Abstract
Contactin-associated protein-like 2 (CNTNAP2) is a large multidomain neuronal adhesion molecule implicated in a number of neurological disorders, including epilepsy, schizophrenia, autism spectrum disorder, intellectual disability, and language delay. We reveal here by electron microscopy that the architecture of CNTNAP2 is composed of a large, medium, and small lobe that flex with respect to each other. Using epitope labeling and fragments, we assign the F58C, L1, and L2 domains to the large lobe, the FBG and L3 domains to the middle lobe, and the L4 domain to the small lobe of the CNTNAP2 molecular envelope. Our data reveal that CNTNAP2 has a very different architecture compared with neurexin 1α, a fellow member of the neurexin superfamily and a prototype, suggesting that CNTNAP2 uses a different strategy to integrate into the synaptic protein network. We show that the ectodomains of CNTNAP2 and contactin 2 (CNTN2) bind directly and specifically, with low nanomolar affinity. We show further that mutations in CNTNAP2 implicated in autism spectrum disorder are not segregated but are distributed over the whole ectodomain. The molecular shape and dimensions of CNTNAP2 place constraints on how CNTNAP2 integrates in the cleft of axo-glial and neuronal contact sites and how it functions as an organizing and adhesive molecule.
Keywords: cell adhesion, cell surface receptor, protein-protein interaction, structural biology, synapse, contactin, contactin-associated protein like, neuropsychiatric disorders, single particle analysis, synaptic organizer
Introduction
Contactin-associated protein-like 2 (CNTNAP2; also known as CASPR2) is a type I trans-membrane cell adhesion molecule. CNTNAP2 is found in the central and peripheral nervous system, where it is highly expressed throughout the brain and spinal cord, particularly in the frontal and temporal lobes, striatum, dorsal thalamus, and specific layers of the cortex (1, 2). In humans, alterations in the CNTNAP2 gene are associated with a variety of neurological disorders, including epilepsy, schizophrenia, autism spectrum disorder (ASD),5 intellectual disability, and language delay, but also obesity (2–5). In addition, in humans, autoantibodies that target the extracellular domain of CNTNAP2 are linked to autoimmune epilepsies, cerebellar ataxia, autoimmune encephalitis, neuromyotonia, Morvan's syndrome, and behavioral abnormalities including amnesia, confusion, and neuropsychiatric features (6–12).
CNTNAP2 carries out multiple functions in the nervous system. In myelinated axons, CNTNAP2 localizes to the juxtaparanodes, unique regions that flank the nodes of Ranvier. Here, CNTNAP2 takes part in an extensive network of proteins that attaches the glial myelin sheath to the axon and that segregates Na+ and K+ channels aiding to propagate nerve impulses efficiently (13). At these axo-glial contact points, the ectodomain of CNTNAP2 binds the adhesion molecule contactin 2 (CNTN2), forming a molecular bridge that spans the extracellular space, whereas the cytoplasmic tail of CNTNAP2 recruits K+ channels (13–17). CNTNAP2 has an emerging role as well at contact points between neurons called synapses, in particular inhibitory synapses, and this role is probably important for its clinical significance (18–20). At synapses, CNTNAP2 localizes to the presynaptic membrane and binds CNTN2 tethered to the postsynaptic membrane, forming a trans-synaptic bridge that spans the synaptic cleft (19). CNTNAP2 knock-out mice develop seizures, hyperactivity, and behavioral abnormalities associated with ASD (21). Knock-out and knockdown studies indicate that CNTNAP2 is essential to maintain normal network activity and synaptic transmission; its loss leads to decreased dendritic arborization and reduced numbers of inhibitory interneurons, excitatory synapses, and inhibitory synapses (21–23). CNTNAP2 also influences the cellular migration of neurons, guiding them to their correct position in the final layered organization of the brain (1, 21, 24). CNTNAP2 thus plays a key role in the formation of neural circuits through its impact on neural connectivity, neural migration, synapse development, and synaptic communication. Through its organizing role at the nodes of Ranvier, it may influence nerve conduction as well.
The extracellular domain of CNTNAP2 contains eight defined domains: a F58C (discoidin) domain, four LNS domains, two EGF-like repeats, and a fibrinogen-like domain (Fig. 1A). Because CNTNAP2 contains a so-called “neurexin repeat” (LNS-EGF-LNS), it has been suggested that CNTNAPs are members of the neurexin family of synaptic cell adhesion molecules (25, 26). CNTN2 consists of six Ig domains followed by four fibronectin domains, and it is tethered to the cell surface by a glycosylphosphatidylinositol anchor (Fig. 1A). At axo-glial contacts, it has been proposed that the ectodomains of CNTNAP2 and CNTN2 form a cis-complex tethered to the axonal membrane, which in turn recruits a second CNTN2 molecule on the opposing glial membrane to form a bridge spanning the axo-glial cleft (13, 16–17, 27). However, a trans-complex consisting of an axonal CNTNAP2 and a glial CNTN2 molecule has also been proposed (28). At synaptic contacts, CNTNAP2 and CNTN2 appear to form a trans-complex spanning the synaptic cleft (19).
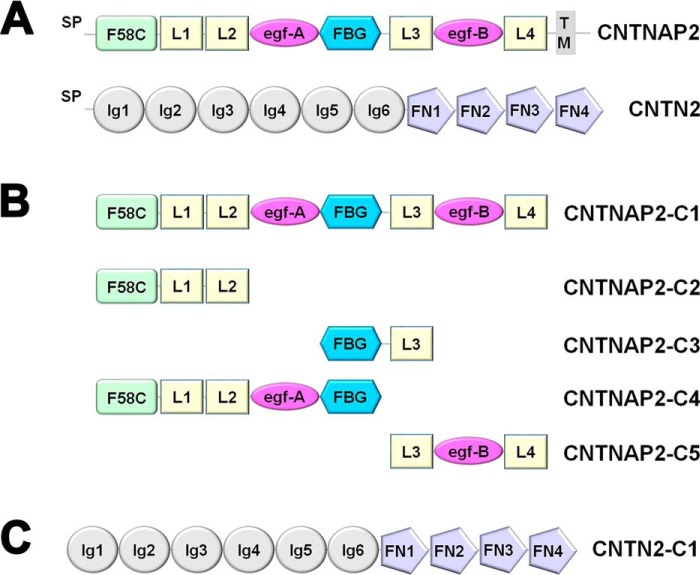
FIGURE 1.
CNTNAP2 and CNTN2. A, domain structure of CNTNAP2 and CNTN2. CNTNAP2 contains a coagulation factor 5/8 type C (F58C) domain; laminin, neurexin, sex hormone binding globulin (LNS or L) domains; egf-like repeats (egf); and a fibrinogen-like (FBG) domain. CNTN2 contains Ig and fibronectin type III domains (FN). Signal peptides (SP) and trans-membrane domain (TM) are indicated. B, CNTNAP2 constructs used in this study. C, CNTN2-C1 construct used in this study. CNTN1-C1 has an analogous domain organization. To improve legibility, CNTNAP2-C1 is abbreviated to CNTNAP2, CNTN2-C1 to CNTN2, and CNTN1-C1 to CNTN1 throughout the text.
The extracellular region of CNTNAP2 is directly linked to disease. A putatively secreted form of the CNTNAP2 ectodomain generated by the homozygous mutation I1253X causes cortical dysplasia-focal epilepsy in humans, a disorder hallmarked by epilepsy as well as cognitive and behavioral deficits; several heterozygous in-frame deletions affecting the N-terminal F58C, L1, and L2 domains are linked to mental retardation, seizures, and speech deficits (24, 29–32). The N-terminal region of CNTNAP2, in particular the F58C domain, is targeted by human autoimmune antibodies associated with encephalitis and/or peripheral nerve hyperexcitability (12, 19). Furthermore, many point mutations in the CNTNAP2 ectodomain have been linked to ASD, although their precise clinical impacts remain to be delineated (18, 33).
To gain insight into the structure and function of CNTNAP2, we overexpressed the extracellular domain of CNTNAP2 and its partner, CNTN2. We established that the ectodomains of CNTNAP2 and CNTN2 interact directly and specifically with each other with low nanomolar affinity. Also, we determined the architecture of the large multidomain extracellular region of CNTNAP2 using electron microscopy. By identifying epitopes and characterizing fragments, we assigned domains within the CNTNAP2 molecular envelope. Our data reveal that CNTNAP2 has a very different architecture compared with neurexin 1α, the prototype for the neurexin superfamily, suggesting that CNTNAP2 uses a different strategy to integrate into the synaptic protein network. Furthermore, the molecular shape and dimensions of CNTNAP2 provide molecular insight into how CNTNAP2 functions in the cleft of axo-glial and neuronal contacts as an organizing and adhesive molecule.
Results
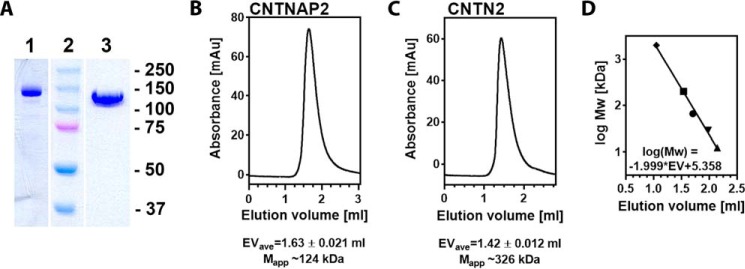
To delineate the architecture of CNTNAP2 and probe its interaction with CNTN2, we produced a panel of purified recombinant proteins in insect cells (Fig. 1, B and C). The extracellular region of CNTNAP2 is observed as a monodisperse protein with an apparent molecular mass of ∼124 kDa by size exclusion chromatography (i.e. close to its calculated molecular mass of 134.1 kDa), suggesting a globular nature (Fig. 2). The CNTN2 ectodomain is also monodisperse in solution, but its apparent molecular mass of ∼326 kDa is much larger than its calculated molecular mass of 108.6 kDa, suggesting that it forms either an elongated or a multimeric species that gel filtration chromatography cannot distinguish between (Fig. 2). To confirm these results, we analyzed the ectodomains of CNTNAP2 and CNTN2 by dynamic light scattering, which revealed a similar difference between the two molecules (i.e. an estimated molecular mass of 168 ± 66 kDa) (polydispersity index 0.154; ∼39% polydispersity) for CNTNAP2 and 444 ± 135 kDa (polydispersity index 0.092; ∼30% polydispersity) for CNTN2, respectively. The monomeric nature of CNTNAP2 was confirmed by electron microscopy (EM) (see below), as it was for CNTN2 as well.6
FIGURE 2.
