Abstract
The Na-H exchanger NHE1 contributes to intracellular pH (pHi) homeostasis in normal cells and the constitutively increased pHi in cancer. NHE1 activity is allosterically regulated by intracellular protons, with greater activity at lower pHi. However, the molecular mechanism for pH-dependent NHE1 activity remains incompletely resolved. We report that an evolutionarily conserved cluster of histidine residues located in the C-terminal cytoplasmic domain between two phosphatidylinositol 4,5-bisphosphate binding sites (PI(4,5)P2) of NHE1 confers pH-dependent PI(4,5)P2 binding and regulates NHE1 activity. A GST fusion of the wild type C-terminal cytoplasmic domain of NHE1 showed increased maximum PI(4,5)P2 binding at pH 7.0 compared with pH 7.5. However, pH-sensitive binding is abolished by substitutions of the His-rich cluster to arginine (RXXR3) or alanine (AXXA3), mimicking protonated and neutral histidine residues, respectively, and the RXXR3 mutant had significantly greater PI(4,5)P2 binding than AXXA3. When expressed in cells, NHE1 activity and pHi were significantly increased with NHE1-RXXR3 and decreased with NHE1-AXXA3 compared with wild type NHE1. Additionally, fibroblasts expressing NHE1-RXXR3 had significantly more contractile actin filaments and focal adhesions compared with fibroblasts expressing wild type NHE1, consistent with increased pHi enabling cytoskeletal remodeling. These data identify a molecular mechanism for pH-sensitive PI(4,5)P2 binding regulating NHE1 activity and suggest that the evolutionarily conserved cluster of four histidines in the proximal cytoplasmic domain of NHE1 may constitute a proton modifier site. Moreover, a constitutively activated NHE1-RXXR3 mutant is a new tool that will be useful for studying how increased pHi contributes to cell behaviors, most notably the biology of cancer cells.
Keywords: actin, focal adhesions, pH regulation, phospholipid, sodium-proton exchange
Introduction
Intracellular pH (pHi) homeostasis is generally maintained near neutral to compensate for metabolic changes and environmental stresses (1). However, dysregulated pHi is seen in a number of diseases. Most cancers have constitutively increased pHi of ∼0.4 units, which enables proliferation and metastasis (2–4). Conversely, neurodegenerative disorders, including Parkinson's and Alzheimer's diseases, are associated with constitutively decreased pHi, which is predicted to enable tau and β-amyloid aggregation as well as cell death (5, 6). Finely tuned pHi homeostasis is maintained by dynamic changes in ion transporter activity that is sensitive to intracellular proton concentrations. However, the molecular mechanisms that mediate pH sensing by ion transporters remain poorly understood.
The ubiquitously expressed Na-H exchanger isoform NHE14 contributes to maintaining pHi homeostasis by generating an electroneutral influx of extracellular Na+ and efflux of intracellular H+ at the plasma membrane. To maintain pHi homeostasis, NHE1 activity increases with acidic pHi and becomes nearly quiescent at neutral pHi. However, NHE1 activity can be increased at neutral pHi in response to growth factor signaling or activated oncogenes by direct phosphorylation of serine residues in the distal C-terminal cytoplasmic domain by kinases such as p90RSK (7), Akt (8), the Rho kinase ROCK (9), and the Nck-interacting kinase NIK (also called MAP4K4) (10). In response to oncogenes, phosphorylation-dependent increases in NHE1 activity override pH-dependent regulation, resulting in the higher pHi in cancer (3). In contrast to how NHE1 activity is regulated by kinases, how it is regulated by acidic pHi remains unclear. More than 30 years ago Aronson and co-workers (11) first proposed a pH-dependent proton modifier site in the C-terminal cytoplasmic domain, but to date this site remains incompletely defined.
Since the first prediction of a proton modifier site regulating NHE1 activity (11), binding to the negatively charged plasma membrane phospholipid phosphatidylinositol 4,5-bisphosphate PI(4,5)P2 emerged as a mechanism for increasing activity of different classes of ion transport proteins (12). Aharonovitz et al. (13) showed the importance of PI(4,5)P2 binding for increased activity for NHE1 that is dependent on two positively charged regions in the membrane-proximal C-terminal cytoplasmic domain (509KKKQETKR515 and 552RFNKKYVKK560 for human NHE1) and subsequently identified electrostatic interactions between PI(4,5)P2 and homologous basic residues in the NHE3 isoform (14). Schelling and co-workers (15) showed that electrostatic interactions between NHE1 and PI(4,5)P2 are dependent on arginine and lysine residues in these two positively charged regions in the C-terminal cytoplasmic domain of NHE1 and also that PI(4,5)P2 binding is pH-dependent, with higher binding affinity at lower pH. However, arginine and lysine with pKa values of 12 and 10.5 in solution, respectively, likely do not titrate with physiological changes in pH to confer pH-dependent binding to PI(4,5)P2.
We report that a cluster of histidine residues between the two positively charged regions in the C-terminal cytoplasmic domain (540HYGHHH545 for human NHE1) regulate pH-dependent PI(4,5)P2 binding and transporter activity. We generated protonated and neutral mimetics by substituting these histidine residues with arginine (NHE1-RXXR3) and alanine (NHE1-AXXA3), respectively, which abolished pH-sensitive PI(4,5)P2 binding in vitro. When expressed in fibroblasts, NHE1-RXXR3 was constitutively active, with greater quiescent activity than wild type NHE1, and conferred higher quiescent pHi as well as increased abundance of contractile actin filaments and cell-substrate adhesions. In contrast, NHE1-AXXA3 had lower quiescent activity than wild type NHE1, and unlike wild type NHE1, its activity did not increase in response to platelet-derived growth factor. Our findings show that an evolutionarily conserved cluster of four His residues in NHE1 regulate transporter activity and suggest that this cluster may constitute an NHE1 proton modifier site first proposed more than 30 years ago.
Results
Cluster of Histidine Residues in Proximal C-terminal Cytoplasmic Domain Confer pH-dependent PI(4,5)P2 Binding
Based on previous findings that the proximal C terminus of NHE1 has pHi-dependent PI(4,5)P2 binding (15), we reasoned that this may be mediated by histidine residues, which can titrate within the physiological pH range and function as a pH sensor site (16). We identified a series of histidine residues located between the arginine- and lysine-rich regions that bind PI(4,5)P2 (Fig. 1A), which are invariant in vertebrate NHE1 orthologs (Fig. 1B). Histidines flanked by arginine/lysine-rich regions in the proximal C terminus are also conserved in other plasma membrane NHE isoforms, although the number of histidine residues varies with isoforms having between one and four (Fig. 1C).
FIGURE 1.
A cluster of histidine residues in the PI(4,5)P2 binding domain of NHE1 is evolutionarily conserved. A, schematic diagram of NHE1 and the location of the two lysine/arginine-rich PI(4,5)P2 binding site (K/R1, K/R2) surrounding a poly-histidine region (HXXHHH). B, sequence alignment of NHE1 orthologs. The His-rich region is shown in red. C, sequence alignment of membrane binding regions of plasma membrane NHEs. Blue, phospholipid binding region KR1. Green, phospholipid binding region KR2. Red, histidine cluster located between KR1 and KR2. *, fully conserved residue; :, strongly conserved residue; ., weakly conserved residue. NCBI accession numbers for sequences: NHE1, NP_003038.2; NHE2, NP_003039.2; NHE3, NP_004165.2; NHE4, NP_001011552.2; NHE5, NP_004585.1.
