. For each concentration of H2O2, the plot of the natural logarithm as a function of time allowed the determination of first-order apparent constants kobs. f, the second-order rate constant was then determined by plotting the first-order apparent constants against [H2O2] (lower panel). The solid lines represent the best linear regression fit of the data to Equations 2 and 3. The calculated kinact for the inhibition of bGP by H2O2 was 185 m−1 min−1. Data represent mean values of three independent experiments ± S.D.
As stated above, cysteine residues in proteins can be oxidized by H2O2 to a different set of reversibly or irreversibly oxidized forms (disulfide, sulfenic, sulfinic, and sulfonic acid forms) that can have an impact on protein functions (15, 20). Fluorescein 5-iodoacetamide (5-IAF) and 5,5′-dithiobis-2-nitrobenzoic acid (DTNB), two alkylating agents that covalently modify free thiol groups, were used in chemical labeling experiments to investigate the oxidation of bGP cysteine residues by H2O2. As shown in Fig. 1d, we observed a strong loss of 5-IAF fluorescence signal in bGP exposed to H2O2, thus confirming the readily oxidative modification of cysteine residues of the enzyme by H2O2 (left panel). DTNB labeling further confirmed the oxidation of bGP cysteine residues by H2O2 and allowed us to determine that most of these residues (10 out of 12 cysteine residues present in human bGP) were oxidized (Fig. 1d, right panel).
We carried out kinetic analysis of the H2O2-dependent inhibition of bGP. As shown in Fig. 1e, semi-logarithmic plots of the percentage of residual bGP activity versus time for different concentrations of H2O2 gave straight lines, suggesting that inhibition obeyed a pseudo first-order reaction. These data were further analyzed by replotting the observed pseudo first-order rate constants (kobs) against H2O2 concentrations (Fig. 1f). The values could be fitted to a straight line passing close to zero. The slope gave a second-order rate constant (kinact) of 185 ± 11 m−1 min−1. This constant is close to values obtained for other enzymes known to be inhibited by reversible oxidation of cysteine residues by H2O2, such as human arylamine N-acetyltransferase 1 (29), caspase 3 (30), protein tyrosine phosphatases (31, 32), or calcineurin (33).
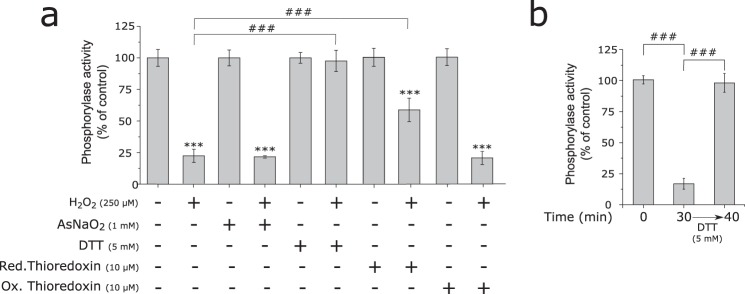
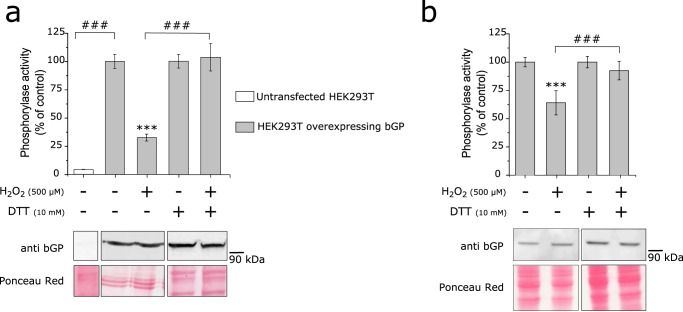
We tested whether the H2O2-dependent inhibition of bGP could be reversed by different reducing agents, including DTT, sodium arsenite, and thioredoxin. As shown in Fig. 2, a and b, a short incubation (10 min) with 5 mm DTT fully reactivated the enzyme inhibited by either bolus addition or flux of H2O2. The ability of DTT to readily restore bGP activity indicated that the inhibition of bGP by H2O2 occurred by the formation of either sulfenic acid and/or a disulfide bond (15, 34). Thioredoxin, a major cellular disulfide reductase (35), significantly reactivated H2O2-inhibited bGP (Fig. 2a). Conversely, sodium arsenite, a reducing agent known to specifically reduce sulfenic acids but not disulfides (36), afforded no significant reactivation of inhibited bGP (Fig. 2a). Taken together, these results suggested the involvement of one or several disulfide bonds in the redox regulation of human bGP. Moreover, Western blotting analyses of H2O2-oxidized and reduced bGP showed that both H2O2-exposed and non-exposed bGP migrated as a single monomer of ∼100 kDa in SDS-PAGE, thus suggesting that inhibition of bGP by H2O2 is due to the reversible formation of intramolecular disulfide bonds (Fig. 1, b and c, lower panels). To ascertain that the reversible inhibition of bGP by H2O2 may occur in a cellular context, an approach based on the transfection of human HEK293T cells (which display negligible GP expression and activity), and previously used to study the regulation of GP activity in cells, was carried out (37). As shown in Fig. 3a, bGP was readily expressed and active in extracts of transfected HEK293T cells when compared with non-transfected cells. We found that exposure of transfected HEK293T cells to H2O2 led to a significant decrease (close to 70% inhibition) in bGP activity. Full bGP activity could be restored upon incubation with DTT. These results were further confirmed in a model human astrocytic cell line (U87MG cells), which expresses bGP (Fig. 3b). Altogether these data are consistent with the experiments carried out with purified bGP (Fig. 1). In addition, our results are in agreement with recent redox proteomics studies indicating that cysteine residues of bGP can be oxidized in cells exposed to H2O2 (23).
FIGURE 2.
Human bGP inhibition by H2O2 is due to the reversible formation of intramolecular disulfide bonds. a, recombinant bGP was first inhibited by H2O2 as in Fig. 1a. Oxidized bGP and non-treated bGP were subsequently and independently incubated with 5 mm DTT, 10 mm sodium arsenite (AsNaO2), 5 mm GSH, or 10 μm thioredoxin for 10 min at 37 °C prior to activity measurement. Results are expressed as the percentage of the control. Red., reduced; Ox., oxidized. Data represent mean values of three independent experiments ± S.D., ***, p < 0.001 when compared with control; ###, p < 0.001 when two non-control groups are compared. b, recombinant bGP was inhibited by H2O2 as in Fig. 1c. Oxidized bGP and non-treated bGP were subsequently incubated with 5 mm DTT for 10 min at 37 °C prior to activity measurement. Reduction restored full activity of bGP. Results are expressed as the percentage of the control. Data represent mean values of three independent experiments ± S.D., ***, p < 0.001 when compared with control; ###, p < 0.001 when two non-control groups are compared.
FIGURE 3.
bGP is inhibited in cells exposed to H2O2. a, HEK293T cells were transfected or not transfected with pCMV6-PYGB plasmid, and cells were exposed to 500 μm H2O2 for 20 min before being harvested. Cell lysis was then performed in the presence or absence of a reducing agent (10 mm DTT), and whole-cell extracts were assayed for endogenous glycogen phosphorylase activity. Data are expressed as the percentage of the control and represent mean values of three independent experiments ± S.D. ***, p < 0.001 when compared with positive control (upper panel); ###, p < 0.001 when two non-control groups are compared. Non-reduced and reduced whole-cell extracts were Western blotted and revealed for brain glycogen phosphorylase using an anti-bGP antibody. Ponceau red stains of the membranes are shown (lower panel). b, U87MG cells were exposed to 500 μm H2O2 for 20 min before being harvested. Cell lysis was then performed in the presence or absence of a reducing agent (10 mm DTT), and whole-cell extracts were assayed for endogenous glycogen phosphorylase activity. Data are expressed as the percentage of the control and represent mean values of three independent experiments ± S.D. ***, p < 0.001 when compared with positive control (upper panel); ###, p < 0.001 when two non-control groups are compared. Western blotting analysis of bGP from cells was revealed for brain glycogen phosphorylase using anti-bGP antibodies. Ponceau red stains of the membranes are shown (lower panel).