Extracellular domains of CNTNAP2 and CNTN2. A, SDS-PAGE analysis of purified recombinant CNTNAP2 (lane 1) and CNTN2 (lane 3). Markers (in kDa) are shown in lane 2. B, size exclusion chromatography of CNTNAP2; C, size exclusion chromatography of CNTN2; D, size exclusion chromatography of standards (2,000, 200, 66, 29, and 12.4 kDa) and resulting calibration line. Samples and standards were run in triplicate. The average elution volume (EVave) and S.D. value are shown. The apparent molecular masses (Mapp) are indicated and reflect large versus small species.
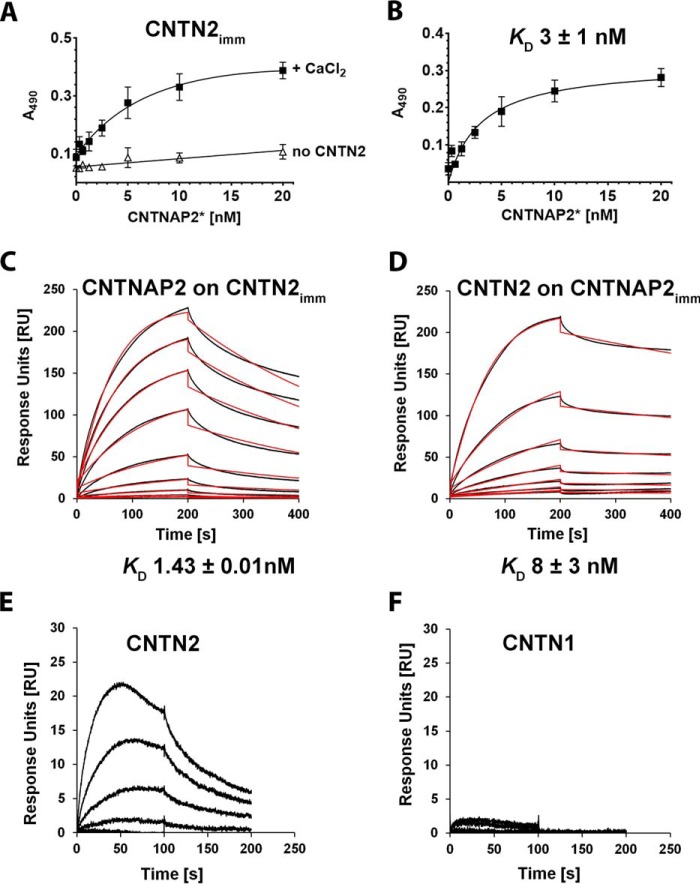
CNTNAP2 has been postulated to interact with CNTN2 on account of cell-based assays (13, 16, 17, 19, 27, 28). To test whether the ectodomains of CNTNAP2 and CNTN2 are sufficient to bind each other directly, we used a solid phase binding assay and showed that CNTNAP2 binds CNTN2 with ∼3 nm affinity (Fig. 3, A and B). We confirmed this interaction with surface plasmon resonance (SPR). CNTN2 immobilized on a biosensor surface bound CNTNAP2 with high affinity (KD ∼1.43 ± 0.01 nm) with kinetic parameters ka ∼(186.0 ± 0.01) × 104 m−1 s−1 and kd ∼(26.50 ± 0.006) × 10−4 s−1 (Fig. 3C). In the reverse assay, immobilized CNTNAP2 also bound soluble CNTN2 with nanomolar affinity (KD ∼8 ± 3 nm) and kinetic parameters ka ∼(8.63 ± 0.03) × 104 m−1 s−1 and kd ∼(6.96 ± 0.02) × 10−4 s−1 (Fig. 3D). The higher affinity observed when CNTN2 is immobilized suggests that constraining the flexibility of CNTN2 may increase its affinity for CNTNAP2. Regardless, both the solid phase and SPR assays demonstrated that the CNTNAP2 and CNTN2 ectodomains bind each other directly with nanomolar affinity. To assess the specificity of this interaction, we tested the binding of CNTN2 and CNTN1, respectively, to a CNTNAP2-coupled biosensor by SPR and revealed that although CNTN2 binds CNTNAP2 readily, CNTN1 does not (Fig. 3, E and F). However, when the contactins were immobilized on the biosensor (the “reverse orientation”), there was little difference between the two, which we believe could be due to the increased affinity gained by constraining the conformation of the long and flexible contactin molecules on the sensor (yielding a ∼6-fold difference in affinity for CNTNAP2 and CNTN2; Fig. 3, C and D).
FIGURE 3.
Binding between CNTNAP2 and CNTN2 ectodomains. A, increasing concentrations of biotinylated CNTNAP2* were incubated in wells with immobilized CNTN2 in presence of 5 mm CaCl2 (■) or in wells lacking CNTN2 (▵). B, specific binding, expressed as the total binding in the presence of Ca2+ minus the binding in the absence of CNTN2. Error bars, S.E. C, binding of soluble CNTNAP2 to a CNTN2-coupled sensor by SPR. Binding curves of CNTNAP2 (0.125–10 nm) (in black) were fit to a 1:1 binding model (red). D, binding of soluble CNTN2 to a CNTNAP2-coupled sensor by SPR. Binding curves of CNTN2 (1.56–200 nm) (black) were fit to a 1:1 binding model (red). E and F, side-by-side comparison of CNTN2 and CNTN1 binding to a single CNTNAP2-coupled sensor by SPR.
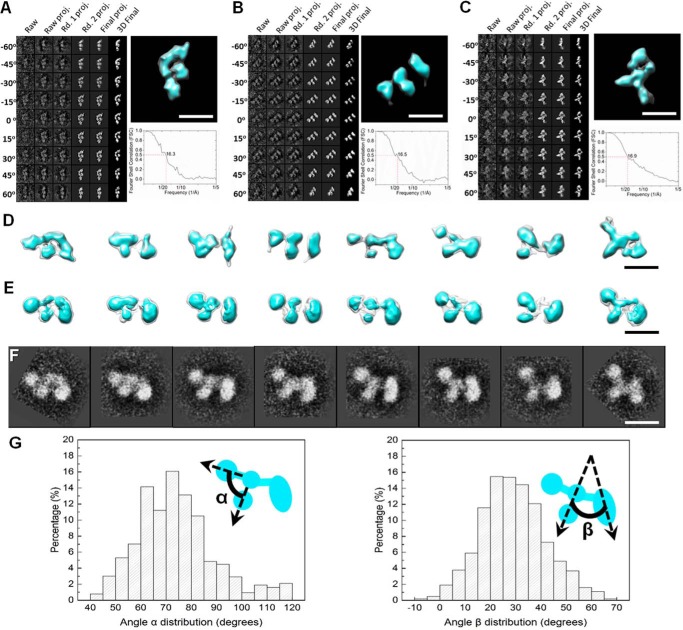
To assess the architecture of CNTNAP2, we used negative staining electron microscopy (NS-EM), because the relatively small molecular mass of CNTNAP2 (134 kDa) makes it challenging to image by cryo-EM. The CNTNAP2 ectodomain is observed as a monomer that is ∼151 Å long and ∼90 Å wide (Fig. 4A). Analysis of 12 representative CNTNAP2 particles revealed a structure composed of four discrete globular densities: a large lobe, a middle lobe clearly composed of two adjacent globules, and a small lobe (Fig. 4B). To reduce the noise, 53,774 particle images were submitted to 200 reference-free two-dimensional (2D) class averaging (Fig. 4C). To highlight the major features of CNTNAP2 and its organization, representative particles from the reference-free class averages were contrasted next to schematic representations, revealing an F-shaped structure (Fig. 4, D and E). To determine the three-dimensional (3D) structure of CNTNAP2, a multireference single-particle reconstruction method was used to refine the particle images (34). To avoid potential bias introduced by initial models during the single-particle 3D refinement and reconstruction stage, we used initial models that were derived from experimental data obtained through electron tomography (ET). In brief, eight representative molecules were selected and imaged from a series of tilt angles. The tilt images from each molecule were aligned and back-projected to produce a corresponding ab initio 3D reconstruction of each molecule using the individual particle electron tomography (IPET) method (35) (Fig. 5, A–D). These eight IPET ab initio 3D reconstructions served as initial models to carry out the multirefinement algorithm with EMAN (34) (Fig. 5E). The 3D reconstructions refined from 53,774 particles also indicated that CNTNAP2 forms an asymmetric, F-shaped molecule composed of three discrete regions: a large lobe (88 × 47 Å), a middle lobe (91 × 40 Å), and a small lobe (54 × 44 Å). The CNTNAP2 ectodomain contains a striking combination of compact lobes that flex with respect to each other via molecular hinges. As shown for the panel of representative particles, the large, middle, and small lobes maintain themselves as well defined entities, whereas the lobes flex with respect to each other (Fig. 5, D and E). This conformational heterogeneity produces a portfolio of F-shaped particles reminiscent of a running dog (Fig. 5F). To examine the impact of the molecular hinges on the conformational freedom of the CNTNAP2 molecule, we carried out a statistical analysis comparing 3,450 CNTNAP2 particle images. Using the middle lobe as a reference point, we determined that the large lobe flexes over a range of ∼65°, whereas the small lobe shows even greater freedom, flexing over a range of ∼74° (Fig. 5G).
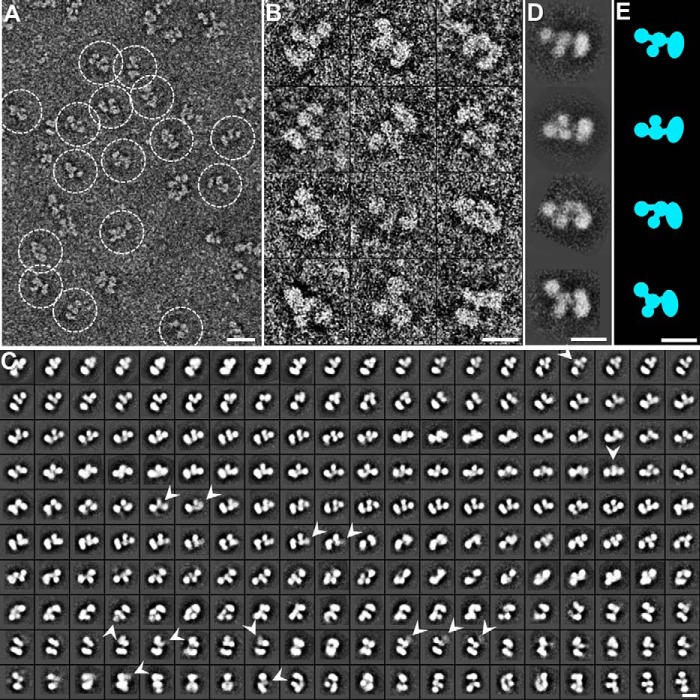
FIGURE 4.