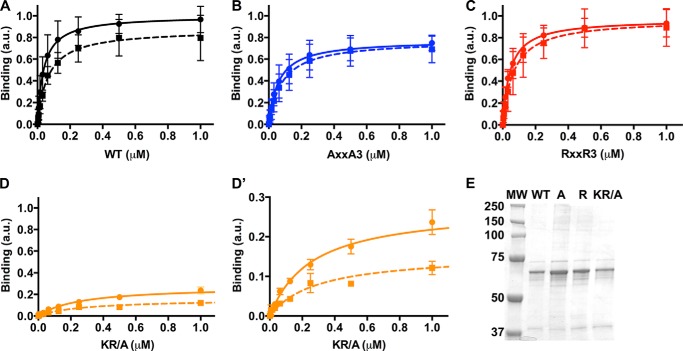
We used an ELISA assay to measure pH-dependent PI(4,5)P2 binding by GST fusion proteins of the C-terminal cytoplasmic domain of human NHE1 (amino acids 503–815; Fig. 2). The assay used PI(4,5)P2-coated multiwell plates and showed that wild type NHE1503–815 (NHE1-WT) had decreased binding at pH 7.5 compared with 7.0, as previously described (15). Maximal binding (Bmax) was 0.87 ± 0.04 arbitrary units (a.u.) at pH 7.5 and 1.0 ± 0.04 a.u. at pH 7.0 (p < 0.05; mean ± S.D.; Fig. 2A and Table 1). Additionally, binding affinity was lower at pH 7.5 compared with pH 7.0, with dissociation constants (kd) of 65 ± 10 nm and 38 ± 6 nm, respectively (p < 0.05; Table 1).
FIGURE 2.
A cluster of histidine residues mediate pH-dependent PI(4,5)P2binding by NHE1. A–D, PI(4,5)P2 binding at pH 7.0 (solid line) and pH 7.5 (dotted line) for wild type (A, black), AXXA3 (B, blue), RXXR3 (C, red) and Arg/Lys residues mutated to alanine (KR/A; D, orange). a.u., arbitrary units. D′ shows the same data in D with a magnified y axis. Data are the mean ± S.D. from six experiments for A–C and three experiments for D. E, Coomassie-stained SDS-PAGE of wild type (WT), RXXR3 (R), AXXA3 (A), and KR/A GST-NHE1 503–815 proteins. The size of the molecular weight marker (MW) is shown in kDa.
TABLE 1.
Binding parameters from ELISA PI(4,5)P2 experiments
The maximal binding, Bmax, is reported in arbitrary units. The dissociation constant, kd, is reported in nm. Data are the mean ± S.D. from 3 (KR/A) and 6 (WT, AXXA3, RXXR3) independent experiments. ND, not determined.
| Construct | Bmax |
kd |
||
|---|---|---|---|---|
| pH 7.5 | pH 7.0 | pH 7.5 | pH 7.0 | |
| Wild type | 0.87 ± 0.04 | 1.00 ± 0.04a | 65 ± 10 | 38 ± 6a |
| AXXA3 | 0.76 ± 0.04b | 0.79 ± 0.03c | 78 ± 14 | 62 ± 9c |
| RXXR3 | 0.97 ± 0.05 | 0.97 ± 0.03 | 67 ± 13 | 44 ± 5 |
| KR/A | 0.16 ± 0.04d | 0.27 ± 0.01d,e | ND | ND |
a p < 0.05 compared to pH 7.5.
b p < 0.05 compared to WT and RXXR3 at pH 7.5.
c p < 0.05 compared to WT and RXXR3 at pH 7.0.
d p < 0.01 compared to WT.
e p < 0.001 compared to pH 7.5.
We next asked whether the cluster of histidines between the arginine and lysine-rich regions (Fig. 1B) is important for pH-dependent PI(4,5)P2 binding. We generated mutant GST fusions of the C-terminal cytoplasmic domain by substituting the four histidines in the cluster with arginines for a protonated mimetic (NHE1-RXXR3) or with alanines for a neutral mimetic (NHE1-AXXA3). Although both mutants bound PI(4,5)P2, pH-dependent binding was abolished, with no significant differences in Bmax or kd for each mutant at pH 7.5 compared with pH 7.0 (Fig. 2, B and C; Table 1). However, Bmax for RXXR3 was significantly greater than AXXA3 at pH 7.5 and pH 7.0 (p < 0.05), and Bmax for AXXA3 at both pH values was significantly lower than WT at both pH values. These results are similar to our previous findings on pH-dependent binding of the focal adhesion protein talin to actin filaments, with pH-insensitive talin mutants having changes in Bmax but not kd (17). Moreover, these data suggest that the protonation state of histidines flanked by the Arg/Lys regions regulates PI(4,5)P2 binding. Consistent with this prediction, the binding of an NHE1 peptide in which the Arg/Lys residues are mutated to alanine (KR/A) had pH-sensitive, but significantly attenuated, PI(4,5)P2 binding (Bmax, Fig. 2D, Table 1).
NHE1-RXXR3 and -AXXA3 Mutants Expressed in Fibroblasts Have Increased and Decreased Activity, Respectively
To determine whether RXXR3 and AXXA3 substitutions affect NHE1 activity, we generated full-length rat NHE1-WT and histidine mutant constructs tagged at the C terminus with an HA epitope and stably expressed each construct in NHE1-deficient PS120 fibroblasts that were previously described (8). Immunoblotting cell lysates with HA antibodies showed that NHE1 migrated as a lower molecular weight band corresponding to the immature, core-glycosylated NHE1 and a higher molecular weight band representing the fully processed, highly glycosylated protein (Fig. 3A) (18). We quantified the total NHE1 expression in NHE1-wild type, -AXXA3, and -RXXR3 cells by densitometry (Fig. 3B). Expression of NHE1-AXXA3 was ∼25% greater than WT and RXXR3 (p < 0.05), which had similar levels of expression. We also determined surface expression by labeling cells with cell-impermeant biotin and HA-immunoblotting of enriched biotin-modified surface proteins from a streptavidin column. This approach revealed a similar abundance of NHE1-WT, -RXXR3, and -AXXA3 at the cell surface, with the majority of NHE1 at the cell surface corresponding to the highly glycosylated form (Fig. 3C).
FIGURE 3.
