Abstract
It is known that there are mechanistic links between circadian clocks and metabolic cycles. Reduced nicotinamide adenine dinucleotide (NADH) is a key metabolic cofactor in all living cells; however, it is not known whether levels of NADH oscillate or not. Here we employed REX, a bacterial NADH-binding protein, fused to the VP16 activator to convert intracellular endogenous redox balance into transcriptional readouts by a reporter gene in mammalian cells. EMSA results show that the DNA binding activity of both T- and S-REX::VP16 fusions is decreased with a reduced-to-oxidized cofactor ratio increase. Transient and stabilized cell lines bearing the REX::VP16 and the REX binding operator (ROP) exhibit two circadian luminescence cycles. Consistent with these results, NADH oscillations are observed in host cells, indicating REX can act as a NADH sensor to report intracellular dynamic redox homeostasis in mammalian cells in real time. NADH oscillations provide another metabolic signal for coupling the circadian clock and cellular metabolic states.
Keywords: biosensor, cell metabolism, circadian, nicotinamide adenine dinucleotide (NADH), oxidation-reduction (redox), Rex, oscillation
Introduction
In the mammalian circadian feedback loop, the CLOCK:BMAL13 (or NPAS2:BMAL1) complex is the positive element that activates expression of its own transcriptional repressors (the negative elements), Period (Per1 and Per2) and Cryptochrome (Cry1 and Cry2) (1). This design of a transcription/translation feedback loop (TTFL) is conserved from bacteria to human. Growing evidence suggests that many metabolic cycles are coupled to circadian transcriptional regulation. For example, in the mammalian nicotinamide adenine dinucleotide (NAD+) synthesis pathway, the activity of nicotinamide phosphoribosyltransferase (NAMPT) is regulated directly by the clock, resulting in circadian rhythmicity of NAD+ concentration (2, 3). Moreover, two NAD+-dependent enzymes, SIRT1 and PARP-1 (poly(ADP-ribose) polymerase 1) are known to modify components of the clock machinery, exemplifying the interplay between circadian and metabolic cycles (4–8). Another NAD+-dependent deacetylase SIRT3 generates rhythmic respiration conveying information from cytoplasmic NAD+ oscillation to mitochondrial oxidative metabolism (9). Recently, it was shown that circadian rhythms of redox state could drive excitability of the master oscillator neurons in mammals (10). More strikingly, circadian reduction-oxidation oscillations of peroxiredoxin (PRX) were uncovered in human red blood cells that are anucleate without the TTFL (11). Furthermore, PRX has been reported to be a conserved marker across phylogenetic kingdoms (12). By in vitro biochemistry experiments, direct evidence showing the redox state on clock protein transcriptional activity is that reduced NADH or NADPH and their oxidized forms have opposite roles for DNA binding of NPAS2:BMAL1 (13). Molecular analysis of clock genes and proteins has been deeply investigated, but how to precisely monitor the intracellular Redox state is not fully developed.
Although the NAD+ oscillation has received much attention, it is inferred that the NAD+/NADH ratio changes could contribute to the redox poise of cells. Likewise, free intracellular NADH is also a redox indicator, similar to its oxidative counterpart. It is thought that the cellular NADH concentration is several orders lower than its oxidative form (14, 15), and changes in free NADH could be a better indicator of the redox poise than NAD+. The conventional method assaying NADH or NAD+ is derived from lactate and pyruvate concentrations (16), which is unsuitable for understanding dynamic changes in intact cells. In addition, the less invasive imaging approach could not distinguish NADH from NADPH as they produce identical autofluorescence signals with ultraviolet excitation (17). Although new strategies (18, 19) have been developed based on two methods mentioned above, it is still pertinent to explore a new NADH biosensor to report the intracellular redox state in living cells in real time.
Among Gram-positive bacteria, the Rex protein is known to act as a redox sensor in response to the cellular NADH/NAD+ ratio changes. Rex functions as a homodimer with an N-terminal DNA-binding domain and a C-terminal NADH-binding domain. When bacteria are growing under an aerobic condition, Rex binds to its operator (ROP) sites upstream of several target respiratory genes, resulting in transcriptional inhibition. At the same time, NAD+ increases the Rex affinity with DNA binding. When oxygen becomes limited, accordingly, NAD+ is reduced back to NADH, which promotes Rex release from DNA (20). Two Rex protein structures, B-Rex from Bacillus subtilis (21) and T-Rex from Thermus aquatics (22, 23), have been resolved. Upon NADH binding, Rex conformation changes from an open to a closed structure of the N-terminal DNA binding domain. Although Rex can sense both NAD+ and NADH in bacteria, its affinity for NADH is much higher than for NAD+ (20, 21). More importantly, Rex does not respond to NADP(H) changes in physiological conditions. Could Rex play a similar role to sense NADH/NAD+ redox state in mammalian cells? Two groups constructed a circularly permuted GFP inserted into a tandem dimer of Rex proteins (24, 25). These fluorescent sensors report intracellular NADH changes independent of their DNA binding function, but this strategy could not observe the redox poise dynamics over time in intact cells.
In this study, the minimal VP16 activation domain from Herpes simplex was fused at the carboxyl terminus of Rex, converting the Rex protein from a repressor in bacteria to a transcriptional activator in mammalian cells. Three Rex proteins were tested for this purpose, B-Rex from Bacillus subtilis, S-Rex from Streptomyces coelicolor A3 (2) as well as T -Rex from Thermus aquaticus. Using an electromobility shift assay (EMSA), the DNA-binding activity of both S- and T-Rex::VP16 decreased with increasing ratios of NADH/NAD+; while B-Rex::VP16 was not effective. With transient and stable expression, we observed two luminescence cycles produced by the reporter gene promoted by the S or T-Rex::VP16 transactivator. Furthermore, the DNA binding activity of Rex::VP16 is far more sensitive to NADH than NAD+. In support of these results, the NADH concentration exhibited about 24 h oscillation in host cells. Thus, we have developed a transcriptional metabolic sensor to detect dynamic changes of free NADH in mammalian cells. NADH oscillations could couple the genetic oscillator together with the metabolic oscillator.
