Abstract
Properly condensed chromosomes are necessary for accurate segregation of the sisters after DNA replication. The Escherichia coli condesin is MukB, a structural maintenance of chromosomes (SMC)-like protein, which forms a complex with MukE and the kleisin MukF. MukB is known to be able to mediate knotting of a DNA ring, an intramolecular reaction. In our investigations of how MukB condenses DNA we discovered that it can also mediate catenation of two DNA rings, an intermolecular reaction. This activity of MukB requires DNA binding by the head domains of the protein but does not require either ATP or its partner proteins MukE or MukF. The ability of MukB to mediate DNA catenation underscores its potential for bringing distal regions of a chromosome together.
Keywords: DNA, DNA binding protein, DNA structure, DNA topology, nucleic acid enzymology, DNA enzymes
Introduction
The Escherichia coli chromosome is condensed about 1000-fold to fit into the nucleoid of the bacterium. A number of factors contribute to this extreme DNA condensation, among them are: DNA supercoiling, the binding of various nucleoid associated proteins like HU, Fis, and H-NS, and the binding of the structural maintenance of chromosomes (SMC)3-like condensin MukBEF (1, 2). SMC proteins act to manage the shape and behavior of chromosomes in both prokaryotes and eukaryotes (3). These proteins dimerize at a hinge region that is flanked by long coiled-coil regions that can be 40–50 nm in length and that end in head domains that bind and hydrolyze ATP, as well as the bridging kleisin protein (MukF for the E. coli condensin (4)). The kleisin is then itself bound by another protein (MukE for the E. coli condensin (4)). There are three versions of the eukaryotic SMC proteins: the condensin, required for packaging of the chromosomes and comprised of SMC2 and SMC4; the cohesin, required for holding sister chromosomes together during mitosis and comprised of SMC1 and SMC3; and the SMC5-SMC6 complex, required for various aspects of DNA repair (3).
It is generally acknowledged that the eukaryotic condensin and cohesin trap DNA topologically in the protein triangle formed by the SMC proteins and the kleisin (5, 6), although an alternative model for DNA binding for the eukaryotic cohesin exists (7). Recent reports suggest that the Bacillus subtilis SMC protein (8) and MukB (9) also trap chromosomes topologically. MukB binds linear and circular DNA in vitro and can induce negative supercoils and knots in relaxed circular DNA in the presence of a topoisomerase (10). Binding of MukB to chromosomal DNA in vivo requires ATP and MukEF (11, 12), although MukB DNA binding in vitro does not (10, 13).
We (14) and the Burger and Oakley (15) labs have shown that the MukB hinge region interacts with the C-terminal β-propeller region of the ParC subunit of the cellular decatenase topoisomerase IV (Topo IV). We have reported (16) that this interaction stimulates the intramolecular activities of Topo IV, negative supercoil relaxation and knotting, but not the intermolecular activities of Topo IV, catenation/decatenation of DNA rings; whereas Berger, Oakley and colleagues (15) have shown a moderate stimulation by MukB of Topo IV-catalyzed decatenation of Crithidia mitochondrial DNA.
Strains deleted for either mukB or the subunits of Topo IV (parE and parC) show chromosome decondensation and segregation defects at the nonpermissive temperature (17, 18). We have proposed that the Topo IV-MukB interaction is necessary for efficient chromosome condensation, with Topo IV possibly stabilizing MukB interactions with distal regions of the chromosome to form loops in the DNA (16). Disruption of this interaction would thereby cause chromosome decondensation that could then disrupt proper chromosome decatenation.
We demonstrate here that MukB can indeed bind two distinct DNA rings and bring them together in a fashion that introduces intermolecular interwindings that can then be catenated by topoisomerase III (Topo III). MukB-mediated DNA catenation did not require and was, in fact, inhibited by ATP. Similarly, the catenation reaction did not require MukEF and was also inhibited in their presence.
Results
MukB Can Mediate Catenation of DNA Rings
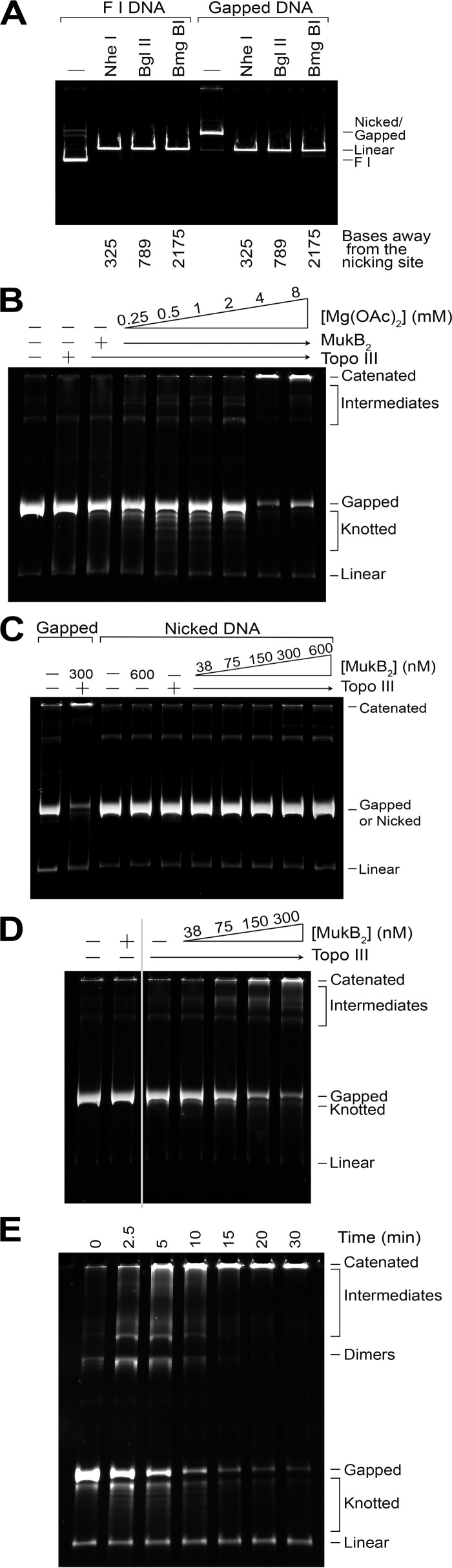
A complicating feature of many studies of topological modulation of DNA by SMC proteins is that the SMC protein and the type II topoisomerase generally used to convert the SMC-induced modulation of the path of the DNA helix to a recordable topological change both use ATP. To allow assessment of the role of ATP in MukB DNA binding, we developed a slightly different assay system. In experiments to be reported elsewhere,4 using a large 11-kbp plasmid DNA, we have developed an assay to score MukB-mediated condensation of DNA. To remain consistent and to address the possibility that the large MukB protein might not interact with small plasmid DNAs in a manner reflective of how it interacts with chromosomal DNA, we used the same large, 11-kbp plasmid DNA in the experiments reported in this manuscript. To eliminate the need for a type II topoisomerase, we formed a small gap (<325 nucleotides) in one of the DNA strands by first nicking the DNA with the nicking restriction endonuclease Nb.BbVCI and then creating a small gap in one strand by digestion with exonuclease III (gapped DNA (gDNA)) (Fig. 1A). The presence of this gap allows Topo III, a type IA topoisomerase that requires a single-stranded DNA site of action but does not require ATP (19), to be used to freeze topological alterations in the DNA.
FIGURE 1.