Negative staining EM images and reference-free class averages of CNTNAP2. A, survey view of CNTNAP2 particles prepared by optimized negative staining. B, 12 representative particles of CNTNAP2. C, all 200 reference-free class averages calculated from 53,774 particles picked from 1,392 micrographs. In some class averages, a domain is fuzzy, probably due to flexibility and the dynamics of the protein (arrowheads). D, four selected reference-free class averages of the particles. E, schematic of particles corresponding to D. Scale bar, 200 Å (A) and 100 Å (B–E).
FIGURE 5.
Three-dimensional reconstruction and conformational variability analysis of CNTNAP2. A–C, process to generate representative 3D density maps, each from an individual CNTNAP2 particle, using IPET (left). Three examples are shown. Final IPET 3D density map of each single CNTNAP2 particle is displayed in the top right panel, and Fourier shell correlation is displayed in the bottom right panel. D, eight 3D density maps each reconstructed from a single molecule from electron tomographic images using the IPET method. E, eight single-particle 3D reconstructions of CNTNAP2. Each reconstruction was refined using an IPET 3D reconstruction as an initial model obtained via a multireference refinement algorithm using the EMAN single-particle reconstruction software. F, selected referenced 2D classifications supporting the range of particle conformational variability seen in D and E. G, histograms of the angles between the small and medium lobe (α) and between the medium and large lobe (β). Envelopes in D and E are displayed at contour levels corresponding to volumes of ∼133 kDa (cyan) and 266 kDa (transparent). Scale bars, 100 Å.
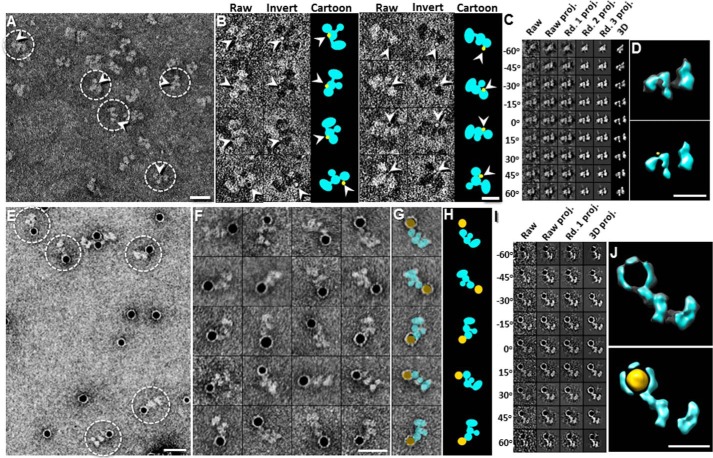
To determine the domain organization within CNTNAP2, we used three independent approaches: nanogold labeling, antibody labeling, and CNTNAP2 fragments. For the nanogold labeling experiment, we labeled the C-terminal hexahistidine tag at the CNTNAP2 L4 domain with two types of Ni-NTA nanogold particles, 1.8 nm (Fig. 6, A–D) and 5.0 nm (Fig. 6, E–J). Survey EM micrographs and representative images of 1.8-nm nanogold-labeled CNTNAP2 showed F-shaped particles with nanogold clusters bound, the visualization of which was enhanced by inverting the contrast to elevate the nanogold above the background noise (Fig. 6B). The 1.8-nm nanogold clusters consistently localized next to the small lobe of CNTNAP2 (Fig. 6B). We confirmed the nanogold location in three dimensions using ET images and IPET 3D reconstruction of a representative CNTNAP2 molecule bound to a 1.8-nm nanogold particle, which enabled us to highlight the protein and the nanogold particle, respectively, by overlaying the 3D map and the contrast-inverted 3D map (Fig. 6, C and D). Survey EM micrographs of 5-nm nanogold-labeled CNTNAP2 also showed dark, round densities corresponding to the nanogold on the surface of CNTNAP2 particles near the small lobe (Fig. 6, E–H). As done for the 1.8-nm nanogold-labeled CNTNAP2, we confirmed the 3D location of the 5-nm gold cluster near the small lobe of a representative CNTNAP2 molecule by overlaying the 3D map and the contrast-inverted 3D map highlighting the protein and the nanogold particle, respectively (Fig. 6, I and J). Both nanogold labeling studies indicated that the small lobe contains the C-terminal CNTNAP2 L4 domain.
FIGURE 6.
Identification of the C-terminal end of CNTNAP2 by nanogold labeling. A, survey NS EM view of CNTNAP2 bound to 1.8-nm Ni-NTA nanogold. B, eight representative images of complexes of CNTNAP2 bound to 1.8-nm Ni-NTA nanogold. Raw particle images are shown in the first column (nanogold in black), contrast-inverted images in the second column (nanogold in white), and schematic representations in the third column (protein in cyan, gold particles in yellow). C, process to generate a representative 3D density map from an individual particle of CNTNAP2 labeled with 1.8-nm nanogold using IPET. D, final IPET 3D density map of a single CNTNAP2 particle labeled with 1.8-nm nanogold (top). To show the nanogold location with respect to the protein, we inverted the final 3D density map (shown in yellow) and overlaid it with the original 3D density map (bottom). E, survey NS-EM of CNTNAP2 bound to 5-nm Ni-NTA nanogold. F, 20 representative images of selected particles. G, the particles shown in the last column of F are overlaid with schematics of CNTNAP2 (cyan) and nanogold (yellow). H, schematic representations of CNTNAP2 bound to nanogold shown in G. I, process to generate a representative 3D density map from a targeted CNTNAP2 particle labeled with 5-nm nanogold using IPET. J, final IPET 3D density map of a single CNTNAP2 particle labeled with 5-nm nanogold (top). Bottom is shown the final 3D density map (cyan) overlaid with its inverted 3D density map (yellow) to visualize the nanogold position bound to CNTNAP2. Scale bar, 200 Å (A, E, and F), 100 Å (B, D, and J).
Second, we then used the monoclonal antibody K67/25 (raised against residues 1124–1265 of the CNTNAP2 L4 domain) to confirm the location of the L4 domain in CNTNAP2 particles. EM micrographs demonstrated a mixture (Fig. 7A) containing F-shaped CNTNAP2 particles, Y-shaped antibody particles, and CNTNAP2-antibody complexes (Fig. 7, B and C). Additionally, because of the flexible nature of the complex and its individual partners, we analyzed single CNTNAP2-antibody complexes using IPET (Fig. 7, D and E). The resolutions of the IPET 3D density maps were sufficient to define the Y-shaped antibody (i.e. three ring- or C-shaped lobes in a characteristic triangular constellation corresponding to the two Fab domains and one Fc domain) (35). We docked the antibody crystal structure (Protein Data Bank code 1IGT) into the Y-shaped 3D density portion, allowing the three domains to flex with respect to each other via their linkers (Fig. 7, D and E). The remaining portion of the 3D map revealed density consistent with the F shape seen for the 3D reconstruction of individual CNTNAP2 molecules, with the small lobe or base of the F shape contacted the antibody (Fig. 5D). Although the complex demonstrated conformational heterogeneity, it still clearly revealed that the antibody bound close to the small lobe of the F-shaped CNTNAP2, confirming that the C-terminal L4 domain carrying the antibody epitope coincides with the small lobe of CNTNAP2.
FIGURE 7.
Identification of the C-terminal end of CNTNAP2 by monoclonal antibody K67/25. A, survey NS-EM view of CNTNAP2 in complex with the monoclonal antibody K67/25. The ratio of particles observed was ∼74 ± 6% CNTNAP2 (rectangles), ∼13 ± 4% antibody (triangles), and ∼13 ± 3% complex (ovals). B, representative CNTNAP2 particles (top), antibody particles (middle), and CNTNAP2-antibody complexes (bottom). The last two columns show the representative reference-free class averages and corresponding schematic. C, series of CNTNAP2-antibody complexes indicating the range of conformational heterogeneity (top) and schematic representations (bottom). D, process to generate a representative 3D density map from a targeted CNTNAP2-antibody complex using IPET. Final IPET 3D density map of a single CNTNAP2-antibody complex shown in the top right panel. Final 3D density map overlaid with an IgG antibody (Protein Data Bank code 1IGT) is shown in the bottom panel. During docking, the linkers between the Fab and Fc domains were allowed to flex. E, process to generate another representative 3D density map from a targeted CNTNAP2-antibody complex using IPET as performed in D. Scale bar, 200 Å (A) and 100 Å (B, C, D, and E).