Cys318 and Cys326 Are Involved in H2O2-dependent Inhibition of bGP through Formation of a Disulfide Bond
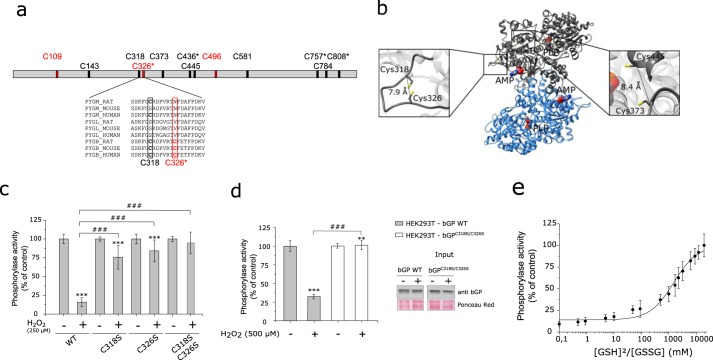
Human bGP has 12 cysteine residues in its primary structure, including four cysteines that are unique to this GP isozyme (Cys326, Cys436, Cys757, and Cys808) (Fig. 4a). Using the structure of human bGP (38), we identified two potential intramolecular disulfide bonds, i.e. Cys318–Cys326 and Cys373–Cys445, the former involving the bGP specific “reactive” cysteine (Cys326), previously identified in a proteomic screen (Fig. 4b and supplemental Table 1) (22, 23). To investigate the involvement of Cys318 and Cys326 in the redox regulation of bGP, these two residues were mutated individually and simultaneously to serine residues to obtain three mutant enzymes: bGPC318S, bGPC326S, and bGPC318S/C326S. As shown in Fig. 4c, contrary to WT bGP, the single mutant enzymes were found to be poorly inhibited by H2O2. The double mutant was not sensitive to H2O2-dependent inhibition. This experiment further indicates that H2O2-dependent inhibition of bGP relies on the oxidation of Cys318 and Cys326 residues. To confirm the redox state of Cys318 and Cys326 residues of bGP exposed to H2O2, we performed differential cysteine labeling and mass spectrometry analysis (39), To this end, the thiol groups of reduced and H2O2-oxidized bGP were first alkylated with iodoacetamide (IAA), reduced with DTT, and then alkylated with N-ethylmaleimide (NEM). The labeled proteins were then trypsin-digested and analyzed by LC-MS/MS (supplemental Fig. 1). Both Cys318 and Cys326 were found to be significantly less labeled by IAA and to display increased NEM labeling (i.e. decrease of the IAA/NEM ratio). In addition, 8 of the 12 cysteines of bGP were found to be oxidized by H2O2 (supplemental Table 1 and supplemental Fig. 1). These results are in agreement with the data obtained with DTNB (Fig. 1d).
FIGURE 4.
Cys318 and Cys326 are critical for H2O2-dependent inhibition of bGP and may form an intramolecular disulfide bond in bGP. a, bGP sequence representation with relative positions of the cysteine residues. Four cysteine residues are specific from the brain isoform (C326*, C436*, C757*, and C808*). In addition, three cysteine residues have been described previously as reactive cysteine (colored in red) (22, 23, 26). Sequences of muscle (PYGM), liver (PYGL), and brain (PYGB) glycogen phosphorylase from rat, mouse, and human containing the three reactive cysteines have been aligned. b, ribbon representation of Cα trace of the dimer of bGP. The AMP-binding site is marked by the allosteric effector AMP (surface representation). The 12 cysteine residues are represented as a yellow stick. The distance separating the Cα of each pair of cysteine residues was measured (supplemental Table 1). Two pairs of cysteines were identified with Cα1-Cα2 <10 Å and are represented (right and left panels): Cys318–Cys326 and Cys373–Cys445. c, single mutation and double mutations of Cys318 and Cys326 were performed and tested for oxidation resistance as described above. Results are expressed as the percentage of the control. Data represent mean values of three independent experiments ± S.D., ***, p < 0.001 when compared with control; ###, p < 0.001 when two non-control groups are compared. d, HEK293T cells were transiently transfected with either WT bGP vector or C318S/C326S mutant bGP vector. Cell medium was gradually replaced by medium without FCS, and cells were then exposed to H2O2 (0, 250, and 500 μm) for 20 min at 37 °C under 5% CO2. Whole-cell extracts were then assayed for endogenous glycogen phosphorylase activity and Western blotting analysis. Activities are expressed as the percentage of the control. Data represent mean values of three independent experiments ± S.D., ***, p < 0.001, **, p < 0.01, when compared with control; ###, p < 0.001 when two non-control groups are compared. e, plot of the relative activity of bGP as a function of the ratio of reduced to oxidized glutathione. bGP activity was measured after incubation of the recombinant bGP with increasing ratios of [GSH]2/[GSSG] (0–20,000 mm) for 18 h at 4 °C. Activity was expressed as the percentage of the control (reduced protein). The solid line is the theoretical fit to Equation 4. The calculated E°′bGP was −267 mV. Data represent mean values of three independent experiments ± S.D.
To further support that Cys318 and Cys326 are indeed involved in the redox regulation of bGP, we transfected wild-type bGP or bGPC318S/C326S mutant in HEK293T cells and exposed them to H2O2. In accordance with the results described above, wild-type bGP was found to be inhibited in cells exposed to H2O2, whereas the bGPC318S/C326S mutant was not (Fig. 4d). Taken together, these results suggest that Cys318 and Cys326 can act as a redox switch through the formation of an intramolecular disulfide bond that impacts the activity of bGP. This disulfide bond is a feature of bGP as the reactive Cys326 residue is unique to the brain isozyme (Fig. 4a), which is consistent with the fact that mGP and lGP are much less sensitive to H2O2 (Fig. 1a) (40).
The standard redox potential of the Cys318–Cys326 disulfide bond was determined using oxidized and reduced glutathione as described previously (Fig. 4e) (41). The activity of the fully reduced bGP was used to calculate the percentage of active bGP, which was plotted as a function of the ratio of reduced to oxidized glutathione. By applying the Nernst equation to the midpoint of the activity and the E°′ value of −240 mV for the GSH/GSSG redox couple, we determined a standard redox potential of −267 mV (E°′bGP) for bGP. This value was consistent with redox potential values found in cytosol and previously reported for other enzymatic systems with functional allosteric disulfide bonds, such as βII-tryptase (Fig. 4e) (42, 43).
