Abstract
Protein phosphatases play vital roles in phosphorylation-mediated cellular signaling. Although there are 11 serine/threonine protein kinases in Mycobacterium tuberculosis, only one serine/threonine phosphatase, PstP, has been identified. Although PstP has been biochemically characterized and multiple in vitro substrates have been identified, its physiological role has not yet been elucidated. In this study, we have investigated the impact of PstP on cell growth and survival of the pathogen in the host. Overexpression of PstP led to elongated cells and partially compromised survival. We find that depletion of PstP is detrimental to cell survival, eventually leading to cell death. PstP depletion results in elongated multiseptate cells, suggesting a role for PstP in regulating cell division events. Complementation experiments performed with PstP deletion mutants revealed marginally compromised survival, suggesting that all of the domains, including the extracellular domain, are necessary for complete rescue. On the other hand, the catalytic activity of PstP is absolutely essential for the in vitro growth. Mice infection experiments establish a definitive role for PstP in pathogen survival within the host. Depletion of PstP from established infections causes pathogen clearance, indicating that the continued presence of PstP is necessary for pathogen survival. Taken together, our data suggest an important role for PstP in establishing and maintaining infection, possibly via the modulation of cell division events.
Keywords: cell division, cell signaling, mycobacteria, Mycobacterium tuberculosis, phosphatase, phosphorylation, protein serine/threonine phosphatase (PSP), PstP, cell division
Introduction
Signal sensing and transduction lead to a wide range of cellular responses and thus must be tightly regulated in pathogenic bacteria to allow optimal survival under variable conditions. A common mode of regulation of the cell's response to external cues is via phosphorylation and dephosphorylation of specific target proteins that lead to cellular responses like altered subcellular localization, protein turnover rates, and protein-protein interactions. Whereas phosphorylation events are mediated by kinases, dephosphorylations are mediated by the action of phosphatases. Bacteria are known to possess the conventional two-component systems involving phosphorylation of His residues in sensor kinases and Asp residues in the corresponding response regulators. These systems are critical for multiple physiological processes and cell survival (1, 2). In most of these systems, kinase-phosphatase activities are possessed by one bifunctional enzyme; for example, in the mycobacterial DevS/DosS system and WalRK system of Bacillus anthracis, DevS and WalK were shown to exhibit both kinase and phosphatase activities (3, 4). There are other known bifunctional kinase/phosphatases that do not belong to two-component systems, such as AceK of Escherichia coli (5) and HPr of Bacillus subtilis (6). Mycobacteria possess 11 two-components systems; besides this, pathogenic bacteria also possess Ser/Thr phosphorylation systems in which a protein that has been phosphorylated by a serine/threonine protein kinase (STPK)6 would be dephosphorylated by a serine/threonine phosphatase. Analysis of the Mycobacterium tuberculosis whole genome sequence identified 11 eukaryotic-like STPKs (PknA–L, except for PknC), one Ser/Thr phosphatase (PstP), one tyrosine kinase (PtkA), and two tyrosine phosphatases (PtpA and PtpB) (7, 8). All of these eukaryotic-like kinases and phosphatases have now been characterized and found to be catalytically active (9–19).
Protein phosphatases belong to distinct families, and members of a family are structurally and functionally conserved enzymes. Phosphatases are classified based on the catalytic domain signature sequence they carry and their substrate preference (the metal-dependent protein phosphatase (PPM) family, the phosphoprotein phosphatase (PPP) family, and the phosphotyrosine phosphatase (PTP) family) (20, 21), and there are examples known from all of these families in bacteria (20, 22, 23). PPP phosphatases are predominantly found in eukaryotes and target Ser/Thr residues in their substrates. The PP2C class of Ser/Thr phosphatases belong to PPM family and require Mg2+/Mn2+ for their activity. In most bacteria, these are usually secreted out of the cell; PTPs are the Tyr phosphatases and are subclassified into class I standard classical PTPs and class II low molecular weight (LMW-PTP), which in many bacteria are secreted into the extracellular milieu (20, 22, 23). M. tuberculosis encodes two tyrosine phosphatases, PtpA and PtpB. Deletion of ptpA, an LMW-PTP, did not impact M. tuberculosis growth either in vitro or in the mouse infection model (24). However, results from competitive co-infection of THP-1 macrophages with H37Rv and MtbΔptpA strains revealed that PtpA is important for long term infection (25). PtpA is secreted from the bacterial cell and dephosphorylates the host's VPS33B (vacuolar protein sorting 33B) protein, a subunit of the VPS-C complex, thus causing the inhibition of phago-lysosomal fusion (25). The interaction of PtpA with the host vacuolar H+ ATPase is necessary for the dephosphorylation of VPS33B and subsequent phagosomal exclusion of V-ATPase (26). PtpB is also secreted into the extracellular milieu, and the disruption of ptpB impairs the pathogen's ability to survive in activated macrophages and guinea pigs (27). Clearance of M. tuberculosis from the lungs occurs via endocytic pathways that require the fusion of phagosome with lysosome, mediated by the VPS-C complex (28). Although no direct substrates of PtpB have been identified to date, the expression of PtpB in activated macrophages attenuated IFN-γ and IL-6 production, possibly through ERK2 and p38 dephosphorylation (29).
The lone serine/threonine phosphatase of M. tuberculosis, PstP, belongs to the PP2C class of PPM family phosphatases and strictly requires the Mn2+ ion for its activity (30). The enzyme localizes to the cell membrane and contains a 240-amino acid intracellular catalytic domain, tethered via a single transmembrane helix to the 196-amino acid-long extracellular domain (18, 31). Although two metal centers are found in the catalytic core in most PP2C phosphatases, the crystal structural of PstP showed the presence of three metal-binding centers (31). The structure of PstP was later refined by the analysis of the trigonal crystal form, which has a similar core structure as the monoclinic crystal but a different flap region (32). The active site residues are conserved between the eukaryotic phosphatase PP2Cα and the PstP (31). The conserved active site residues Asp-38 and Asp-229 are involved in metal binding, whereas Arg-20 is involved in binding with the phosphate moiety in the target proteins, thus bridging together to stabilize the active structure of the complex (31).
The PstP-encoding gene is located in an operon that encodes for essential kinases PknA and PknB, which show similar expression profiles (7, 33, 34). Previous data from our laboratory have shown that PknA and PknB phosphorylate PstP on specific residues in its cytosolic domain, and this phosphorylation is influenced by the presence of Zn2+ ions and inorganic phosphate (Pi). The phosphatase-dead PstP mutants (PstPcD38G, PstPcD229G, and PstPcR20G) are more efficiently phosphorylated by PknA and PknB. Importantly, we found that the phosphorylated PstP is more active compared with its unphosphorylated counterpart (35), suggesting a possible reverse regulation mediated through phosphorylation cascades by STPKs. Investigations carried out thus far are in vitro studies using purified PstP. Initial high throughput data suggested that under in vitro culture, pstP is nonessential, but the same group using Himar-based transposon mutagenesis indicated that it might be essential for growth (36, 37). The present study aims to investigate the impact of PstP overexpression or depletion on the growth of the pathogen and its survival within the host. To examine these aspects, conditional gene replacement mutants of pstP have been generated in both Mycobacterium smegmatis and M. tuberculosis. Using these mutants, we investigated the role of various domains of PstP in modulating cellular events, cell division, and host adaptation.
Results
Impact of Overexpression of PstP
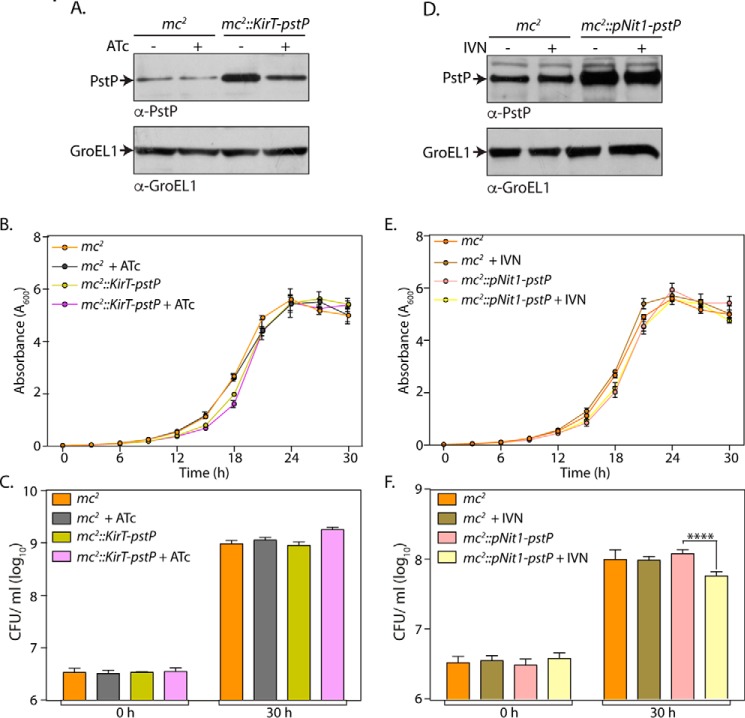
Our laboratory has previously shown that overexpression of PknA or PknB results in cell death, probably due to hyperphosphorylation of substrates critical in modulating cell survival. Because PstP is the sole phosphatase in M. tuberculosis, we speculated that overexpression of PstP would also be likely to impact cell survival. To test this hypothesis, PstP was cloned into the IVN-inducible episomal vector pNit-1 (36) and tet-regulatable integrative vector pST-KirT (37). The constructs were suitably prepared and electroporated into the M. smegmatis mc2155 strain, and the recombinants were analyzed for PstP expression in the presence or absence of inducer. In the case of mc2::KirT-pstP strain, we noticed moderate levels of overexpression in the absence of anhydrotetracycline (ATc) (Fig. 1A), which was consistently observed in four independent experiments. The expression was reduced significantly upon the addition of ATc (albeit not to the endogenous levels), indicating that the promoter could be turned off upon the addition of inducer (Fig. 1A). Based on the growth pattern analysis, we concluded that mild overexpression of PstP from the integrative pST-KirT-pstP does not alter the growth profile (Fig. 1B). To quantitate the differences, cfu at 0 and 30 h were enumerated (Fig. 1C). The presence or absence of ATc did not alter the cfu obtained in the case of mc2, suggesting that the presence of ATc is not toxic to the cells. Analysis suggested that the mild overexpression of PstP in mc2::KirT-pstP strain did not impact the number of cfu (Fig. 1E). Interestingly, even this level of expression led to a statistically significant increase in the cell length, which reverted back upon the addition of ATc (Fig. 2, A and B).