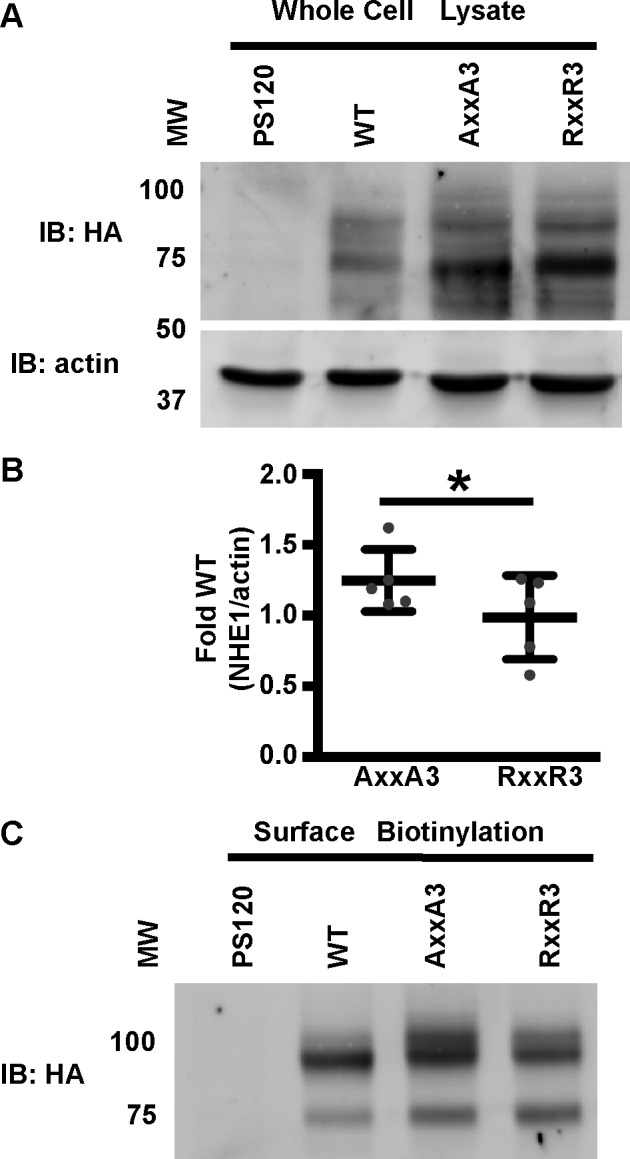
Expression of NHE1-WT and histidine mutants in fibroblasts. A, HA and actin immunoblots (IB) of total lysates from NHE1-deficient PS120 cells and cells stably expressing NHE1-WT, -RXXR3, and -AXXA3 tagged with an HA epitope. B, relative NHE1 expression in AXXA3 and RXXR3 cells was quantified from Western blots by densitometry. The total NHE1 signal, representing the immature and processed forms of NHE1, was normalized to actin and expressed as -fold wild type. Bars represent the mean ± S.D., and gray dots represent relative NHE/actin ratio from five independent preparations of cells. *, p < 0.05. C, HA immunoblot of enriched biotinylated cell surface proteins. Data are representative of experiments from two separate cell preparations. The molecular weight marker (MW) is shown in kDa.
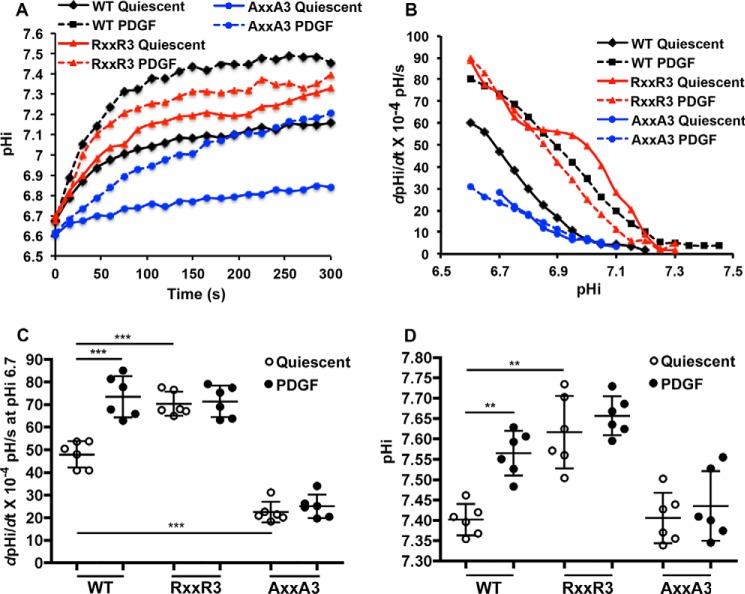
Based on Bmax values for PI(4,5)P2 binding, we tested the prediction that NHE1-RXXR3 and NHE1-AXXA3 would have constitutively increased and decreased activity, respectively. We measured time-dependent pHi recoveries from an NH4Cl-induced acid load in a nominally HCO3−-free buffer as previously described (8) (Fig. 4A). We also determined NHE1 activity as the pHi-dependent rate of pHi recovery (dpHi/dt) (Fig. 4, B and C). For NHE1-WT, quiescent activity increased with platelet-derived growth factor (PDGF; 50 ng/ml), similar to our previous findings (8, 10). At pHi 6.7, quiescent activity of 47.99 ± 5.78 dpHi/dt × 10−4 pH/s (mean ± S.D.) significantly increased with PDGF to 73.24 ± 9.10 dpHi/dt × 10−4 pH/s. For cells expressing NHE1-RXXR3, quiescent activity at pHi 6.7 was 70.34 ± 5.38 dpHi/dt × 10−4 pH/s and significantly greater than quiescent NHE1-WT but not different than NHE1-WT activity with PDGF. Additionally, NHE1-RXXR3 activity did not increase with PDGF, with activity at pH 6.7 of 71.35 ± 6.98 dpHi/dt × 10−4 pH/s not significantly different than quiescent. Moreover, quiescent pHi, determined in the presence of 25 mm HCO3−, was 7.39 ± 0.04 in cells expressing NHE1-WT and significantly increased with PDGF to 7.56 ± 0.05. Quiescent pHi with RXXR3 was 7.61 ± 0.08, which was significantly higher than with NHE1-WT and also did not increase with PDGF (Fig. 4D). These data indicate that NHE1-RXXR3 is constitutively more active than NHE1-WT, including generating a higher quiescent pHi.
FIGURE 4.
Histidine mutants have distinct quiescent activities that do not increase with PDGF. NHE1 activity was determined as the rate of pHi recovery from an NH4Cl-induced acid load in cells loaded with the pH-sensitive dye BCECF. A, means of time-dependent pHi recoveries in the absence (quiescent; solid lines) and presence of PDGF (50 ng/ml; dotted lines) from five independent cell preparations. B, means of pHi-dependent recovery rates (dpHi/dt) in the absence (quiescent; solid lines) and presence of PDGF (50 ng/ml; dotted lines) from six independent cell preparations. C, dpHi/dt at pHi 6.6 in quiescent and PDGF-treated cells from data shown in B. Data are the means ± S.D. of six independent cell preparations. **, p < 0.001; ***, p < 0.001. D, the pHi of indicated cell types at quiescence and with PDGF (50 ng/ml PDGF for 5 min). Data are the means ± S.D. of six independent cell preparations.
Activity of NHE1-AXXA3 was significantly lower than NHE1-WT (Fig. 4, A–C) and did not increase with PDGF. Activity was 21.43 ± 4.52 dpHi/dt × 10−4 pH/s and 25.09 ± 5.15 dpHi/dt × 10−4 pH/s in the absence and presence of PDGF, respectively. Notably, NHE1-AXXA3 retained pHi-dependent activity that decreased at higher pHi. Despite the lower activity of NHE1-AXXA3, quiescent pHi in cells expressing this mutant was 7.40 ± 0.06 and not different than that with NHE1-WT. However, pHi with NHE1-AXXA3 did not increase with PDGF (Fig. 4D). Decreased NHE1 activity and, in contrast to our findings, decreased pHi are seen with alanine substitutions in arginine and lysine residues in the charged 513–520 and 556–564 regions with the predicted mechanisms of decreased PI(4,5)P2 binding (13, 15).