Results
Generation of the Rex Transactivator and Its DNA-binding ROPs
To test if Rex could sense NADH/NAD+ in mammalian cells, we adopted the strategy based on the tetracycline-controlled transactivator (tTA) system which was developed as a genetic switch in eukaryotes (26). The tTA is a fusion between the Escherichia coli tetracycline repressor and the minimal transcriptional activation domain of the Herpes simplex virus VP16 gene. The tet operator (tetO) is the target sequence of the tTA, which drives downstream gene expression. However, a tetracycline derivative, doxycycline (Dox), prevents binding of tTA to the tetO sequence. Using this system with the administration of Dox, inducible and reversible Clock gene expression in brain was achieved (27). As there are mechanistic similarities between the tetracycline repressor and the Rex protein in bacteria, we expect that the fusion of Rex and the VP16 activation domain could bind to its ROP, activating the downstream luciferase gene expression. On the contrary, NADH associates with Rex and releases it from DNA binding (Fig. 1a). The intracellular redox state could then be converted into transcriptional luminescence readouts, and recorded by a LumiCycle instrument.
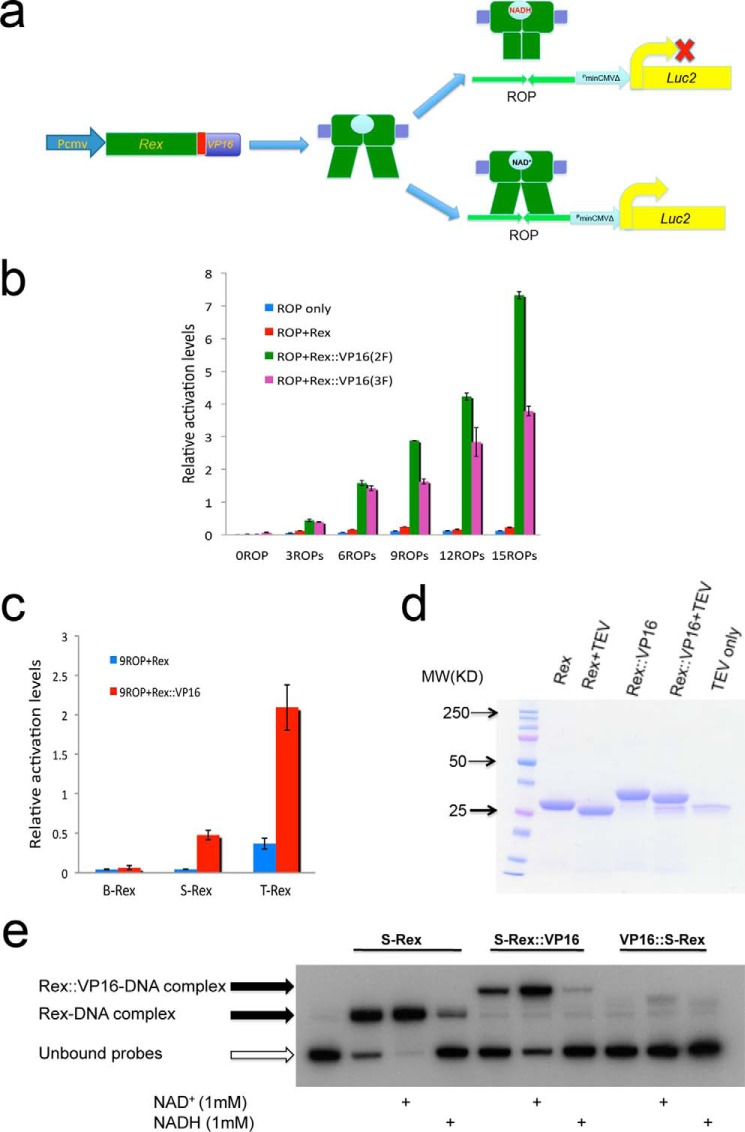
FIGURE 1.
The Rex::VP16/ROPs is a newly developed approach sensing endogenous dynamic redox changes in mammalian cells. a, schematic representation of the Rex::VP16/ROPs working diagram. b, different VP16 repeats and ROP repeats showed respective activations in HEK293T cells. c, one VP16 fused by T-Rex or S-Rex showed the report gene activations. The pGL4.10.9ROPs was used for co-transfection in HEK293T cells. d, partially purified reconstituted Rex proteins were treated by TEV to remove the N-terminal His tag. e, EMSA results showed that only C-terminal VP16-fused S-Rex exhibited DNA binding activity and responded to NADH/NAD+ treatments.