FIGURE 1.
Impact of overexpression of PstP. M. smegmatis mc2155 strain was electroporated with pKirT-pstP and pNit1-pstP to generate mc2::pKirT-pstP and mc2::pNit1-pstP strains, respectively. A, mc2 or mc2::pKirT-pstP cultures were initiated at an A600 of 0.1 and grown in the absence or the presence of 50 ng/ml ATc for 12 h. Whole cell lysates (WCLs) were prepared, and 35 μg (for PstP) and 20 μg (for GroEL1) of lysates were resolved on SDS-PAGE and probed with α-PstP and α-GroEL1 antibodies. B, mc2 or mc2::pKirT-pstP cultures were seeded at an initial A600 of 0.02 and grown in the absence or presence of 50 ng/ml ATc. Absorbance was measured every 3 h for 30 h. The experiment was performed in triplicate, the mean was plotted with the help of GraphPad Prism version 6 software, and the error bar represents S.D. C, cfu were enumerated at 0 and 30 h from the growth curve experiment in B. D, mc2 or mc2::pNit1-pstP cultures were seeded at an initial A600 of 0.1 and grown in the absence or presence of 5 μm IVN. WCLs were prepared and analyzed for expression of PstP and GroEL1 as described in A. E and F, mc2 or mc2::pNit1-pstP cultures were seeded at an initial A600 of 0.02 and grown in the absence or presence of 5 μm IVN. The growth curve was performed, and corresponding cfu data were analyzed and plotted as described above. ****, p < 0.0001, two-way ANOVA test, mean with S.D.
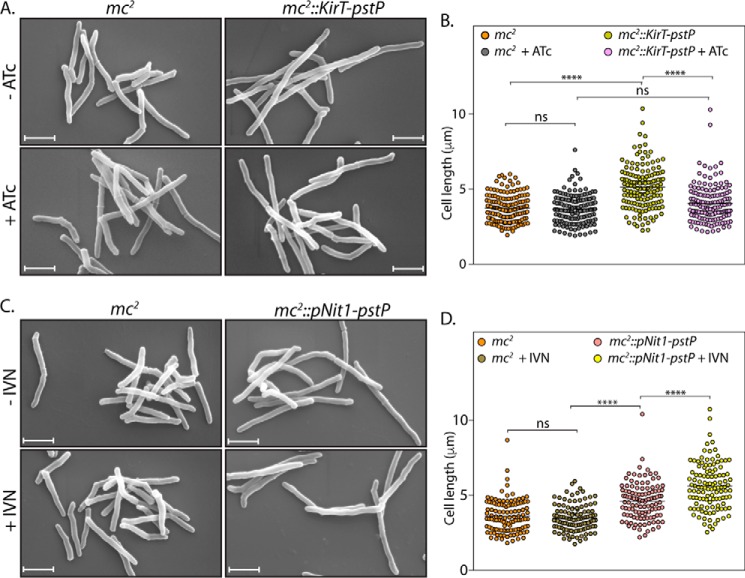
FIGURE 2.
Impact of overexpression of PstP on morphology. A, mc2 or mc2::KirT-pstP cultures were seeded at 0.1 and allowed to grow in the presence and absence of 50 ng/ml ATc inducer for 12 h. Cells were processed for SEM as described under “Materials and Methods.” Scale bar, 1 μm. B, cell lengths of 150 individual cells from different SEM images were measured using Smart Tiff software, and data were analyzed using GraphPad Prism version 6. Mean lengths are plotted, and significance is calculated using ordinary one-way ANOVA with p < 0.0001 (****). Mean lengths of mc2, mc2 +ATc, mc2::KirT-pstP −ATc, and mc2::KirT-pstP +ATc were 3.74, 3.68, 5.13, and 4.03 μm, respectively. C, mc2 or mc2::pNit1-pstP cultures were initiated at an A600 of 0.1 in the absence or presence of 5 μm IVN and grown for 12 h. SEM was performed as described. Scale bar, 1 μm. D, cell lengths of 115 individual cells from SEM images were measured by Smart Tiff software and analyzed as mentioned in B. Mean cell lengths of mc2, mc2 +IVN, mc2::pNit-pstP −IVN, and mc2::pNit1-pstP +IVN samples were 3.61, 3.49, 4.60, and 5.61 μm, respectively. ****, p < 0.0001, ordinary one-way ANOVA. ns, not significant.
The results obtained for mc2::pNit1-pstP varied between two phenotypes. At times, we observed significant cell death upon the addition of IVN. Western blot analysis from such an experiment exhibited significant overexpression of PstP upon the addition of IVN (data not shown). However, on the majority of occasions, we observed a marginal difference in the growth in the absence and presence of IVN (Fig. 1E). Correspondingly, the Western blot analysis indicated marginal differences in the expression pattern of PstP in the absence and presence of IVN (Fig. 1D). To quantitate the impact of PstP overexpression on cell survival, cfu analysis was performed with mc2 and mc2::pNit1-pstP cultures grown in the presence or absence IVN (Fig. 1F). We observed a marginal but statistically significant decrease in the cfu upon the addition of IVN (Fig. 1F). Scanning electron microscopy revealed that overexpression of PstP resulted in elongation of cells (Fig. 2, C and D). The presence or absence of ATc or IVN did not alter the cell length of mc2. Interestingly, we observed slight but statistically significant change in mean cell length upon the addition of IVN (Fig. 2D). These results suggest a role for PstP in maintaining normal cell lengths.
Depletion of PstP in M. smegmatis Leads to Cell Death
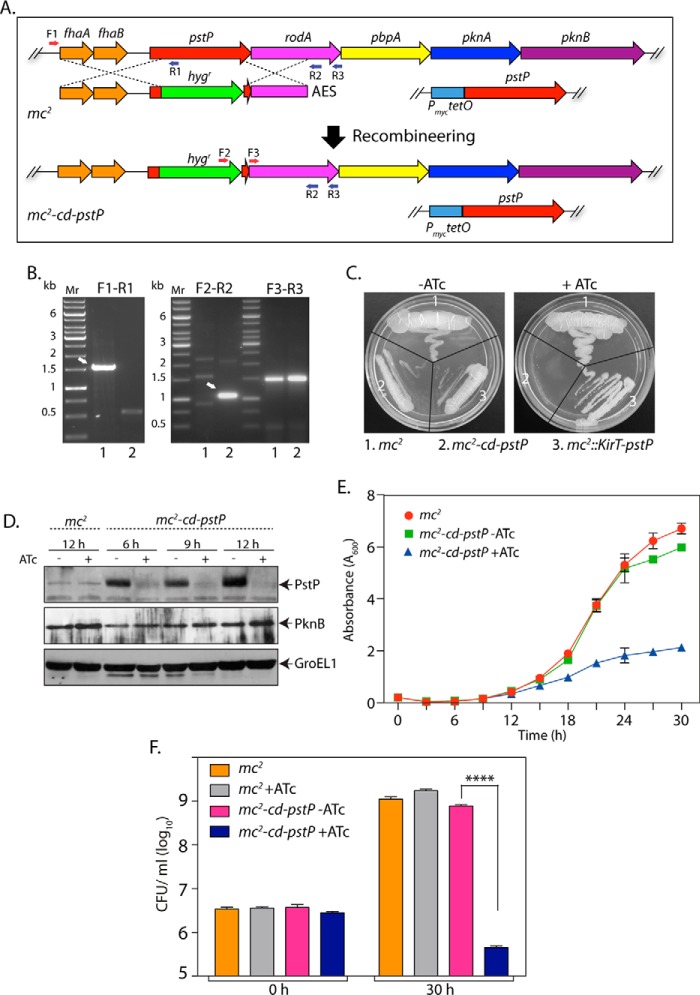
To decipher the role of PstP in regulating cellular events, we began with creating a conditional gene replacement pstP mutant in M. smegmatis. Previous attempts to generate mycobacterial pstP gene replacement mutants in our laboratory were unsuccessful, possibly due to polarity effects on the expression of the downstream genes in the operon. We therefore modified the strategy for generating the allelic exchange substrate (AES), wherein the carboxyl terminus of the hygr gene was fused with a spacer (encoding four glycine residues and one serine residue) followed by the last 21 nucleotides of the pstP gene (encoding the last seven amino acids of PstP). Because the translational stop codon of PstP overlaps with the translational start codon of RodA, we reasoned that the above strategy would minimally disrupt both transcription and translation of the downstream genes. The mc2-KirT-pstP strain was electroporated with pNitA-ET to generate recombineering-proficient merodiploid strain. This strain was electroporated with blunt-ended linearized AES to replace the pstP at the native locus with a modified hygr gene. Fidelity of recombination at the native pstP locus was confirmed by performing PCR amplification with specific primers across the replacement junctions (Fig. 3, A and B). To determine the impact of PstP depletion on cell growth, we streaked M. smegmatis wild type (mc2), merodiploid (mc2::KirT-pstP), and the mutant (mc2-cd-pstP) on plates in the presence or absence of ATc (Fig. 3C). As expected, both wild type and merodiploid strains grew regardless of the presence of ATc. However, the conditional mutant grew well only in the absence of ATc, because in the presence of ATc we did not detect any growth on plates (Fig. 3C). Western blot analysis of the whole cell lysates, isolated at different time points post-ATc addition, confirmed the depletion of PstP (Fig. 3D). To examine the possibility of polarity effects on the expression of downstream genes of the operon, the whole cell lysates were analyzed with anti-PknB antibody. The expression of PknB was found to be similar regardless of the presence of ATc, indicating that the replacement of pstP at its native locus did not alter expression of the downstream genes. A comparison of the growth pattern analysis of the mutant versus the wild type strain revealed that the depletion of PstP significantly reduced the growth (Fig. 3E). The reduced growth observed in the presence of ATc could be due to either bacteriostatic or bactericidal phenotypes. To resolve this question, we enumerated cfu at the 0 and 30 h time points in the presence and absence of ATc. The depletion of PstP resulted in ∼3 log-fold decrease in the survival of bacteria, suggesting that depletion of PstP was detrimental to cell survival (Fig. 3F).