Cys318–Cys326 Disulfide Bond Impairs AMP-dependent Activation of bGP
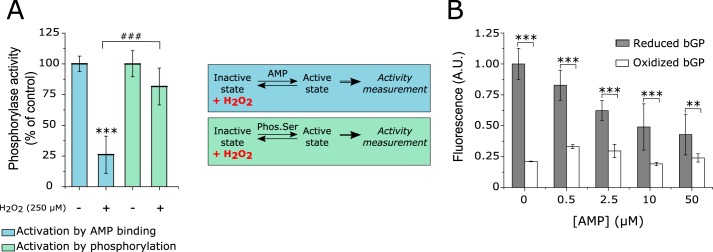
bGP is an allosteric enzyme found in equilibrium between an inactive (or T-state) and an active (or R-state). The transition between these two states is under the control of 1) the phosphorylation of Ser14 and 2) the binding of allosteric effectors (such as AMP) in the AMP-binding site. The latter is composed of several secondary structures including two helices (helices 2 and 8) and three loops (adenine, β4/β5, and cap′ loops) (38). Interestingly, the Cys318 and Cys326 residues belong to the adenine loop present in the AMP-binding site (supplemental Fig. 2). In addition, in mGP, this conserved loop interacts with AMP, flanking the AMP-binding site and stabilizing the active state of the enzyme (44). Therefore, the formation of the Cys318–Cys326 disulfide bond upon exposure to H2O2 may thus impact the activation of bGP by AMP through the structural modification of the adenine loop, but not the activation of the enzyme by phosphorylation on Ser14. To further analyze this point, we carried out experiments where bGP was first oxidized by H2O2, and the remaining activity of the enzyme was monitored either after activation by AMP or after phosphorylation by phosphorylase kinase (Fig. 5A). Although H2O2-inhibited bGP was not activated by AMP, the oxidized enzyme was readily activated upon phosphorylation by phosphorylase kinase (Fig. 5A). These results strongly suggested that the formation of the Cys318–Cys326 disulfide bond by H2O2 impairs the AMP-dependent activation of bGP but not its activation by phosphorylation.
FIGURE 5.
H2O2 exposure and Cys318–Cys326 disulfide bond formation impacts the AMP-dependent activation of bGP. A, bGP was incubated with 250 μm H2O2 for 30 min at 37 °C. The oxidized enzyme was then incubated with AMP or phosphorylase kinase prior to activity measurement. Phos. Ser, phosphorylated serine. Results are expressed as the percentage of the control. Data represent mean values of three independent experiments ± S.D., ***, p < 0.001 when compared with control; ###, p < 0.001 when two non-control groups are compared. B, analysis of the binding of AMP to reduced and oxidized bGP was performed by incubating the treated or untreated enzyme with 5 μm fluorescent mant-AMP. To ascertain that the fluorescence was due to binding of mant-AMP to the enzyme, increasing concentrations of AMP were added. The addition of increasing concentrations of AMP resulted in a loss of fluorescence. A. U., arbitrary units.
The fluorescent analogue of AMP, mant-AMP, was used to test the ability of H2O2-oxidized bGP to bind AMP. This analogue displays increased fluorescence when bound to proteins (45, 46). As expected, significant fluorescence of mant-AMP at 448 nm (λex: 355 nm) was observed in the presence of reduced bGP. The addition of increasing concentrations of AMP induced a significant decrease in fluorescence, thus confirming that mant-AMP fluorescence was due to the binding of this AMP analogue in the AMP-binding site of bGP (Fig. 5B). In contrast, when the experiments were carried out with H2O2-inhibited bGP, only a slight increase in mant-AMP fluorescence (close to 25% of the control reduced enzyme) was observed. These results strongly suggest that the formation of the Cys318–Cys326 disulfide bond in the adenine loop alters the binding of AMP and the subsequent activation of the bGP. Molecular dynamics simulations of bGP with or without the Cys318–Cys326 disulfide bond further supported these data (supplemental Fig. 3 and supplemental Table 3). Indeed, the presence of the Cys318–Cys326 disulfide bond was found to rigidify the adenine loop, thus impairing the flanking of the AMP-binding site (supplemental Fig. 3 and supplemental Table 3).
Discussion
Brain glycogen has long been considered as an emergency glucose store that provides neuronal protection during stress episodes, including hypoglycemia and hypoxic stress (3–5, 7). Over the past years, increasing interest has been paid to brain glycogen as it is also critical in normal brain functions and high cognitive processes, in particular by providing lactate and energy for the astrocyte-neuron lactate shuttle (8, 9). In mammalian cells, the mobilization of the glycogen store depends on GP, the rate-limiting enzyme of glycogenolysis that exists as three distinct isozymes: the liver, the muscle, and the brain GP (1, 14). GPs are allosteric enzymes, regulated by both phosphorylation (by phosphorylase kinase) and the binding of allosteric effectors (such as AMP), allowing the respective regulation of GP by extracellular signals and the local energy demand (1, 14). In addition, recent redox proteomics studies suggest that bGP activity might be regulated by ROS, through the modification of critical reactive cysteine residues (22, 23, 26). In this study, we report that bGP is reversibly controlled by an intramolecular disulfide bond between Cys318 and Cys326, the latter cysteine residue being found only in the brain form of GP. The critical role of these two cysteines (in particular Cys326) was confirmed by mutational analysis (Fig. 4, c and d). Our work suggests that this disulfide bond acts as a redox switch of the AMP-binding site (allosteric site) and controls the activation of the brain form of GP by AMP, without altering the regulation of the enzyme by phosphorylation.
Several proteins and enzymes, such as PKA, are redox-regulated through reversible cysteine oxidation. In particular, this redox regulation can occur through the formation of reversible disulfide bonds, as observed in Hsp33, bII tryptase, or Lon protease (41, 42, 47). These functional disulfide bonds regulate protein functions. They form in the cytosol and present higher redox potentials comprised between −120 and −270 mV (48). In our case, the redox potential of human bGP, −267 mV, is consistent with the regulation of this enzyme by a functional disulfide bond (Fig. 2b). The functional role of these disulfide bonds relies on the reversibility of their formation by reductants. We used several reducing agents to determine the reversibility of the inhibition. Redox-maintaining molecules, such as GSH and glutaredoxin 1, present higher redox potential (−240 and −233 mV, respectively) when compared with bGP, suggesting that these antioxidant systems may not be involved in the redox regulation of bGP in vivo (49). However, oxidoreductases, including thioredoxin, display a redox potential of −270 mV, indicating that the reduction reaction of bGP by thioredoxin is thermodynamically favorable and suggesting that thioredoxin could be a primary reductant of bGP in cells (49). These observations were supported by the low reactivation of the oxidized bGP by GSH and the ability of DTT (redox potential of −330 mV) and thioredoxin to better reverse the bGP oxidation in vitro (Fig. 2a). Interestingly, thioredoxin is known to interact with oxidized GP, in particular in cardiac cells (50).
GPs are enzyme-regulated by phosphorylation and the binding of allosteric effectors. In bGP, the binding of AMP is critical for the enzyme activation as this isoform is mainly regulated by allosteric effectors and to a lesser extent by phosphorylation (51, 52). As Cys318 and Cys326 are located in the adenine loop, which belongs to the AMP-binding site, the Cys318–Cys326 disulfide bond is likely to alter the structural properties of the loop, resulting in the alteration of the binding of AMP (Fig. 5B, supplemental Fig. 3 and supplemental Table 3). These findings emphasize the critical role of AMP and the AMP-binding site for the regulation of bGP. Recent data suggest that the AMP-dependent activation of bGP is critical for the regulation of glycogen metabolism and functions in the brain (52).
The reversible redox regulation of bGP was also observed in U87MG astrocytes and HEK293T cells transfected with wild-type bGP, but not with HEK293T cells transfected with bGP mutated for Cys318 and Cys326 (supplemental Fig. 1). As astrocytes express both the mGP and the bGP isozymes (13), biochemical and cellular studies suggested that bGP is mainly regulated by the binding of allosteric effectors, whereas mGP is tailored to respond to phosphorylation (51, 52). Our findings indicate that the redox regulation of bGP may allow a preferential regulation of glycogenolysis by phosphorylation under oxidative conditions. It has been shown that neurons exclusively express the brain isozyme of GP and not the muscle one (3). This observation suggested that glycogen degradation in these cells is designed mainly to respond to their own energy needs (3). The selective regulation of oxidized bGP by phosphorylation, and hence extracellular signals, indicates that ROS may significantly impact glycogen degradation in neurons. This may lead to a shift toward the phosphorylation-dependent regulation of bGP instead of the effector-dependent regulation of bGP. Further studies are needed to better understand the differential regulation of glycogenolysis in neurons and astrocytes under oxidative conditions and its impact on astrocytic and neuronal glycogen. Furthermore, allosteric and phosphorylation activation mechanisms are interrelated in cells. However, the relative contribution of phosphorylation versus allosteric control of GP in brain cells remains to be elucidated. The selective regulation of bGP under oxidative conditions provides additional insights about how the two activation mechanisms may independently influence glycogenolysis in these cells.