MukB-mediated catenation of DNA. A, the gapped DNA substrate has gaps that are less than 325 nucleotides in length. Either supercoiled pCG09 form I DNA (F1 DNA) or gapped pCG09 DNA prepared as described under “Experimental Procedures,” were treated with the indicated restriction enzymes that cut the plasmid DNA only once at the distance indicated from the nicking site. Complete digestion to linear product indicates that the DNA must be double-stranded at the indicated site. B, in the presence of Topo III, MukB mediates knotting of single DNA rings at low Mg2+ concentrations, but mediates catenation of DNA rings at moderate Mg2+ concentrations. Reaction mixtures containing 0.7 nm gapped DNA, 25 nm Topo III (a plus above the lane followed by an arrow indicates that all reactions analyzed in lanes covered by the arrow contained the indicated component), 300 nm MukB (as dimer), as indicated, and the indicated concentrations of Mg(OAc)2, were incubated for 30 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” C, a gap is required to observe DNA catenation. Standard reaction mixtures containing either gapped DNA or DNA that had only been nicked with Nb.BbVCI (nicked DNA), 25 nm Topo III, as indicated, and the indicated concentration of MukB were incubated for 30 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” D, MukB-mediated catenation of DNA in the presence of Topo III as a function of MukB concentration. Standard reaction mixtures containing 25 nm Topo III and the indicated concentrations of MukB were incubated for 5 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” E, time course of DNA catenation at 300 nm MukB. A standard reaction mixture was increased 8-fold in size in volume and incubated at 37 °C. Aliquots (20 μl) were removed at the indicated times and processed and analyzed as described under “Experimental Procedures.” Note that the zero time aliquot was removed after mixing all components of the reaction together, accounting for the slight catenation and knotting observed. Catenated, large networks of catenated DNA rings that do not enter the gel, some of this material will wash off the top of the gels during handling. Intermediates, less complicated catenated networks of DNA rings were probably made of chains of DNA rings linked once. Gapped, gDNA starting material. Knotted, single DNA rings that have become knotted. Linear, a trace amount of linear pCG09 DNA present in the gDNA preparation. All assays were repeated at least three times, representative gels are shown.
The products formed when gDNA was incubated in the presence of MukB and Topo III varied depending on the concentration of Mg2+ in the reaction mixture (Fig. 1B). At low concentrations of Mg2+, bands that moved faster than the gDNA were observed that represented the formation of knots in the DNA, as has been observed previously (10). At higher concentrations of Mg2+, DNA product accumulated at the top of the gel (labeled Catenated in Fig. 1). Accumulation of these large products that did not enter the gel was dependent on the presence of a gap in the DNA (Fig. 1C), increased with increasing concentrations of MukB (Fig. 1D), and with time of incubation (Fig. 1E).
These large products were reminiscent of where catenated DNA rings migrate on agarose gels (20). The large size of the network of interlocked DNA rings generated prevents the products from entering the gel. The reaction mixtures were digested routinely with Proteinase K in the presence of SDS, eliminating the possibility that MukB aggregation on the DNA was preventing the DNA products from entering the gel. Furthermore, product bands could be observed at early times in the reaction that migrated between the gDNA and the top of the gel (labeled Intermediates in Fig. 1) that were apparently chased into larger products (Fig. 1C). These bands likely represented small strings of DNA rings catenated together (i.e. dimer, trimer, etc.), supporting the idea that the large products were catenated DNA.
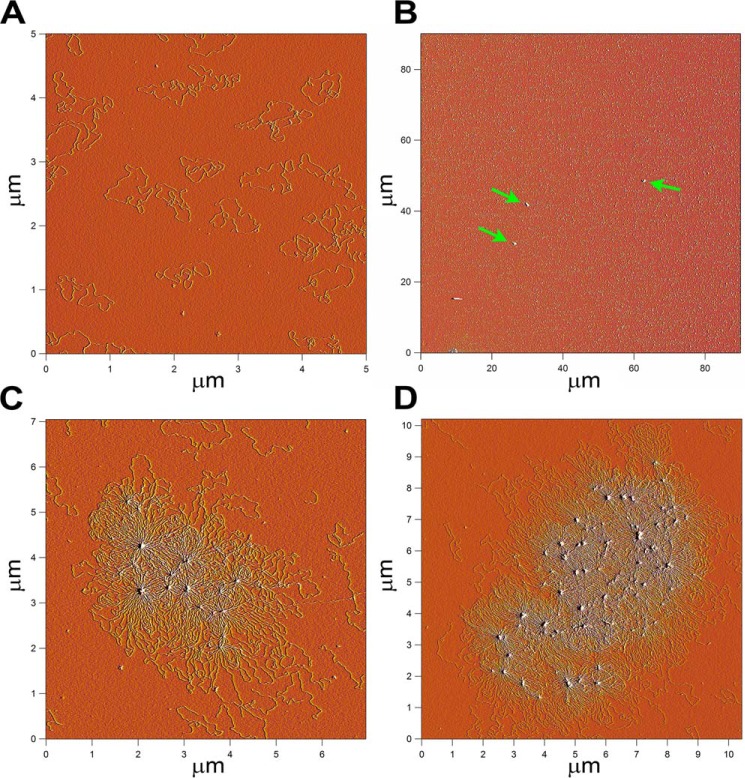
To observe directly whether the large DNA products were catenated DNA rings, they were examined by atomic force microscopy (AFM). The preparation of gDNA appeared as single DNA rings in the AFM fields at high density as expected (Fig. 2A). On the other hand, a preparation of catenated DNA spread on mica for AFM at the same molar concentration (per DNA monomer) as the gDNA displayed very few features. Some small, dense objects could be discerned (arrows in Fig. 2B) when large fields were examined. Closer inspection of these small, dense objects revealed them to be large networks of catenated DNA (Fig. 2, C and D). It was not possible to determine from the AFM images whether the networks represented DNA rings that were each catenated only once or multiple times to another DNA ring or a mixture of such.
FIGURE 2.
Atomic force microscopic images of catenated DNA. A, monomer pCG09 DNA rings. B, low resolution image of catenated DNA spread at the same total concentration of monomer DNA as in panel A. The green arrows point to the catenated DNA. C and D, high resolution images of the catenated DNA called out in panel B. Microscopy was as described under “Experimental Procedures.”
To further establish that the large products were catenated DNA rings, a preparation of catenated DNA was treated with Topo IV, which is a very active decatenating enzyme (21, 22). In the presence of very low concentrations of Topo IV (125 pm), the large DNA products were partially resolved into intermediate bands and the monomer gDNA (Fig. 3). Incubating with a 4-fold higher concentration of Topo IV resolved all of the large DNA products into monomer gDNA (Fig. 3).
FIGURE 3.