Two repeats of the minimal VP16 activation domain (VP16(2F)) or three repeats (VP16(3F)) were fused at the 3′-end of the B-Rex. Different ROP repeats were inserted in the upstream region of the luciferase gene in the pGL4.10 plasmid. After co-transfection in HEK293T cells, the Rex fusion could activate the target gene expression, and luminescence was increased with more ROP repeats, suggesting Rex::VP16 is a functional transactivator binding to its ROP. Surprisingly, the VP16(3F) did not enhance, however, it decreased Rex transcriptional activity compared with the VP16 (2F) (Fig. 1b). This indicates that the C-terminal fusion affects the N-terminal DNA binding domain of Rex. As such, only one minimal VP16 peptide was fused at the 3′-end of the three Rex proteins. Interestingly, B-Rex::VP16 did not show much activation, but both S-Rex:: VP16 and T-Rex ::VP16 exhibited transcriptional activity (Fig. 1c). We then asked whether Rex::VP16 still corresponds to NAD(H) changes. The reconstituted Rex protein was expressed and partially purified from bacteria, and the N-terminal His tag was removed with the TEV protease treatment (Fig. 1d) because the fusion with an N-terminal peptide of Rex interfered with DNA binding (28). EMSA experiments were performed to investigate Rex::VP16 DNA binding activity. In terms of S-Rex, it showed that most proteins were bound with DNA, NAD+ promoted Rex DNA binding activity; on the contrary, NADH decreased association between Rex and DNA, consistent with previous results (20). For S-Rex::VP16, similar to that seen on the S-Rex, NAD+ and NADH treatment had an opposite role upon DNA binding, indicating the fusion has same function as its original counterpart (Fig. 1e). If VP16 was fused at the 5′-end of Rex, DNA binding and NADH/NAD+ responses were abolished (Fig. 1e) (28). Thus, Rex::VP16 could act as a transactivator in vivo and sense NAD(H) in vitro.
The DNA Binding Activity of Rex::VP16 Corresponds to NADH/NAD+ Ratio Changes
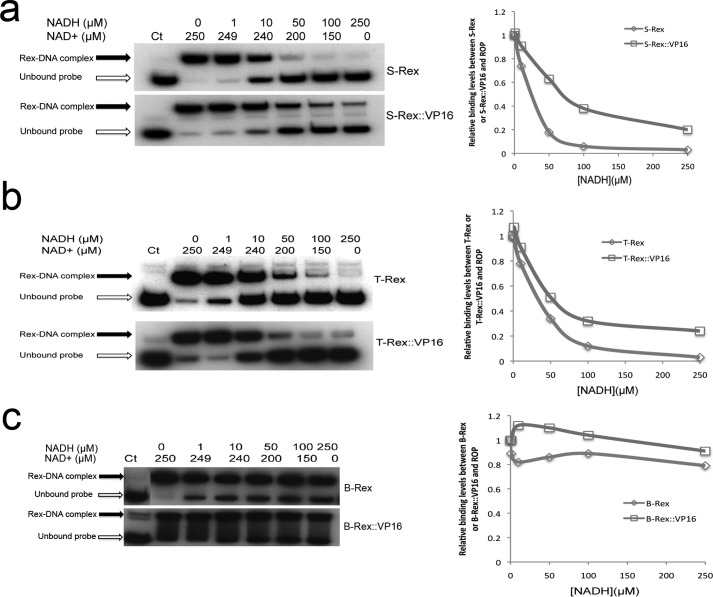
Total intracellular NAD cofactors are unlikely to change over time, but the redox state fluctuation might be due to NADH/NAD+ ratio variation in mammalian cells. We tested whether Rex::VP16 could sense NADH/NAD+ ratio changes in vitro under a constant level of total NAD cofactors. The EMSA results revealed that both S-Rex:: VP16 and T-Rex ::VP16 DNA binding activity was gradually decreased with increased NADH ratios, consistent with changes of their respective original Rex without the VP16 peptide (Fig. 2, a and b). Of note, both B-Rex::VP16 and its original protein were not sensitive to NADH/NAD+ ratio changes in vitro (Fig. 2c), suggesting there are differences in Rex redox sensing among bacterium species.
FIGURE 2.
EMSA results showed that the DNA binding activity of Rex and Rex::VP16 was differently corresponding to NADH/NAD+ ratio changes among three Rex proteins and densitometries were shown below. a, S-Rex (100 nm) and S-Rex::VP16 (350 nm); b, T-Rex (100 nm) and T-Rex::VP16 (100 nm); c, B-Rex (500 nm) and B-Rex::VP16 (100 nm).