Glycogen store and glycogenolysis are critical for high cognitive processes, in particular learning and memory consolidation. In this context, astrocytic mGP and bGP are phosphorylated by phosphorylase kinase in response to neurotransmitters such as serotonin and noradrenaline (53). In addition, ROS are also essential to synaptic plasticity and memory formation (54). Our results suggest that the redox regulation of bGP may be involved in cognitive processes that rely on glycogenolysis. In addition, these results further support the hypothesis that glycogen performs a broad range of functions in the brain (52).
Finally, glycogen has been found to accumulate in the brain in several pathological conditions, including amyotrophic lateral sclerosis, Alzheimer's disease, ischemic stroke, and aging, and is a direct cause of neurodegeneration in Lafora disease (7, 10–12, 55). Lafora disease is an autosomal recessive pathology caused by the mutation of either laforin or malin. These two proteins are involved in the regulation of glycogen accumulation through the proteasome-dependent degradation of glycogen-related proteins including glycogen synthase. The resulting over-synthesis of glycogen in neurons results in the accumulation of poorly branched glycogen, forming inclusions called Lafora bodies and causing neuronal loss and epilepsies (10). These diseases are also characterized by an increased production of ROS and oxidative stress (56, 57). Here, we demonstrate that bGP displays a high sensitivity to oxidative conditions and suggest that its activity may be altered by oxidative stress, resulting in the impairment of brain energy metabolism that could contribute to neurodegenerative disease pathogenesis, as well as the deleterious accumulation of glycogen in neurons and astrocytes.
Conclusion
In conclusion, our study provides evidence that the brain isoform of GP can be regulated by ROS (in particular H2O2), through the formation of an intramolecular disulfide bond between Cys318 and Cys326, a reactive cysteine residue found only in the brain form of GP. This disulfide bond acts as a redox switch of the AMP-binding site and selectively precludes bGP regulation by the allosteric effector AMP, which binds in the AMP-binding site. Our findings suggest that, under oxidative conditions, bGP could be selectively regulated by phosphorylation/dephosphorylation events but not by allosteric effectors. It is known that ROS have a dual role in the brain, as mediators for brain function and as a source of oxidative stress in brain diseases, including neurodegenerative diseases. Our study shows the isoform-selective regulation of bGP by oxidants and provides a molecular basis for the regulation of glycogenolysis by ROS, which may influence brain function that relies on glycogen store.
Experimental Procedures
Materials
The oligonucleotides used for the site-directed mutagenesis were purchased from Eurofins. pCMV6 carrying bGP cDNA (pCMV6-PYGB) was purchased from OriGene Technologies, Inc.
Antibodies raised against brain glycogen phosphorylase were obtained from Santa Cruz Biotechnology (SC-81751). Anti-histidine tag (Sigma) antibodies, l-arabinose, isopropyl-1-thio-β-d-galactopyranoside, protease inhibitor cocktail, nickel-nitrilotriacetic acid Superflow resin, imidazole, DTT, BSA, phosphorylase kinase from rabbit muscle, ATP, AMP, phosphoglucomutase, EDTA, glucose 1,6-diphosphate, Pi, glycogen phosphorylase from rabbit muscle, H2O2, glucose, glucose oxidase, catalase, N-ethylmaleimide, iodoacetamide, 5-IAF, DTNB, GSH, GSSG, FCS, and glucose 6-phosphate were purchased from Sigma-Aldrich. ECL Western blotting detection reagent and buffer exchange columns were purchased from GE Healthcare. Glucose-6-phosphate dehydrogenase was purchased from Roche Applied Science. NADP was purchased from Apollo Scientific. U87-MG cells were purchased from ATCC. HEK293T were kindly provided by Dr. Ait-Si-Ali (CNRS UMR7216, Université Paris Diderot). DMEM, RPMI medium, non-essential amino acids, and glutamine supplements were purchased from Life Technologies. Mant-AMP was purchased from Jena Bioscience.
Methods
Alignment of Glycogen Phosphorylase Sequences
Muscle (PYGM), liver (PYGL), and brain (PYGB) glycogen phosphorylase sequences were obtained from UniProt/Swiss-Prot. The following sequences were analyzed: P11217, P06737, and P11216 (Homo sapiens, human); P09812, P09811, and P53534 (Rattus norvegicus, rat); P00489 (Oryctolagus cuniculus, rabbit); and Q9WUB3, Q9ET01, and Q8CI94 (Mus musculus, mouse). The multiple sequence alignment was performed using Clustal Omega server (European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI)).
cDNA Constructs
The human bGP cDNA was obtained in the eukaryotic pCMV6 vector (provided by OriGene Technologies, Inc.) and subcloned into the pET28a vector for expression and purification of recombinant bGPs. The resulting construct encoded for His6-tagged fusion recombinant proteins (His-bGP). Escherichia coli C41(DE3)/pGro7 (encoding the GroEL/GroES chaperonin protein complex) strains were transformed with plasmid pET28a carrying His-bGP and used to express and purify the recombinant proteins.
Site-directed Mutagenesis of bGP
Cysteine residues 318 and 326 were mutated into serine residues using the Agilent QuikChange site-directed mutagenesis kit according to the manufacturer's instructions. Briefly, the mutagenesis of each cysteine residue was performed by amplification of the whole plasmids (pCMV6 and pET28a) carrying bGP, using 5′ and 3′ primers containing single point mutations of interest. The sequence of each oligonucleotide pair used is presented in supplemental Table 4.
Expression and Purification of Recombinant Human bGP
E. coli bacteria were cultured at 37 °C. Expression of the GroEL/GroES chaperonin protein complex was first induced by the addition of 1 mm l-arabinose to culture. Expression of recombinant His-bGP was then induced by the addition of 500 μm isopropyl-1-thio-β-d-galactopyranoside to culture. The bacteria were further cultured at 16 °C overnight. The bacteria were pelleted by centrifugation (4,000 × g, 10 min), washed with cold PBS, and harvested by centrifugation (4,000 × g, 10 min). The pellets were stored at −80 °C until required.
The bacteria were resuspended in 35 ml of lysis buffer (PBS, pH 8, 300 mm NaCl, 0.5% Triton X-100, 1 mg/ml lysozyme, protease inhibitor cocktail) and incubated for 1 h at 4 °C. The lysate was sonicated on ice (8-s pulses for up to 7 min) and centrifuged (17,000 × g, 30 min, 4 °C). The supernatant was collected and incubated with 1 ml of nickel-nitrilotriacetic acid Superflow resin in the presence of 10 mm imidazole (final concentration) for a minimum of 2 h at 4 °C. The resin was then poured into a column and washed successively with washing buffer (PBS, pH 8, 300 mm NaCl) containing 0.1% Triton X-100 and a stepwise gradient of imidazole in washing buffer until a concentration of 20 mm imidazole (final concentration) was achieved. His-tagged proteins were eluted with washing buffer containing 300 mm imidazole. Purified proteins were incubated for 10 min with 10 mm DTT and protease inhibitor cocktail. The purified protein was then exchanged against PBS, pH 7.1, using a PD 10 desalting column. The protein concentration was measured using the standard Bradford assay with BSA as standard and by absorbance measurement at 280 nm, using a theoretical ϵM: 115,170 m−1 cm−1. The purity of the protein was assessed by SDS-PAGE analysis.