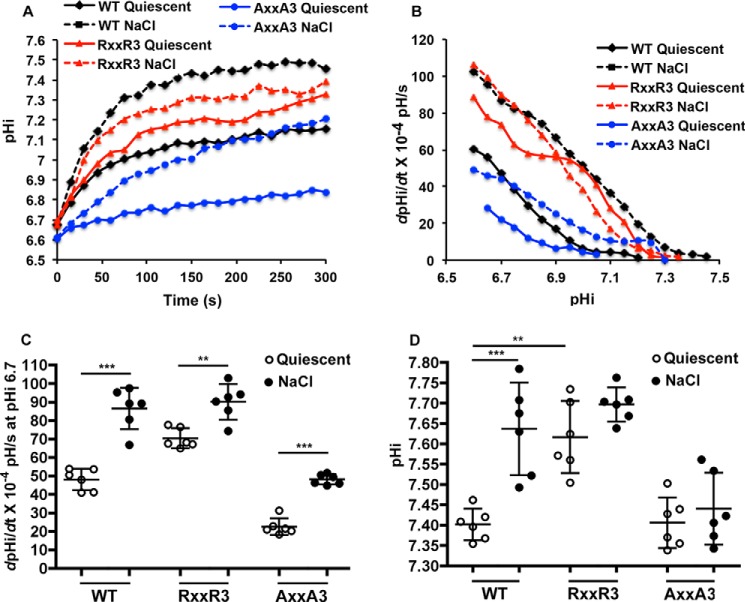
In contrast to the lack of response to PDGF, activity of both NHE1-RXXR3 and -AXXA3 increased with hyperosmolarity (Fig. 5A), determined in a nominally HCO3−-free buffer supplemented with 100 mm NaCl. Compared with their respective quiescent activities indicated above, with hyperosmolarity NHE1-WT activity increased to 86.25 ± 11.22 dpHi/dt × 10−4 pH/s, NHE1-RXXR3 increased to 90.36 ± 9.56 dpHi/dt × 10−4 pH/s, and NHE1-Axx3 increased to 48.11 ± 2.77 dpHi/dt × 10−4 pH/s (Fig. 5, B and C). However, hyperosmolarity only increased pHi with expression of NHE1-WT but not NHE1-RXXR3 or NHE1-Axx3 (Fig. 5D). These data suggest that NHE1 activity with hyperosmolarity is mostly independent of PI(4,5)P2 binding, unlike growth factor-regulated activity.
FIGURE 5.
Histidine mutants retain increased activity with hyperosmolarity. A, means of time-dependent pHi recoveries in the absence (quiescent; solid line) and presence of excess 100 mm NaCl (dotted lines) from 4 independent cell preparations. B, means of pHi-dependent recoveries (dpHi/dt) in the absence (quiescent; solid lines) and presence of excess 100 mm NaCl (dotted lines) from 4 independent cell preparations. C, dpHi/dt) at pHi 6.6 for data shown in B. Data are the means ± S.D. of six independent cell preparations. D, the pHi of indicated cell types at quiescence and with 100 mm NaCl (5 min). Data are the means ± S.D. of six independent cell preparations. **, p < 0.01; ***, p < 0.001
Differences in Contractile Actin Filaments and Focal Adhesions with NHE1-RXXR3 and -AXXA3
Increased NHE1 activity and pHi promote the assembly of actin filaments (8, 19–21). Consistent with these findings, we observed more contractile actin stress fibers in quiescent cells expressing NHE1-RXXR3 compared with NHE1-WT and -AXXA3 (Fig. 6A). Quantifying the intensity of rhodamine-phalloidin fluorescence confirmed significant differences between the three cell types (Fig. 6B). Additionally, immunolabeling for phosphorylated myosin light chain (pMLC) showed abundant and thick bundled actin filaments traversing the cell length highly decorated by pMLC with RXXR3. In contrast, actin stress fibers were fewer and appeared thinner and shorter with NHE1-WT and NHE1-AXXA3. With NHE1-WT, pMLC predominantly decorated the distal ends of actin filaments in contrast to the entire length with NHE1-RXXR3. With NHE1-AXXA3, pMLC labeling was mostly punctate with minimal labeling along actin filaments. Of note, actin aggregates, likely Hirano bodies, were seen in all three cell lines and is a feature of the parental PS120 fibroblasts (22).
FIGURE 6.
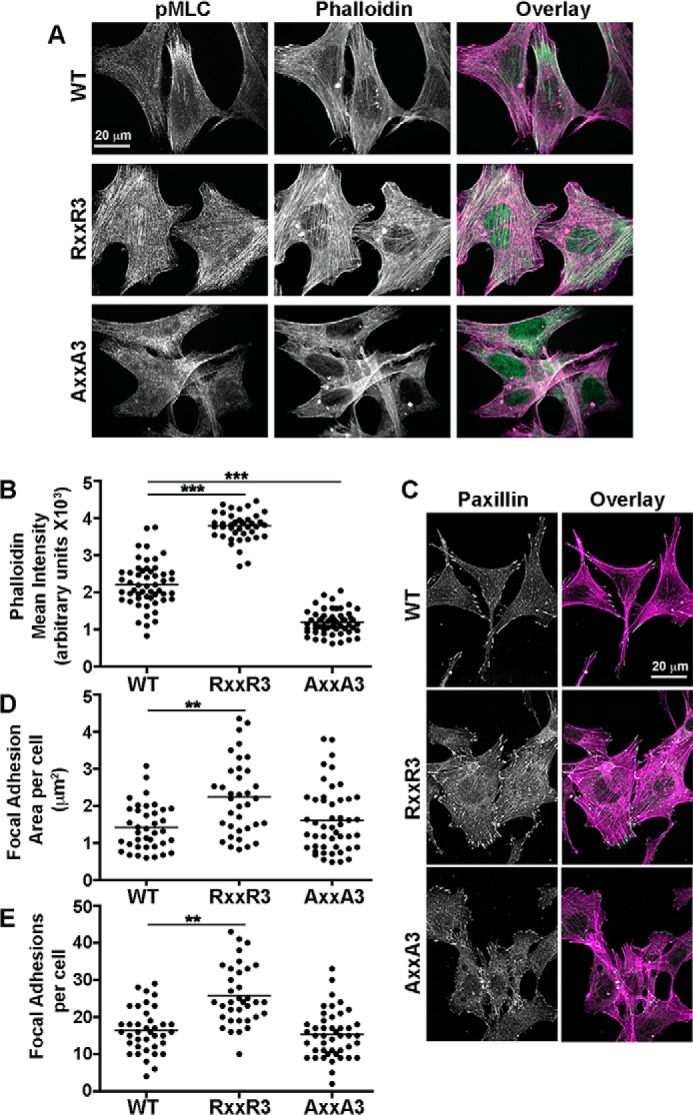
Histidine mutants have differences in abundance of contractile actin filaments and focal adhesions. A, representative confocal images of the indicated cell types at quiescence labeled with pMLC (green) and rhodamine-phalloidin (magenta). B, mean fluorescence intensities of rhodamine-phalloidin for WT (53 cells), RXXR3 (42 cells), and AXXA3 (58 cells) in three separate cell preparations. C, representative confocal images of the indicated cell types at quiescence labeled with paxillin (green) and rhodamine-phalloidin (magenta). Paxillin labeling is also shown in monochromatic LUT. D and E, quantification of the peripheral focal adhesion area per cell (D) and total number per cell (E). Shown are WT (43 cells), RXXR3 (41 cells), and AXXA3 (49 cells) in three independent cell preparations. **, p < 0.01; *** p < 0.001.