FIGURE 3.
Depletion of PstP in M. smegmatis leads to poor cell survival. A, schematic depiction of generation of gene replacement mutant using the recombineering method. Primers used for screening the mutant are shown. B, agarose gels showing PCR products obtained using different primer pairs with genomic DNA obtained from mc2 (lane 1) and putative mc2-cd-pstP mutant (lane 2). F3-R3 amplifying a region of the rodA gene (∼1.5 kb) was used as a control. F1-R1, ∼1.5-kb fragment for mc2 and no amplicon expected for mc2-cd-pstP. F2-R2, no amplicon expected for mc2 and ∼1.1 kb for mc2-cd-pstP. C, mc2, mc2::pKirT-pstP, and mc2-cd-pstP cultures were grown until A600 reached ∼0.6, and the cultures were diluted to an A600 of 0.1. Diluted cultures were streaked on 7H10 agar plates not containing or containing 50 ng/ml ATc. D, mc2 and mc2-cd-pstP cultures were initiated at an A600 of 0.1 and grown in the absence or presence of ATc for 6, 9, or 12 h. Whole cell lysates were isolated, resolved on SDS-PAGE, transferred to nitrocellulose membrane, and probed with α-PstP, α-PknB, and α-GroEL1 antibodies. E, mc2 and mc2-cd-pstP cultures were initiated at an initial A600 of 0.02 and grown in the absence or presence of ATc for 30 h. The experiment was performed in triplicates, and the error bar represents S.E. F, cfu were enumerated at 0 and 30 h for both mc2 and mc2-cd-pstP in the presence and absence of ATc. cfu data were analyzed and plotted as mean with S.D. (error bars) using GraphPad Prism version 6, and significance was calculated using two-way ANOVA with p < 0.0001 (****).
Decrease in PstP Expression Compromises M. tuberculosis Growth
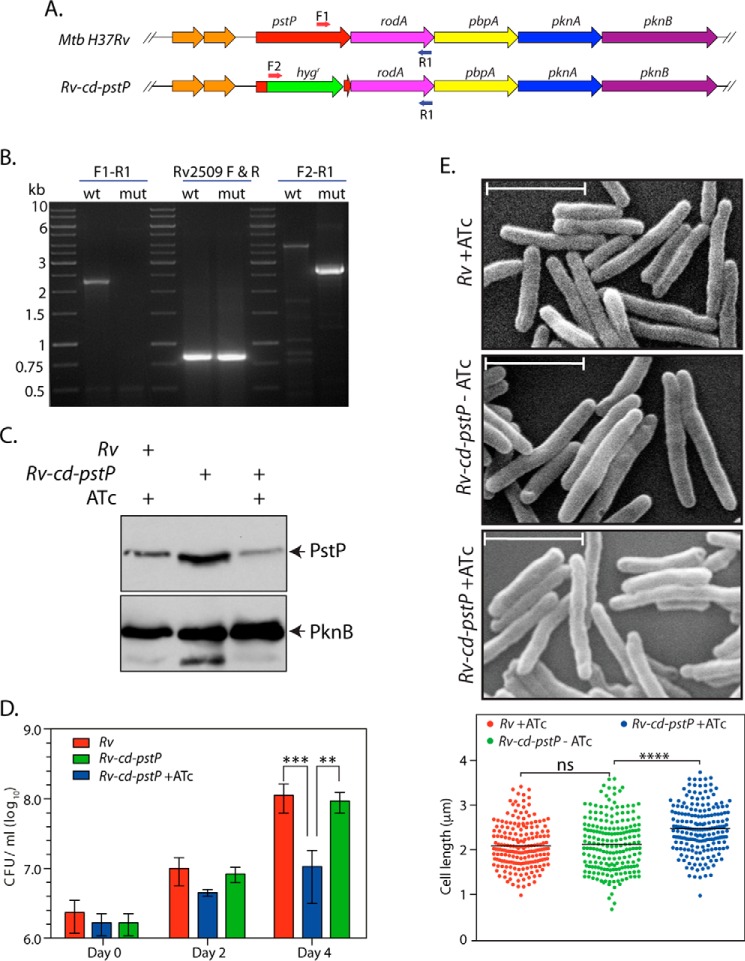
A conditional gene replacement of pstP in M. tuberculosis was created with the help of recombineering as described under “Materials and Methods.” Replacement of pstP at the native locus was confirmed by performing PCRs with appropriate primers (Fig. 4A), using the genomic DNA isolated from Rv and Rv-cd-pstP (Fig. 4B). Although the expression of PknB (from the last gene in the operon; Fig. 4A) was similar in the absence/presence of ATc, there was a significant decrease in PstP expression in the presence of ATc (Fig. 4C). Although this decrease in PstP expression was significant when compared with expression in the absence of ATc (compare lanes 2 and 3 of Fig. 4C), the levels of PstP expression in the presence of ATc were only ∼1.9-fold lower than PstP expression levels detected in wild type cells (from the native locus; compare lane 3 with lane 1 in Fig. 4C). Nevertheless, we analyzed the mutant for growth defects by determining the cfu/ml of culture grown in the absence or presence of ATc for 0, 2, and 4 days. We observed the impact of PstP decreased expression to be ∼1 log-fold (10-fold) compared with the wild type (Fig. 4D). Thus, a tight regulation of PstP expression appears to be critical for optimal growth and survival. Moreover, scanning electron microscopy analysis indicated marginal elongation of cells upon partial depletion of PstP (Fig. 4E).
FIGURE 4.
Decrease in PstP expression compromises M. tuberculosis growth. A, schematic representation of Rv and Rv-cd-pstP mutant. The primers used for PCR-based confirmation of the generated mutant are depicted by arrows. B, agarose gels showing the PCR amplicons obtained using genomic DNA from Rv (wt) and Rv-cd-pstP (mut). The left panel depicts PCR using primer F1-R1 wt (2.265 kb) and F1-R1 mut (no amplicon). The middle panel depicts control PCR using Rv2509 F-R primers. The right panel depicts PCR with F2-R1 wt (no amplicon) and F2-R1 mut (2.61 kb). C, Rv and Rv::cd-pstP cultures were initiated at an A600 of 0.1 and grown in the absence or presence of ATc (1 μg/ml) for 4 days. WCLs were prepared and analyzed by Western blot with α-PstP and α-PknB antibodies. D, Rv and Rv::cd-pstP cultures initiated at an A600 of 0.1 were grown in the presence or absence of ATc for 0, 2, or 4 days. Cultures were serially diluted and plated on 7H10 agar plates supplemented with OADC without ATc. Mean log10 cfu for cultures on day 4 were 8.04 cfu for Rv, 7.9 cfu for Rv::cd-pstP −ATc, and 6.0 cfu for Rv::cd-pstP +ATc. The experiment was performed in triplicate, and mean values with S.D. were been plotted using GraphPad Prism version 6. Significance was calculated using two-way ANOVA with p < 0.001 (***) and p < 0.01 (**). E, SEM analysis of Rv, Rv::cd-pstP −ATc, and Rv::cd-pstP +ATc cultures grown for 4 days. Scale bar, 1 μm. F, 198 cells of each cell type were visualized by SEM and measured per sample with the help of Smart Tiff software. Mean cell lengths for Rv, Rv-cd-PstP −ATc, and Rv-cd-PstP +ATc samples were 2.09, 2.12, and 2.47 μm, respectively. Data were plotted using GraphPad Prism version 6, and significance was calculated using two-way ANOVA with p < 0.0001 (****). ns, not significant.
The Phosphatase Activity of PstP Is Essential for in Vitro Growth
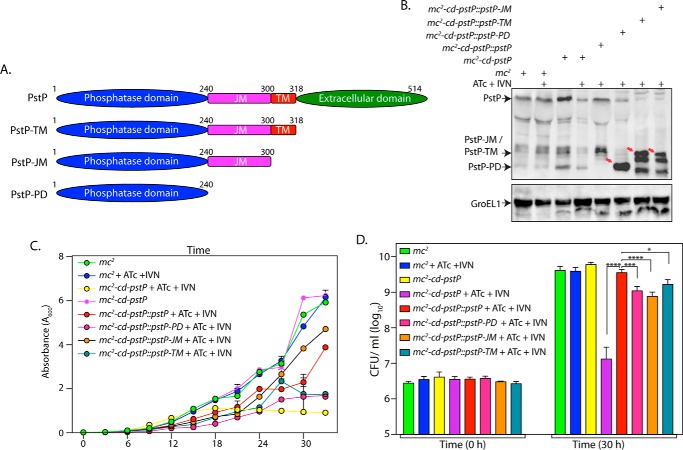
PstP has four distinct domains: an intracellular catalytic domain (amino acids 1–240), a 60-amino acid-long juxtamembrane domain, a transmembrane domain that carries 18 amino acids, and an extracellular domain of 196 amino acids (Fig. 5A). Although the catalytic domain was shown to be functional in vitro (18), the roles played by the remaining domains have not been elucidated either in vitro or in vivo. To decipher the roles of the different domains and the active site residues in the functioning of PstP in vivo, wild type PstP, PstP deletion mutants, and PstP point mutants were cloned into pNitA. The constructs were electroporated into the M. smegmatis PstP conditional mutant mc2-cd-pstP, and the transformants were assessed for the ability of the mutated PstP proteins (expressed from the plasmids) to complement PstP depletion (in the presence of ATc). We standardized the conditions for appropriate expression of the PstP deletion fragments by inducing with different concentrations of IVN. Western blot analysis clearly demonstrated the expression of all three deletion fragments (Fig. 5B, indicated by arrows). Growth analysis in liquid culture as well as cfu analysis suggested effective complementation by episomally expressed wild type PstP in the presence of ATc (Fig. 5, C and D). Importantly, we observed compromised growth with all three deletion mutants, and the cfu analyses are in agreement with the observed growth patterns (Fig. 5, C and D). Together, analysis suggests that although the deletion fragments could complement, the ability to complement is not equivalent to that of full-length PstP protein.