SDS-PAGE Electrophoresis and Western Blotting
Proteins were loaded onto 7.5% polyacrylamide gels, and electrophoretic protein separation was carried out at 110 V (constant voltage) under non-reducing conditions. For SDS-PAGE, the presence of protein was revealed using Coomassie Blue R250. After migration by SDS-PAGE, the proteins were transferred onto a nitrocellulose membrane at a constant current of 200 mA at 4 °C for 1 h. Membranes were incubated at 4 °C overnight with appropriately diluted primary antibody. After washing, the membranes were incubated for 2 h at room temperature with peroxidase-coupled secondary antibody. The proteins were visualized by chemiluminescence detection using ECL substrate and LAS 4000 (Fujifilm). The images were analyzed using Gimp 2 software.
Phosphorylation of Ser14
bGP was phosphorylated using phosphorylase kinase from rabbit muscle (1 unit/mg of bGP), activated by preincubation for 1 h in phosphorylation buffer (20 mm Tris-HCl buffer, pH 7.7, 0.22 mm ATP, 3.3 mm MgCl2, 0.5 mm CaCl2, 0.5 mm NaF) for activation. Phosphorylation was performed by the addition of activated phosphorylase kinase to the bGP solution, in phosphorylation buffer, and incubation for 2 h at room temperature. The resulting activity was assessed with or without AMP. Phosphorylation of rabbit mGP was used as a control. Phosphorylated bGP was then subjected to buffer exchange against PBS buffer, pH 6.9, using a PD MiniTrap G25 (GE Healthcare) desalting column.
GP Activity Assay
GP activity was measured in the direction of glycogenolysis as described elsewhere (58). Briefly, the formation of glucose 1-phosphate was determined using a coupled assay system containing phosphoglucomutase, glucose-6-phosphate dehydrogenase, and NADP, by measuring NADPH(H)+ formation at 340 nm. The phosphorylase activity assay was carried out at 37 °C in PBS, pH 6.9. The mixture consisted of bGP (0.1 μm final concentration) with or without 1 mm AMP, 0.25% glycogen, 2 mm EDTA, 0.8 mm NADP, 10 mm magnesium acetate, 5 μm glucose 1,6-diphosphate, 5 units of glucose-6-phosphate dehydrogenase, and 5 units of phosphoglucomutase in a final volume of 250 μl. Each measurement was performed in triplicate. GP activity was expressed as the percentage of the control.
Effect of H2O2 on GP Activity
Purified GP was reduced using 1 mm DTT for 20 min on ice. DTT was then removed using a PD SpinTrap G25 (GE Healthcare) desalting column, and the protein concentration was determined by Bradford assay using BSA as standard. To test the impact of H2O2 on muscle, liver, and brain GP, 0.25 μm purified enzyme was incubated with or without H2O2 (250 μm final) in PBS, pH 6.9, for 30 min at 37 °C in a final volume of 25 μl. The reaction was stopped by the addition of catalase (300 units/ml of final concentration). The residual activity of GP was assayed as described above. GP activity in the absence of H2O2 was considered as 100% activity. Each measurement was performed in triplicate.
To mimic the cellular generation of H2O2, 0.25 μm recombinant bGP was incubated with 1.2 units of glucose oxidase in the presence of 5 mm glucose in PBS buffer, pH 6.9. In our conditions, the addition of glucose to glucose oxidase allowed the formation of H2O2 at a rate of 6 μm H2O2/min, which is considered to be physiologically relevant. The aliquot was removed every 5 min until 30 min (27–29). Catalase (300 units/ml) was added to counteract any further generation of H2O2. Residual activity was then assayed as described previously (38, 40). bGP activity at t0 was considered as 100% activity. The same experiment was performed in the presence of catalase (300 units/ml) as a control. Glucose oxidase and catalase independently had no effect on bGP activity and bGP activity measurement. In addition, glucose, an allosteric inhibitor of bGP, which binds in the catalytic site of the enzyme (59), did not impact bGP inhibition by H2O2 or bGP activity assay. Prior to experiments, the production rate of H2O2 by glucose oxidase was assessed spectrophotometrically at 240 nm, by measuring H2O2 formation over time in PBS buffer, pH 6.9, containing 5 mm glucose (ϵM(H2O2) = 43.6 m−1 cm−1).
To test whether the oxidation of bGP was reverted by reducing agent, bGP was first inhibited as described previously (40). 5 mm DTT was then added to the mixture and incubated 10 min at 37 °C prior to the enzyme activity assay. Results are expressed as the percentage of the control (bGP incubated in the absence of H2O2).
To test whether oxidized bGP could be activated by phosphorylation, bGP (without ligands or non-phosphorylated) was incubated with H2O2 as described above. The oxidized enzyme was then phosphorylated by phosphorylase kinase as described previously (38) and assayed for activity in the absence of AMP. Results are expressed as the percentage of each positive control.
Modification of bGP Cysteine Residues by H2O2
To test whether bGP oxidation was concomitant with the modification of cysteine residues, we performed cysteine labeling experiments using 5-IAF and DTNB (Ellman's reagent).
5-IAF Labeling
Inhibition of bGP by H2O2 was performed as described previously (29). After inhibition, 20 μm 5-IAF was added to the mixture and incubated for 10 min at 37 °C. Then, 0.5 μg of each sample was run on SDS-PAGE and blotted on a nitrocellulose membrane, in the dark. Fluorescein was detected by fluorescence measurement (λex: 492 nm; λem: 520 nm), and bGP was revealed using specific antibody as described previously (29).
DTNB Labeling
bGP was inhibited by H2O2 as described under “Results.” Reaction was stopped by dilution (10×) in PBS buffer, pH 7.5. Proteins were denaturated and labeled with DTNB by the addition of 100 mm Tris-HCl solution containing 6.4 m guanidine HCl and 5 mm DTNB. Controls containing only PBS buffer, protein, or H2O2 were also performed. Absorbance was measured at 405 nm. Theoretical absorbance corresponding to the labeling of all cysteine in bGP was calculated, compared with the control, and used to determine the number of oxidized cysteines.
Mass Spectrometry
Prior to treatment, bGP was fully reduced and precipitated using TCA to stabilize cysteine residues. Proteins were then centrifuged, and pellets were successively washed using 1) acetone containing 5 mm HCl and 2) acetone. We employed mass spectrometry with differential cysteine labeling to identify the redox active disulfide bond in the enzyme. With that aim, 1 μm precipitated bGP was resuspended in PBS buffer containing 250 mm IAA or incubated with 25 mm H2O2 for 10 min at room temperature. Proteins were precipitated again using TCA and washed as described under “Experimental Procedures.” Pellets were finally resuspended in loading buffer containing 250 mm IAA to alkylate any unpaired cysteine residues. Samples were first purified on a 7.5% SDS-polyacrylamide gel stained with Coomassie Blue R250. After in-gel reduction (5 mm DTT final concentration, 30 min, 56 °C) and alkylation (200 mm NEM, 20 min, room temperature in the dark), gel slices were recovered by trypsin (Roche Applied Science) at 12.5 ng/μl in 25 mm ammonium bicarbonate, 0.05% CaCl2 and incubated overnight at 37 °C. Then, the reaction was stopped with 100 μl of 5% formic acid. Peptides were extracted from gel and incubated twice in 5% formic acid prior to sonication and incubation on acetonitrile. Then, supernatants from the all fractions of the same sample were pooled and dried using a SpeedVac.