Increased NHE1 activity and pHi also increase the assembly and maturation of cell-substrate focal adhesions (9, 23). Accordingly, immunolabeling of the focal adhesion-associated protein paxillin showed more and larger peripheral focal adhesions with NHE1-RXXR3 compared with NHE1-WT (Fig. 6, C–E). With NHE1-RXXR3 there were pronounced clusters of focal adhesions at cell margins as well as numerous focal contacts along the dorsal cell surface. With NHE1-WT, focal adhesions appeared to be predominantly at cell protrusions, with a limited number of focal contacts at the dorsal cell surface. With NHE1-AXXA3, focal adhesions were similar in size and number to those with NHE1-WT, although punctate paxillin labeling at the dorsal surface was more pronounced. Consistent with differences in focal adhesion abundance, cells were more spread with NHE1-RXXR3 and more fusiform and elongated with NHE1-AXXA3, compared with NHE1-WT. Together, these findings suggest that increased activity with RXXR3 is associated with substantial changes in cell morphology with a contractile phenotype indicated by abundant pMLC-decorated actin stress fibers and focal adhesions.
Discussion
NHE1 is an acid extruder controlling H+ efflux at the plasma membrane and a major regulator of pHi in many cells. For normal pHi homeostasis, NHE1 activity increases in response to acidic pHi and becomes quiescent at neutral pHi. The pHi regulation of NHE1 activity was proposed over 30 years ago to be mediated by an internal H+ modifier site independent of the proton binding site used for substrate transport (11); however, the precise identity of this site remains unclear. Subsequent studies suggested that a proton modifier site located in the C-terminal cytoplasmic domain modulates a set point for NHE1 activity (24, 25) and found that binding PI(4,5)P2 is necessary for maximal NHE1 activity (13, 15), with increased binding at lower pH. However, studies on PI(4,5)P2 binding focused on arginines and lysines in a putative modifier site, which likely do not change charge within the physiological pH range to regulate binding affinity for anionic phospholipids. We found that a cluster of four histidines in the putative modifier site confer pH-dependent PI(4,5)P2 binding. When these histidines are substituted to arginines, NHE1 has constitutively increased activity, and pHi is higher, whereas alanine substitutions result in significantly decreased activity, although quiescent pHi is not different compared with wild type NHE1. Although RXXR3 and AXXA3 mutants lack pH-dependent PI(4,5)P2 binding and their activity in cells is increased and decreased, respectively, they retain pH sensitivity with greater activity at lower pHi. The pHi-dependent activity of these mutants could be maintained by alternative proton modifier sites that have been proposed in the NHE1 distal C terminus and transmembrane domain (26).
A pH-dependent “His switch” for binding membrane phospholipids is emerging as a regulatory mechanism for many proteins. However, His switches have predominantly been identified in cytosolic rather than integral membrane proteins. We previously showed that activation of the actin-binding protein cofilin at higher pH is mediated by deprotonation of cofilin-His-133, which decreases affinity for binding PI(4,5)P2 to release cofilin from its inactive, plasma membrane-bound state (19). We also showed a similar pH-sensitive His switch for releasing membrane-associated Dbs, a guanine nucleotide exchange factor for Cdc42 (27). Additionally, the FYVE domain that targets many proteins, including endosome antigen 1 and guanine nucleotide exchange factors, to intracellular vesicle membranes has two conserved His residues that confer a pH-sensitive His switch for binding phosphatidylinositol 3-phosphate (28, 29). Intracellular membrane targeting of proteins containing ENTH and ANTH domains is also regulated by His switches for binding membrane phospholipids (30). However, for integral membrane ion transport proteins, the role of pH-dependent His switches for phospholipid binding and activity is less clear. Fliegel and co-workers (31) recognized the histidine cluster in NHE1 we studied, but did not test PI(4,5)P2 binding, and found that human NHE1-540HYGAAA and -HYGRRR mutants have no effect on activity, which compared with our results suggests the possibility that His-540 may be a critical residue for pH sensing. In the NHE isoform NHE3, His-499, which is equivalent to His-543 of NHE1 (Fig. 1C), confers pHi sensitive activity (32); however, the role of His-499 in PI(4,5)P2 binding was not reported. Additionally, Na-HCO3 exchangers in the SLC4A family contain multiple histidines within and surrounding a PI(4,5)P2 binding site (33), although to our knowledge the importance of these histidines for PI(4,5)P2 binding has not been reported. Hence, our findings are significant in showing a pH-dependent His switch for PI(4,5)P2 binding that regulates activity of an integral membrane ion transport protein.
Our findings also suggest substantial differences in how growth factors and hyperosmolarity increase NHE1 activity, as activity of NHE1-RXXR3 and -AXXA3 increases with hyperosmolarity but not with PDGF. Hyperosmolarity is proposed to activate NHE1 through a mechanical, cell-shrinkage-dependent mechanism (34), and a predicted volume-sensitive site on NHE1 is different than sites required to mediate the stimulatory effects of growth factors (35). Many growth factors and hormones activate NHE1 by increasing the phosphorylation of distinct serine residues in the distal C terminus of NHE1, with direct phosphorylation confirmed by the kinases p90RSK (7), Akt (8), ROCK (9), and Nck-interacting kinase (NIK)/MAP4K4 (10). Ligand-increased NHE1 activity is inhibited by alanine substitutions in these serine residues. However, activity of the NHE1-AXXA3 mutant is significantly lower compared with wild type and does not increase with PDGF, which suggests that for wild type NHE1, PI(4,5)P2 binding by the membrane-proximal C terminus and phosphorylation of the distal C terminus are integrated. If the set point or pH dependence of a proton modifier site is changed by ligand-dependent NHE1 phosphorylation, it remains to be determined whether the pKa values of histidine residues regulating PI(4,5)P2 binding might be upshifted. Wakabayashi et al. (36, 37) proposed that growth factors and hormones increasing diacylglycerol activate NHE1 through a lipid-interacting domain (LID) encompassing amino acids 542–598 in human NHE1 that includes the PI(4,5)P2 binding region. They found that the LID domain binds phorbol esters and used FRET reporters to show increased association of the NHE1 C terminus with the plasma membrane in cells treated with phorbol esters. However, phorbol ester and PI(4,5)P2 binding within the LID may be distinct because alanine substitutions for Leu-573 and Ile-574 were identified as mediators of phorbol ester binding (36). Recent work from Schelling and co-workers (38) suggests that decreased NHE1 activity from disrupted PI(4,5)P2 binding contributes to apoptosis in renal tubular atrophy. They found that amphipathic, long chain acyl-CoA metabolites, which accumulate with glomerular injury, decrease NHE1 activity by competing with PI(4,5)P2 binding. Future studies could address whether a similar competitive inhibition of PI(4,5)P2 binding might contribute to the proposed NHE1-dependent apoptosis in neurodegenerative diseases (5).