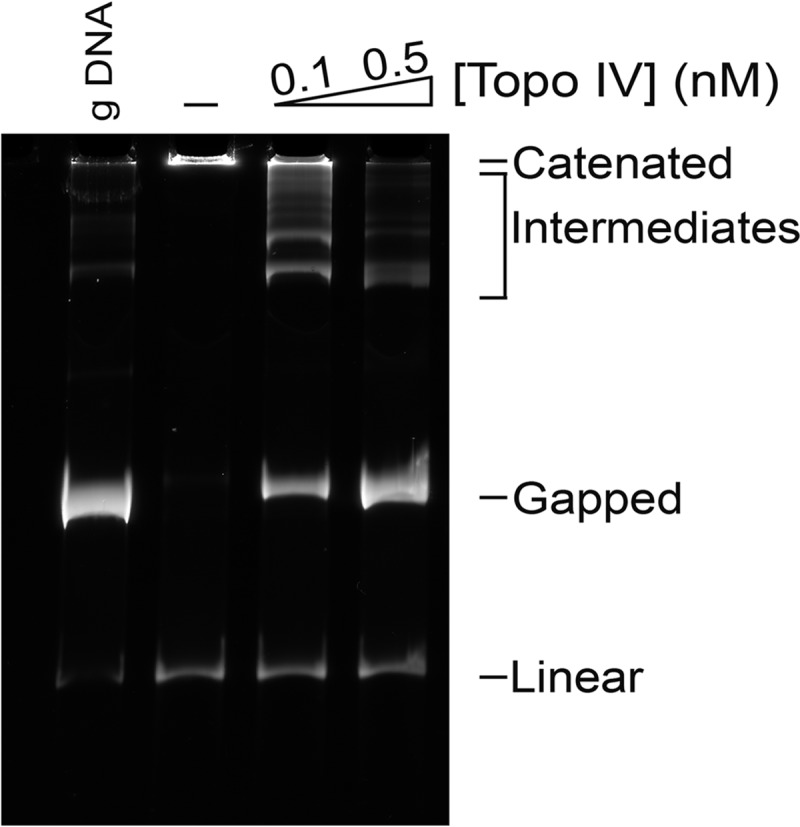
Topo IV decatenates catenated DNA formed by MukB and Topo III. Catenated DNA formed in the presence of MukB and Topo III was isolated and purified as described under “Experimental Procedures.” Reaction mixtures (20 μl) containing catenated DNA (∼100 ng), and either no or the indicated concentrations of Topo IV were incubated for 30 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” The assay was repeated at least three times, a representative gel is shown.
The data reported in Figs. 2 and 3 prove that the large DNA products observed in Fig. 1 were networks of catenated DNA and that MukB had to be present in the reaction mixture for this product to be formed. However, it was still possible that MukB was merely acting in some way to neutralize the negative charges on the DNA molecules to such an extent that Topo III itself was capable of catenating the DNA molecules. The condensation of DNA that occurs when the negative charges of the phosphates are neutralized by greater than 90% allows catenation of DNA rings by topoisomerases (20). Under our assay conditions, Topo III alone could not catenate the DNA rings, even at Mg2+ concentrations 4-fold greater than what was used routinely in the MukB-mediated DNA catenation reaction (Fig. 4A). This observation indicated that the Mg2+ concentrations used in the standard reaction condition was itself insufficient to neutralize the negative charge of the DNA to the extent that Topo III could catenate the DNA. To address whether MukB was playing an active role in distorting the path of the DNA helix leading to catenation, we used a variant MukB protein that was deficient in DNA-binding activity and assessed its ability to mediate DNA catenation.
FIGURE 4.
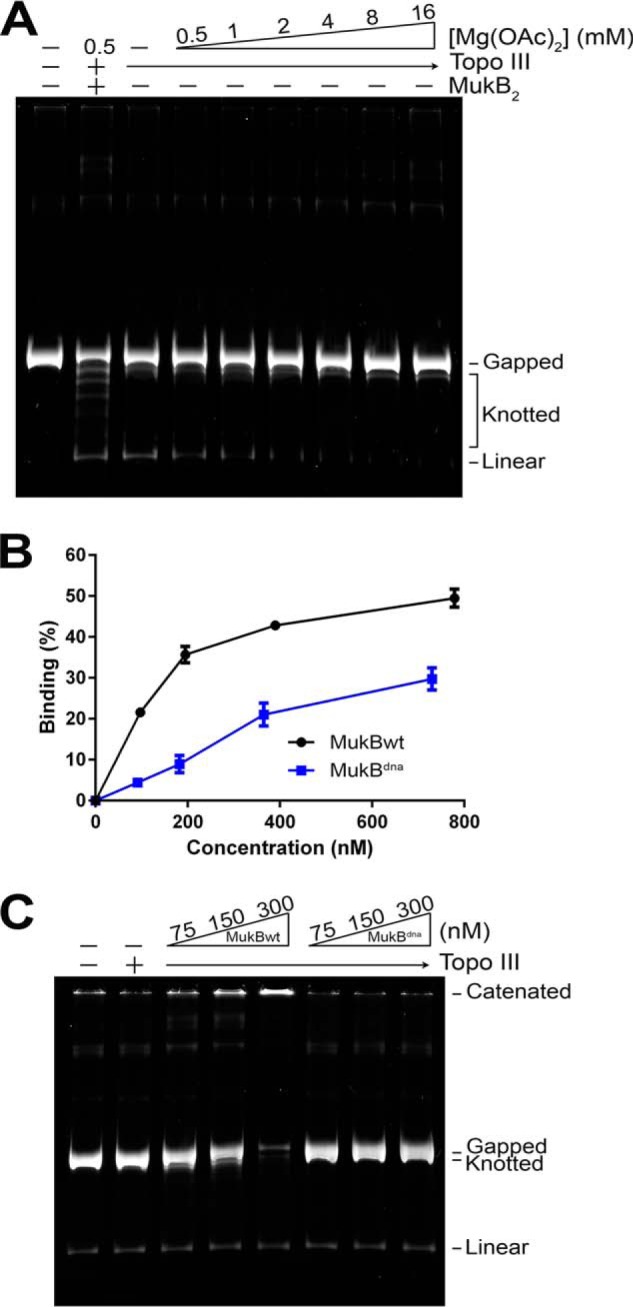
MukB DNA-binding activity is required for catenation. A, Topo III alone does not catenate the gDNA. Standard reaction mixtures containing gDNA and 25 nm Topo III with the indicated concentrations of Mg(OAc)2 were incubated for 30 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” The assay was repeated at least three times, a representative gel is shown. B, nitrocellulose filter DNA binding assays of wild type MukB and MukBdna. Binding to a duplex 5′-32P 50-bp long oligonucleotide was measured as described under “Experimental Procedures.” The mean ± S.D. of three experiments is plotted. C, MukB DNA-binding activity is required for DNA catenation. Standard reaction mixtures containing 25 nm Topo III and either no MukB or the indicated concentrations of either MukB or MukBdna were incubated for 30 min at 37 °C processed and analyzed as described under “Experimental Procedures.” The assay was repeated at least three times, a representative gel is shown.
We had previously engineered and purified a MukB variant that was defective in DNA binding (MukBdna (16), R187E/R189E). These two amino acid substitutions are the equivalent of the R216E and R218E mutations made in Haemophilus ducreyi MukB that elicited DNA binding deficiency (13). These two mutations sit on the head domain of the MukB dimer.
The DNA-binding activity of MukBdna was about one-fourth that of the wild type protein in a nitrocellulose filter binding assay using a duplex 50-bp long oligonucleotide as the substrate (Fig. 4B) (based on a non-linear regression curve fit for saturation binding of one site using total ligand). MukBdna failed to mediate catenation of DNA rings when compared with equivalent concentrations of wild type protein (Fig. 4C), indicating that the DNA-binding function of MukB was necessary to observe the catenation activity. Taken together, we conclude from the data presented in this section that MukB mediates DNA catenation by binding two distinct DNA rings and bringing them in close enough proximity that Topo III can catenate them. A possible alternate explanation, that binding of MukB to the DNA provides a preferred binding site for Topo III action, seems unlikely because Topo III acts only on single-stranded DNA (19).
ATP Inhibits MukB-mediated Catenation of DNA
MukB, as all other SMC proteins (3), is an ATPase, albeit not a very active one on its own (10). Biochemical and structural studies have demonstrated that the binding of two ATPs is shared by the two head domains in the dimeric protein, bringing them together in a closed conformation (23). ATPase activity is stimulated by binding of the kleisin, MukF, and its associated protein, MukE (24), suggesting that the cycle of ATP binding and hydrolysis could regulate the opening and closing of the tripartite ring of the SMC complexes. This action has been invoked as a necessary part of the ability of the eukaryotic cohesin and condensin to trap DNA and allow it to exit (3). In both B. subtilis and E. coli, the ability of SMC and MukB, respectively, to bind and hydrolyze ATP has been shown to be necessary for their binding to chromosomal DNA (8, 12, 25, 26). However, binding of DNA to MukB in vitro does not require ATP (Fig. 4) (10).