Samples were purified by ZipTip (Millipore) before the LC-MS/MS analysis eluting in a solution containing 60% ACN, 0.1% formic acid. Desalted samples were then diluted 10 times prior to fractionation on a capillary reverse phase column (nano C18 Dionex Acclaim PepMap100, 75-μm inner diameter × 50 cm) at a constant flow rate of 220 nl/min, with a gradient of 2–40% buffer B containing 90% ACN, 10% water, 0.1% formic acid (buffer A: 98% water, 2% ACN, 0.1% formic acid). LC was directly coupled to a QqOrbitrap mass spectrometer (Q Exactive, Thermo Fisher Scientific). MS experiments consisted of a survey MS scan (400–2,000 m/z; resolution 70,000) followed by an MS/MS analysis of the 10 most intense precursors, with a dynamic exclusion time of 30 s of the previously fragmented precursors.
After processing raw files with the in-house-developed software MaxQuant 1.5.3.8 (60), data were searched against the PYGB sequence with Andromeda (61). Carbamidomethylated cysteines and NEM cysteines were set as variables similar to modifications such as oxidation of methionine, and N-terminal acetylation. Mass deviation of 0.5 Da was set as maximum allowed for MS/MS peaks, and a maximum of two missed cleavages was allowed. Maximum false discovery rates were set to 0.01 on both peptide and protein levels. Minimum required peptide length was 7 amino acids.
Perseus version.1.5.1.6 was used for data analysis and processing. We used the evidence.txt file to compare the abundance for each peptide modified with carbamidomethyl or NEM. Results are expressed as the ratio of NEM/CAM labeling (where CAM is carbamidomethyl). A decrease in the NEM/CAM ratio suggests that the cysteine residue is oxidized upon the addition of H2O2.
Kinetic Analysis and Determination of the Second-order Rate Inhibition Constant (kinact)
The determination of the second-order rate inhibition constant (kinact) was determined using the pseudo first-order method. Recombinant bGP (0.25 μm) was incubated with various concentrations of H2O2 (0–400 μm) at 37 °C in PBS buffer, pH 6.9. Aliquots were removed at various time points and assayed for residual bGP activity. Inactivation of bGP by H2O2 can be represented by the equation
| (Eq. 1) |
where [bGP] is the concentration of active enzyme and kinact is the second-order rate constant. Considering that H2O2 is in excess, Equation 1 can be simplified as
| (Eq. 2) |
with
| (Eq. 3) |
The apparent first-order inactivation rate constants kobs were measured for each concentration of H2O2 from the slope of the natural logarithm (ln) of the percentage of residual activity plotted against time. The second-order rate constant kinact was determined from the slope of kobs plotted against H2O2 concentrations (Equation 3) and equal to 185 m−1 min−1.
Redox Potential E°′ of the bGP Disulfide Bond
The standard redox potential of the Cys318–Cys326 disulfide bond was determined using reduced and oxidized glutathione. Recombinant bGP (0.25 μm) was incubated with increasing ratios of [GSH]2/[GSSG] (0–20,000 mm) for 18 h at 4 °C. bGP activity was then assayed as described previously (38). The thiol disulfide exchange between bGP and glutathione is described as follows
where bGPRED and bGPOX are the reduced and oxidized forms of bGP, respectively, and n is the number of subunits reduced/oxidized. Considering Reaction 1 and the result that the activity of the oxidized form is 14.04% relative to the reduced form, in the presence of AMP, the theoretical fit was realized using the data-fitting program Origin Pro 8, according to Equation 4.
| (Eq. 4) |
The extracted equilibrium constant Keq, which corresponds to the midpoint of the activity, was applied to the Nernst equation to calculate the standard state redox potential of the enzyme (E°′bGP)
| (Eq. 5) |
where E°′GSH is the standard state redox potential of GSH (−240 mV), R is the gas constant, T is the temperature, and F is the Faraday constant.
Binding of AMP
AMP binding experiments were carried out using fluorescent mant-AMP, an analogue of AMP that fluoresces when bound to proteins. bGP (0.25 μm, either reduced or oxidized) was incubated with mant-AMP (5 μm) and increasing concentrations of AMP (0–50 μm) in Tris-HCl buffer (pH 7.5) for 30 min at 25 °C. Fluorescence of bound mant-AMP was determined by spectrofluorometry using λex = 355 nm, λem = 448 nm (Flex Station 3, Molecular Devices).
Cell Culture and H2O2 Exposure
Astrocytic U-87MG cells were purchased from ATCC and grown in DMEM low glucose containing 10% FCS, 1% GlutaMAX, and non-essential amino acids at 37 °C in 5% CO2 incubator. HEK293T cells were grown in RPMI medium containing 10% FCS and 1% GlutaMAX at 37 °C in a 5% CO2 incubator.
U-87MG cells were serum-starved 18 h before the experiments. Cells were then exposed to the indicated concentrations of H2O2 (0–500 μm) for 20 min at 37 °C in a 5% CO2 incubator. After treatment, cells were washed three times with PBS, pH 7, and lysed in PBS, pH 7, containing 0.1% Triton and protease inhibitors. Cells were submitted to a slight sonication before centrifugation, and the supernatant was collected for assay of bGP activity and Western blotting analysis.
HEK293T cells were transiently transfected with wild-type or C318S/C326S mutant bGP carrying plasmid pCMV6. Cells were exposed to H2O2 in RPMI medium and assayed for phosphorylase activity.
Structural Analysis
Structural analyses were performed using the crystal structures of human bGP (Protein Data Bank (PDB): 5IKP). The missing loops (including the adenine loop 319–326) were constructed using the WinCoot software. Structural analyses were performed using Chimera software, and distances between cysteine residues were measured between Cα. A cut-off distance of 10 Å between Cα was used to identify the potential disulfide bonds. All the distances are reported in supplemental Table 2.
Molecular Modeling and Molecular Dynamics Simulations
Four dimeric systems were examined that correspond to (i) the native brain X-ray dimeric structure with and without an AMP molecule (PDB ID: 5IKP) and (ii) modeled structures with a disulfide bond between Cys318 and Cys326, with and without an AMP molecule. For X-ray structures, regions with missing residues were completed with the Modeler9.14 software (62). Ten models were initially constructed and then refined with the fast loop Modeler procedure. The model with the best DOPE score was selected as a starting structure for molecular dynamics simulations. This model was also used as a template structure to build models with disulfide bond (DISU patch option in Modeler). The model with the best DOPE score among 10 models was selected as a starting structure for the molecular dynamics simulations. The position of AMP molecules was obtained from the X-ray structure (PDB ID: 5IKP).
Molecular dynamics stimulations were carried out using Gromacs5.1.1 (63), choosing the amber99sb-ildn force field for the protein and AMP molecules. AMP topology and parameters were retrieved from REDDB-F90 (q4md Force Field Tools, available from the R.E.D. Force Field Tools page). The systems were solvated with TIP3 water molecules and neutralized with counter ions. A classical protocol (64) was followed, i.e. minimization, thermalization, and equilibration with position restraints on protein heavy atoms during 1 ns. Restraints were finally removed, and the systems were simulated for 100 ns. For each system, a replicate simulation of 100 ns was performed by changing initial velocities. All bond lengths were constrained with LINCS (Linear Constraint Solver). Temperature was maintained at 300 K with the velocity-rescale method and a time constant of 0.1 ps. Pressure was maintained isotropically at 1 bar using the Parrinello-Rahman algorithm with a time constant of 2.0 ps. The neighbor list was updated every fifth step with a cut-off of 1.0 nm. Lennard-Jones interactions were switched to zero at 1.0 nm. Particle Mesh Ewald (PME) with real space cut-off at 1.0 nm was used for electrostatics. Gromacs tools and home-made Python scripts were used for the analyses.
Statistical Analysis
Results are presented as mean ± S.D. of three independent experiments. Statistical analysis was performed using one-way analysis of variance followed by Bonferroni's post hoc test, using the OriginPro 8 software. If only two groups were compared, the mean comparison t test was used. A p < 0.05 was considered as significant. p < 0.05, p < 0.01, and p < 0.001 were indicated by one, two, and three asterisks, respectively. p ≥ 0.05 were considered as not significant and not marked.