FIGURE 5.
Deletion of different PstP domains leads to mildly compromised rescue. A, schematic depiction of domain structure of PstP. PD, phosphatase domain; JM, juxtamembrane; TM, transmembrane. B, M. smegmatis mc2, mc2-cd-pstP, mc2-cd-pstP::pstP-TM, mc2-cd-pstP::pstP-JM, and mc2-cd-pstP::pstP-PD cultures were initiated at an A600 of 0.1 in the absence or presence of ATc + IVN (50 ng/ml ATc + 5 μm IVN) for 12 h. WCLs were prepared, and 40 μg (for PstP) and 20 μg (for GroEL1) of lysates were resolved on SDS-PAGE, transferred to nitrocellulose membrane, and probed with α-PstP and α-GroEL1 antibodies. C, mc2, mc2-cd-pstP, mc2-cd-pstP::pstP-TM, mc2-cd-pstP::pstP-JM, and mc2-cd-pstP::pstP-PD cultures were initiated at an A600 of 0.02, and growth was monitored in the absence or presence of ATc + IVN every 3 h for 30 h. Growth analysis is plotted as the mean with error bars representing S.D. D, cfu enumeration was performed for mc2, mc2-cd-pstP, mc2-cd-pstP::pstP-TM, mc2-cd-pstP::pstP-JM, and mc2-cd-pstP::pstP-PD from the cultures in C at 0 and 30 h. Data were plotted as the mean with S.D., and significance was calculated using two-way ANOVA with p < 0.0001 (****), p < 0.001 (***), and p < 0.05 (*).
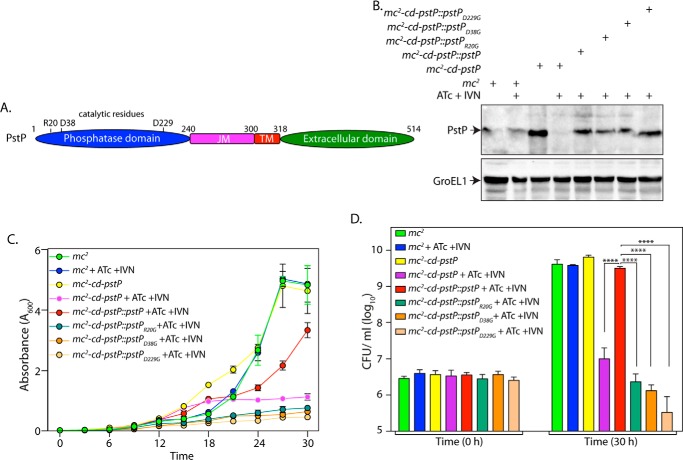
It has previously been shown that mutating Asp-38 and Asp-229 to glycine resulted in ∼90% loss of the dephosphorylation activity, whereas mutating Arg-20 to Gly resulted in ∼60% loss of its activity (35). We sought to investigate the role of phosphatase activity of PstP in regulating growth (Fig. 6A). Western blot analysis of lysates prepared from the transformants confirmed the expression of PstP and its active site mutants (Fig. 6B). Growth pattern and cfu analysis suggested that none of the point mutants (D38G, R20G, and D229G) could complement PstP depletion (Fig. 6, C and D). Although these mutants display varying levels of residual phosphatase activity (35), it appears that a 60% loss in phosphatase activity is sufficient to compromise in vitro growth. Thus, our results suggest that the activity of PstP is essential for mycobacterial growth.
FIGURE 6.
The catalytic activity of PstP is essential for in vitro growth. A, schematic depiction of domain structure of PstP. Residues critical for catalysis are indicated. B, mc2, mc2-cd-pstP, mc2-cd-pstP::pstPR20G, mc2-cd-pstP::pstPD38G, and mc2-cd-pstP::pstPD229G strains were initiated at A600 of 0.1 in the absence or presence of ATc + IVN (50 ng/ml ATc + 5 μm IVN) for 12 h. WCLs were prepared, and 40 μg (for PstP) and 20 μg (for GroEL1) of lysates were resolved on SDS-PAGE and probed with α-PstP and α-GroEL1 antibodies. C, mc2, mc2-cd-pstP, mc2-cd-pstP::pstPR20G, mc2-cd-pstP::pstPD38G, and mc2-cd-pstP::pstPD229G strains were seeded at an A600 of 0.02, and growth was monitored in the absence or presence of ATc + IVN every 3 h for 30 h. Growth analysis is plotted as the mean with error bars representing S.D. D, cfu enumeration was performed for mc2, mc2-cd-pstP, mc2-cd-pstP::pstPR20G, mc2-cd-pstP::pstPD38G, and mc2-cd-pstP::pstPD229G strains from the cultures in C at 0 and 30 h. Data were plotted as the mean with S.D., and significance was calculated using two-way ANOVA with p < 0.0001 (****).
Depletion of PstP Results in Multiseptum Phenotype
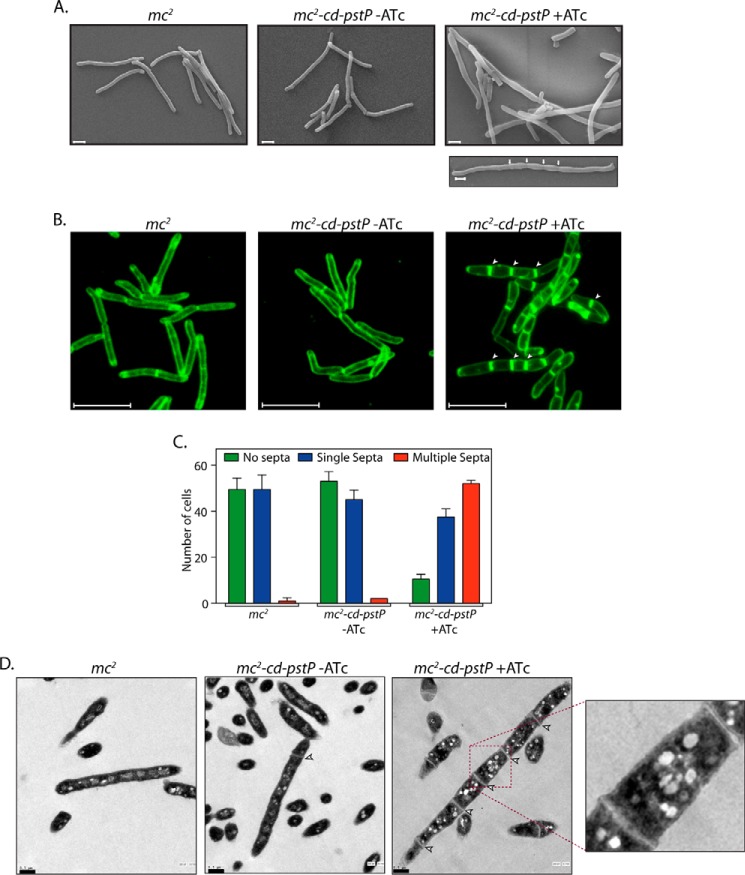
The pstP gene is located in an operon that encodes for PknA, PknB, RodA, and PbpA. These four genes have all been suggested to play a critical role in cell shape maintenance, cell division, and cell wall synthesis processes (34, 38). PknA and PknB modulate the activities of a number of proteins that participate in cell division and cell wall synthesis by mediating their phosphorylation (39–41). Given the fact that PstP is the sole serine/threonine phosphatase in mycobacterium and is carried by an operon that carries genes controlling cell shape and cell division (including two kinases), we investigated any possible role that PstP might play in modulating cell shape and cell division by examining the impact of PstP depletion on these parameters using scanning electron microscopy (SEM). Interestingly, significantly elongated and bulged cells with multiple septa were observed in PstP-depleted samples (ATc treatment-mediated 12-h depletion), suggesting a possible role for PstP in modulating cell division (Fig. 7A). To confirm this phenotype, we labeled the cells with FM4–64 dye and seeded on a soft agar stage. This lipophilic dye exhibits fluorescence upon binding with the outer layer of the cell membrane, thus serving as a tool to stain and visualize plasma membranes and septa. Although the FM4-64 fluorescence pattern of mc2-cd-pstP (−ATc) strain was observed to be similar to that of mc2, we observed distinctly elongated and wider cells with multiple septa in mc2-cd-pstP upon the addition of ATc (Fig. 7B). 198 cells of each strain were scored for their number of septa, and it was observed that there was a ∼6-fold increase in multiseptate mc2-cd-pstP (+ATc) cells as compared with wild type cells or mc2-cd-pstP (−ATc) cells. TEM analysis revealed that the distance between two septa was much shorter than the average normal cell length in multiseptate cells (Fig. 7D). These results suggest the possibility that in the absence of PstP-mediated dephosphorylation of target substrates, the cells display division defects.
FIGURE 7.