Although we currently lack a unified mechanistic description for the complex interplay of regulated NHE1 activity, many publications described here indicate the importance of PI(4,5)P2 binding. Our findings reveal the critical significance of a histidine cluster for PI(4,5)P2 binding, which contributes to the established pHi dependence of NHE1 activity. Beyond insights on mechanisms regulating NHE1 activity, the constitutively active NHE1-RXXR3 mutant we report has promise as a new tool for determining functional consequences of increased pHi, particularly in cancer cell biology, based on increased pHi with activated oncogene and our recent finding (4) that increased pHi in the absence of activated oncogenes is sufficient to induce hyperproliferation and dysplasia.
Experimental Procedures
Sequence Alignment of NHE1 C-terminal Juxtamembrane Region
The sequence surrounding the His-rich region of human NHE1 was aligned to NHE1 orthologs from other species and to other human plasma membrane NHE family members (NHE2–5) using Clustal Omega (39). NCBI accession numbers used for alignment were: Homo sapiens NHE1, NP_003038.2; H. sapiens NHE2, NP_003039.2; H. sapiens NHE3, NP_004165.2; H. sapiens NHE4, NP_001011552.2; H. sapiens NHE5, NP_004585.1; Mus musculus NHE1, NP_058677.1; Rattus norvegicus NHE1, NP_036784.1; Gallus gallus NHE1, NP_001038108.1; Xenopus laevis NHE1, NP_001081553.1; Danio rerio NHE1, NP_001106952.1.
Expression and Purification of GST-NHE1 503–815
The C-terminal cytoplasmic domain of human NHE1 (amino acids 503–815) in the pGEX vector was previously described (10). Site-directed mutagenesis was used to mutate His-540, -543, -544, and -545 to alanines or arginines (QuikChange Lightning, Agilent, Santa Clara, CA). DNA primers were designed using an online primer design tool (QuikChange® Primer Design Program) and purchased from Elim Biopharmaceuticals (Hayward, CA). DNA sequencing was performed to verify mutations. Expression and affinity purification of wild type and mutant GST-NHE1 was performed as previously described (22). Protein was concentrated using an Amicon Ultracel-20K Centrifugal Filter Unit and stored at −80 °C. Protein integrity and purity were determined by Coomassie Blue-stained SDS-PAGE gels.
ELISA PI(4,5)P2 Binding Assay
Pre-blocked Cova PI(4,5)P2 screening plates (Echelon Biosciences, Salt Lake City, UT, catalogue number 1278H-6) were used for binding assays. Wild type and mutant GST-NHE1 were incubated at concentrations from 0 to 1 μm in binding buffer (10 mm HEPES, 150 mm NaCl, 1% mouse serum) of pH 7.5 or 7.0 for 1 h at room temperature. Wells were washed 3 times (3 min each) with wash buffer (binding buffer + 0.05% Tween 20) and incubated for 1 h at room temperature with peroxidase-conjugated anti-GST antibody (Abcam, Cambridge, MA, catalogue number ab137840, lot number GR169731–2) diluted 1:5000 in binding buffer. After washing 3 times, 100 μl of 3,3′,5,5′-tetramethylbenzidine (Sigma, catalogue number T8665, lot number SLBM6117V) was added to each well for 15 min, and the optical density was read at 370 nm on a SpectraMax M5 plate reader (Molecular Dynamics, Sunnyvale, CA). Statistical analyses were performed using GraphPad software.
Cell Expression of Recombinant NHE1
The mammalian expression construct, pCMV-NHE1, containing the full-length rat NHE1 sequence and a 3′ HA epitope tag was previously described (22, 40, 41). Site-directed mutagenesis was used to mutate His-544, -547, -548, and -549 to alanines or arginines as described above. Wild type and mutant NHE1-HA were stably expressed in NHE1-deficient PS120 cells, derived from CCL39 hamster lung fibroblasts as described (22). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% FBS and penicillin/streptomycin (complete media). The expression of wild type and mutant NHE1 was determined by immunoblotting of cell lysates, whereas the cell surface expression was determined by the selective biotinylation and isolation of cell surface proteins using a commercially available kit (Pierce Cell Surface Protein Isolation kit, Thermo Fisher Scientific, catalogue number 89881). Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes for immunoblotting. The membranes were probed with mouse anti-HA (12CA5) epitope tag (1:1000, ThermoFisher Scientific, catalogue number MA1–12429) for NHE1-HA or mouse anti-actin clone C4 (1:5,000, ED Millipore MAB1501) as a loading control. Peroxidase-conjugated goat anti-mouse (Bio-Rad, catalogue number 172-6516, lot number 350003069) and goat anti-rabbit secondary antibodies (Bio-Rad, catalogue number 172-1019, lot number 350003011) were used at a 1:10,000 dilution. Bound antibodies were detected by enhanced chemiluminescence.
NHE1 Activity
NHE1 activity was determined as the rate of pHi recovery (dpHi/dt) from an NH4Cl-induced acid load, as previously described (8). Briefly, cells were plated in triplicate for each condition and maintained in complete medium for 24 h before incubating with DMEM containing 0.2% FBS for 20 h to induce quiescence. Cells loaded with 1 μm 2,7-biscarboxyethyl-5(6)-carboxyfluorescein (BCECF; Invitrogen) in a nominally HCO3−-free HEPES buffer were incubated for 10 min in HEPES buffer containing 40 mm NH4Cl, which was rapidly aspirated and replaced with HEPES buffer in the absence (control) or presence of PDGF (50 ng/ml) or NaCl (100 mm). Ratios of BCECF fluorescence at Ex490/Em530 and Ex440/Em530 were acquired every 15 s for 5 min. Fluorescence ratios were converted to pHi by calibrating each well with 10 μm nigericin (Invitrogen) at pH 7.4 and pH 6.6. NHE1 activity was expressed as dpHi/dt by evaluating the derivative of the slope of the time-dependent pHi recovery at intervals of 0.05 pH units, and statistical analyses were performed using GraphPad software. Steady-state pHi was determined as previously described (8). In brief, quiescent cells loaded with BCECF in buffer containing 25 mm HCO3− were washed and incubated in HCO3− buffer in the absence or presence of PDGF or NaCl for 5 min before measuring fluorescence ratios and calibrating pHi as described above.