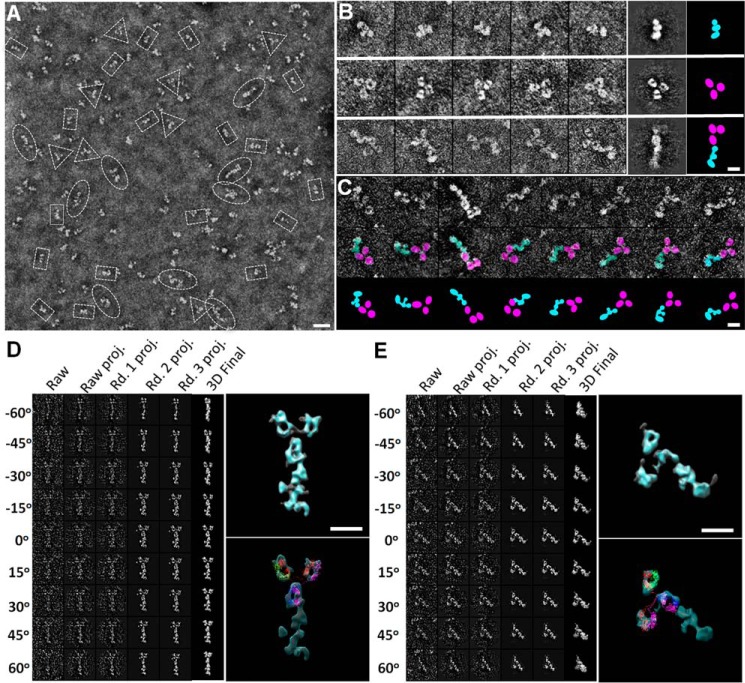
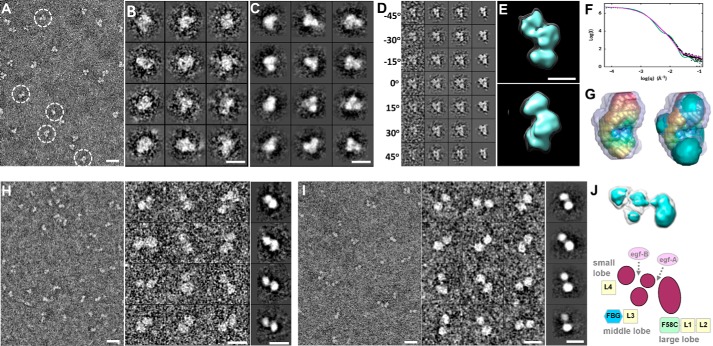
Third, we examined three fragments of CNTNAP2 (F58C-L1-L2, FBG-L3, and L3-egf-B-L4) using NS-EM, ET, and small angle x-ray scattering (SAXS). The N-terminal fragment CNTNAP2-C2 (F58C-L1-L2) was seen as a compact moiety with dimensions 101 × 67 Å (i.e. similar to those of the large lobe in NS-EM images) (Fig. 8, A–C). The CNTNAP2-C2 fragment resolved into three similarly sized domains in raw particles and class averages, consistent with it containing one F58C and two LNS domains; subsequent careful inspection of NS-EM images of the full-length CNTNAP2 ectodomain revealed that the large lobe could also be resolved into three individual globules in some particles. The size and shape of CNTNAP2-C2 were further confirmed with ET images by reconstructing a representative IPET 3D density map from an individual molecule (Fig. 8, D and E), which matched the size and shape determined by SAXS (Fig. 8, F and G). The fragment FBG-L3 (CNTNAP2-C3) was observed as two globular domains connected by a flexible linker (dimensions 96 × 43 Å) (Fig. 8H). The fragment L3-egf-B-L4 (CNTNAP2-C5) was also seen as two connected globular domains with dimensions 108 × 50 Å, although these were separated by a larger distance, consistent with the presence of an EGF-like repeat (Fig. 8I). Taken together, our results suggest that the large lobe in CNTNAP2 contains the domains F58C, L1, and L2; the medium lobe contains the FBG and L3 domains; and the small lobe contains the C-terminal L4 domain, leading to a putative assignment for the domain organization of CNTNAP2 (Fig. 8J). Our conformational variability analysis (Fig. 5G) and our domain assignment for CNTNAP2 (Fig. 8J) suggest that CNTNAP2 has molecular hinges that coincide with the EGF-like repeats, permitting the lobes to flex with respect to each other.
FIGURE 8.
Deconstruction of the CNTNAP2 extracellular domain using fragments. A, survey NS-EM view of the CNTNAP2-C2 fragment (F58C-L1-L2). B, selected raw images of CNTNAP2-C2 particles. C, selected reference-free 2D class averages of CNTNAP2-C2. D, process to generate a representative 3D density map from a targeted CNTNAP2-C2 particle using IPET. E, final IPET 3D reconstruction viewed from two perpendicular angles. F, log-log plot of the SAXS data for CNTNAP2-C2 (●), the fit from the averaged ab initio SAXS bead model (magenta line), and the calculated scattering from the CNTNAP2-C2 EM envelope contoured at 5.19σ (green line). G, left, averaged ab initio SAXS shape (rainbow-colored) calculated for 25 bead models and the range of all 25 bead models (gray). Right, superposition of the averaged SAXS shape (rainbow-colored) with the CNTNAP2-C2 EM envelope (cyan) contoured at 5.19σ. H, survey NS-EM view, selected particle images, and selected reference-free 2D class averages of CNTNAP2-C3 (FBG-L3). I, survey NS-EM view, selected particle images, and reference-free 2D class averages of CNTNAP2-C5 (L3-egf-B-L4). J, tentative domain assignment of the large, middle, and small lobes of CNTNAP2. Scale bar, 200 Å (A, H (left), and I (left)), 100 Å (B, C, H (middle and right), and I (middle and right)), and 50 Å (E).
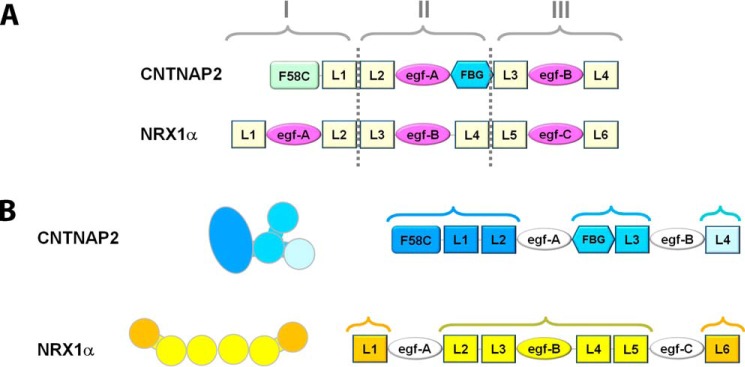
CNTNAP2 and neurexin 1α possess a similar domain composition consisting of LNS domains interspersed with EGF-like repeats (Fig. 9A), and it has widely been assumed that they share similar architectures. Crystal structures and EM studies have shown that the ectodomain of neurexin 1α forms a rod-shaped assembly made up of domains L2–L5 (36–38). Although the EGF-like repeats are not visible in the EM images for CNTNAP2 or neurexin 1α (Fig. 4) (38), the location of these small, ∼40-amino acid domains could be determined via the crystal structures (36, 37). The domains L1 and L6 are flexibly tethered on either side via egf-A and egf-C, yielding a molecule that spans ∼200 Å (36–38). Whereas CNTNAP2 and neurexin 1α contain EGF-like repeats adjacent to molecular hinges, neurexin 1α contains an additional EGF-like repeat (egf-B) that works as a lock, packing the central domains L3 and L4 side-by-side into a horseshoe-shaped reelin-like repeat forming the core of the rod-shaped assembly (36), a configuration not seen in CNTNAP2. Thus, the locations of the molecular hinges in the extracellular region of CNTNAP2 and neurexin 1α are different. The two proteins have a fundamentally different architecture (i.e. CNTNAP2 adopts an F-shaped molecule segregated into three major lobes, whereas neurexin 1α adopts a rod-shaped core with terminal domains flexibly tethered on either side (Figs. 4, 5, and 9B). Consequently, our data suggest that CNTNAP2 and α-neurexins may possess fundamentally different structure-function relationships and molecular mechanisms through which they recruit partners and carry out their function at neuronal contact sites (further detailed under “Discussion”).
FIGURE 9.
CNTNAP2 and neurexin 1α possess different three-dimensional architectures. A, CNTNAP2 and neurexin 1α ectodomains color-coded according to their domain identities reveal a similar composition. Based on amino acid sequence alone, neurexins can be divided into three repeats, I, II, and III; CNTNAP2 shares common aspects. B, composition of CNTNAP2 and neurexin 1α ectodomains color-coded to indicate their three-dimensional architectural organization (see also the “Results” section).
Discussion
We have investigated structure-function relationships of CNTNAP2, a neuronal cell adhesion molecule at axo-glial and synaptic contacts that is implicated in a variety of neurological disorders, including epilepsies and autism spectrum disorder. Our results indicate that 1) the extracellular domains of CNTNAP2 and CNTN2 bind each other tightly and specifically with low nanomolar affinity; 2) CNTNAP2 forms a relatively globular F-shaped molecule that is divided into three distinct lobes; 3) the lobes flex with respect to each other at hinge points near the EGF-like repeats; 4) the N-terminal large lobe is composed of F58C, L1, and L2, the middle lobe contains the FBG and L3 domains, and the C-terminal small lobe contains L4; and 5) the structural organization of CNTNAP2 is profoundly different from that of neurexin 1α, the prototype for the neurexin superfamily. Our results have implications for how CNTNAP2 stabilizes synaptic and axo-glial contacts, because the architecture and dimensions of CNTNAP2 not only determine how CNTNAP2 fits into the narrow extracellular clefts at contact sites, but also how it binds protein partners. Our results differ drastically from those of a recent study examining the structure of CNTNAP2 by EM, where the domains were assigned in the opposite order in the CNTNAP2 molecular envelope compared with our experimentally validated orientation (39). In addition, in that study, no interaction was detected between CNTNAP2 and CNTN2, although CNTNAP2 was found to bind CNTN1 (39), unlike the results presented here. Key differences in the experimental approach for the two biolayer interferometry studies are that we used as bait and ligand highly purified monomeric CNTNAP2 and CNTN2 carrying only a small hexahistidine affinity tag, which we produced in insect cells, whereas the other study used unpurified Fc fusion proteins immobilized on an Fc capture biosensor captured from conditioned medium of transfected cells, and the proteins were produced in glycosylation-deficient HEK293 GnTI cells.