Author Contributions
C. M. and F. R. L. designed experiments, managed the project, and wrote the manuscript. C. M., R. D., A. C., E. P., L. C. B., I. H., J. V., C. E., J. M. D., and F. R. L. contributed reagents, performed experiments, and/or analyzed the data. All the authors discussed the results and commented on the manuscript.
Supplementary Material
This work was supported by the University Paris Diderot and CNRS. This work was also supported by Ph.D. fellowships from the University Paris Diderot (Ecole Doctorale BioSPC) (to C. M.), Région Ile de France (to R. D.), and University Paris Diderot (to A. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as a Paper of the Week.

This article contains supplemental Figs. 1–3 and supplemental Tables 1–4.
- GP
- glycogen phosphorylase
- bGP
- brain GP
- mGP
- muscle GP
- lGP
- liver GP
- ROS
- reactive oxygen species
- 5-IAF
- fluorescein 5-iodoacetamide
- DTNB
- 5,5′-dithiobis-2-nitrobenzoic acid
- NEM
- N-ethylmaleimide
- ACN
- acetonitrile
- FA
- formic acid
- mant-AMP
- 2′/3′-(N-methyl-anthraniloyl)-adenosine-5′-monophosphate, triethylammonium salt
- DOPE
- discrete optimized protein energy.
References
- 1. Roach P. J. (2002) Glycogen and its metabolism. Curr. Mol. Med. 2, 101–120 [DOI] [PubMed] [Google Scholar]
- 2. Cataldo A. M., and Broadwell R. D. (1986) Cytochemical identification of cerebral glycogen and glucose-6-phosphatase activity under normal and experimental conditions. II. Choroid plexus and ependymal epithelia, endothelia and pericytes. J. Neurocytol. 15, 511–524 [DOI] [PubMed] [Google Scholar]
- 3. Saez I., Duran J., Sinadinos C., Beltran A., Yanes O., Tevy M. F., Martínez-Pons C., Milán M., and Guinovart J. J. (2014) Neurons have an active glycogen metabolism that contributes to tolerance to hypoxia. J. Cereb. Blood Flow Metab. 34, 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choi I.-Y., Seaquist E. R., and Gruetter R. (2003) Effect of hypoglycemia on brain glycogen metabolism in vivo. J. Neurosci. Res. 72, 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown A. M., Sickmann H. M., Fosgerau K., Lund T. M., Schousboe A., Waagepetersen H. S., and Ransom B. R. (2005) Astrocyte glycogen metabolism is required for neural activity during aglycemia or intense stimulation in mouse white matter. J. Neurosci. Res. 79, 74–80 [DOI] [PubMed] [Google Scholar]
- 6. Swanson R. A., Sagar S. M., and Sharp F. R. (1989) Regional brain glycogen stores and metabolism during complete global ischaemia. Neurol. Res. 11, 24–28 [DOI] [PubMed] [Google Scholar]
- 7. Hossain M. I., Roulston C. L., and Stapleton D. I. (2014) Molecular basis of impaired glycogen metabolism during ischemic stroke and hypoxia. PloS One 9, e97570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suzuki A., Stern S. A., Bozdagi O., Huntley G. W., Walker R. H., Magistretti P. J., and Alberini C. M. (2011) Astrocyte-neuron lactate transport is required for long-term memory formation. Cell 144, 810–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gibbs M. E., Anderson D. G., and Hertz L. (2006) Inhibition of glycogenolysis in astrocytes interrupts memory consolidation in young chickens. Glia 54, 214–222 [DOI] [PubMed] [Google Scholar]
- 10. Duran J., Gruart A., García-Rocha M., Delgado-García J. M., and Guinovart J. J. (2014) Glycogen accumulation underlies neurodegeneration and autophagy impairment in Lafora disease. Hum. Mol. Genet. 23, 3147–3156 [DOI] [PubMed] [Google Scholar]
- 11. Sato N., and Morishita R. (2015) The roles of lipid and glucose metabolism in modulation of β-amyloid, tau, and neurodegeneration in the pathogenesis of Alzheimer disease. Front. Aging Neurosci. 7, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dodge J. C., Treleaven C. M., Fidler J. A., Tamsett T. J., Bao C., Searles M., Taksir T. V., Misra K., Sidman R. L., Cheng S. H., and Shihabuddin L. S. (2013) Metabolic signatures of amyotrophic lateral sclerosis reveal insights into disease pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 10812–10817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pfeiffer-Guglielmi B., Fleckenstein B., Jung G., and Hamprecht B. (2003) Immunocytochemical localization of glycogen phosphorylase isozymes in rat nervous tissues by using isozyme-specific antibodies. J. Neurochem. 85, 73–81 [DOI] [PubMed] [Google Scholar]
- 14. Newgard C. B., Hwang P. K., and Fletterick R. J. (1989) The family of glycogen phosphorylases: structure and function. Crit. Rev. Biochem. Mol. Biol. 24, 69–99 [DOI] [PubMed] [Google Scholar]
- 15. D'Autréaux B., and Toledano M. B. (2007) ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824 [DOI] [PubMed] [Google Scholar]
- 16. Veal E. A., Day A. M., and Morgan B. A. (2007) Hydrogen peroxide sensing and signaling. Mol. Cell 26, 1–14 [DOI] [PubMed] [Google Scholar]
- 17. Dickinson B. C., and Chang C. J. (2011) Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 7, 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Friguet B. (2006) Oxidized protein degradation and repair in ageing and oxidative stress. FEBS Lett. 580, 2910–2916 [DOI] [PubMed] [Google Scholar]
- 19. Go Y.-M., Chandler J. D., and Jones D. P. (2015) The cysteine proteome. Free Radic. Biol. Med. 84, 227–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brandes N., Schmitt S., and Jakob U. (2009) Thiol-based redox switches in eukaryotic proteins. Antioxid. Redox Signal. 11, 997–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wouters M. A., Fan S. W., and Haworth N. L. (2010) Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxid. Redox Signal. 12, 53–91 [DOI] [PubMed] [Google Scholar]
- 22. Wang C., Weerapana E., Blewett M. M., and Cravatt B. F. (2014) A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 11, 79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weerapana E., Wang C., Simon G. M., Richter F., Khare S., Dillon M. B. D., Bachovchin D. A., Mowen K., Baker D., and Cravatt B. F. (2010) Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gould N. S., Evans P., Martínez-Acedo P., Marino S. M., Gladyshev V. N., Carroll K. S., and Ischiropoulos H. (2015) Site-specific proteomic mapping identifies selectively modified regulatory cysteine residues in functionally distinct protein networks. Chem. Biol. 22, 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lo Conte M., Lin J., Wilson M. A., and Carroll K. S. (2015) A chemical approach for the detection of protein sulfinylation. ACS Chem. Biol. 10, 1825–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang J., Gupta V., Tallman K. A., Porter N. A., Carroll K. S., and Liebler D. C. (2015) Global, in situ, site-specific analysis of protein S-sulfenylation. Nat. Protoc. 10, 1022–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Giulivi C., and Davies K. J. (2001) Mechanism of the formation and proteolytic release of H2O2-induced dityrosine and tyrosine oxidation products in hemoglobin and red blood cells. J. Biol. Chem. 276, 24129–24136 [DOI] [PubMed] [Google Scholar]
- 28. Mueller S., Pantopoulos K., Hübner C. A., Stremmel W., and Hentze M. W. (2001) IRP1 activation by extracellular oxidative stress in the perfused rat liver. J. Biol. Chem. 276, 23192–23196 [DOI] [PubMed] [Google Scholar]