The Rex::VP16/ROPs System Reveals Intracellular NADH Oscillations in Mammalian Cells
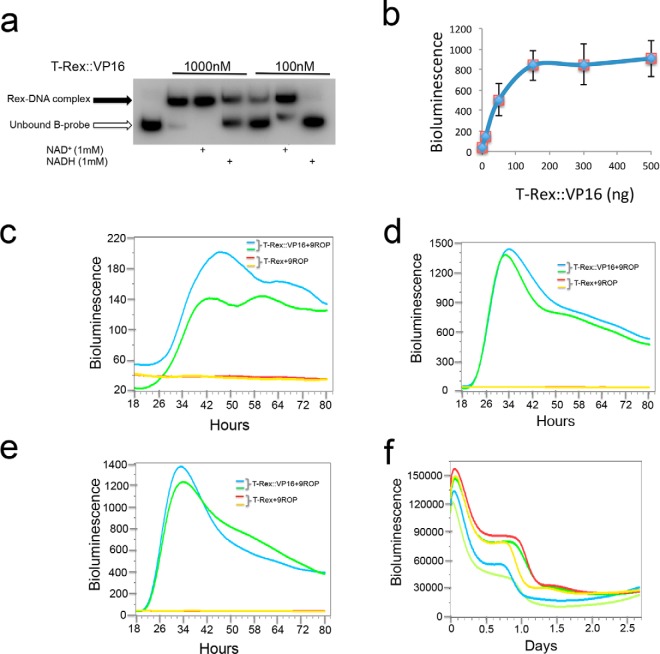
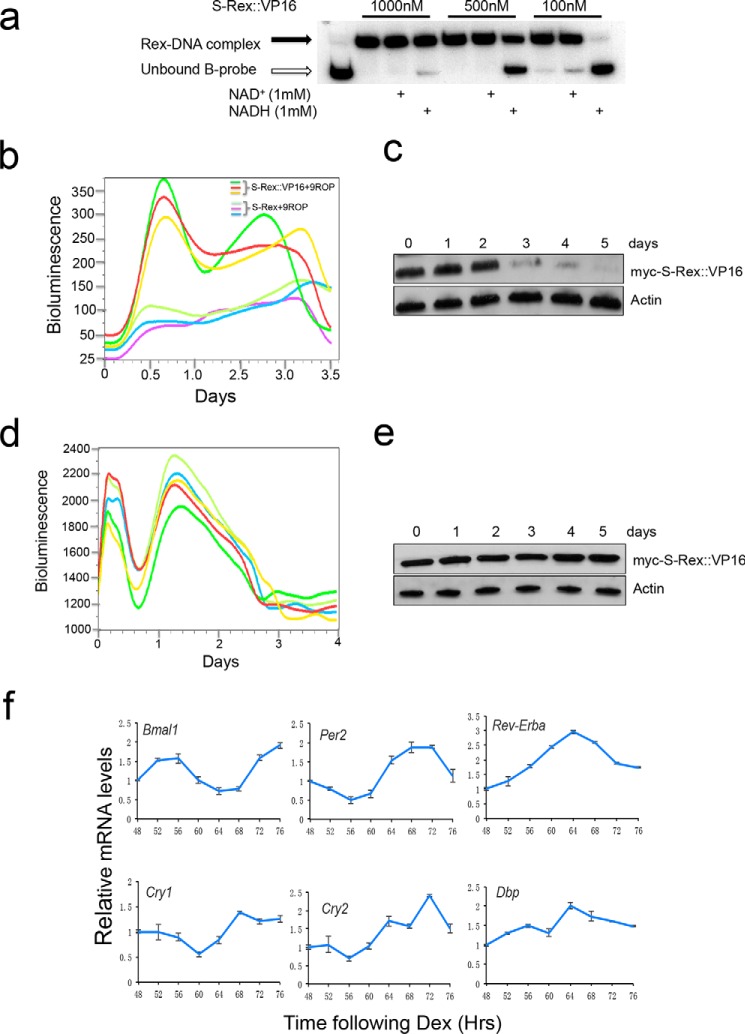
As both T-Rex:: VP16 and S-Rex ::VP16 are responsive to NADH/NAD+ ratio changes in vitro, they potentially could act as redox sensors in vivo. We noticed that the Rex sensitivity to NADH was dependent on the ratio between the Rex protein amount and DNA quantities. When T-Rex::VP16 was 1000 nm, NADH treatment resulted in partial dissociation from DNA. However, the same NADH treatment abolished total Rex DNA binding when the protein amount was ten times less (Fig. 3a) This indicates that the ratio between Rex and ROPs is important for Rex to sense cellular redox changes in vivo. Then we performed an activation titration assay. In this co-transfection with constant ROPs plasmids, the reporter gene activation was gradually saturated with more and more Rex plasmids (Fig. 3b). Based on these results, we anticipate that the T-Rex::VP16 fusion is not sensitive to NADH changes if transfected plasmids are beyond a threshold, otherwise, appropriately expressed Rex proteins could efficiently sense redox changes. Supporting this speculation, two activation oscillations were observed when 10 ng of Rex plasmids were transfected (Fig. 3c); however, the second cycle was not seen when using 150 ng (Fig. 3d) or 300 ng (Fig. 3e) plasmids due to Rex protein saturation. The stabilized cell line transfected by both plasmids expressing T-Rex::VP16 and bearing 9ROPs also showed two activation cycles (Fig. 3f). Similar to that seen in the T-Rex::VP16, different S-Rex::VP16 protein amounts also showed dissociation from DNA under NADH treatments (Fig. 4a). Optimized transient co-transfection exhibited two activation cycles (Fig. 4b). In terms of the first cycle, three individual dishes showed the peaks at the same time. However, the second cycles displayed different peaks and periods were not exactly 24 h. These suggested that the Rex protein levels could be different in individual transfection on the 2.5∼3.0 day.
FIGURE 3.
The ratio between plasmids expressing T-Rex::VP16 and plasmids bearing 9ROPs was important for activation cycles. a, T-Rex::VP16 protein amount was important for corresponding to NADH treatments. b, activation titration of T-Rex::VP16 was shown in U2OS cells. After co-transfection with the same amount of 20 ng plasmids bearing 9ROPs and different amount of plasmids expressing T-Rex::VP16, luminescence was measured at 24 h in U2OS cells. c, two cycles with low amplitudes were produced when 10-ng plasmids expressing T-Rex::VP16 were co-transfected with plasmids bearing 9ROPs in U2OS cells. d and e, second cycle was not produced when 150 ng (d) and 300 ng (e) plasmids expressing T-Rex::VP16 were co-transfected with plasmids bearing 9ROPs in U2OS cells. f, two activation cycles were shown in the stabilized U2OS cell line expressing T-Rex::VP16 proteins and bearing 9ROPs (in the pGL4.28 vector).
FIGURE 4.