Depletion of PstP results in multiseptum phenotype. A, SEM analysis of mc2 and mc2::cd-pstP strains grown in the absence or presence of ATc (50 ng/ml) for 12 h was performed as described (50). White arrows in the bottom right panel indicate septa. Scale bar, 1 μm. B, FM4-64 labeling and microscopy of mc2 and mc2::cd-pstP strains grown in the absence or presence of ATc was performed as described under “Materials and Methods.” The arrows indicate the presence of septa. Scale bar, 5 μm. C, 100 individual cells per cell type per FM4-64 labeling experiment were scored for aseptate, uniseptate, or multiseptate phenotypes. The experiment was performed twice, and the average is represented in the graph. D, TEM analysis of mc2 and mc2::cd-pstP strains grown in the absence or presence of ATc for 12 h. Processed samples were visualized on grids under FEI G2 Spirit. Scale bar, 0.5 μm. Error bars, S.D.
Depletion of PstP in M. tuberculosis Negatively Impacts Its Survival in Mice
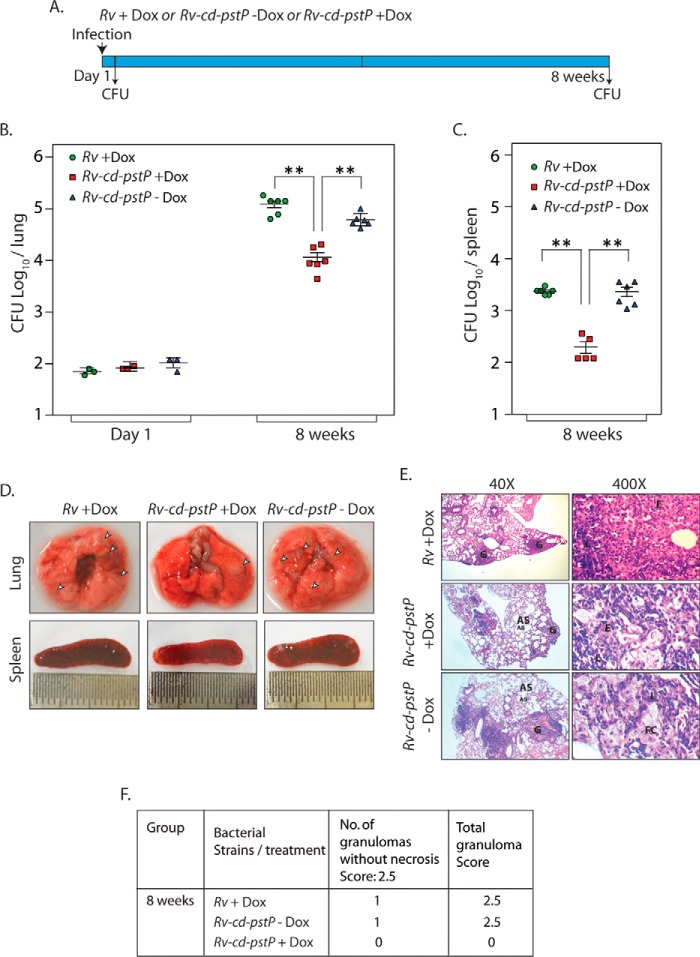
To investigate whether stringent regulation of PstP expression is critical for the survival of pathogen within the host, we infected mice with Rv or Rv-cd-pstP, and the mice were either provided or not provided doxycycline (Fig. 8A). Analysis of cfu determined 1 day post-infection showed equivalent implantation of wild type and mutant bacilli in mice lungs (Fig. 8B). To determine the impact of PstP depletion on the survival, cfu were enumerated both in lungs and in the spleen of mice 8 weeks post-infection (Fig. 8, B and C). Gross pathological assessment of lungs at 8 weeks post-infection affirmed the presence of distinct tubercles in the lung tissue of mice infected with Rv and Rv-cd-pstP without ATc, whereas a lesser number of tubercles were observed in the case of mice infected with Rv-cd-pstP that were given doxycycline (Fig. 8D). This was also reflected in the histopathological data and scores, wherein no distinct granulomas were detected in the lungs of mice infected with Rv-cd-pstP +doxycycline hydrochloride (Dox) (Fig. 8, E and F). These data corroborated the cfu counts obtained in the lungs as well as the spleen 8 weeks post-infection, which were 1 log-fold (4.0 versus 5.1 and 4.8 for lungs and 2.1 versus 3.3 and 3.3 for spleen cfu) lower in mice infected with Rv-cd-pstP +Dox samples compared with those obtained in Rv and Rv-cd-pstP −Dox-infected mice (Fig. 8, B and C). Taken together, these results underline the importance of PstP for the survival of the pathogen in the host.
FIGURE 8.
Depletion of PstP in M. tuberculosis negatively impacts its survival in mice. A, schematic representation of the mouse infection experiment. B and C, BALB/c mice were infected with Rv (9 mice) and Rv-cd-PstP (18 mice) as described previously (50). Rv-cd-PstP-infected mice were divided into two groups, and one group was provided Dox (1 mg/kg) in their drinking water. Rv-infected mice (9 mice) were also provided Dox in their drinking water. The water was changed every third day. Three mice from each group were sacrificed 1 day post-infection, and lung homogenates were plated in duplicates to determine mycobacterial implantation efficiency. The six remaining mice per group were sacrificed 56 days post-infection, and lung and spleen homogenates were similarly plated. B, the mean log10 cfu obtained for lung homogenates from Rv, Rv-cd-PstP +Dox, and Rv-cd-PstP −Dox at day 1 and day 56 were 5.1, 4.0, and 4.8 cfu, respectively. C, the mean log10 cfu obtained for spleen homogenates from Rv +Dox, Rv-cd-PstP +Dox, and Rv-cd-PstP at day 56 were 3.3, 2.1, and 3.3, respectively. Error bars, S.E. **, p < 0.01, two-tailed nonparametric t test. D, gross lung and spleen pathology of Rv +Dox-, Rv-cd-PstP +Dox-, and Rv-cd-PstP-infected mice 8 weeks post-infection. Prominent tubercles are indicated by white arrowheads. E, ×40 and ×400 images of H&E-stained lung sections of the infected mice. G, granuloma; FC, foamy cells; AS, alveolar space; E, epithelioid cells. F, granuloma score of the histopathology sections shown in E.
Depletion of PstP Decreases the Bacillary Load Even in an Established Infection
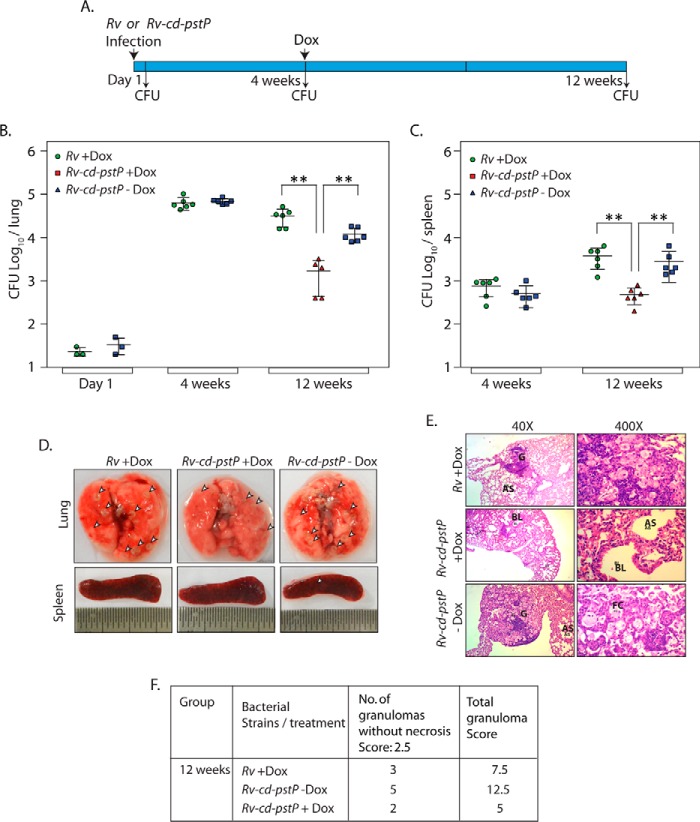
To determine the impact of PstP depletion on the survival of bacilli in a well established infection, we infected mice with Rv or Rv-cd-pstP and allowed the infection to progress over 4 weeks. The Rv-cd-pstP-infected mice were then divided into two groups, and only one group was provided Dox. Dox was also provided to Rv infected mice to serve as a control. The bacillary loads in the lungs and spleens of the mice were determined after 1 day, 4 weeks, and 12 weeks post-infection (Fig. 9A, time line diagram). It is apparent from the data in Fig. 9, B and C, that the bacillary load was equivalent after 1 day and 4 weeks in the case of both mice infected with Rv and mice infected with Rv-cd-pstP. Interestingly, depletion of PstP led to ∼1 log-fold (3.14 versus 4.49 and 4.08 in lungs and 2.54 versus 3.57 and 3.45 in spleen cfu) decrease in the cfu both in lungs and spleen even in established infection (Fig. 9, B and C). In agreement with this observation, we noticed that the number of distinct tubercles in the lungs were lower in PstP-depleted samples (Fig. 9D). Histopathological analysis of lungs 12 weeks post-infection revealed the presence of a higher number of granulomas in the case of Rv and Rv-cd-PstP. However, in the presence of Dox, we observed a lower number of granulomas and correspondingly lower score in case of Rv-cd-PstP infection (Fig. 9, E and F). These data emphasize the need for controlled expression of PstP not only for establishment of infection (Fig. 8), but also for continued maintenance of an established infection.
FIGURE 9.