Immunolabeling
Immunocytochemistry was performed as previously described (8). In brief cells plated at a density of 50 × 104 cells/well of a 6-well plate containing 100-mm glass coverslips were maintained in complete medium for 24 h, washed, and maintained in DMEM containing 0.2% FBS for 20 h. After washing with 100 mm phosphate buffer (PB), cells were fixed in 4% paraformaldehyde in PB for 10 min, permeabilized in 0.1% Triton X-100 in PB for 10 min, and incubated in blocking PB containing 2% BSA for 60 min. Cells were incubated overnight at 4 °C with primary antibodies to pMLC (T18/S19, rabbit, Cell Signaling Technologies, catalogue number 3674, lot number 5) and paxillin (mouse, BD Transduction Laboratories, catalogue number 610620, lot number 66614) diluted 1:200 in PB containing 0.01% Triton X-100. After washing with PB, cells were incubated for 1 h with PB containing secondary antibodies Alexa Fluor 488 goat anti-rabbit (Thermo Fisher Scientific, catalogue number A11008, lot number 458548) or goat anti-mouse (Thermo Fisher Scientific, catalogue number A11001, lot number 455497) (1:200) and rhodamine-phalloidin (Thermo Fisher Scientific, catalogue number R415, lot number 1616945) (1:400). Fluorescence images were acquired using a spinning disk confocal (Yokogawa; CSU-10) on a microscope (Nikon; TE-2000) with a 60X objective (Plan TIRF 1.49 NA) equipped with a cooled CCD camera and analyzed using NIS-Elements software (Nikon). To quantify rhodamine-phalloidin fluorescence, background subtraction was first performed from an ROI that did not include cells, and then mean background-subtracted rhodamine fluorescence was determined as the average of three uniform regions of interest for each of the indicated number of cells in three separate preparations. Quantitative analysis of focal adhesions was performed on maximum intensity projections generated from a z-stack. Background subtraction was performed from an ROI that did not include cells. Focal adhesions were selected for analysis if they were located near or touching plasma membrane and had actin filaments terminating within. The “Auto detect ROI” tool with a graphical user interface tuning parameters setting of 5 was used to select ROIs. Area and mean intensity of each focal adhesion was quantified and exported to Microsoft Excel for analysis; graphs were generated using Prism. Outliers greater than two standard deviations from the mean were excluded from the final analysis.
Author Contributions
B. A. W. and A. S. conceived the initial studies. B. A. W. generated and characterized expression in cell lines, including surface biotinylation. K. A. W. performed and analyzed the in vitro PI(4,5)P2 binding assays. B. K. G.-H. performed quantification of focal adhesions. A. S. generated GST constructs used in in vitro binding assays and performed the initial characterization of lipid binding. C. C. performed the Western blot analysis. D. L. B. performed the NHE1 activity assays, pHi determinations, and immunolabeling studies. B. A. W., K. A. W., B. K. G.-H., and D. L. B. contributed to the writing of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health F32 NRSA (National Research Service Award) Postdoctoral Fellowship CA177085 (to K. A. W.) and Grant R01 GM116384 (to D. L. B.). This work was also supported in whole or part by Canadian Institutes of Health research postdoctoral fellowship (to B. A. W.) and Deutsche Forschungsgemeinschaft (DFG) Postdoctoral Fellowship SCHO 1410/1-1 (to A. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institutes of Health.
- NHE1
- sodium hydrogen exchanger 1
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- pHi
- intracellular pH
- PDGF
- platelet-derived growth factor
- pMLC
- phosphorylated myosin light chain
- LID
- lipid-interacting domain
- BCECF
- 2,7-biscarboxyethyl-5(6)-carboxyfluorescein
- PB
- phosphate buffer
- ROI
- region(s) of interest.
References
- 1. Boron W. (2004) Regulation of intracellular pH. Adv. Physiol. Educ. 28, 160–179 [DOI] [PubMed] [Google Scholar]
- 2. Cardone R. A., Casavola V., and Reshkin S. J. (2005) The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat. Rev. Cancer 5, 786–795 [DOI] [PubMed] [Google Scholar]
- 3. Webb B. A., Chimenti M., Jacobson M. P., and Barber D. L. (2011) Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer 11, 671–677 [DOI] [PubMed] [Google Scholar]
- 4. Grillo-Hill B. K., Choi C., Jimenez-Vidal M., and Barber D. L. (2015) Increased H+ efflux is sufficient to induce dysplasia and necessary for viability with oncogene expression. eLife 10.7554/eLife.03270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harguindey S., Reshkin S. J., Orive G., Arranz J. L., and Anitua E. (2007) Growth and trophic factors, pH, and the Na+/H+ exchanger in Alzheimer's disease, other neurodegenerative diseases, and cancer: new therapeutic possibilities and potential dangers. Curr. Alzheimer Res. 4, 53–65 [DOI] [PubMed] [Google Scholar]
- 6. Fang B., Wang D., Huang M., Yu G., and Li H. (2010) Hypothesis on the relationship between the change in intracellular pH and incidence of sporadic Alzheimer's disease or vascular dementia. Int. J. Neurosci. 120, 591–595 [DOI] [PubMed] [Google Scholar]
- 7. Takahashi E., Abe J., Gallis B., Aebersold R., Spring D. J., Krebs E. G., and Berk B. C. (1999) p90(RSK) is a serum-stimulated Na+/H+ exchanger isoform-1 kinase: regulatory phosphorylation of serine 703 of Na+/H+ exchanger isoform-1. J. Biol. Chem. 274, 20206–20214 [DOI] [PubMed] [Google Scholar]
- 8. Meima M. E., Webb B. A., Witkowska H. E., and Barber D. L. (2009) The sodium-hydrogen exchanger NHE1 is an Akt substrate necessary for actin filament reorganization by growth factors. J. Biol. Chem. 284, 26666–26675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tominaga T., Ishizaki T., Narumiya S., and Barber D. L. (1998) p160ROCK mediates RhoA activation of Na-H exchange. EMBO J. 17, 4712–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yan W., Nehrke K., Choi J., and Barber D. L. (2001) The Nck-interacting kinase (NIK) phosphorylates the Na+-H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J. Biol. Chem. 276, 31349–31356 [DOI] [PubMed] [Google Scholar]
- 11. Aronson P. S., Nee J., and Suhm M. A. (1982) Modifier role of internal H+ in activating the Na+-H+ exchanger in renal microvillus membrane vesicles. Nature 299, 161–163 [DOI] [PubMed] [Google Scholar]
- 12. Hilgemann D. W., Feng S., and Nasuhoglu C. (2001) The complex and intriguing lives of PIP2 with ion channels and transporters. Sci. STKE 2001, re19. [DOI] [PubMed] [Google Scholar]