Both transient and stabilized transfection showed two activation cycles converted by S-Rex::VP16. a, EMSA results showed that correspondence to NADH treatments in S-Rex::VP16 DNA binding was dependent on the protein amount. b, two activation cycles were observed in the transient co-transfection of S-Rex::VP16 and 9ROPs plasmids in U2OS cells. c, Western blot results showed that levels of S-Rex protein were stable in the first 2 days, then reduced quickly in transiently transfected cells. d, stabilized cell line showed two activation cycles converted by S-Rex::VP16. Different curves represent repeats of the same cell line. e, Western blot results showed that levels of S-Rex protein were stable in the stabilized cell line. f, qPCR results were shown that six clock genes oscillated over 48–76 h. U2OS cells transfected by plasmids expressing S-Rex::VP16 and plasmids bearing 9ROPs were synchronized by dexamethasone (Dex), and mRNAs were extracted at 4-h intervals starting at 48 h after Dex treatments.
To rule out the bioluminescence cycles are due to Rex protein oscillations, levels of the S-Rex::VP16 protein were analyzed. Interestingly, in the transient transfected cells, the Rex protein quantity was constant in the starting 2 days, afterward, it was reduced dramatically (Fig. 4c). This result suggested that the bioluminescence signal indeed comes from the Rex protein expression, and bioluminescence cycles could be due to intracellular redox changes. Furthermore, the stabilized cell line expressing both Rex::VP16 and ROPs also showed two oscillation cycles (Fig. 4d), then lost rhythms completely. However, the Rex protein levels were constant in these cells (Fig. 4e), suggesting associations between Rex and NADH were affected by the pH changes or nutrient consumption of cell culture media. Interestingly, six clock genes including Bmal1, Per2, Rev-Erba, Cry1, Cry2, and Dbp still had good rhythms over 48–76 h in the same condition (Fig. 4f).
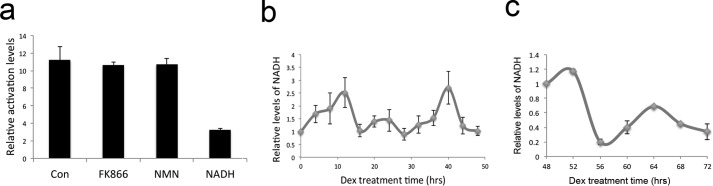
Next, we asked whether the S-Rex::VP16 activation activity is affected by endogenous NAD+ or NADH levels. FK866, a selective inhibitor of NAMPT, which is a rate-limiting step in the NAD+ salvage synthesis pathway in mammals, and nicotinamide mononucleotide (NMN), which is a product of the NAMPT enzymatic reaction, were employed to decrease or increase NAD+ levels in vivo (29). Interestingly, both FK866 and NMN administration could not change S-Rex::VP16 transcriptional activity. However, NADH treatment significantly reduced its activation levels (Fig. 5a). This indicated that S-Rex::VP16 is more sensitive to NADH than NAD+, consistent with the Rex activity regulation in prokaryotes. We hypothesize that the activation cycles by Rex::VP16 could be a readout of endogenous NADH level changes. In accordance with our observation, the NADH level displays circadian oscillations in the host cells (Fig. 5b). In comparison with the redox state and the clock gene expression, we found that both cellular NADH levels (Fig. 5c) and clock gene transcription, such as Rev-Erba and Dbp, peaked around at 64 h (Fig. 4f), suggesting the reduced redox state could promote the expression of clock genes.
FIGURE 5.
Rex::VP16 could report cellular endogenous NADH oscillations. a, NADH treatments resulted in repression of the S-Rex::VP16 transcriptional activation. NADH (1 mm), FMN (100 μm), or FK866 (10 nm) treatments 1 day after transfection 24 h in U2OS cells, then bioluminescence was measured. b, cellular NADH levels showed circadian oscillations. Data are mean ± S.D. n = 3. c, endogenous NADH levels also oscillated over 48–72 h. U2OS cells expressing both S-Rex::VP16 and 9ROPs were synchronized by Dex, and NADH was analyzed at 4-h intervals starting at 48 h after Dex treatments. Data are mean ± S.D. n = 3.
Discussion
Although levels of NAD+/NADH can be estimated based on the concentrations of cellular steady-state metabolites following cell disruption (16), this approach cannot be applied in dynamic studies in intact cells. Furthermore, the old method measures total NAD+ and NADH amounts, which are protein bound and free in the cytosol and in the mitochondria. Nevertheless, only free NAD(H) in the cytosol contributes to the cellular redox potential. How to examine the free NAD+ or NADH is not fully understood.
A less invasive strategy to monitor cellular NAD(H) is based on autofluorescence. NADH and NADPH are fluorescent under ultraviolet excitation, while NAD+ and NADP+ are not, but this approach cannot distinguish between NADH and NADPH (17).
In addition, it was reported that NAD+ levels exhibit circadian oscillations in animals (2, 3), but it was not clear whether NADH concentration changes over time. In the present study, using a heterologous fusion protein from bacteria, we revealed that Rex was a NADH sensor and our constructed genetic tool could measure endogenous cellular redox dynamic changes. Similar to NAD+ oscillations, the NADH level converted by the Rex sensor shows circadian changes in mammalian cells, consistent with the result by the conventional NADH assay method. Although Rex could sense both NAD+ and NADH in prokaryotes, it binds NAD+ 20,000 times less than NADH (21). Our results also showed that it was far more sensitive to NADH than NAD+ in mammalian cells (Fig. 5a), strongly suggesting that Rex is a NADH sensor.
More recently, by combining a circular permuted GFP with the Rex protein, two groups reported that this genetically encoded sensor could examine NADH/NAD+ changes in living cells (24, 25). However, this fluorescent sensor could not measure cellular redox dynamic changes over time. In our system, NADH amounts were converted into transcriptional luminescence signals, which were recorded by a LumiCycle instrument in intact cells.