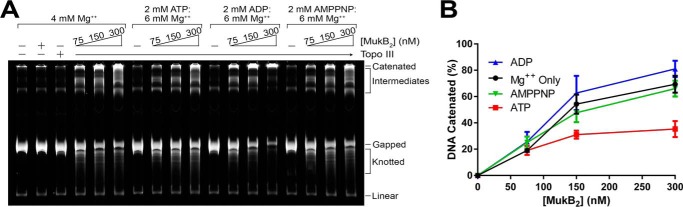
We investigated the influence of ATP, ADP, and the non-hydrolyzable ATP analog, AMP-PNP on the MukB-mediated DNA catenation reaction (Fig. 5). In general, DNA catenation reactions are difficult to quantify because of the likelihood that some of the catenated DNA network washes off the top of the gel during electrophoresis and staining, leaving quantification of the relative extent of catenation in any particular lane on the gel problematic. Nevertheless, it was clear from assessing the amount of gDNA remaining in any particular lane compared with the control lane that ATP inhibited the MukB-mediated catenation reaction, whereas ADP stimulated slightly and AMP-PNP inhibited to a lesser extent compared with ATP (Fig. 5, A and B). These data are consistent with previous observations and indicate that even when the head domains spend the vast majority of the time in the closed conformation (in the presence of AMP-PNP, which has been shown to lock the head domains together as ATP does (23)) MukB is able to bind DNA and modify topology.
FIGURE 5.
Effect of ATP moieties on MukB-mediated DNA catenation. A, MukB-mediated DNA catenation reactions containing the indicated concentrations of MukB, Mg(OAc)2, and ATP, ADP, or AMP-PNP were incubated for 30 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” The experiment was repeated three times, a representative gel is shown. B, quantification of the extent of DNA catenation in the reactions. The fraction of DNA catenated in any particular lane was calculated as 1 − the ratio of the gDNA remaining compared with the starting material. Because the networks of catenated DNA wash off the top of the gel during electrophoresis and handling, using a ratio of the catenated DNA at the top of the gel to the gDNA remaining in any particular lane will not give accurate values of the extent of catenation. The plot shows the mean ± S.D. of three experiments.
MukEF Inhibits MukB-mediated Catenation of DNA
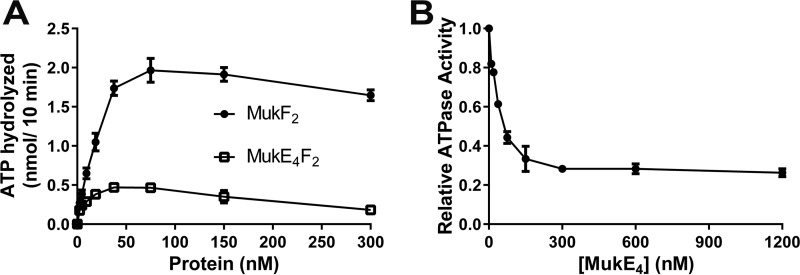
Binding of MukEF to MukB stimulates turnover of ATP that is bound to MukB, i.e. the ATPase activity of MukB is enhanced (24). Thus, we asked if the addition of MukEF to MukB-mediated catenation reactions that were inhibited by ATP led to a recovery of the catenation activity. First, based on observations communicated to us by David Sherratt (University of Oxford),5 we assessed the requirement for MukE in stimulation of MukB ATPase (Fig. 6). Interestingly, the presence of MukE actually inhibited stimulation of the MukB ATPase by MukF (Fig. 6B). Maximum stimulation of MukB ATPase by MukF occurred at the substoichiometric ratio of 1 MukF dimer to 4 MukB dimers (Fig. 6A), suggesting that the MukF was acting catalytically with respect to MukB dimers and that turnover of the MukB-MukF complex was rapid and modulated by ATP. On the other hand, MukE inhibition of the MukF-stimulated MukB ATPase suggested that the presence of MukE tempered the rate of MukF-MukB turnover.
FIGURE 6.
Effect of MukF, MukE, and the MukEF complex on MukB ATPase activity. A, MukF alone is sufficient to stimulate MukB ATPase activity. Standard ATPase reaction mixtures containing 300 nm MukB2 and the indicated concentration of either MukF or the MukEF complex were incubated for 10 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” The plot shows the mean ± S.D. from three experiments. The very low ATPase activity of MukB alone (0.11 nmol/10 min ± 0.1 nmol) was subtracted from all values. B, MukE inhibits MukF-stimulated MukB ATPase activity. Standard reaction mixtures containing 300 nm MukB2, 75 nm MukF2, and the indicated concentrations of MukE were incubated, processed, and analyzed as described under “Experimental Procedures.” The plot shows the mean ± S.D. from three experiments. Even though MukE is a dimer, the concentration is given as a tetramer because the final stoichiometry in a MukBEF complex is MukB2(MukE2MukF)2 (23).
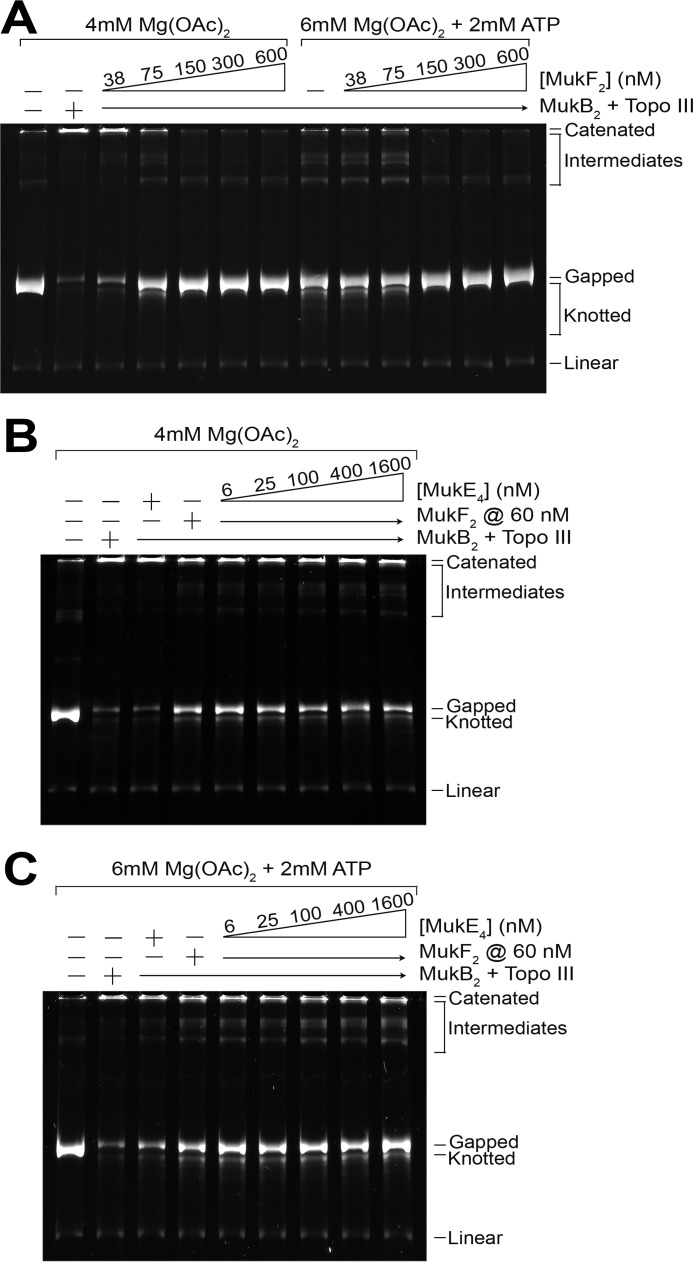
MukF inhibited MukB-mediated catenation of DNA (Fig. 7A) at molar ratios to MukB where it stimulated the MukB ATPase activity maximally (Fig. 6A). Because both the binding of ATP and the binding of MukF to MukB act to form a closed protein loop (23), it was not surprising that the addition of ATP to catenation reactions containing both MukF and MukB did not alter the observed inhibition. Furthermore, the addition of MukE did not overcome the inhibition by MukF either in the presence or absence of ATP (Fig. 7, B and C). We conclude from these data that the head domains of MukB must be conformationally free for catenation to occur.