- 13. Aharonovitz O., Zaun H. C., Balla T., York J. D., Orlowski J., and Grinstein S. (2000) Intracellular pH regulation by Na+/H+ exchange requires phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 150, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alexander R. T., Jaumouillé V., Yeung T., Furuya W., Peltekova I., Boucher A., Zasloff M., Orlowski J., and Grinstein S. (2011) Membrane surface charge dictates the structure and function of the epithelial Na+/H+ exchanger. EMBO J. 30, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abu Jawdeh B. G., Khan S., Deschênes I., Hoshi M., Goel M., Lock J. T., Shinlapawittayatorn K., Babcock G., Lakhe-Reddy S., DeCaro G., Yadav S. P., Mohan M. L., Naga Prasad S. V., Schilling W. P., Ficker E., and Schelling J. R. (2011) Phosphoinositide binding differentially regulates NHE1 Na+/H+ exchanger-dependent proximal tubule cell survival. J. Biol. Chem. 286, 42435–42445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schönichen A., Webb B. A., Jacobson M. P., and Barber D. L. (2013) Considering protonation as a posttranslational modification regulating protein structure and function. Annu. Rev. Biophys. 42, 289–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Srivastava J., Barreiro G., Groscurth S., Gingras A. R., Goult B. T., Critchley D. R., Kelly M. J., Jacobson M. P., and Barber D. L. (2008) Structural model and functional significance of pH-dependent talin-actin binding for focal adhesion remodeling. Proc. Natl. Acad. Sci. U.S.A. 105, 14436–14441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Counillon L., Pouysségur J., and Reithmeier R. A. (1994) The Na+/H+ exchanger NHE-1 possesses N- and O-linked glycosylation restricted to the first N-terminal extracellular domain. Biochemistry 33, 10463–10469 [DOI] [PubMed] [Google Scholar]
- 19. Frantz C., Barreiro G., Dominguez L., Chen X., Eddy R., Condeelis J., Kelly M. J., Jacobson M. P., and Barber D. L. (2008) Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J. Cell Biol. 183, 865–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Magalhaes M. A., Larson D. R., Mader C. C., Bravo-Cordero J. J., Gil-Henn H., Oser M., Chen X., Koleske A. J., and Condeelis J. (2011) Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J. Cell Biol. 195, 903–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clement D. L., Mally S., Stock C., Lethan M., Satir P., Schwab A., Pedersen S. F., and Christensen S. T. (2013) PDGFRα signaling in the primary cilium regulates NHE1-dependent fibroblast migration via coordinated differential activity of MEK1/2-ERK1/2-p90RSK and AKT signaling pathways. J. Cell Sci. 126, 953–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Denker S. P., Huang D. C., Orlowski J., Furthmayr H., and Barber D. L. (2000) Direct binding of the Na-H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H+ translocation. Mol. Cell 6, 1425–1436 [DOI] [PubMed] [Google Scholar]
- 23. Choi C.-H., Webb B. A., Chimenti M. S., Jacobson M. P., and Barber D. L. (2013) pH sensing by FAK-His58 regulates focal adhesion remodeling. J. Cell Biol. 202, 849–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wakabayashi S., Bertrand B., Shigekawa M., Fafournoux P., and Pouysségur J. (1994) Growth factor activation and “H+-sensing” of the Na+/H+ exchanger isoform 1 (NHE1). Evidence for an additional mechanism not requiring direct phosphorylation. J. Biol. Chem. 269, 5583–5588 [PubMed] [Google Scholar]
- 25. Orlowski J., and Grinstein S. (1997) Na+/H+ exchangers of mammalian cells. J. Biol. Chem. 272, 22373–22376 [DOI] [PubMed] [Google Scholar]
- 26. Wakabayashi S., Ikeda T., Iwamoto T., Pouysségur J., and Shigekawa M. (1997) Calmodulin-binding autoinhibitory domain controls “pH-sensing” in the Na+/H+ exchanger NHE1 through sequence-specific interaction. Biochemistry 36, 12854–12861 [DOI] [PubMed] [Google Scholar]
- 27. Frantz C., Karydis A., Nalbant P., Hahn K. M., and Barber D. L. (2007) Positive feedback between Cdc42 activity and H+ efflux by the Na-H exchanger NHE1 for polarity of migrating cells. J. Cell Biol. 179, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee S. A., Eyeson R., Cheever M. L., Geng J., Verkhusha V. V., Burd C., Overduin M., and Kutateladze T. G. (2005) Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proc. Natl. Acad. Sci. U.S.A. 102, 13052–13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He J., Vora M., Haney R. M., Filonov G. S., Musselman C. A., Burd C. G., Kutateladze A. G., Verkhusha V. V., Stahelin R. V., and Kutateladze T. G. (2009) Membrane insertion of the FYVE domain is modulated by pH. Proteins 76, 852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hom R. A., Vora M., Regner M., Subach O. M., Cho W., Verkhusha V. V., Stahelin R. V., and Kutateladze T. G. (2007) pH-dependent binding of the Epsin ENTH domain and the AP180 ANTH domain to PI(4,5)P2-containing bilayers. J. Mol. Biol. 373, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dibrov P., Murtazina R., Kinsella J., and Fliegel L. (2000) Characterization of a histidine rich cluster of amino acids in the cytoplasmic domain of the Na+/H+ exchanger. Biosci. Rep. 20, 185–197 [DOI] [PubMed] [Google Scholar]
- 32. Cha B., Oh S., Shanmugaratnam J., Donowitz M., and Yun C. C. (2003) Two histidine residues in the juxta-membrane cytoplasmic domain of Na+/H+ exchanger isoform 3 (NHE3) determine the set point. J. Membr. Biol. 191, 49–58 [DOI] [PubMed] [Google Scholar]
- 33. Hong J. H., Yang D., Shcheynikov N., Ohana E., Shin D. M., and Muallem S. (2013) Convergence of IRBIT, phosphatidylinositol (4,5) bisphosphate, and WNK/SPAK kinases in regulation of the Na+-HCO3− cotransporters family. Proc. Natl. Acad. Sci. U.S.A. 110, 4105–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krump E., Nikitas K., and Grinstein S. (1997) Induction of tyrosine phosphorylation and Na+/H+ exchanger activation during shrinkage of human neutrophils. J. Biol. Chem. 272, 17303–17311 [DOI] [PubMed] [Google Scholar]
- 35. Bianchini L., Kapus A., Lukacs G., Wasan S., Wakabayashi S., Pouysségur J., Yu F. H., Orlowski J., and Grinstein S. (1995) Responsiveness of mutants of NHE1 isoform of Na+/H+ antiport to osmotic stress. Am. J. Physiol. 269, C998–C1007 [DOI] [PubMed] [Google Scholar]
- 36. Wakabayashi S., Nakamura T. Y., Kobayashi S., and Hisamitsu T. (2010) Novel phorbol ester binding motif mediates hormonal activation of Na+/H+ exchanger. J. Biol. Chem. 285, 26652–26661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shimada-Shimizu N., Hisamitsu T., Nakamura T. Y., Hirayama N., and Wakabayashi S. (2014) Na+/H+ exchanger 1 is regulated via its lipid-interacting domain, which functions as a molecular switch: a pharmacological approach using indolocarbazole compounds. Mol. Pharmacol. 85, 18–28 [DOI] [PubMed] [Google Scholar]
- 38. Khan S., Abu Jawdeh B. G., Goel M., Schilling W. P., Parker M. D., Puchowicz M. A., Yadav S. P., Harris R. C., El-Meanawy A., Hoshi M., Shinlapawittayatorn K., Deschênes I., Ficker E., and Schelling J. R. (2014) Lipotoxic disruption of NHE1 interaction with PI(4,5)P2 expedites proximal tubule apoptosis. J. Clin. Invest. 124, 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kapus A., Grinstein S., Wasan S., Kandasamy R., and Orlowski J. (1994) Functional characterization of three isoforms of the Na+/H+ exchanger stably expressed in Chinese hamster ovary cells: ATP dependence, osmotic sensitivity, and role in cell proliferation. J. Biol. Chem. 269, 23544–23552 [PubMed] [Google Scholar]
- 41. Orlowski J. (1993) Heterologous expression and functional properties of amiloride high affinity (NHE-1) and low affinity (NHE-3) isoforms of the rat Na/H exchanger. J. Biol. Chem. 268, 16369–16377 [PubMed] [Google Scholar]