The NAD+/NADH ratio calculated by the cellular lactate and pyruvate amount is about 700 (14), illustrating that free NAD+ levels are dramatically higher than those of NADH. The free NAD+ level could not be affected significantly by the conversion of NAD+ to NADH. We could predict that NADH changes are far more effective than NAD+ changes in affecting the intracellular NAD+/NADH ratio, indicating that NADH plays a more important role than NAD+ in regulating the cellular redox state. In addition, the Rex::VP16 fusion is more sensitive to NADH than NAD+ changes, reminiscent of the endogenous corepressor CtBP (carboxyl-terminal-binding protein). CtBP senses levels of free NADH to inhibit expression of target genes as a redox sensor. Similar to the Rex protein, NADP(H) does not function on the CtBP transcription activity (14). CtBP binding to its partner is promoted by increasing NADH at concentrations in the nm range, in accordance with the physiological level of NADH around 110 nm. Of note, binding of both T-Rex:: VP16 and S-Rex ::VP16 to their DNA was regulated by NADH concentrations in the μm range in vitro, similar to their original Rex proteins (Fig. 2, a and b). This is probably due to differences between in vivo and in vitro conditions. Because the original Rex protein can sense physiological levels of NAD+/NADH in E. coli, it is conceivable that the Rex::VP16 fusion plays a similar role in mammalian cells.
Among the three Rex proteins tested in our study, both T- and S-Rex could bind to the ROPs site and sense NADH changes in vivo and in vitro, but B-Rex failed to show much activation and was not sensitive to NADH treatment (Fig. 2). This demonstrated that there are differences among Rex family proteins in our experimental conditions. However, the fluorescent sensor developed based on the same B-Rex is highly and specifically responsive to NADH in mammalian cells. Using this approach, Zhao et al. examined the cytosolic and nuclear NADH level of 130 nm (25), which was close to the result of 110 nm measured by the conventional method (14). Also, they reported that the free NADH level is much higher in the mitochondrial matrix than in the nucleus and cytosol. This indicates that Rex is a metabolic sensor regulated by the physiological NADH level in mammalian cells. Even though both S-Rex and T-Rex could act as redox sensors in our experiment, their induced cycles were not stable and robust. Future work would be focused to modify these Rex proteins by various mutations for optimization.
It has been known that the NAD+ level displays circadian oscillations, and both NAD+-dependent SIRT1 and PARP1 connect the cellular redox state with circadian systems (6, 8). We show here that the NADH level converted by Rex displays rhythmic cycles in mammalian cells. Consistent with these data, it was reported that the free NADH concentration within epidermal stem cells showed a daily fluctuation and was controlled by the circadian clock (30). How does the NADH oscillation affect the circadian clock? One possibility is that, as NADH or NADPH enhances DNA binding of the CLOCK:BMAL1 (13), NADH oscillations could drive the heterodimer rhythmically entering and exiting in the promoter regions of the clock-controlled genes. The qPCR results showed that transcripts of two clock-controlled genes, Rev-Erba and Dbp, peaked at the same time as NADH levels at 64 h, supporting the idea that high levels of NADH promote clock gene transcription (Figs. 4f and 5c). Another possibility is that changes in the NADH level could be directly detected by endogenous specific redox sensors and transmitted into transcriptional signals. For instance, CtBP associates with NADH and acts as a corepressor for a few target genes. Of these, the SIRT1 transcription is regulated by this redox sensor. Hypoxia induces cellular high levels of NADH, which increases recruitment of CtBP, thus, inhibits SIRT1 gene expression (15). In this context, note that NAD+/NADH ratio is dramatically high, the interconversion between NAD+ and NADH leads to a large change in levels of NADH, not NAD+. It is tempting to speculate that NADH might play a more important role than NAD+ in SIRT1 regulation of circadian rhythms. Intriguingly, CtBP regulates the E-box clock gene expression as an activator of CLOCK/CYCLE in Drosophila (31). These results indicate that NADH tightly couples the cellular redox to transcriptional oscillations via CtBP and SIRT1.
Interestingly, one genetic biosensor comprising a circularly permutated Venus fluorescent protein and a NAD+-binding domain from a bacterial protein could be used to measure free subcellular NAD+ concentrations (32). We report that the Rex::VP16/ROPs system could detect free NADH oscillations in mammalian cells.
Experimental Procedures
Materials
Both pGL4.10 and pGL4.28 plasmids were purchased from Promega. The pcDNATM3.1/myc-His B vector was from Invitrogen. All other primary reagents, such as NADH, NAD+, and FK866, etc., were purchased from Sigma-Aldrich or as otherwise indicated.
Cell Cultures
HEK293T cells were grown in the Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 100 units ml−1 penicillin and 100 μg ml−1 streptomycin. U2OS cells were grown in the McCoy's 5A medium with the same supplements as HEK293T cells. Cell cultures were regularly maintained in a 5% CO2 incubator. In terms of cells for luminescence recording, they were grown in the DMEM supplemented with 0.035% sodium bicarbonate, 10 mm HEPES, 1% fetal bovine serum, 2 mm l-glutamine, 1 mm sodium pyruvate, 100 units ml−1 penicillin, 100 μg ml−1 streptomycin, 100 μm luciferin, and 100 nm dexamethasone (Dex).