FIGURE 7.
Effect of MukF and MukE on MukB-mediated catenation of DNA. A, MukF inhibits MukB-mediated catenation of DNA. Standard DNA catenation reactions containing 300 nm MukB2 either in the presence or absence of ATP and either in the presence or absence of the indicated concentrations of MukF were incubated for 30 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” The assay was repeated at least three times, a representative gel is shown. B and C, MukE does not overcome MukF inhibition of MukB-mediated DNA catenation. Standard DNA catenation reaction mixtures containing 300 nm MukB2, 75 nm MukF2, and the indicated concentrations of MukE4 either in the absence (panel B) or presence (panel C) of ATP were incubated for 30 min at 37 °C, processed, and analyzed as described under “Experimental Procedures.” The assay was repeated at least three times, a representative gel is shown.
Discussion
We have described a reaction mediated by the bacterial condensin MukB that requires the intertwining of two separate molecules of DNA for them to be catenated by the action of Topo III.
Possible Mechanisms of the MukB-mediated Catenation Reaction
Catenation of DNA rings can occur in the absence of any protein other than the topoisomerase necessary to pass DNA strands to lock the topological linkage in place. In these reactions, about 90% of the negative charges on ColE1 plasmid DNA molecules had to be neutralized for the DNA to aggregate and be catenated by DNA gyrase. Charge neutralization could be effected by the presence of mono-, di-, and trivalent cations (20). The E. coli type I enzyme topoisomerase I worked as well as DNA gyrase as long as there was some nicked DNA present in the reactions. In that case, as with the reaction we describe here with Topo III, the type I enzyme is passing DNA strands through a double strand break created by gating the DNA opposite the nick (or on the single-stranded gap in our case). And, indeed, Topo III alone could readily catenate the DNA rings if sufficient spermidine was added to the catenation reactions to neutralize charge (data not shown). However, our standard reaction conditions contain only 4 mm Mg2+, an insufficient amount to account for extensive charge neutralization, as shown by the failure of Topo III alone to catenate the DNA (Fig. 4A).
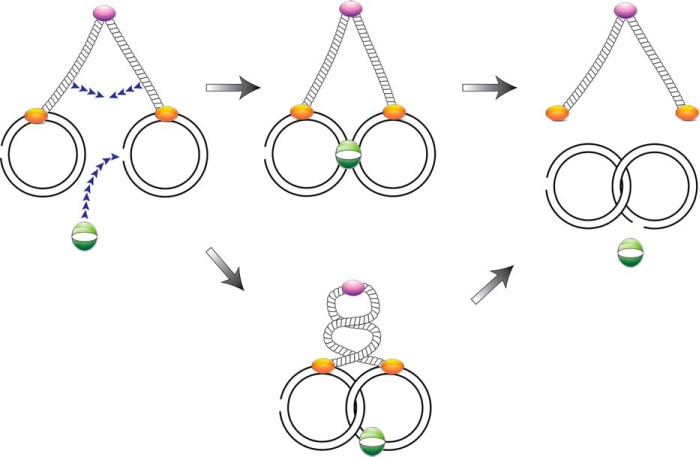
It is conceivable that MukB-mediated catenation resulted from MukB itself acting as the agent of charge neutralization. Catenation could be observed at 75 nm MukB (Fig. 1B). The DNA substrate is 11 kbp in length and present at 0.7 nm in the reaction mixture. Neutralization of 90% of all the negative charges by direct binding of all the MukB in the reaction mixture to the DNA would therefore require that the MukB head dimer bind about 90 bp of DNA (and completely neutralize the charge). This seems quite unlikely. The presumptive DNA-binding site on the head domain dimer is only about 80 Å across at its widest point (23). Instead, we argue that MukB is playing an active role by binding two separate DNA rings and bringing them together in such a way that the helices either become intertwined or are brought close enough together that when Topo III passes strands catenation occurs (Fig. 8).
FIGURE 8.
Model for MukB-mediated DNA catenation. MukB (purple, hinge domain, stippled tubes, as coiled-coils, and orange, head domains) likely mediates catenation by bringing two molecules of gapped DNA close enough together that the probability increases when Topo III (green oval) gates on the ssDNA in the gap to pass a duplex segment from the other molecule of DNA through the gap. MukB is likely to bind two DNA molecules in the process, one via each head domain in the MukB dimer. Proximity of the DNA molecules might be achieved by either movement of the coiled-coil regions bringing the head domains closer together (top pathway) or perhaps intertwining of the coiled-coil regions might lead to intertwining of the two DNA molecules (bottom pathway). The model is discussed in the text.
Topological Alteration of the DNA Observed in the Reactions Varied as a Function of the Mg2+ Concentration
At Mg2+ concentrations less than 2 mm, the intramolecular reaction of knotting was observed; whereas at Mg2+ concentrations of 4 mm or greater, the intermolecular reaction of catenation occurred. It has previously been demonstrated that MukB can bridge two DNA helices and that in doing so it favored binding to right-handed crossings of the helices (27). Indeed, knots induced by MukB in circular DNA are almost exclusively right-handed (10). Although we tried to visualize the handedness of the crossovers between DNA rings in the catenated product by coating the DNA with RecA and imaging in the AFM, we were unable to accumulate sufficient data that would allow us to make a definitive statement. Those crossovers we did observe in dimeric catenanes were right-handed. In general, the binding affinity of MukB to DNA decreases with increasing Mg2+ concentrations. Also ATP is not required for the catenation reaction, suggesting that MukB head domains spend most of the time in the reaction in monomeric form. Variation in Mg2+ concentration might alter the likelihood that when one head domain in a MukB dimer is bound to a segment of DNA the second one will also bind to a segment in the same DNA molecule. At low Mg2+ concentrations, each monomer of a head domain dimer binds DNA segments on the same DNA ring, whereas at higher Mg2+ concentrations, each monomer of a head domain dimer binds DNA segments on different DNA rings. Perhaps the change in Mg2+ concentration might alter the conformational disposition of the coiled-coil domains. Of course, it is also possible that catenation involves binding of DNA to a region of MukB other than the head domain.
It is interesting to consider the sharp transition that we observe between knotting and catenation as a function of Mg2+ concentration in vitro in light of the observations that the total Mg2+ concentration in E. coli is about 100 mm (28), whereas free intracellular Mg2+ is much lower, less than 5 mm (29) and more likely about 1 mm (30). At the most extreme, one might then conclude that MukB-mediated DNA catenation does not occur at all in vivo, or that it happens under rare circumstances. However, the equilibrium between the bound and free forms of Mg2+ intracellularly and with free Mg2+ extracellularly is a complex one that is further complicated, with respect to the catenation reaction described here, by the presence of significant intracellular concentrations of polyamines such as spermidine (6 mm (31)), which can bind the DNA substrate and effect charge neutralization (20).