Depletion of PstP decreases the bacillary load even in an established infection. A, schematic outline depicting the experiment. B and C, BALB/c mice groups were infected with Rv and Rv-cd-pst. Three mice each from Rv- and Rv-cd-pstP-infected groups were sacrificed 1 day post-infection, and cfu were determined. Six mice each from Rv and Rv-cd-pstP infected groups were sacrificed 4 weeks post-infection, and the bacillary loads in the lungs and spleen were determined. Log10 cfu for Rv and Rv-cd-pstP in the lungs were 4.8 and 4.84, and values for the spleen were 2.66 and 2.56, respectively. Rv-cd-pstP-infected mice were divided into two groups (n = 6) 4 weeks post-infection, and one group was provided Dox (1 mg/kg) for 8 weeks. cfu were enumerated in lungs and spleen 12 weeks post-infection. The bacillary loads (log10 cfu) in the lungs for Rv + Dox, Rv-cd-pstP +Dox and Rv-cd-pstP -Dox were 4.49, 3.14, and 4.08, and bacillary loads in the spleen were 3.57, 2.54, and 3.45, respectively. Error bars, S.E. **, p < 0.01, two-tailed nonparametric t test. D, representative images of lungs and spleen after 12 weeks of infection. Prominent granulomas are indicated by arrowheads. E, ×40 and ×400 histopathology images of lung sections at 12 weeks post-infection. G, granuloma; FC, foamy cells; AS, alveolar space; E, epithelioid cells; BL, bronchial lumen. F, granuloma score of the histopathology sections of E.
Discussion
Although the eukaryotic-like phosphosignaling systems in prokaryotes are emerging as regulatory systems that are as important to the cell as their more prominent eukaryotic counterparts, the study of their functions is still in its infancy. Sensing the host environment is key to the appropriate execution of pathogen developmental programs, and mycobacterial phosphatases are a major family of sensor/signaling proteins. Dephosphorylation by phosphatases is critical for regulation of serine/threonine/tyrosine protein kinase-mediated cellular signaling (42, 43). In most prokaryotes, the genes encoding STPKs and corresponding protein phosphatases are carried by the same operon, seemingly following the ratio of one phosphatase per kinase (20, 43). For example, in Streptococcus pneumoniae, the phosphatase PhpP interacts with the kinase StkP (the genes encoding both lying in the same operon), and the presence of the kinase is essential for appropriate phosphatase localization. Additionally, cells could only survive deletion of phosphatase gene when the kinase gene stkP was also deleted (44). Mycobacteria and corynebacteria, however, encode multiple kinases, but they encode for only one serine/threonine phosphatase belonging to the PPM family (7, 45). The physiological purpose of such an arrangement remains undiscovered. The present study targets the investigation of the physiological impact of overexpressing and depleting PstP on growth and survival of the pathogen both in vitro and in vivo.
Among the criteria used to classify bacteria is cell shape (46). Multiple bacterial proteins are cell shape regulators. The pstP is the first gene of an operon that also carries rodA, pbpA, pknA, and pknB genes. RodA, named for its role in conferring bacteria with a rodlike shape, is a putative lipid II flippase (47, 48). PbpA, although not yet characterized, is predicted to be a transpeptidase required for cross-linking peptidoglycans in the periplasm space (49). Previous studies have shown that overexpression of PknA or PknB in M. smegmatis or Mycobacterium bovis BCG leads to altered cellular morphology. Whereas the overexpression of PknB results in bulged cells, overexpression of PknA results in elongated cells (34). Moreover, overexpression of PknB is detrimental to in vitro growth and cell survival (50). Our efforts to overexpress PstP using inducible nitrile promoter have not been very fruitful. Although on two occasions, we observed significant overexpression that led to cell death, in most of the experiments, we observed marginally higher expression. We do not have a good explanation for this phenomenon. We speculate that the organism found a way to make the nitrile promoter refractory to inducer concentrations. Interestingly, even with this marginal higher expression of PstP, we observed a statistically significant increase in the mean cell length (Fig. 2). Thus, it appears that the entire operon is involved in the modulation and maintenance of cell morphology and cell division.
To decipher the importance of PstP in regulating cellular events, we started with generating M. smegmatis and M. tuberculosis pstP conditional mutant strains by carrying out gene replacements (Figs. 3 and 4). The M. smegmatis pstP conditional mutant (mc2-cd-pstP) displayed almost complete depletion of PstP in the presence of ATc. This depletion of PstP was detrimental to cell survival (Fig. 3). Only partial depletion of PstP was observed in the M. tuberculosis pstP conditional mutant (Fig. 4), but despite this, we observed ∼1 log-fold difference in cell survival, suggesting that stringent modulation of PstP expression is critical for optimal growth.
PstP and 9 of the 11 STPKs in M. tuberculosis contain a single transmembrane helix, connected to an extracytoplasmic domain (7, 38). The extracellular domain of PknD has been shown to be involved in sensing osmotic stress (51). The extracellular PASTA domains of PknB interact with muropeptides in the periplasmic space and are essential for in vitro growth of M. tuberculosis (50, 52). The membrane-anchoring transmembrane domain, but not the extracytoplasmic domain of PknA, is found to be essential for in vitro growth (53). For most mycobacterial kinases and the sole phosphatase PstP, very little is known about the functional significance of the membrane-anchoring or the extracytoplasmic domains. The phosphatase domain of PstP has been found to be necessary and sufficient for efficient catalytic activity in vitro (18, 35). We observed that active site mutants of PstP completely failed to rescue growth defects of the M. smegmatis pstP conditional mutant in vitro, indicating the importance of catalytically active PstP phosphatase for cell survival (Fig. 6). Similar experimental analyses with multiple other PstP mutants (Fig. 5) revealed that deletion mutants show marginally compromised survival (0.4–0.6 log-fold lower compared with the wild type). Thus, it appears that all of the domains, including the extracellular domain, are necessary for effective functional complementation.
Cell division is a complex process comprising cell elongation, septum formation, and subsequent cytokinesis, involving a myriad of proteins. Interestingly, several proteins involved in cell division and cell wall synthesis processes have been identified to be phosphorylated in phosphoproteomic studies (54–57). Among the 301 proteins identified in a high throughput phosphoproteomic study from M. tuberculosis H37Rv, 69 proteins were linked to cell division and cell wall synthesis processes (54). In a recent study, 398 M. bovis BCG proteins were identified to be phosphorylated, and 19.5% of these phosphoproteins were involved in various cellular processes (57). Similarly, phosphoproteomic analysis of M. smegmatis mc2 and M. bovis BCG has revealed the phosphorylation of a number of cell division proteins, such as FtsQ, FtsW, CwsA, FtsK, and FtsY (56). Although all these proteins are important for cell division, FtsZ, a homolog of eukaryotic tubulin harboring GTPase activity, acts as a primer for divisome assembly (58). PknA phosphorylates FtsZ, thus impairing its GTPase activity and polymerization functions (59). Phosphorylation at the carboxyl terminus of FtsZ also impairs its interaction with other divisome proteins, FipA and FtsQ (59). Taken together, these studies confirm the significant role of phosphorylation in modulating the cell division process. Hence, we hypothesized that the depletion of PstP would have an impact on the cell division process. Data from scanning electron microscopy, fluorescence microscopy, and transmission electron microscopy experiments presented in Fig. 7 show the formation of multiple septa upon PstP depletion. Thus, it appears that appropriate dephosphorylation of cell division proteins is critical to orderly cell elongation and cytokinesis.
Phosphatases play an important role in determining the virulence of the pathogen. PtpA and PtpB, secretory tyrosine phosphatases of M. tuberculosis, have been shown to be crucial for intracellular survival of the pathogen (25, 27). However, to date, the impact of PstP depletion on mycobacterial survival in the host has not been assessed. Data from mouse infection experiments that we have carried out indicate that even partial depletion of PstP compromises pathogen survival by ∼10-fold (Fig. 8). Interestingly, partial depletion of PstP from an established infection (4 weeks post-infection) also led to a ∼10-fold decrease in the survival, signifying the need for continued expression of PstP for the maintenance of M. tuberculosis infection (Fig. 9). Specific inhibitors of PtpA and PtpB with useful IC50 values have been recently identified (60, 61). Standard inhibitors of PP2C phosphatases, such as cyclosporine and sodium fluoride, are not very effective against PstP. Taken together, these findings underline the importance of PstP both for in vitro growth and in vivo survival and suggest that PstP could be an effective target for therapeutic intervention.
Materials and Methods
Bacterial Strains and Reagents
A list of constructs generated and strains used in the study is presented in Table 1. Sequences and descriptions of the oligonucleotides used in the study are provided in supplemental Table 1. All medium components were purchased from BD Biosciences. Restriction endonucleases and DNA-modifying enzymes were procured from New England Biolabs and MBI Fermentas. Oligonucleotide primers and analytical grade chemicals were procured from Sigma-Aldrich or GE Healthcare. α-PstP, α-PknB, and α-GroEL-1 antibodies for immunoblotting were raised in the laboratory. Electron microscopy reagents were procured from Electron Microscopy Sciences. Doxycycline hydrochloride was purchased from Biochem Pharmaceutical (Mumbai, India). The pENTR/directional TOPO cloning kit was purchased from Invitrogen. Mycobacterial shuttle plasmids pNit-1 (Kanr) (36), pJV53 (Kanr) (62), and pNit-ET (Kanr) (63) were kind gifts from Dr. Christopher Sassetti, Dr. Graham Hatfull, and Dr. Eric Rubin, respectively.
TABLE 1.