CNTNAP2 at Synaptic and Axo-glial Contacts
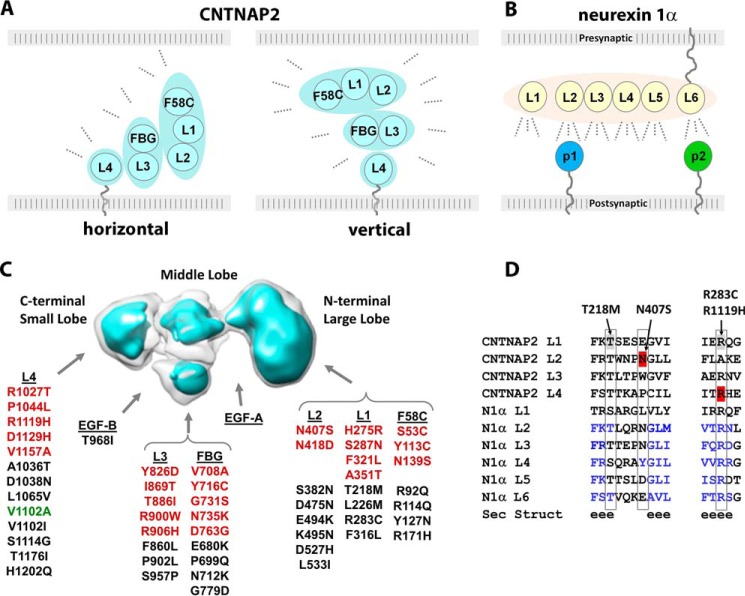
Adhesion molecules like CNTNAP2 shape protein networks at synaptic and axo-glial contacts by binding protein partners. Their ability to recruit partners is heavily influenced by how these molecules are positioned in the extracellular space between the cells at the contact site (i.e. how their overall dimensions, domains, and molecular hinges fit in the cleft). In the CNS, synaptic clefts are estimated to span ∼200–240 Å at excitatory synapses (40–43) but only ∼120 Å at inhibitory synapses (44), although even narrower gaps were recently suggested for excitatory (∼160 Å) and inhibitory (100 Å) synapses, respectively (44). The axo-glial cleft at juxtaparanodes putatively spans ∼74–150 Å (i.e. an intermediate distance between the paranodal and internodal clefts for which more accurate measurements are known) (45, 46). Therefore, given the dimensions of the CNTNAP2 ectodomain (∼145 Å long × ∼90 Å wide × ∼50 Å thick with a ∼50-residue membrane tether), the long axis of the molecule probably fits horizontally in the narrow cleft of inhibitory synapses and juxtaparanodes, primary locations for CNTNAP2 (Fig. 10A). Likewise, CNTNAP2 is also easily accommodated in a horizontal orientation at excitatory synaptic contacts, although a vertical orientation (i.e. the long axis orthogonal to the membranes) cannot be ruled out in these wider clefts (Fig. 10A). Our results indicate that the lobes of CNTNAP2 flex with respect to each other; they may also change upon protein partner binding so that the molecule could fit in alternative ways in the cleft. If CNTNAP2 seeks out the periphery of the cleft where the two membranes widen from each other, then a vertical orientation would be feasible as well. Intriguingly, the synaptic organizer synCAM1 localizes to the periphery of synaptic contact sites, and its distribution further changes in response to synaptic activity (43). In the case of neurexin 1α, its ∼200-Å-long, rodlike shape most certainly restricts it to a horizontal orientation in the synaptic cleft, facilitating the recruitment of its postsynaptically tethered partners along its length (Fig. 10B). The orientation of the CNTNAP2 ectodomain in the cleft of synaptic and axo-glial contact sites therefore is important, because it can fundamentally impact how CNTNAP2 interacts with its protein partners. The architecture and dimensions of CNTNAP2 provided in this study therefore place limits on how CNTNAP2 recruits partners such as CNTN2 to stabilize axo-glial and synaptic contact sites. Of course, the conformation and oligomerization state of molecules such as CNTNAP2 and CNTN2 could become altered in the synaptic cleft (e.g. in response to synaptic activity).
FIGURE 10.
Structural architecture and functional relationships of CNTNAP2. A, possible orientations of the CNTNAP2 ectodomain in the cleft of synaptic and axo-glial contacts. B, a horizontal orientation of the presynaptic neurexin 1α ectodomain at synaptic clefts promotes binding of protein partners tethered to the postsynaptic membrane (p1 and p2). C, location of amino acid substitutions in CNTNAP2 found in patient (red) and control (black) groups or both (green) as described in the “Results” section. D, sequence alignment of LNS domains from CNTNAP2 and neurexin 1α. Secondary structure prediction is shown (e is β-strand). Mutations discussed are indicated.
Multiple CNTNAP2 molecules could easily fit in a synaptic cleft given that the surface areas of postsynaptic densities for synapses on dendritic spines typically span ∼0.04–0.15 μm2 in adult mice, corresponding to a ∼2,250–4,370-Å-wide circular patch (42, 47). Surface areas for inhibitory synaptic contact sites are much larger than excitatory postsynaptic densities (6,500–14,000 Å in length) (48). Accurate dimensions for juxtaparanodal regions have not been obtained yet. Efforts to estimate the number of CNTNAP2 molecules per contact site, however, are complicated because it is not known whether the distribution of CNTNAP2 throughout synaptic contacts or axo-glial contacts is uniform.
Interaction of CNTNAP2 with CNTN2
Presynaptic CNTNAP2 and postsynaptic CNTN2 were recently shown to engage each other directly in a macromolecular complex at synaptic contacts (19). However, at axo-glial contacts, it has been proposed that CNTNAP2 binds CNTN2 in a side-by-side complex tethered to the axonal membrane (i.e. in cis); this cis-complex reaches across the extracellular space to bind a second CNTN2 molecule on the opposing glial membrane (i.e. in trans), forming a tripartite complex that spans the axo-glial cleft (13, 16, 27). Whether CNTNAP2 alone is sufficient to form the trans-complex with a bridging CNTN2 molecule or whether a cis-complex of CNTNAP2-CNT2 is required to form a tripartite complex is controversial (28). Our data indicate that the CNTNAP2 and CNTN2 ectodomains are sufficient to bind each other directly with high affinity in the low nanomolar range, although in the context of the contact site cleft, their affinity may be different. It will be important to investigate the structure of the CNTNAP2-CNTN2 complex as well as experimentally determine whether CNTNAP2 uses similar mechanisms to bind other putative partners (16, 20, 22, 49).
CNTNAP2 and Disease
Many alterations in the CNTNAP2 gene have been found; these include SNPs, deletions, point mutations, and defects at splice donor/acceptor splice sites (2, 18, 21, 29–31, 33, 50–52). Homozygous deletion of CNTNAP2 results in epilepsy, intellectual disability, and ASD, but it is unclear to what extent heterozygous mutations of CNTNAP2 confer appreciable disease risk (33, 52). Two large scale sequencing studies identified ∼66 point mutations, of which 24 were found uniquely in ASD patients and not in control subjects (18, 33). Mapping the point mutations on the CNTNAP2 envelope shows that they distribute over the entire extracellular region, and neither the disease nor the control group mutations preferentially locate to a particular lobe of the ectodomain (Fig. 10C). In contrast, human pathogenic autoantibodies targeting CNTNAP2 appear to predominantly target the N-terminal region of CNTNAP2, in particular the F58C and L1 domains found in the large lobe (12, 19), suggesting that they might disrupt a particular function or have efficient access to only a limited portion of CNTNAP2 in the cleft of contact sites.
Closer examination of the CNTNAP2 point mutations identified in the disease and control groups reveals complex structure-function relationships. Some point mutations in the disease group appear to be mild (substituting similar residues) and would not be expected to disrupt the protein fold, whereas other mutations in the control group would be expected to be deleterious. Although no structures are known for CNTNAP2 LNS domains, they are structurally homologous to LNS domains in neurexin 1α, enabling structural predictions to be made (Fig. 10D). For example, the disease mutation N407S maps to the L2 domain in CNTNAP2 and is expected to have a mild effect on the protein structure; this residue is expected to be solvent-exposed, and the LNS domain fold tolerates many different residues (Glu, Asp, Asn, Trp, Tyr, Leu, and Pro) at this position (Fig. 10D). N407S is not aberrantly retained in the ER or aberrantly trafficked (53). It is possible that this mutation disrupts protein function (e.g. protein partner binding) or destabilizes interactions between the group F58C, L1, and L2 or alters mRNA stability. In contrast, the control group mutation T218M in the CNTNAP2 L1 domain appears more severe because it probably replaces the terminal residue of a β-strand, where normally exclusively a Ser or Thr is found (Fig. 10D); the side chain hydroxyl plays an important structural role in the LNS domain fold by forming hydrogen bonds with the backbone of an adjacent loop and β-strand. The much larger, hydrophobic Met would not be able to carry out this structural role and would be expected to destabilize the protein fold despite the benign clinical manifestation of the T218M mutation. Further underscoring the complexity of interpreting disease mutations, R283C and R1119H are found in a very structurally conserved region of the LNS domain fold, replacing a virtually conserved Arg residue in the CNTNAP2 L1 and L4 domains, respectively. In structural analogues, this Arg is completely buried inside the protein and forms key hydrogen bonds with residues from three β-stands. Curiously, whereas R1119H is found in the disease group, the potentially structurally more damaging R283C is found in the control group. Higher resolution structural information will disclose whether mutations in CNTNAP2 have the potential to negatively impact structure-function relationships, but the impact of each mutation will probably have to be assessed by evaluating an endophenotype rather than a clinical contribution. Together, the studies presented here form the basis to pursue the molecular mechanisms of CNTNAP2 and its partners, to further understand its role in the formation and stabilization of synaptic and axo-glial contacts.
Experimental Procedures
Protein Expression and Purification
The human contactin-associated protein-like 2 (CNTNAP2) ectodomain (S32QK … CGAS1217; accession number BC093780) or fragments followed by a C-terminal ASTSHHHHHH tag were produced using baculovirus-mediated overexpression in HighFive cells with Insect-XPRESS+L-Glutamine medium (Lonza). Briefly, medium containing the secreted proteins was concentrated after protease inhibitors were added, dialyzed overnight (25 mm sodium phosphate, pH 8.0, 250 mm NaCl), and purified with an Ni-NTA column (Invitrogen; 25 mm sodium phosphate, pH 8.0, 500 mm NaCl, eluted with an imidazole gradient). Subsequently, the protein was dialyzed into 50 mm MES, pH 7.0, 50 mm NaCl, 3% glycerol overnight; incubated with 5 mm CaCl2 for 0.5 h; applied to a MonoQ column (GE Healthcare) equilibrated with 50 mm MES, pH 7.0; and subsequently eluted with an NaCl gradient. Last, proteins were applied to a HiLoad Superdex-200 16/60 size exclusion column (GE Healthcare) equilibrated with 50 mm MES, pH 7.0, 100 mm NaCl. Human contactin 2 (CNTN2) ectodomain (L29ESQ … VRNG1004; accession number BC BC129986.1) followed by a C-terminal SASTSHHHHHH tag was produced using baculovirus-mediated overexpression as well. Human contactin 1 (CNTN1) ectodomain (V32SEE … KISGA995; accession number NM_001843.2) followed by a C-terminal GSASTSHHHHHH tag was produced using baculovirus-mediated overexpression also. Briefly, medium containing the secreted proteins was concentrated after protease inhibitors were added, dialyzed overnight (25 mm sodium phosphate, pH 8.0, 250 mm NaCl), and purified with an Ni-NTA column (Invitrogen; 25 mm sodium phosphate, pH 8.0, 500 mm NaCl, eluted with an imidazole gradient). Subsequently, the protein was dialyzed into 25 mm Tris, pH 8.0, 100 mm NaCl, applied to a Mono Q column (GE Healthcare) equilibrated with 25 mm Tris, pH 8.0, 50 mm NaCl, and subsequently eluted with an NaCl gradient. Last, proteins were applied to a HiLoad Superdex-200 16/60 size exclusion column (GE Healthcare; 10 mm HEPES, pH 8.0, 50 mm NaCl). Purified proteins were stored in in flash-frozen aliquots. For analytical size exclusion chromatography, proteins were loaded on a Superdex 200 PC 3.2/30 column in 25 mm HEPES, pH 8.0, 100 mm NaCl, 5 mm CaCl2 in a 50-μl sample volume and run at 0.08 ml/min (in triplicate). Protein standards from Sigma (200, 66, 29, and 12.4 kDa) and blue dextran (2,000 kDa) were used to calibrate the column, loaded in a 50-μl sample volume and run at 0.08 ml/min. In parallel, samples were analyzed by dynamic light scattering using a Malvern Zetasizer Nano at 1 mg/ml in 10 mm HEPES, pH 8.0, 50 mm NaCl, 20 mm EDTA at 25 °C following centrifugation for 10 min at 13,000 rpm (3-μl sample volume, in triplicate). Relevant theoretical molecular masses are as follows: CNTNAP2-C1, 133,658 Da; CNTNAP2-C2, 59,720 Da; CNTNAP2-C3, 43,420 Da; CNTNAP2-C4, 86,624 Da; CNTNAP-C5, 48,684 Da; CNTN2-C1, 108,116 Da; CNTN1-C1, 108,469 Da.