The Effect of MukEF on MukB ATPase Activity
It had been shown previously that MukEF stimulated the MukB ATPase activity (24). As communicated to us by David Sherratt,5 only MukF is required to stimulate the MukB ATPase. MukF stimulation of the MukB ATPase was maximal at the substoichiometric ratio of MukF to MukB of 1:4. This observation seems somewhat at odds with structural studies showing that the MukF dimer binds at the bottom (compared with the orientation of the coiled-coil regions) of the MukB head domain dimer and that ATP hydrolysis causes the head domain dimer to come apart, with one MukF monomer becoming largely dissociated from the complex, leaving only a flexible linker bound to one of the monomers of the MukB head domain (23).
It is difficult to assign biological significance to this observation at the moment. It is possible that MukF acts catalytically with respect to MukB, dissociating completely after ATP hydrolysis. Another possibility is that the actual agent of MukB ATPase activity is a conglomerate of MukB dimers associated head-to-head and that the MukF dimer need only bind to one end of the chain to trigger ATP hydrolysis throughout. A recent study of DNA release by the yeast cohesin showed that asymmetric ATP hydrolysis by the head domains of Smc1 and Smc3 was involved (32). Perhaps the head domains in a MukB dimer are also non-equivalent.
The presence of MukE inhibited MukF-stimulated MukB ATPase activity, indicating that MukE takes an active role in modulating the ATPase cycle of MukB despite not being bound directly to MukB. If the MukB ATPase activity is a direct measure of the distribution of MukB between closed (head domains bound together) and open conformations, then it is likely that the presence of MukE favors the closed conformation.
The Effect of ATP and MukEF on MukB-mediated Catenation
In the absence of ATP, MukF inhibited MukB-mediated DNA catenation, as did the presence of ATP alone. The addition of MukE did not relieve inhibition by MukF. Because both the binding of ATP and MukF to the head domains serve to bring those domains together, prima facie, these observations indicate that an open conformation of MukB favors DNA catenation. Previous studies have shown: (i) that MukB does not require ATP to bind to plasmid DNA and alter its topology (10) or bridge two DNAs (27), (ii) that the pre-formed MukBEF complex is defective in binding plasmid DNA (13), and (iii) that adding MukEF to a MukB-DNA complex appears to displace MukB from the DNA (13). Our observations are consistent with these previous observations; however, we observe distinct inhibition of the MukB-mediated catenation reaction by the addition of ATP, with less inhibition observed with AMP-PNP, and no inhibition observed with ADP. The binding of AMP-PNP is known to bring the head domains together (23). Thus, the observation that AMP-PNP does not inhibit the catenation reaction to the same extent as ATP suggests that it is not the closing of the MukB head domains per se that is inhibitory, but the cycling between the closed and open conformations supported by the presence of ATP. It is possible that such cycling, which presumably entails changes to the disposition of the hinge domain and the coiled-coil arms of the protein, prevents selectivity of MukB binding to two different DNAs.
The preponderance of biochemical evidence outlined above from this laboratory and others argues strongly that MukB requires neither ATP nor MukEF to act on DNA. How then to reconcile the biochemical observations to those in vivo that indicate mukE and mukF mutations have the same phenotype as mukB mutations (33, 34), and that MukB ATP-binding and MukEF are required for MukB localization in the cell (11, 12)? These observations are not necessarily contradictory. We still do not understand the cycle of ATP and MukEF binding to MukB completely, and it could just as well be the case that the presence of ATP and MukEF are required for either loading MukB to DNA or unloading it from DNA in vivo.
Possible Role of the Catenation Reaction in DNA Condensation by MukB
The mechanism by which MukB condenses DNA is unknown. Topological trapping of DNA in a tripartite complex formed between the SMC protein, the kleisin, and accessory protein has been proposed as a possible mechanism. Presumably, by trapping distal segments of the chromosome, DNA condensation is achieved by forming loops between these DNA segments. Trapping of DNA by MukB alone was not affected by ATP (9), although the effect of AMP-PNP was not examined, and was reported to be dependent on MukB binding to single-stranded DNA. The fact that MukB can mediate catenation of DNA rings does not argue one way or another for any particular mode of DNA binding by the protein. However, we have proposed that MukB stabilizes loops between distal segments of a chromosome by intertwining them (16) with the topological alteration being stabilized by the interacting Topo IV. MukB-mediated DNA catenation clearly fits in with this model.
Experimental Procedures
DNAs and Proteins
MukB and MukBdna (MukB R187E/R189E) were prepared as described (14, 16). Topo III was prepared as described (35). Topo IV was prepared as described (36). Plasmid pCG09 is 11,088 bp long and was described in Ref. 16. Gapped pCG09 DNA was prepared as follows. Reaction mixtures (600 μl) containing supercoiled pCG09 (60 μg), 1× concentration of New England Biolabs Smart Cut buffer, the nicking restriction endonuclease Nb.BbVCI (New England Biolabs, 8.5 units), and exonuclease III (New England Biolabs, 2 units) were incubated at 37 °C for 30 min. The reaction was terminated by the addition of EDTA to 45 mm followed by extraction with phenol-chloroform, the DNA was recovered by ethanol precipitation, resuspended in 10 mm Tris-HCl (pH 8.0 at 4 °C), 1 mm EDTA, divided into small aliquots, and stored at −80 °C. As measured by the ability of the gDNA to be digested with restriction enzymes, the gaps were less than 100 nucleotides in length.
Purification of MukF
A freshly transformed BL21(DE3) (pET11a-mukF) colony was inoculated to 500 ml of LB medium containing 200 μg/ml of ampicillin and grown at 37 °C overnight. This culture was then used to inoculate 20 liters of LB medium containing 100 μg/ml of ampicillin in a New Brunswick Scientific Fermentor to an A600 of 0.05. The cells were grown at 37 °C to an A600 of 0.4 and MukF overproduction was induced by the addition of 0.4 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 25 °C. Cells were harvested by centrifugation, resuspended at 1:1 (w/v) in Tris buffer B (50 mm Tris-HCl (pH 7.5 at 4 °C), 0.1 mm EDTA, 2 mm DTT, 20% glycerol) with 150 mm NaCl, frozen in liquid nitrogen, and stored at −80 °C. After gradually thawing the frozen cells on ice and then at room temperature, the cell suspension was adjusted to 50 mm Tris-HCl (pH 8.0 at 4 °C), 10 mm DTT, 20 mm EDTA, and 0.2 mg/ml of lysozyme. The suspension was incubated on ice for 10 min, at room temperature for 10 min, and then on ice for another 10 min, followed by centrifugation at 100,000 × g for 1 h at 4 °C. The supernatant (fraction 1) was collected and kept on ice. Fraction 1 was mixed slowly with ground (NH4)2SO4 powder to 35% saturation, stirred for 1 h at 4 °C, and precipitated protein was collected by centrifugation in an SS34 rotor at 19,000 rpm for 30 min at 4 °C. The pellet was resuspended in Tris buffer B to a conductivity equivalent to 50 mm NaCl in Tris buffer B (fraction 2, 256 mg of protein). Fraction 2 was loaded onto a 19-ml Q-Sepharose column that had been equilibrated previously with 3 column volumes of Tris buffer A (50 mm Tris-HCl (pH 7.5 at 4 °C), 1 mm EDTA, 5 mm DTT, and 10% glycerol) containing 50 mm NaCl. The column was washed with 3 column volumes of Tris buffer B with 50 mm NaCl and protein was eluted with a 10 column volume gradient of 50 to 750 mm NaCl in Tris buffer B. Peak fractions were pooled according to SDS-PAGE analysis for the MukF protein band (fraction 3). Fraction 3 was mixed slowly with ground (NH4)2SO4 powder to 60% saturation, stirred for 1 h at 4 °C, and precipitated protein was collected by centrifugation in an SS34 rotor at 19,000 rpm for 30 min at 4 °C. The pellet was resuspended in Tris buffer A with 0.5 m NaCl (fraction 3′). Fraction 3′ was gel filtered through a 125-ml Superdex 200 column (GE Healthcare, 2.3 × 58 cm) that had been equilibrated with Tris buffer A with 0.5 m NaCl. The column was developed at 1 ml/min. Peak fractions were pooled (fraction 4, 36.3 mg), dialyzed against storage buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.1 mm EDTA, 5 mm DTT, 40% glycerol), frozen in liquid nitrogen, and stored at −80 °C.