Plasmids and Generation of Stabilized Cell Lines
Three Rex genes were individually cloned into the pcDNATM3.1/myc-His B vector between HindIII and BamHI sites for the mammalian expression. The single VP16 sequence (sense 5′-ccggccgacgccctggacgacttcgacctggacatgctg-3′) or more repeats (26) was inserted between BamHI and XbalI sites downstream of Rex genes. Then six amino acids (GAGAGA) were added as a linker between Rex and VP16 to ensure the activator functional. In terms of reconstituted Rex proteins, they were cloned into pHIS.Parallel1 vector for expression in E. coli. One or more ROP repeats (sense 5′-ttaattgtgaaatacttcacaatatcgt-3′) sequences (21) were inserted into the upstream region of the luciferase gene in pGL4.10 or pGL4.28 between KpnI and NheI sites. The minimal CMV promoter was inserted at the HindIII site of the pGL4.10 plasmid. All cell transfections were performed by the Effectene Transfection Reagent from Qiagen per the manufacturer's instructions. In terms of stabilized U2OS cell lines carrying Rex genes and ROP sequences, they were developed from single colonies with 50 μg ml−1 hygromycin (pGL4.28) and 100 μg ml−1 G418 (pcDNATM3.1).
Luciferase Assay
HEK293T cells (5 × 104 per well, in 24-well plates) were lysed in 100 μl of lysis buffer after 24 h transfections. Luminescence was measured following the kit protocol from Promega.
Real-time Luminescence Monitoring of Circadian Oscillations
U2OS cells were transfected by plasmids expressing Rex::VP16 and plasmids bearing ROPs. After 24 h, cells were cultured on the DMEM recording medium containing Dex for synchronization. Luciferase activity was recorded in real time using a 32-channel luminometer (LumiCycle, Actimetrics) that was maintained at 37 °C in a walk-in warm room. Stabilized cell lines were also synchronized by Dex before luminescence recording.
Electrophoretic Mobility Shift Assays (EMSAs)
PCR products (∼200 bp) that included the ROP palindromic sequence in the ywcJ promoter region from Bacillus subtilis were produced from primers: sense 5′-attggtaccgaatacgctttcatcata-3′, antisense 5′-attgctagccaaagcatactgttcaac-3′, then they were labeled with [γ-32P]ATP using polynucleotide kinase. Binding reactions were performed in a final volume 10 μl (1 μg of the nonspecific competitor poly(dI-dC), 20 mm Tris-HCl, pH 8.0, 10% glycerol, 1 mm MgCl2, 40 mm KCl). After 15 min of incubation of Rex or Rex::VP16 proteins and probes (7.6 nm) at room temperature, then reactions were loaded on a 6% polyacrylamide gel in TBE. Dried gels were radiographed on the films in the dark.
NADH Assays
NADH concentrations were measured based on the protocol from Abcam. U2OS cells were harvested and processed following the instructions in the kit. Colorimetric measurements were performed at room temperature at 450 nm on a Spectramax M2 instrument.
Western Blot Analysis
Anti-myc antibody was from Invitrogen. Actin was used as an internal loading control.
Quantitative RT-PCR
RT-PCR was performed using the following primers, and the GAPDH gene was used as an internal control for normalization. Bmal1 sense 5′-acaactttggtatcgtggaagg-3′, antisense 5′-ggacattgcgttgcatgttgg-3′; Per2 sense 5′-gacatgagaccaacgaaaactgc-3′, antisense 5′-aggctaaaggtatctggactctg-3′; Rev-Erba sense 5′-ccagagaggagaacggattcc-3′, antisense 5′-tggctgctaagcccattgc-3′; Cry1 sense 5′-ttggaaaggaacgagacgcag-3′, antisense 5′-cggttgtccaccattgagtt-3′; Cry2 sense 5′-tcccaaggctgttcaaggaat-3′, antisense 5′-tgcatcccgttctttcccaaa-3′; Dbp sense 5′-ctgatcttgccctatcaagcatt-3′, antisense 5′-cgatgtcttcgagggtcaaag-3′.
Author Contributions
J. S. T. and G. H. conceived and designed the study and wrote the paper. Y. Z. performed the qPCR of clock genes. Y. S. and S. Y. cloned Rex genes and made plasmid constructions. Y. C. purified the Rex proteins. H. W. makes contributions to analysis and interpretation of data.
Acknowledgments
We thank Jiaxi Wu (rotation graduate in UT Southwestern Medical Center) for technical support. Dr. Wachenfeldt in Lund University kindly provided the plasmid bearing the Rex gene from Bacillus subtilis in the early stage of this project. The bacterial genomic DNA samples for Rex cloning were contributed by different laboratories.
This work was supported by National Natural Science Foundation of China Grant 31571206 (to G. H.). The authors declare that they have no conflicts of interest with the contents of this article.
- CLOCK
- circadian locomoter output cycle kaput
- BMAL1
- brain and muscle Arnt-like protein 1
- NAD(H)
- nicotinamide adenine dinucleotide
- NPAS2
- neuronal PAS domain-containing protein 2
- Per
- period
- Cry
- cryptochrome
- SIRT1
- sirtuin 1
- NAMPT
- nicotinamide phosphoribosyltransferase
- PARP-1
- poly(ADP-ribose) polymerase 1
- TTFL
- transcription/translation feedback loop
- ROP
- Rex-binding operator
- EMSA
- electrophoretic mobility shift assay
- CtBP
- C-terminal-binding protein 1
- NMN
- nicotinamide mononucleotide
- DBP
- D element-binding protein.