Plasmids and strains used in the study
| Description | Source | |
|---|---|---|
| Plasmid constructs | ||
| pST-KirT | Integrative Mtb expression vector with N-terminal FLAG tag and r-tetR cloned in SnaBI site; Kanr | Ref. 64 |
| pNit-1 | IVN inducible Mtb expression vector; Kanr | Ref. 36 |
| pNit-ET | Construct containing gp60 and gp61 genes from Che9c phase under nitrile-inducible promoter; Kanr | Ref. 63 |
| pJV3 | Construct containing gp60 and gp61 genes from Che9c phase under acetamide inducible promoter; Kanr | Ref. 62 |
| pMV-261A | E. coli-mycobacterium shuttle vector, Aprar | Kind gift from Dr. Jacobs |
| pNitA | pNit-1 construct wherein kanr gene with the aprar gene from pMV261A; Aprar | This study |
| pNitA-ET | pNit-ET construct wherein kanr gene was replaced with the aprar gene from pMV261A; Aprar | This study |
| pJV53A | pJV53 construct wherein kanr gene was replaced with the aprar gene from pMV261A; Aprar | This study |
| pKirT-pstP | pstP cloned into NdeI-HindIII sites of pST-KirT; Kanr | This study |
| pNit1-pstP | pstP cloned into NdeI-HindIII sites of pNit-1; Kanr | This study |
| pNitA-pstP | pstP cloned into NdeI-HindIII sites of pNitA; Aprar | This study |
| pNitA-pstP-TM | pstP-TM cloned into NdeI-HindIII sites of pNitA; Aprar | This study |
| pNitA-pstP-JM | pstP-JM cloned into NdeI-HindIII sites of pNitA; Aprar | This study |
| pNitA-pstP-PD | pstP-PD cloned into NdeI-HindIII sites of pNitA; Aprar | This study |
| pNitA-pstPR20G | pstPR20G cloned into NdeI-HindIII sites of pNitA; Aprar | This study |
| pNitA-pstPD38G | pstPD38G cloned into NdeI-HindIII sites of pNitA; Aprar | This study |
| pNitA-pstPD229G | pstPD229G cloned into NdeI-HindIII sites of pNitA; Aprar | This study |
| Strains | ||
| DH5α | E. coli strain used for cloning experiment | Invitrogen |
| mc2 | M. smegmatis mc2155 strain | ATCC, 700084 |
| Rv | M. tuberculosis H37Rv strain | ATCC |
| Rv::pNitA-ET | Rv strain electroporated with pNitA-ET construct | This study |
| mc2::pJV53A | mc2 strain electroporated with pJV53A construct | This study |
| mc2::KirT-pstP | mc2 strain electroporated with pST-KirT-pstP construct | This study |
| Rv::KirT-pstP | Rv strain electroporated with pST-KirT-pstP construct | This study |
| mc2-cd-pstP | PstP conditional mutant in mc2::KirT-pstP merodiploid strain | This study |
| Rv-cd-pstP | PstP conditional mutant in Rv::KirT-pstP merodiploid strain | This study |
| mc2-cd-pstP::pstP | mc2-cd-pstP strain complemented with pNitA-pstP construct | This study |
| mc2-cd-pstP::pstP-TM | mc2-cd-pstP strain complemented with pNitA-pstP-TM construct | This study |
| mc2-cd-pstP::pstP-JM | mc2-cd-pstP strain complemented with pNitA-pstP-JM construct | This study |
| mc2-cd-pstP::pstP-PD | mc2-cd-pstP strain complemented with pNitA-pstP-PD construct | This study |
| mc2-cd-pstP::pstPR20G | mc2-cd-pstP strain complemented with pNitA-pstPR20G construct | This study |
| mc2-cd-pstP::pstPD38G | mc2-cd-pstP strain complemented with pNitA-pstPD38G construct | This study |
| mc2-cd-pstP::pstPD229G | mc2-cd-pstP strain complemented with pNitA-pstPD229G construct | This study |
Generation of Plasmid Constructs
The pstP gene was amplified from M. tuberculosis genomic DNA with adaptor primers carrying NdeI and HindIII sites at the 5′-ends of the forward and reverse primers, respectively, using Phusion DNA polymerase (New England Biolabs). The amplicon was digested with NdeI-HindIII and cloned into the corresponding sites in pNit1 and pST-KirT (64) to generate pNit1-pstP and pST-KirT-pstP constructs, respectively. pNit1, pJV53, and pNit-ET constructs were modified by replacing the kanr gene with the aprar gene from pMV261-apra (a kind gift from Dr. William Jacobs), to generate pNitA, pJV53A, and pNitA-ET, respectively. PstP deletion mutants were created by amplifying specific fragments of the gene with adaptor primers carrying NdeI-HindIII sites and cloning the amplicons into pENTR vector. This was followed by subcloning the NdeI-HindIII fragments into the corresponding sites in pNit-apra vector. PstP point mutants created in a previous study (35) were subcloned into pNitA vector. Details of constructs used in the study are provided in Table 1.
Generation of M. smegmatis and M. tuberculosis Conditional Gene Replacement Mutants
M. tuberculosis H37Rv and M. smegmatis mc2155 strains were electroporated with integration-proficient pST-KirT-pstP (with PstP containing an amino-terminal FLAG tag) construct, to generate merodiploid strains Rv::KirT-pstP and mc2::KirT-pstP. These strains were then electroporated with pNitA-ET and pJV53A, respectively, to generate recombineering proficient strains. These strains were electroporated. The hygromycin resistance gene (hygr) along with its promoter was amplified from pYUB1474 construct (65) (a kind gift from Dr. William Jacobs) using adaptor primers carrying PflMI sites compatible with those found in pYUB1474. The reverse primer of hygr was designed such that the stop codon would be replaced with an amino acid spacer (GGSGG). The amplicon obtained was cloned into pENTR vector to generate pENTR-hygr. The apramycin resistance gene (aprar) was amplified from pMV261A and cloned into pENTR vector to generate pENTR-aprar. Upstream and downstream flanks (∼1 kb on either side) of pstP were amplified from genomic DNA of M. tuberculosis and M. smegmatis. The forward primer of the downstream (3′) flank was designed to carry a PflMI site along with the last seven amino acids of PstP, such that these amino acids would be in frame with the hygr gene. We also introduced an EcoRV site in the 5′-flank forward and 3′-flank reverse primers after the PflMI site. Upstream, downstream flanks digested with PflMI were ligated with the hygr gene from pENTR-hygr, the apramycin resistance gene from pENTR-aprar, and oriE and λcos fragment from pYUB1474 to generate allelic exchange substrates. The AES cassette containing 5′-flank-hygr-3′-flank was released with EcoRV digestion, and the cassette was electroporated into recombineering-proficient merodiploid strains. Induction of recombineering genes before competent cell preparation was performed as described earlier (62). Conditional mutants mc2-cd-pstP and Rv-cd-pstP were screened by PCR amplification using specific primer sets to confirm that genuine recombination had occurred at the native locus (Figs. 3 and 4).
Growth Pattern Analysis
M. smegmatis mc2 wild type and mc2-cd-pstP mutant strains were electroporated with pST-KirT-pstP, pNit1-pstP, pNitA-pstP, and pNitA-pstPmutant (deletion mutants or point mutants) constructs to generate recombinant strains. The cultures were grown in 7H9 medium supplemented with 10% ADC (albumin, dextrose, and catalase) or in 7H10 agar medium supplemented with 10% OADC (oleic acid, albumin, dextrose, and catalase). To analyze growth on plates, cultures were diluted to an A600 of ∼0.1 and streaked on 7H10 agar plates containing either no antibiotics or ATc (50 ng/ml). To analyze growth patterns in liquid culture, cultures were initiated in triplicates at an A600 of ∼0.02, either in the presence or absence of ATc, and absorbance at 600 nm was measured every 3 h for 30 h. The data were plotted using GraphPad Prism version 6 software. To enumerate cfu at 0 and 30 h, cultures were harvested, washed twice with PBST (PBS containing 0.05% Tween 80), resuspended, and serially diluted, and different dilutions were plated on 7H10 agar plates containing OADC. In the case of M. tuberculosis (Rv) or M. tuberculosis mutant (Rv-cd-pstP), cultures were seeded at an A600 of ∼0.1, either in the presence or absence of ATc, and cultures were grown for 4 days. cfu were determined on day 0, day 2, and day 4 on 7H10 agar plates in the absence of all antibiotics.
Western Blotting Analysis
Cultures of M. smegmatis mc2, mc2::KirT-pstP, mc2::pNit1-pstP, mc2-cd-pstP, or mc2-cd-pstP::pstPwt/mutant strains seeded at an A600 of ∼0.1 were grown for different times in the absence or presence of ATc or IVN or ATc + IVN. In the case of M. tuberculosis strains Rv or Rv-cd-pstP, cultures seeded at an A600 of 0.1 were grown in the absence or presence of ATc for 4 days. Whole cell lysates were isolated from harvested cells and analyzed by Western blotting as described earlier (53).
Scanning Electron Microscopy
Cultures of M. smegmatis mc2 or mc2-cd-pstP strains were initiated at an A600 of 0.1 and grown for 12 h in the absence or presence of ATc in filtered 7H9 medium. Similarly, Rv and Rv-cd-PstP cultures were initiated at an A600 of ∼0.1 and grown for 4 days in the absence or presence of ATc in filtered 7H9 medium. Cells were harvested, and SEM analysis was performed as described earlier (50).
Transmission Electron Microscopy
M. smegmatis mc2 and mc2-cd-pstP strains were grown until mid-log phase in the absence or presence of ATc. The cells were harvested at 4,300 × g and washed three times with 100 mm sodium phosphate buffer (pH 7.4). Transmission electron microscopy was performed as described earlier (66).
Immunofluorescence Analysis
FM4-64 labeling methodology was used to visualize mycobacteria by fluorescence. For this, 7H9-agarose pads were prepared on frosted slides (Corning Microslide Frosted; 75 × 25 mm) using an AB gene frame (Thermo Scientific; 17 × 54 mm). The 7H9-agarose pads comprised high resolution low melting agarose (Sigma; 1.5%) in 7H9 medium supplemented with ADC. mc2 or mc2-cd-pstP cultures at an A600 of ∼0.6 were diluted to an A600 of ∼0.1, and 1 μl of diluted culture of either mc2 or mc2-cd-PstP was spread on the agarose pad (either with or without ATc (50 ng/ml)). The agarose pads were supplemented with FM4-64 (2 μg/ml) (67). Image acquisition was performed on a Leica TCS SP8 confocal laser scanning microscope at ×63 oil immersion with 3× optical zoom (68).