Purification of MukE
MukE was purified essentially as for MukF above with the following differences. Strain BL21(DE3) (pET11a-mukE) was used. Protease inhibitor mixture (Sigma) was added at 29 μl/ml of thawed cell suspension before lysis. Fraction 1 (1.24 g) was fractionated by chromatography on a 60-ml Q-Sepharose column equilibrated with Tris buffer A containing 50 mm NaCl using a gradient of 50 to 600 mm NaCl. Peak fractions were pooled according to SDS-PAGE analysis for the MukE protein band (fraction 2). Fraction 2 (273 mg) was concentrated by precipitation with (NH4)2SO4 and gel filtered through Superdex 200 equilibrated in buffer A with 50 mm NaCl as above to give fraction 3 (75 mg). Fraction 3 was loaded onto a 7-ml hydroxyapatite HTP (Bio-Rad) column equilibrated with Tris buffer A containing 50 mm NaCl. The column was washed with 2 column volumes of Tris buffer A with 50 mm NaCl and protein was eluted with a 10-column volume gradient of 20 to 250 mm sodium phosphate buffer (pH 7.2) in Tris buffer A. Peak fractions were pooled (fraction 4, 30 mg) according to SDS-PAGE analysis, dialyzed against storage buffer, frozen in liquid nitrogen, and stored at −80 °C.
Purification of the MukEF Complex
The MukEF complex was purified essentially as for MukF above with the following differences. Strain BL21(DE3)(pET11a-mukFE) was used. Nucleic acid in fraction 1 was precipitated by adding Polymin P slowly to 0.07%. After stirring for 10 min at 4 °C. The mixture was centrifuged in a SS34 rotor at 19,000 rpm for 10 min at 4 °C and the supernatant was collected (fraction 1′). Fraction 1′ was then treated with (NH4)2SO4 powder. Fraction 2 (900 mg) was fractionated by chromatography on a 46-ml Q-Sepharose column equilibrated with Tris buffer A containing 50 mm NaCl using a gradient of 50 to 600 mm NaCl. Peak fractions were pooled according to SDS-PAGE analysis for the MukE and MukF protein bands (fraction 3). Fraction 3 was concentrated by precipitation with (NH4)2SO4 and gel filtered through Superdex 200 as above to give fraction 4 (25.5 mg) that was stored as for MukF.
DNA Catenation Assay
Standard reaction mixtures (20 μl) containing 50 mm HEPES-KOH (pH 7.5), 4 mm Mg(OAc)2, 20 mm KCl, 10 mm DTT, 100 μg/ml of BSA, 0.7 nm gDNA, 25 nm Topo III, and 300 nm MukB2 were incubated at 37 °C for either 5 or 30 min. EDTA and NaCl were then added to 20 and 150 mm, respectively, and the incubation continued for 5 min. Proteinase K and SDS were then added to 0.2 mg/ml and 0.5%, respectively, and the incubation continued for 30 min. Reaction products were assayed by electrophoresis through vertical 0.9% agarose gels at 2 V/cm for 17 h at room temperature using 50 mm Tris-HCl (pH 7.8 at 23 °C), 40 mm NaOAc, 1 mm EDTA as the electrophoresis buffer. The buffer was recirculated during the electrophoresis. Gels were stained with SYBR Gold (Invitrogen) and scanned using a GE Typhoon 9500 laser scanner at 350 V.
Isolation of Catenated DNA
Standard DNA catenation reaction mixtures were increased in volume by 10-fold and incubated for 30 min at 37 °C. Incubation time in the presence of Proteinase K and SDS was increased to 1 h. The DNA was purified by dialysis using Biotech Cellulose Ester Dialysis Membranes with a molecular mass cut-off of 300 kDa. Samples were first dialyzed for 5 min against H2O (1 liter) and then for 3 × 24 h against 1 liter of 10 mm Tris-HCl (pH 8.0 at 4 °C), 1 mm EDTA. The catenated DNA was divided into small aliquots and stored frozen at −80 °C.
Decatenation of Catenated DNA
Reaction mixtures (20 μl) containing 50 mm Tris-HCl (pH 7.5 at 37 °C), 6 mm Mg(OAc)2, 10 mm DTT, 50 μg/ml of BSA, 100 mm K-glutamate, 0.4 mm ATP, catenated DNA (∼100 ng), and Topo IV as indicated, were incubated for 30 min at 37 °C. Reactions were then processed and analyzed as described for the catenation assay.
Nitrocellulose DNA Binding Assay
Binding to a 5′-end labeled duplex DNA formed by annealing the oligonucleotide 5′-ATC TCG GGC TAT TCT TTT GAT TTA TAA GGG ATT TTG CCG ATT TCG GAA CC and its complement was measured in reaction mixtures (20 μl) containing 50 mm HEPES-KOH (pH 7.5), 20 mm KCl, 10 mm DTT, 0.5 mm Mg(OAc)2, 5 nm labeled duplex DNA, and the indicated concentrations of either MukB or MukBdna. Reactions were incubated for 15 min at 37 °C, diluted with 100 μl of wash buffer (50 mm HEPES-KOH, 20 mm KCl, and 0.5 mm Mg(OAc)2) equilibrated to 37 °C and immediately filtered through Millipore HAWP filters that had been soaked in wash buffer. The filters were washed three times with 1 ml of wash buffer at room temperature, dried, and counted in a liquid scintillation counter.
ATPase Assay
Reaction mixtures (10 μl) containing 50 mm HEPES-KOH (pH 7.5), 20 mm KCl, 10 mm DTT, 2 mm Mg(OAc)2, 100 μg/ml of BSA, 2 mm ATP, [γ-32P]ATP (2 μCi), 300 nm MukB2, and the indicated concentrations of MukF, MukE, and MukEF were incubated for 10 min at 37 °C. EDTA was then added to 100 mm and 1 μl of the reaction mixture was spotted on a PEI cellulose plate (Millipore). The plate was then developed in an enclosed glass chamber using 0.5 m LiCl, 1 m HCOOH. The plate was dried at 60 °C, exposed to a phosphorimager screen, and read in a GE Typhoon FLA7000 Scanner. The fraction of ATP hydrolyzed to Pi was determined using Image Gauge software (Fuji).
Atomic Force Microscopy
Catenated DNA was diluted to ∼1 ng/ml in 20 mm HEPES-KOH (pH 7.0), 5 mm MgCl2 and a drop was placed on freshly cleaved mica and incubated for 30 min at room temperature. Monomeric pCG09 DNA was incubated with the mica for 2 min. Samples were washed gently with ∼10 ml of Molecular Biology Grade H2O (Fisher) and then dried under a stream of ultra-pure dry N2. The mica was then loaded onto the stage of an Asylum Research (Goleta CA) MFP-3D-BIO Atomic Force Microscope and the samples imaged using an Olympus AC240 probe (k = ∼2 N/m) at an amplitude and gain that was adjusted to produce the best results. The free amplitude was always <10 nm. Images were acquired and processed using Igor Pro Software (Wave Metrics) with the Asylum Research software loaded.