Negative Stain EM Specimen Preparation
All samples were prepared by optimized negative staining with dilution buffer 25 mm Tris, pH 8.0, 100 mm NaCl, 3 mm CaCl2. CNTNAP2 sample was diluted to 0.005 mg/ml. For labeling studies, CNTNAP2 and Ni-NTA nanogold (Nanoprobes) were mixed with a molar ratio of ∼1:5, incubated for 1 h at room temperature, and then diluted to 0.005 mg/ml with respect to CNTNAP2. The CNTNAP2 monoclonal antibody K67/25 (NeuroMab) was diluted to 0.005 mg/ml. CNTNAP2 and K67/25 were mixed with a molar ratio of 1:1, incubated for 1 h at room temperature, and then diluted to end concentrations of 0.005 mg/ml CNTNAP2 and 0.0068 mg/ml K67/25, respectively. When making grids, an aliquot (∼4 μl) of sample was placed on a 5-nm thin carbon-coated 200-mesh copper grid (CF200-Cu, EMS) that had been glow-discharged for 15 s. After ∼1 min of incubation, excess solution was blotted with filter paper, and the grid was stained for ∼10–60 s by submersion in two drops (∼35 μl) of 1% (w/v) uranyl formate (54, 55) on Parafilm before being dried with nitrogen at room temperature.
Electron Microscopy Data Acquisition and Image Preprocessing
The NS micrographs were acquired at room temperature on a Gatan UltraScan 4Kx4K CCD by a Zeiss Libra 120 transmission electron microscope (Carl Zeiss NTS) operating at 120 kV at ×80,000 to ×125,000 magnification under near Scherzer focus (0.1 μm) and a defocus of 0.6 μm. Each pixel of the micrographs corresponded to 1.48 Å for ×80,000 magnification and 0.94 Å for ×125,000 magnification. Micrographs were processed with the EMAN, SPIDER, and FREALIGN software packages (34, 56, 57). The defocus and astigmatism of each micrograph were examined by fitting the contrast transfer function (CTF) parameters with its power spectrum by ctffind3 in the FREALIGN software package (57). Micrographs with distinguishable drift effects were excluded, and the CTF was corrected with SPIDER software (56). Only isolated particles from the NS-EM images were initially selected and windowed using the boxer program in EMAN and then manually adjusted. A total of 1,392 micrographs from CNTNAP2 samples were acquired, in which a total of 53,774 particles were windowed and selected. A total of 105 micrographs from CNTNAP2 and CNTNAP2-antibody complex samples were acquired, in which a total of 945 particles were windowed and selected. Particles were aligned and classified by reference-free class averaging with refine2d.py in the EMAN software package.
Electron Tomography Data Acquisition and Image Preprocessing
Electron tomography data of CNTNAP2, CNTNAP2-nanogold, and CNTNAP2-antibody K67/25 complex were acquired under ×125,000 magnification (each pixel of the micrograph corresponds to 0.94 Å in the specimens) and ×80,000 magnification (each pixel of the micrograph corresponds to 1.48 Å in the specimens) with 50- and 600-nm defocus by a Gatan Ultrascan 4,096 × 4,096-pixel CCD equipped in a Zeiss Libra 120 Plus transmission electron microscope operated under 120 kV. The specimens mounted on a Gatan room temperature high-tilt holder were tilted at angles ranging from −66 to 66° in steps of 1.5°. The total electron dose was ∼200 e−/Å2. The tilt series of tomographic data were controlled and imaged by manual operation and Gatan tomography software. Tilt series were initially aligned together with the IMOD software package (58). The CTF of each tilt micrograph was determined by ctffind3 in the FREALIGN software package and then corrected by CTF correction software, TOMOCTF (59). The tilt series of each particle image were semiautomatically tracked and selected using IPET software (35).
IPET 3D Reconstruction
Ab initio 3D reconstructions were conducted using the IPET reconstruction method described previously (35). In brief, the small image containing only a single targeted particle was selected and windowed from a series of tilted whole micrographs after CTF correction. An initial model was obtained by directly back-projecting these small images into a 3D map according to their tilt angles. The 3D reconstruction refinements were performed with three rounds of refinement using an electron tomography reconstruction algorithm. Each round contained several iterations. In the first round, circular Gaussian edge soft masks were used. In the second round, particle-shaped soft masks were used. In the third round, the last particle-shaped soft mask of the second round was used in association with an additional interpolation method during the determination of the translational parameters. In this last round, translational searching was carried out to sub-pixel accuracy by interpolating the images five times in each dimension using the triangular interpolation technique.
IPET Fourier Shell Correlation (FSC) Analysis
The resolution of the IPET 3D reconstructions was analyzed using the FSC criterion, in which center-refined raw ET images were split into two groups based on having an odd- or even-numbered index in the order of tilt angles. Each group was used independently to generate its 3D reconstruction by IPET; these two IPET 3D reconstructions were then used to compute the FSC curve over their corresponding spatial frequency shells in Fourier space (using the RF 3 command in SPIDER) (56). The frequency at which the FSC curve falls to a value of 0.5 was used to assess the resolution of the final IPET 3D density map.
Single Particle 3D Reconstruction
Eight IPET ab initio 3D density maps of CNTNAP2 generated through IPET reconstruction were low pass-filtered to 40 Å and then used as initial models for single-particle multireference refinement by using multirefine in EMAN (34). The final single-particle 3D maps have resolutions from 13.7 to 17.0 Å based on the 0.5 Fourier shell correlation criterion (34). The maps were then low pass-filtered to 16 Å for structural manipulation. To visualize molecular envelopes, the “hide dust” function was applied in Chimera (60).
Antibody Docking and Interpretation of IPET 3D Density Maps for Individual Antibody-CNTNAP2 Complexes
The resolution of the IPET 3D reconstructions (∼1–3 nm) of the antibody-CNTNAP2 complex was sufficient to first locate the antibody in the density map. We docked the crystal structure of an antibody (Protein Data Bank code 1IGT (61)). Using our previously published procedure (62), Fab and Fc domains were rigid body-docked into a density map envelope by Chimera (60), allowing the three domains to flex with respect to each other via their linkers. The remaining unoccupied density corresponded to the CNTNAP2 molecule in the complex.
CNTNAP2 Angle Statistical Analysis
The angle between the small and medium lobe (α) was defined by the angle between line P1P2 and line P2P3, in which P1, P2, and P3 are characteristic points on the small and medium lobes. The angle between the medium and large lobe (β) was defined by the angle between line P3P4 and P5P6, in which P3, P4, P5, and P6 are characteristic points on medium and large lobes. Although the lobes in 3D maps would be related by a dihedral angle, in 2D class averages, we observe a projection of the dihedral angle. A total of 3,450 CNTNAP2 uniformly oriented particles were selected for measuring angles α and β. Particles with significantly different orientations on the grid were excluded to eliminate the influence of particle orientation on angle distributions. The results of angle α and β distributions are shown in Fig. 5G.
Molecular Ratio Calculation
The particle ratios of CNTNAP2, antibody K67/25, and their complex were obtained by calculating the average and S.D. of particle ratios in 10 micrographs of ×80,000 magnification. A total of 1,136 particles were counted.
Solid Phase Binding Assays
For solid phase binding assays with immobilized CNTN2, 200 ng of CNTN2 in Binding Buffer/Ca2+ (20 mm Tris, pH 8.0, 100 mm NaCl, 5 mm CaCl2) was coated in 96-well plates at room temperature (for 2 h), blocked with Blocking Buffer (1% gelatin, 20 mm Tris, pH 8.0, 100 mm NaCl, 5 mm CaCl2) for 2 h, and then incubated for 1 h with increasing concentrations of biotinylated CNTNAP2 C1* (0–20 nm) in Binding Buffer/Ca2+ (in triplicate). To assess nonspecific background binding, wells without CNTN2 were also incubated with biotinylated CNTNAP2 C1* (0–20 nm) in Binding Buffer/Ca2+ (in duplicate). Wells were then emptied and washed three times. To develop the signal, all wells were incubated with the anti-Streptavidin HRP conjugate (1:5,000) for 45 min followed by the addition of the substrate o-phenylenediamine for 10 min. The reaction was then stopped by adding 50 μl/well of 2 m H2SO4, and the plate was read at 490 nm. The KD value was calculated by fitting the data after subtraction of the background to a one-site total binding equation using the non-linear regression model in GraphPad Prism. Error bars show S.E.