- 29. Atmane N., Dairou J., Paul A., Dupret J.-M., and Rodrigues-Lima F. (2003) Redox regulation of the human xenobiotic metabolizing enzyme arylamine N-acetyltransferase 1 (NAT1): reversible inactivation by hydrogen peroxide. J. Biol. Chem. 278, 35086–35092 [DOI] [PubMed] [Google Scholar]
- 30. Hampton M. B., Stamenkovic I., and Winterbourn C. C. (2002) Interaction with substrate sensitises caspase-3 to inactivation by hydrogen peroxide. FEBS Lett. 517, 229–232 [DOI] [PubMed] [Google Scholar]
- 31. Lee S.-R., Yang K.-S., Kwon J., Lee C., Jeong W., and Rhee S. G. (2002) Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 277, 20336–20342 [DOI] [PubMed] [Google Scholar]
- 32. Bhattacharya S., LaButti J. N., Seiner D. R., and Gates K. S. (2008) Oxidative inactivation of protein tyrosine phosphatase 1B by organic hydroperoxides. Bioorg. Med. Chem. Lett. 18, 5856–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bogumil R., Namgaladze D., Schaarschmidt D., Schmachtel T., Hellstern S., Mutzel R., and Ullrich V. (2000) Inactivation of calcineurin by hydrogen peroxide and phenylarsine oxide: evidence for a dithiol-disulfide equilibrium and implications for redox regulation. Eur. J. Biochem. 267, 1407–1415 [DOI] [PubMed] [Google Scholar]
- 34. Poole L. B. (2015) The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 80, 148–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu J., and Holmgren A. (2014) The thioredoxin antioxidant system. Free Radic. Biol. Med. 66, 75–87 [DOI] [PubMed] [Google Scholar]
- 36. Gupta V., and Carroll K. S. (2014) Sulfenic acid chemistry, detection and cellular lifetime. Biochim. Biophys. Acta 1840, 847–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang T., Wang S., Lin Y., Xu W., Ye D., Xiong Y., Zhao S., and Guan K.-L. (2012) Acetylation negatively regulates glycogen phosphorylase by recruiting protein phosphatase 1. Cell Metab. 15, 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mathieu C., Li de la Sierra-Gallay I. L., Duval R., Xu X., Cocaign A., Léger T., Woffendin G., Camadro J.-M., Etchebest C., Haouz A., Dupret J.-M., and Rodrigues-Lima F. (2016) Insights into brain glycogen metabolism: the structure of human brain glycogen phosphorylase. J. Biol. Chem. 291, 18072–18083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seiwert B., Hayen H., and Karst U. (2008) Differential labeling of free and disulfide-bound thiol functions in proteins. J. Am. Soc. Mass Spectrom. 19, 1–7 [DOI] [PubMed] [Google Scholar]
- 40. Dairou J., Pluvinage B., Noiran J., Petit E., Vinh J., Haddad I., Mary J., Dupret J.-M., and Rodrigues-Lima F. (2007) Nitration of a critical tyrosine residue in the allosteric inhibitor site of muscle glycogen phosphorylase impairs its catalytic activity. J. Mol. Biol. 372, 1009–1021 [DOI] [PubMed] [Google Scholar]
- 41. Nishii W., Kukimoto-Niino M., Terada T., Shirouzu M., Muramatsu T., Kojima M., Kihara H., and Yokoyama S. (2015) A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis. Nat. Chem. Biol. 11, 46–51 [DOI] [PubMed] [Google Scholar]
- 42. Cook K. M., McNeil H. P., and Hogg P. J. (2013) Allosteric control of βII-tryptase by a redox active disulfide bond. J. Biol. Chem. 288, 34920–34929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmidt B., Ho L., and Hogg P. J. (2006) Allosteric disulfide bonds. Biochemistry 45, 7429–7433 [DOI] [PubMed] [Google Scholar]
- 44. Sprang S. R., Withers S. G., Goldsmith E. J., Fletterick R. J., and Madsen N. B. (1991) Structural basis for the activation of glycogen phosphorylase b by adenosine monophosphate. Science 254, 1367–1371 [DOI] [PubMed] [Google Scholar]
- 45. Jämsen J., Baykov A. A., and Lahti R. (2010) Nucleotide- and substrate-induced conformational transitions in the CBS domain-containing pyrophosphatase of Moorella thermoacetica. Biochemistry 49, 1005–1013 [DOI] [PubMed] [Google Scholar]
- 46. Hiratsuka T. (1983) New ribose-modified fluorescent analogs of adenine and guanine nucleotides available as substrates for various enzymes. Biochim. Biophys. Acta 742, 496–508 [DOI] [PubMed] [Google Scholar]
- 47. Jakob U., Eser M., and Bardwell J. C. (2000) Redox switch of Hsp33 has a novel zinc-binding motif. J. Biol. Chem. 275, 38302–38310 [DOI] [PubMed] [Google Scholar]
- 48. Cook K. M., and Hogg P. J. (2013) Post-translational control of protein function by disulfide bond cleavage. Antioxid. Redox Signal. 18, 1987–2015 [DOI] [PubMed] [Google Scholar]
- 49. Aslund F., Berndt K. D., and Holmgren A. (1997) Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J. Biol. Chem. 272, 30780–30786 [DOI] [PubMed] [Google Scholar]
- 50. Park E. M., and Thomas J. A. (1989) Reduction of protein mixed disulfides (dethiolation) by Escherichia coli thioredoxin: a study with glycogen phosphorylase b and creatine kinase. Arch. Biochem. Biophys. 272, 25–31 [DOI] [PubMed] [Google Scholar]
- 51. Crerar M. M., Karlsson O., Fletterick R. J., and Hwang P. K. (1995) Chimeric muscle and brain glycogen phosphorylases define protein domains governing isozyme-specific responses to allosteric activation. J. Biol. Chem. 270, 13748–13756 [DOI] [PubMed] [Google Scholar]
- 52. Müller M. S., Pedersen S. E., Walls A. B., Waagepetersen H. S., and Bak L. K. (2015) Isoform-selective regulation of glycogen phosphorylase by energy deprivation and phosphorylation in astrocytes. Glia 63, 154–162 [DOI] [PubMed] [Google Scholar]
- 53. Gibbs M. E. (2015) Role of glycogenolysis in memory and learning: regulation by noradrenaline, serotonin and ATP. Front. Integr. Neurosci. 9, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Massaad C. A., and Klann E. (2011) Reactive oxygen species in the regulation of synaptic plasticity and memory. Antioxid. Redox Signal. 14, 2013–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gertz H. J., Cervos-Navarro J., Frydl V., and Schultz F. (1985) Glycogen accumulation of the aging human brain. Mech. Ageing Dev. 31, 25–35 [DOI] [PubMed] [Google Scholar]
- 56. Kim G. H., Kim J. E., Rhie S. J., and Yoon S. (2015) The role of oxidative stress in neurodegenerative diseases. Exp. Neurobiol. 24, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de la Cruz V. P., Korrapati S. V., and Chaverrí J. P. (2015) Redox Status and Aging Link in Neurodegenerative Diseases 2015. Oxid. Med. Cell. Longev. 2015, 494316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maddaiah V. T., and Madsen N. B. (1966) Kinetics of purified liver phosphorylase. J. Biol. Chem. 241, 3873–3881 [PubMed] [Google Scholar]
- 59. Martin J. L., Johnson L. N., and Withers S. G. (1990) Comparison of the binding of glucose and glucose 1-phosphate derivatives to T-state glycogen phosphorylase b. Biochemistry 29, 10745–10757 [DOI] [PubMed] [Google Scholar]
- 60. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 62. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 63. van Drunen R., berendsen H. J. (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comp. Phys. Comm. 91, 43–56 [Google Scholar]
- 64. Lindahl E. (2015) Molecular dynamics simulations. Methods Mol. Biol. 1215, 3–26 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.