Author Contributions
S. B. and K. M. designed the experiments, S. B. performed all the experiments, S. B. and K. M. analyzed the data, R. H. purified MukF, MukE, and the MukEF complex, K. M. wrote the manuscript
Acknowledgments
We thank Navid Paknejad and the Memorial Sloan Kettering Molecular Cytology Core Facility for the AFM images.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34558 (to K. J. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
R. Kumar, M. Grosbart, P. Nurse, S. Bahng, and C. Wyman, unpublished data.
D. Sherratt, personal communication.
- SMC
- structural maintenance of chromosome protein
- Topo IV
- topoisomerase IV
- Topo III
- topoisomerase III
- gDNA
- gapped DNA
- AFM
- atomic force microscopy
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate.
References
- 1. Thanbichler M., and Shapiro L. (2006) Chromosome organization and segregation in bacteria. J. Struct. Biol. 156, 292–303 [DOI] [PubMed] [Google Scholar]
- 2. Luijsterburg M. S., White M. F., van Driel R., and Dame R. T. (2008) The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol. 43, 393–418 [DOI] [PubMed] [Google Scholar]
- 3. Uhlmann F. (2016) SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 [DOI] [PubMed] [Google Scholar]
- 4. Yamazoe M., Onogi T., Sunako Y., Niki H., Yamanaka K., Ichimura T., and Hiraga S. (1999) Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 18, 5873–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haering C. H., Farcas A. M., Arumugam P., Metson J., and Nasmyth K. (2008) The cohesin ring concatenates sister DNA molecules. Nature 454, 297–301 [DOI] [PubMed] [Google Scholar]
- 6. Cuylen S., Metz J., and Haering C. H. (2011) Condensin structures chromosoma DNA through topological links. Nat. Struct. Mol. Biol. 18, 894–901 [DOI] [PubMed] [Google Scholar]
- 7. Sun M., Nishino T., and Marko J. F. (2013) The SMC1-SMC3 cohesin heterodimer structures DNA through supercoiling-dependent loop formation. Nucleic Acids Res. 41, 6149–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wilhelm L., Burmann F., Minnen A., Shin H. C., Toseland C. P., Oh B. H., and Gruber S. (2015) SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife 4, 10.7554/eLife.06659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Niki H., and Yano K. (2016) In vitro topological loading of bacterial condensin MukB on DNA, preferentially single-stranded DNA rather than double-stranded DNA. Sci. Rep. 6, 29469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Petrushenko Z. M., Lai C. H., Rai R., and Rybenkov V. V. (2006) DNA reshaping by MukB: right-handed knotting, left-handed supercoiling. J. Biol. Chem. 281, 4606–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. She W., Wang Q., Mordukhova E. A., and Rybenkov V. V. (2007) MukEF Is required for stable association of MukB with the chromosome. J. Bacteriol. 189, 7062–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Badrinarayanan A., Reyes-Lamothe R., Uphoff S., Leake M. C., and Sherratt D. J. (2012) In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science 338, 528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Petrushenko Z. M., Lai C. H., and Rybenkov V. V. (2006) Antagonistic interactions of kleisins and DNA with bacterial Condensin MukB. J. Biol. Chem. 281, 34208–34217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hayama R., and Marians K. J. (2010) Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 18826–18831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y., Stewart N. K., Berger A. J., Vos S., Schoeffler A. J., Berger J. M., Chait B. T., and Oakley M. G. (2010) Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc. Natl. Acad. Sci. U.S.A. 107, 18832–18837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayama R., Bahng S., Karasu M. E., and Marians K. J. (2013) The MukB-ParC interaction affects the intramolecular, not intermolecular, activities of topoisomerase IV. J. Biol. Chem. 288, 7653–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., and Suzuki H. (1990) New topoisomerase essential for chromosome segregation in E. coli. Cell 63, 393–404 [DOI] [PubMed] [Google Scholar]
- 18. Hiraga S., Niki H., Imamura R., Ogura T., Yamanaka K., Feng J., Ezaki B., and Jaffé A. (1991) Mutants defective in chromosome partitioning in E. coli. Res. Microbiol. 142, 189–194 [DOI] [PubMed] [Google Scholar]
- 19. DiGate R. J., and Marians K. J. (1988) Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 263, 13366–13373 [PubMed] [Google Scholar]
- 20. Krasnow M. A., and Cozzarelli N. R. (1982) Catenation of DNA rings by topoisomerases: mechanism of control by spermidine. J. Biol. Chem. 257, 2687–2693 [PubMed] [Google Scholar]
- 21. Hiasa H., and Marians K. J. (1996) Two distinct modes of strand unlinking during θ-type DNA replication. J. Biol. Chem. 271, 21529–21535 [DOI] [PubMed] [Google Scholar]
- 22. Ullsperger C., and Cozzarelli N. R. (1996) Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J. Biol. Chem. 271, 31549–31555 [DOI] [PubMed] [Google Scholar]
- 23. Woo J. S., Lim J. H., Shin H. C., Suh M. K., Ku B., Lee K. H., Joo K., Robinson H., Lee J., Park S. Y., Ha N. C., and Oh B. H. (2009) Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell 136, 85–96 [DOI] [PubMed] [Google Scholar]
- 24. Chen N., Zinchenko A. A., Yoshikawa Y., Araki S., Adachi S., Yamazoe M., Hiraga S., and Yoshikawa K. (2008) ATP-induced shrinkage of DNA with MukB protein and the MukBEF complex of Escherichia coli. J. Bacteriol. 190, 3731–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mascarenhas J., Volkov A. V., Rinn C., Schiener J., Guckenberger R., and Graumann P. L. (2005) Dynamic assembly, localization and proteolysis of the Bacillus subtilis SMC complex. BMC Cell Biol. 6, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shin H. C., Lim J. H., Woo J. S., and Oh B. H. (2009) Focal localization of MukBEF condensin on the chromosome requires the flexible linker region of MukF. FEBS J. 276, 5101–5110 [DOI] [PubMed] [Google Scholar]
- 27. Petrushenko Z. M., Cui Y., She W., and Rybenkov V. V. (2010) Mechanics of DNA bridging by bacterial condensin MukBEF in vitro and in singulo. EMBO J. 29, 1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moncany M. L., and Kellenberger E. (1981) High magnesium content of Escherichia coli B. Experientia 37, 846–847 [DOI] [PubMed] [Google Scholar]
- 29. Alatossava T., Jütte H., Kuhn A., and Kellenberger E. (1985) Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 162, 413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Froschauer E. M., Kolisek M., Dieterich F., Schweigel M., and Schweyen R. J. (2004) Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol. Lett. 237, 49–55 [DOI] [PubMed] [Google Scholar]
- 31. Tabor C. W., and Tabor H. (1985) Polyamines in microorganisms. Microbiol. Rev. 49, 81–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elbatsh A. M., Haarhuis J. H., Petela N., Chapard C., Fish A., Celie P. H., Stadnik M., Ristic D., Wyman C., Medema R. H., Nasmyth K., and Rowland B. D. (2016) Cohesin releases DNA through asymmetric ATPase-driven ring opening. Mol. Cell 61, 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Niki H., Jaffé A., Imamura R., Ogura T., and Hiraga S. (1991) The new gene mukB codes for a 177 kDa protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamanaka K., Ogura T., Niki H., and Hiraga S. (1996) Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol. Gen. Genet. 250, 241–251 [DOI] [PubMed] [Google Scholar]
- 35. Hiasa H., DiGate R. J., and Marians K. J. (1994) Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 269, 2093–2099 [PubMed] [Google Scholar]
- 36. Peng H., and Marians K. J. (1999) Overexpression and purification of bacterial topoisomerase IV. Methods Mol. Biol. 94, 163–169 [DOI] [PubMed] [Google Scholar]