Surface Plasmon Resonance
Binding of CNTNAP2 to CNTN2 was assessed in Running Buffer (25 mm HEPES, pH 8.0, 150 mm NaCl, 5 mm CaCl2, and 0.05% Tween 20) at 25 °C with a Biacore T100 system. CNTN2 (217 RU) and CNTNAP2 (1,009 RU) were immobilized separately on C1 sensor chips (matrix-free carboxymethylated sensors optimized for large molecules; GE Healthcare). Specific binding data were obtained by injecting a series of CNTNAP2 concentrations over a ligand-coupled sensor and subtracting from the signal that was obtained from a series flowing CNTNAP2 simultaneously over a sensor with no ligand immobilized. The following CNTNAP2 concentrations were used: CNTNAP2 (0, 0.125, 0.25, 0.5, 1.0, 2.0, 4.0, 6.0, 8.0, and 10 nm) and CNTN2 (0, 1.565, 3.125, 6.25, 12.5, 25, 50, 100, and 200 nm) flowed at 30 μl/min for 200 s (association step) followed by Running Buffer for 200 s (dissociation step).The sensor was regenerated after each protein injection with 3 mm NaOH. The data were processed using a kinetic analysis, and the KD was calculated from sensorgram data fit to a ∼1:1 stoichiometric model. The curves were fit using a local fitting method (Rmax local fitting). Global fitting was also possible but resulted in slightly worse fits; however, the calculated KD values did not differ significantly. The KD values for CNTNAP2/CNTN2 binding for two independent experiments were averaged (the average and error (S.D.) are given). For CNTNAP2/CNTN2 binding, the S.E. values for kd and ka calculated by the Biacore T100 software were used to calculate the error on the KD. To assess the interaction of CNTNAP2 with CNTN1 versus CNTN2, CNTNAP2 was immobilized on a C1 sensor chip (1664 RU; GE Healthcare); specific binding data were obtained by injecting a series of CNTN1 or CNTN2 concentrations (0, 1.56, 3.12, 6.25, 12.5, 25, 50, 100, and 200 nm) over the same CNTNAP2 biosensor at a flow rate of 30 μl/min for 100 s (association step), followed by Running Buffer for 100 s (dissociation step) as described above. The experiment was repeated twice, yielding similar results. The reverse experiment immobilizing contactins and flowing CNTNAP2 was performed as well. It is unknown whether immobilizing CNTN2 and/or CNTNAP2 on the biosensor induces conformational changes or changes in the oligomeric state.
SAXS
All SAXS data were collected using a Rigaku BioSAXS-1000 camera on a FR-E++ x-ray source. The CNTNAP2 C2 samples were measured at concentrations of 2.5, 1.25, and 0.62 mg/ml. For each concentration 70 μl of buffer and sample were manually pipetted into separate tubes of an eight-tube PCR strip capillary cell and sealed. These were loaded into an aligned quartz flow cell under vacuum in the BioSAXS camera using an automatic sample changer. Series of 1-h exposures were collected and averaged in SAXLab to produce separate sample and buffer curves ranging from 12 to 16 h of total exposure. Buffer subtraction, absorption correction, and molecular weight calibration were performed using the SAXNS-ES server. Data analysis, including zero concentration extrapolation, was performed with the Primus program, and the P(r) was calculated using GNOM, both from the ATSAS suite (63). The ab initio molecular shape was generated from an average of 25 DAMMIF (64) runs, using the saxns_dammif utility. Calculation of the fit to the SAXS data was performed using CRYSOL. The EM2DAM utility was used to find the optimum EM map contour level for fitting to the SAXS data.
Author Contributions
G. Rudenko and G. Ren designed the study and wrote the paper. Z. L., Jianfang Liu, Jiankang Liu, L. Z., and G. Ren designed, performed, and analyzed the experiments involving EM studies. F. C. and X. Z. designed and constructed vectors for the expression of proteins. S. R., A. K., F. C., Y. W., S. S., and X. Z. purified proteins. S. R. and L. H. performed and interpreted SPR experiments. M. W. performed and interpreted the SAXS experiments. S. R., A. K., and S. S. performed biochemical and biophysical characterization of CNTNAP2, CNTN2, and CNTN1. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We gratefully thank Elior Peles for the gift of the CNTNAP2 cDNA.
This work was supported by NIMH, National Institutes of Health, Grant R01MH077303 with additional support provided by the Sealy Center for Structural Biology and Molecular Biophysics (University of Texas Medical Branch) and the Brain and Behavior Research Foundation (to G. Rudenko). Work at the Molecular Foundry (G. Ren) was supported by the United States Department of Energy under Contract DE-AC02-05CH11231. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
3D maps have been deposited in the EM Data Bank with the following accession codes: CNTNAP2 IPET 3D reconstructions: EMD-9556 to -9563; Single particle 3D reconstructions: EMD-9545, -9546, -9550–9555; 1.8 nm nanogold labelled CNTNAP2 IPET 3D reconstruction: EMD-9547; 5 nm nanogold labelled CNTNAP2 IPET 3D reconstruction: EMD-9548; Antibody-CNTNAP2 complex IPET 3D reconstructions: EMD-9543, -9544; and CNTNAP2-C2 IPET 3D reconstruction: EMD-9549.
Z. Lu, M. V. V. V. S. Reddy, Jianfang Liu, A. Kalichava, Jiankang Liu, L. Zhang, S. Seshadrinathan, X. Zhong, G. Ren, and G. Rudenko, unpublished data.
- ASD
- autism spectrum disorder
- LNS
- laminin, neurexin, sex hormone binding globulin
- SPR
- surface plasmon resonance
- NS
- negative staining
- ET
- electron tomography
- IPET
- individual particle electron tomography
- Ni-NTA
- nickel-nitrilotriacetic acid
- SAXS
- small angle x-ray scattering
- CTF
- contrast transfer function
- FSC
- Fourier shell correlation
- RU
- resonance units.
References
- 1. Scott-Van Zeeland A. A., Abrahams B. S., Alvarez-Retuerto A. I., Sonnenblick L. I., Rudie J. D., Ghahremani D., Mumford J. A., Poldrack R. A., Dapretto M., Geschwind D. H., and Bookheimer S. Y. (2010) Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci. Transl. Med. 2, 56ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rodenas-Cuadrado P., Ho J., and Vernes S. C. (2014) Shining a light on CNTNAP2: complex functions to complex disorders. Eur. J. Hum. Genet. 22, 171–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lancaster E., and Dalmau J. (2012) Neuronal autoantigens-pathogenesis, associated disorders and antibody testing. Nat. Rev. Neurol. 8, 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zweier C. (2012) Severe intellectual disability associated with recessive defects in CNTNAP2 and NRXN1. Mol. Syndromol. 2, 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchner D. A., Geisinger J. M., Glazebrook P. A., Morgan M. G., Spiezio S. H., Kaiyala K. J., Schwartz M. W., Sakurai T., Furley A. J., Kunze D. L., Croniger C. M., and Nadeau J. H. (2012) The juxtaparanodal proteins CNTNAP2 and TAG1 regulate diet-induced obesity. Mamm. Genome 23, 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Irani S. R., Alexander S., Waters P., Kleopa K. A., Pettingill P., Zuliani L., Peles E., Buckley C., Lang B., and Vincent A. (2010) Antibodies to Kv1 potassium channel-complex proteins leucine-rich, glioma inactivated 1 protein and contactin-associated protein-2 in limbic encephalitis, Morvan's syndrome and acquired neuromyotonia. Brain 133, 2734–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lancaster E., Huijbers M. G., Bar V., Boronat A., Wong A., Martinez-Hernandez E., Wilson C., Jacobs D., Lai M., Walker R. W., Graus F., Bataller L., Illa I., Markx S., Strauss K. A., Peles E., Scherer S. S., and Dalmau J. (2011) Investigations of caspr2, an autoantigen of encephalitis and neuromyotonia. Ann. Neurol. 69, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Melzer N., Golombeck K. S., Gross C. C., Meuth S. G., and Wiendl H. (2012) Cytotoxic CD8+ T cells and CD138+ plasma cells prevail in cerebrospinal fluid in non-paraneoplastic cerebellar ataxia with contactin-associated protein-2 antibodies. J. Neuroinflammation 9, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Becker E. B., Zuliani L., Pettingill R., Lang B., Waters P., Dulneva A., Sobott F., Wardle M., Graus F., Bataller L., Robertson N. P., and Vincent A. (2012) Contactin-associated protein-2 antibodies in non-paraneoplastic cerebellar ataxia. J. Neurol. Neurosurg. Psychiatry 83, 437–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Irani S. R., and Vincent A. (2012) The expanding spectrum of clinically-distinctive, immunotherapy-responsive autoimmune encephalopathies. Arq. Neuropsiquiatr. 70, 300–304 [DOI] [PubMed] [Google Scholar]
- 11. Irani S. R., Pettingill P., Kleopa K. A., Schiza N., Waters P., Mazia C., Zuliani L., Watanabe O., Lang B., Buckley C., and Vincent A. (2012) Morvan syndrome: clinical and serological observations in 29 cases. Ann. Neurol. 72, 241–255 [DOI] [PubMed] [Google Scholar]
- 12. Olsen A. L., Lai Y., Dalmau J., Scherer S. S., and Lancaster E. (2015) Caspr2 autoantibodies target multiple epitopes. Neurol. Neuroimmunol. Neuroinflamm. 2, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Faivre-Sarrailh C., and Devaux J. J. (2013) Neuro-glial interactions at the nodes of Ranvier: implication in health and diseases. Front. Cell Neurosci. 7, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poliak S., Gollan L., Martinez R., Custer A., Einheber S., Salzer J. L., Trimmer J. S., Shrager P., and Peles E. (1999) Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron 24, 1037–1047 [DOI] [PubMed] [Google Scholar]
- 15. Arroyo E. J., Xu T., Poliak S., Watson M., Peles E., and Scherer S. S. (2001) Internodal specializations of myelinated axons in the central nervous system. Cell Tissue Res. 305, 53–66 [DOI] [PubMed] [Google Scholar]
- 16. Poliak S., Salomon D., Elhanany H., Sabanay H., Kiernan B., Pevny L., Stewart C. L., Xu X., Chiu S. Y., Shrager P., Furley A. J., and Peles E. (2003) Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J. Cell Biol. 162, 1149–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poliak S., and Peles E. (2003) The local differentiation of myelinated axons at nodes of Ranvier. Nat. Rev. Neurosci. 4, 968–980 [DOI] [PubMed] [Google Scholar]