References
- 1. Mohawk J. A., Green C. B., and Takahashi J. S. (2012) Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35, 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ramsey K. M., Yoshino J., Brace C. S., Abrassart D., Kobayashi Y., Marcheva B., Hong H. K., Chong J. L., Buhr E. D., Lee C., Takahashi J. S., Imai S., and Bass J. (2009) Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324, 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakahata Y., Sahar S., Astarita G., Kaluzova M., and Sassone-Corsi P. (2009) Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Asher G., Gatfield D., Stratmann M., Reinke H., Dibner C., Kreppel F., Mostoslavsky R., Alt F. W., and Schibler U. (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134, 317–328 [DOI] [PubMed] [Google Scholar]
- 5. Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., Guarente L. P., and Sassone-Corsi P. (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belden W. J., and Dunlap J. C. (2008) SIRT1 is a circadian deacetylase for core clock components. Cell 134, 212–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Asher G., Reinke H., Altmeyer M., Gutierrez-Arcelus M., Hottiger M. O., and Schibler U. (2010) Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142, 943–953 [DOI] [PubMed] [Google Scholar]
- 8. Kumar V., and Takahashi J. S. (2010) PARP around the clock. Cell 142, 841–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peek C. B., Affinati A. H., Ramsey K. M., Kuo H. Y., Yu W., Sena L. A., Ilkayeva O., Marcheva B., Kobayashi Y., Omura C., Levine D. C., Bacsik D. J., Gius D., Newgard C. B., Goetzman E., Chandel N. S., Denu J. M., Mrksich M., and Bass J. (2013) Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342, 1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang T. A., Yu Y. V., Govindaiah G., Ye X., Artinian L., Coleman T. P., Sweedler J. V., Cox C. L., and Gillette M. U. (2012) Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science 337, 839–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Neill J. S., and Reddy A. B. (2011) Circadian clocks in human red blood cells. Nature 469, 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edgar R. S., Green E. W., Zhao Y., van Ooijen G., Olmedo M., Qin X., Xu Y., Pan M., Valekunja U. K., Feeney K. A., Maywood E. S., Hastings M. H., Baliga N. S., Merrow M., Millar A. J., Johnson C. H., Kyriacou C. P., O'Neill J. S., and Reddy A. B. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rutter J., Reick M., Wu L. C., and McKnight S. L. (2001) Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293, 510–514 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Q., Piston D. W., and Goodman R. H. (2002) Regulation of corepressor function by nuclear NADH. Science 295, 1895–1897 [DOI] [PubMed] [Google Scholar]
- 15. Zhang Q., Wang S. Y., Fleuriel C., Leprince D., Rocheleau J. V., Piston D. W., and Goodman R. H. (2007) Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc. Natl. Acad. Sci. U. S.A. 104, 829–833 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16. Williamson D. H., Lund P., and Krebs H. A. (1967) The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem. J. 103, 514–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Skala M. C., Riching K. M., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K. W., White J. G., and Ramanujam N. (2007) In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. U. S.A. 104, 19494–19499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blacker T. S., Mann Z. F., Gale J. E., Ziegler M., Bain A. J., Szabadkai G., and Duchen M. R. (2014) Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat. Commun. 5, 3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Christensen C. E., Karlsson M., Winther J. R., Jensen P. R., and Lerche M. H. (2014) Non-invasive in-cell determination of free cytosolic [NAD+]/[NADH] ratios using hyperpolarized glucose show large variations in metabolic phenotypes. J. Biol. Chem. 289, 2344–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brekasis D., and Paget M. S. (2003) A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J. 22, 4856–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang E., Bauer M. C., Rogstam A., Linse S., Logan D. T., and von Wachenfeldt C. (2008) Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol. Microbiol. 69, 466–478 [DOI] [PubMed] [Google Scholar]
- 22. Sickmier E. A., Brekasis D., Paranawithana S., Bonanno J. B., Paget M. S., Burley S. K., and Kielkopf C. L. (2005) X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure 13, 43–54 [DOI] [PubMed] [Google Scholar]
- 23. McLaughlin K. J., Strain-Damerell C. M., Xie K., Brekasis D., Soares A. S., Paget M. S., and Kielkopf C. L. (2010) Structural basis for NADH/NAD+ redox sensing by a Rex family repressor. Mol. Cell 38, 563–575 [DOI] [PubMed] [Google Scholar]
- 24. Hung Y. P., Albeck J. G., Tantama M., and Yellen G. (2011) Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 14, 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao Y., Jin J., Hu Q., Zhou H. M., Yi J., Yu Z., Xu L., Wang X., Yang Y., and Loscalzo J. (2011) Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab. 14, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baron U., Gossen M., and Bujard H. (1997) Tetracycline-controlled transcription in eukaryotes: novel transactivators with graded transactivation potential. Nucleic Acids Res. 25, 2723–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong H. K., Chong J. L., Song W., Song E. J., Jyawook A. A., Schook A. C., Ko C. H., and Takahashi J. S. (2007) Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS genetics 3, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Du X., and Pène J. J. (1999) Identification, cloning and expression of p25, an AT-rich DNA-binding protein from the extreme thermophile, Thermus aquaticus YT-1. Nucleic Acids Res. 27, 1690–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Revollo J. R., Körner A., Mills K. F., Satoh A., Wang T., Garten A., Dasgupta B., Sasaki Y., Wolberger C., Townsend R. R., Milbrandt J., Kiess W., and Imai S. (2007) Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 6, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stringari C., Wang H., Geyfman M., Crosignani V., Kumar V., Takahashi J. S., Andersen B., and Gratton E. (2015) In vivo single-cell detection of metabolic oscillations in stem cells. Cell Reports 10, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Itoh T. Q., Matsumoto A., and Tanimura T. (2013) C-terminal binding protein (CtBP) activates the expression of E-box clock genes with CLOCK/CYCLE in Drosophila. PloS one 8, e63113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cambronne X. A., Stewart M. L., Kim D., Jones-Brunette A. M., Morgan R. K., Farrens D. L., Cohen M. S., and Goodman R. H. (2016) Biosensor reveals multiple sources for mitochondrial NAD(+). Science 352, 1474–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]