- 18. Bakkaloglu B., O'Roak B. J., Louvi A., Gupta A. R., Abelson J. F., Morgan T. M., Chawarska K., Klin A., Ercan-Sencicek A. G., Stillman A. A., Tanriover G., Abrahams B. S., Duvall J. A., Robbins E. M., Geschwind D. H., et al. (2008) Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 82, 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pinatel D., Hivert B., Boucraut J., Saint-Martin M., Rogemond V., Zoupi L., Karagogeos D., Honnorat J., and Faivre-Sarrailh C. (2015) Inhibitory axons are targeted in hippocampal cell culture by anti-Caspr2 autoantibodies associated with limbic encephalitis. Front. Cell Neurosci. 9, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varea O., Martin-de-Saavedra M. D., Kopeikina K. J., Schürmann B., Fleming H. J., Fawcett-Patel J. M., Bach A., Jang S., Peles E., Kim E., and Penzes P. (2015) Synaptic abnormalities and cytoplasmic glutamate receptor aggregates in contactin associated protein-like 2/Caspr2 knockout neurons. Proc. Natl. Acad. Sci. U.S.A. 112, 6176–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peñagarikano O., Abrahams B. S., Herman E. I., Winden K. D., Gdalyahu A., Dong H., Sonnenblick L. I., Gruver R., Almajano J., Bragin A., Golshani P., Trachtenberg J. T., Peles E., and Geschwind D. H. (2011) Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 147, 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anderson G. R., Galfin T., Xu W., Aoto J., Malenka R. C., and Südhof T. C. (2012) Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proc. Natl. Acad. Sci. U.S.A. 109, 18120–18125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gdalyahu A., Lazaro M., Penagarikano O., Golshani P., Trachtenberg J. T., and Gescwind D. H. (2015) The autism related protein contactin-associated protein-like 2 (CNTNAP2) stabilizes new spines: an in vivo mouse study. PLoS One 10, e0125633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strauss K. A., Puffenberger E. G., Huentelman M. J., Gottlieb S., Dobrin S. E., Parod J. M., Stephan D. A., and Morton D. H. (2006) Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N. Engl. J. Med. 354, 1370–1377 [DOI] [PubMed] [Google Scholar]
- 25. Bellen H. J., Lu Y., Beckstead R., and Bhat M. A. (1998) Neurexin IV, caspr and paranodin: novel members of the neurexin family: encounters of axons and glia. Trends Neurosci. 21, 444–449 [DOI] [PubMed] [Google Scholar]
- 26. Banerjee S., Paik R., Mino R. E., Blauth K., Fisher E. S., Madden V. J., Fanning A. S., and Bhat M. A. (2011) A laminin G-EGF-laminin G module in neurexin IV is essential for the apico-lateral localization of contactin and organization of septate junctions. PLoS One 6, e25926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Traka M., Goutebroze L., Denisenko N., Bessa M., Nifli A., Havaki S., Iwakura Y., Fukamauchi F., Watanabe K., Soliven B., Girault J. A., and Karagogeos D. (2003) Association of TAG-1 with Caspr2 is essential for the molecular organization of juxtaparanodal regions of myelinated fibers. J. Cell Biol. 162, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Savvaki M., Theodorakis K., Zoupi L., Stamatakis A., Tivodar S., Kyriacou K., Stylianopoulou F., and Karagogeos D. (2010) The expression of TAG-1 in glial cells is sufficient for the formation of the juxtaparanodal complex and the phenotypic rescue of tag-1 homozygous mutants in the CNS. J. Neurosci. 30, 13943–13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zweier C., de Jong E. K., Zweier M., Orrico A., Ousager L. B., Collins A. L., Bijlsma E. K., Oortveld M. A., Ekici A. B., Reis A., Schenck A., and Rauch A. (2009) CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am. J. Hum. Genet. 85, 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gregor A., Albrecht B., Bader I., Bijlsma E. K., Ekici A. B., Engels H., Hackmann K., Horn D., Hoyer J., Klapecki J., Kohlhase J., Maystadt I., Nagl S., Prott E., Tinschert S., et al. (2011) Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1. BMC Med. Genet. 12, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mefford H. C., Muhle H., Ostertag P., von Spiczak S., Buysse K., Baker C., Franke A., Malafosse A., Genton P., Thomas P., Gurnett C. A., Schreiber S., Bassuk A. G., Guipponi M., Stephani U., et al. (2010) Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 6, e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Al-Murrani A., Ashton F., Aftimos S., George A. M., and Love D. R. (2012) Amino-terminal microdeletion within the CNTNAP2 gene associated with variable expressivity of speech delay. Case Rep. Genet. 2012, 172408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murdoch J. D., Gupta A. R., Sanders S. J., Walker M. F., Keaney J., Fernandez T. V., Murtha M. T., Anyanwu S., Ober G. T., Raubeson M. J., DiLullo N. M., Villa N., Waqar Z., Sullivan C., Gonzalez L., et al. (2015) No evidence for association of autism with rare heterozygous point mutations in contactin-associated protein-like 2 (CNTNAP2), or in other contactin-associated proteins or contactins. PLoS Genet. 11, e1004852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ludtke S. J., Baldwin P. R., and Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 35. Zhang L., and Ren G. (2012) IPET and FETR: experimental approach for studying molecular structure dynamics by cryo-electron tomography of a single-molecule structure. PLoS One 7, e30249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen F., Venugopal V., Murray B., and Rudenko G. (2011) The structure of neurexin 1α reveals features promoting a role as synaptic organizer. Structure 19, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller M. T., Mileni M., Comoletti D., Stevens R. C., Harel M., and Taylor P. (2011) The crystal structure of the α-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function. Structure 19, 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Comoletti D., Miller M. T., Jeffries C. M., Wilson J., Demeler B., Taylor P., Trewhella J., and Nakagawa T. (2010) The macromolecular architecture of extracellular domain of αNRXN1: domain organization, flexibility, and insights into trans-synaptic disposition. Structure 18, 1044–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rubio-Marrero E. N., Vincelli G., Jeffries C. M., Shaikh T. R., Pakos I. S., Ranaivoson F. M., von Daake S., Demeler B., De Jaco A., Perkins G., Ellisman M. H., Trewhella J., and Comoletti D. (2016) Structural characterization of the extracellular domain of CASPR2 and insights into its association with the novel ligand contactin1. J. Biol. Chem. 291, 5788–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lucić V., Yang T., Schweikert G., Förster F., and Baumeister W. (2005) Morphological characterization of molecular complexes present in the synaptic cleft. Structure 13, 423–434 [DOI] [PubMed] [Google Scholar]
- 41. Zuber B., Nikonenko I., Klauser P., Muller D., and Dubochet J. (2005) The mammalian central nervous synaptic cleft contains a high density of periodically organized complexes. Proc. Natl. Acad. Sci. U.S.A. 102, 19192–19197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harris K. M., and Weinberg R. J. (2012) Ultrastructure of synapses in the mammalian brain. Cold Spring Harb. Perspect. Biol. 4, a005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perez de Arce K., Schrod N., Metzbower S. W., Allgeyer E., Kong G. K., Tang A. H., Krupp A. J., Stein V., Liu X., Bewersdorf J., Blanpied T. A., Lucić V., and Biederer T. (2015) Topographic mapping of the synaptic cleft into adhesive nanodomains. Neuron 88, 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. High B., Cole A. A., Chen X., and Reese T. S. (2015) Electron microscopic tomography reveals discrete transcleft elements at excitatory and inhibitory synapses. Front. Synaptic Neurosci. 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salzer J. L., Brophy P. J., and Peles E. (2008) Molecular domains of myelinated axons in the peripheral nervous system. Glia 56, 1532–1540 [DOI] [PubMed] [Google Scholar]
- 46. Nans A., Einheber S., Salzer J. L., and Stokes D. L. (2011) Electron tomography of paranodal septate-like junctions and the associated axonal and glial cytoskeletons in the central nervous system. J. Neurosci. Res. 89, 310–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dani A., Huang B., Bergan J., Dulac C., and Zhuang X. (2010) Superresolution imaging of chemical synapses in the brain. Neuron 68, 843–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Linsalata A. E., Chen X., Winters C. A., and Reese T. S. (2014) Electron tomography on γ-aminobutyric acid-ergic synapses reveals a discontinuous postsynaptic network of filaments. J. Comp. Neurol. 522, 921–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen N., Koopmans F., Gordon A., Paliukhovich I., Klaassen R. V., van der Schors R. C., Peles E., Verhage M., Smit A. B., and Li K. W. (2015) Interaction proteomics of canonical Caspr2 (CNTNAP2) reveals the presence of two Caspr2 isoforms with overlapping interactomes. Biochim. Biophys. Acta 1854, 827–833 [DOI] [PubMed] [Google Scholar]
- 50. Alarcón M., Abrahams B. S., Stone J. L., Duvall J. A., Perederiy J. V., Bomar J. M., Sebat J., Wigler M., Martin C. L., Ledbetter D. H., Nelson S. F., Cantor R. M., and Geschwind D. H. (2008) Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 82, 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. O'Roak B. J., Deriziotis P., Lee C., Vives L., Schwartz J. J., Girirajan S., Karakoc E., Mackenzie A. P., Ng S. B., Baker C., Rieder M. J., Nickerson D. A., Bernier R., Fisher S. E., Shendure J., and Eichler E. E. (2011) Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 43, 585–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sampath S., Bhat S., Gupta S., O'Connor A., West A. B., Arking D. E., and Chakravarti A. (2013) Defining the contribution of CNTNAP2 to autism susceptibility. PLoS One 8, e77906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Falivelli G., De Jaco A., Favaloro F. L., Kim H., Wilson J., Dubi N., Ellisman M. H., Abrahams B. S., Taylor P., and Comoletti D. (2012) Inherited genetic variants in autism-related CNTNAP2 show perturbed trafficking and ATF6 activation. Hum. Mol. Genet. 21, 4761–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang L., Song J., Newhouse Y., Zhang S., Weisgraber K. H., and Ren G. (2010) An optimized negative-staining protocol of electron microscopy for apoE4.POPC lipoprotein. J. Lipid Res. 51, 1228–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang L., Song J., Cavigiolio G., Ishida B. Y., Zhang S., Kane J. P., Weisgraber K. H., Oda M. N., Rye K. A., Pownall H. J., and Ren G. (2011) Morphology and structure of lipoproteins revealed by an optimized negative-staining protocol of electron microscopy. J. Lipid Res. 52, 175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., and Leith A. (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 57. Grigorieff N. (2007) FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 157, 117–125 [DOI] [PubMed] [Google Scholar]
- 58. Kremer J. R., Mastronarde D. N., and McIntosh J. R. (1996) Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 [DOI] [PubMed] [Google Scholar]
- 59. Fernández J. J., Li S., and Crowther R. A. (2006) CTF determination and correction in electron cryotomography. Ultramicroscopy 106, 587–596 [DOI] [PubMed] [Google Scholar]
- 60. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 61. Harris L. J., Larson S. B., Hasel K. W., and McPherson A. (1997) Refined structure of an intact Ig G2a monoclonal antibody. Biochemistry 36, 1581–1597 [DOI] [PubMed] [Google Scholar]
- 62. Zhang X., Zhang L., Tong H., Peng B., Rames M. J., Zhang S., and Ren G. (2015) 3D structural fluctuation of IgG1 antibody revealed by individual particle electron tomography. Sci. Rep. 5, 9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D. T., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]