Infection of Mice with Mycobacteria
Rv and Rv-cd-PstP cells were grown to mid-log phase and processed, and mice were infected with these cells, as described (50). BALB/c mice of either sex that were 6–8 weeks old (obtained from the National Institute of Immunology breeding facility) were infected by the aerosol route. The lung bacillary loads were enumerated 24 h post-infection to determine the implantation dosage. For reducing PstP levels in mice infected with Rv-cd-PstP, Dox was supplied at a concentration of 1 mg/kg with 5% sucrose in the drinking water. Mice were dissected at the desired time point, bacillary loads were determined, and histopathology of lung sections was performed as described earlier (50).
Author Contributions
A. K. S., V. K. N., and Y. S. conceived and designed the experiments. A. K. S., D. A., A. G., and L. K. S. performed the experiments. V. K. N., Y. S., A. K. S., D. A., V. M., and A. S. analyzed the data. V. K. N., Y. S., A. S., and A. K. S. wrote the paper.
Supplementary Material
Acknowledgments
We thank the Tuberculosis Aerosol Challenge Facility (TACF) of International Center for Genetic Engineering and Biotechnology, New Delhi (ICGEB) for support in all animal experiments. We thank Dr. Dhiraj Kumar for helpful discussions and for allowing use of the TACF facility. We thank the scanning electron microscopy facility and the biocontainment facility at the National Institute of Immunology. We thank the confocal microscope facility and transmission electron microscopy facility at CSIR-Institute of Genomic and Integrative Biology. We thank Rekha Rani, Mahendra Pratap Singh, and Manish Kumar for support in SEM/TEM and confocal imaging. We thank Dr. Christopher Sassetti, Dr. Graham Hatfull, Dr. Eric Rubin, and Dr. William Jacobs for providing the plasmids.
This work was supported by Indo French Centre for the Promotion of Advanced Research Grant 5303-1 (to V. K. N., Y. S., and V. M.) and BSC0104 (CSIR) (to Y. S.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table 1.
- STPK
- serine/threonine protein kinase
- Dox
- doxycycline hydrochloride
- PPM
- metal-dependent protein phosphatase
- PPP
- phosphoprotein phosphatase
- PTP
- phosphotyrosine phosphatase
- LMW
- low molecular weight
- ATc
- anhydrotetracycline
- IVN
- isovaleronitrile
- AES
- allelic exchange substrate
- SEM
- scanning electron microscopy
- WCL
- whole cell lysate
- ANOVA
- analysis of variance.
References
- 1. Bourret R. B., and Silversmith R. E. (2010) Two-component signal transduction. Curr. Opin. Microbiol. 13, 113–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stock A. M., Robinson V. L., and Goudreau P. N. (2000) Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 [DOI] [PubMed] [Google Scholar]
- 3. Kaur K., Kumari P., Sharma S., Sehgal S., and Tyagi J. S. (2016) DevS/DosS sensor is bifunctional and its phosphatase activity precludes aerobic DevR/DosR regulon expression in Mycobacterium tuberculosis. FEBS J. 283, 2949–2962 [DOI] [PubMed] [Google Scholar]
- 4. Dhiman A., Bhatnagar S., Kulshreshtha P., and Bhatnagar R. (2014) Functional characterization of WalRK: a two-component signal transduction system from Bacillus anthracis. FEBS Open Bio 4, 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. LaPorte D. C., and Chung T. (1985) A single gene codes for the kinase and phosphatase which regulate isocitrate dehydrogenase. J. Biol. Chem. 260, 15291–15297 [PubMed] [Google Scholar]
- 6. Galinier A., Kravanja M., Engelmann R., Hengstenberg W., Kilhoffer M. C., Deutscher J., and Haiech J. (1998) New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc. Natl. Acad. Sci. U.S.A. 95, 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E. 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 8. Wong D., Chao J. D., and Av-Gay Y. (2013) Mycobacterium tuberculosis-secreted phosphatases: from pathogenesis to targets for TB drug development. Trends Microbiol. 21, 100–109 [DOI] [PubMed] [Google Scholar]
- 9. Chaba R., Raje M., and Chakraborti P. K. (2002) Evidence that a eukaryotic-type serine/threonine protein kinase from Mycobacterium tuberculosis regulates morphological changes associated with cell division. Eur. J. Biochem. 269, 1078–1085 [DOI] [PubMed] [Google Scholar]
- 10. Peirs P., Parmentier B., De Wit L., and Content J. (2000) The Mycobacterium bovis homologous protein of the Mycobacterium tuberculosis serine/threonine protein kinase Mbk (PknD) is truncated. FEMS Microbiol. Lett. 188, 135–139 [DOI] [PubMed] [Google Scholar]
- 11. Molle V., Girard-Blanc C., Kremer L., Doublet P., Cozzone A. J., and Prost J. F. (2003) Protein PknE, a novel transmembrane eukaryotic-like serine/threonine kinase from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 308, 820–825 [DOI] [PubMed] [Google Scholar]
- 12. Koul A., Choidas A., Tyagi A. K., Drlica K., Singh Y., and Ullrich A. (2001) Serine/threonine protein kinases PknF and PknG of Mycobacterium tuberculosis: characterization and localization. Microbiology 147, 2307–2314 [DOI] [PubMed] [Google Scholar]
- 13. Molle V., Kremer L., Girard-Blanc C., Besra G. S., Cozzone A. J., and Prost J. F. (2003) An FHA phosphoprotein recognition domain mediates protein EmbR phosphorylation by PknH, a Ser/Thr protein kinase from Mycobacterium tuberculosis. Biochemistry 42, 15300–15309 [DOI] [PubMed] [Google Scholar]
- 14. Gopalaswamy R., Narayanan P. R., and Narayanan S. (2004) Cloning, overexpression, and characterization of a serine/threonine protein kinase pknI from Mycobacterium tuberculosis H37Rv. Protein Expr. Purif. 36, 82–89 [DOI] [PubMed] [Google Scholar]
- 15. Arora G., Sajid A., Gupta M., Bhaduri A., Kumar P., Basu-Modak S., and Singh Y. (2010) Understanding the role of PknJ in Mycobacterium tuberculosis: biochemical characterization and identification of novel substrate pyruvate kinase A. PLoS One 5, e10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumar P., Kumar D., Parikh A., Rananaware D., Gupta M., Singh Y., and Nandicoori V. K. (2009) The Mycobacterium tuberculosis protein kinase K modulates activation of transcription from the promoter of mycobacterial monooxygenase operon through phosphorylation of the transcriptional regulator VirS. J. Biol. Chem. 284, 11090–11099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lakshminarayan H., Narayanan S., Bach H., Sundaram K. G., and Av-Gay Y. (2008) Molecular cloning and biochemical characterization of a serine threonine protein kinase, PknL, from Mycobacterium tuberculosis. Protein Expr. Purif. 58, 309–317 [DOI] [PubMed] [Google Scholar]
- 18. Boitel B., Ortiz-Lombardía M., Durán R., Pompeo F., Cole S. T., Cerveñansky C., and Alzari P. M. (2003) PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol. Microbiol. 49, 1493–1508 [DOI] [PubMed] [Google Scholar]
- 19. Koul A., Choidas A., Treder M., Tyagi A. K., Drlica K., Singh Y., and Ullrich A. (2000) Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J. Bacteriol. 182, 5425–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sajid A., Arora G., Singhal A., Kalia V. C., and Singh Y. (2015) Protein phosphatases of pathogenic bacteria: role in physiology and virulence. Annu. Rev. Microbiol. 69, 527–547 [DOI] [PubMed] [Google Scholar]
- 21. Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 22. Shi L., Potts M., and Kennelly P. J. (1998) The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 22, 229–253 [DOI] [PubMed] [Google Scholar]
- 23. Shi L. (2004) Manganese-dependent protein O-phosphatases in prokaryotes and their biological functions. Front. Biosci. 9, 1382–1397 [DOI] [PubMed] [Google Scholar]
- 24. Grundner C., Cox J. S., and Alber T. (2008) Protein tyrosine phosphatase PtpA is not required for Mycobacterium tuberculosis growth in mice. FEMS Microbiol. Lett. 287, 181–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bach H., Papavinasasundaram K. G., Wong D., Hmama Z., and Av-Gay Y. (2008) Mycobacterium tuberculosis virulence is mediated by PtpA dephosphorylation of human vacuolar protein sorting 33B. Cell Host Microbe 3, 316–322 [DOI] [PubMed] [Google Scholar]
- 26. Wong D., Bach H., Sun J., Hmama Z., and Av-Gay Y. (2011) Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc. Natl. Acad. Sci. U.S.A. 108, 19371–19376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh R., Rao V., Shakila H., Gupta R., Khera A., Dhar N., Singh A., Koul A., Singh Y., Naseema M., Narayanan P. R., Paramasivan C. N., Ramanathan V. D., and Tyagi A. K. (2003) Disruption of mptpB impairs the ability of Mycobacterium tuberculosis to survive in guinea pigs. Mol. Microbiol. 50, 751–762 [DOI] [PubMed] [Google Scholar]
- 28. Clarke M., Maddera L., Engel U., and Gerisch G. (2010) Retrieval of the vacuolar H-ATPase from phagosomes revealed by live cell imaging. PLoS One 5, e8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou B., He Y., Zhang X., Xu J., Luo Y., Wang Y., Franzblau S. G., Yang Z., Chan R. J., Liu Y., Zheng J., and Zhang Z. Y. (2010) Targeting mycobacterium protein tyrosine phosphatase B for antituberculosis agents. Proc. Natl. Acad. Sci. U.S.A. 107, 4573–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chopra P., Singh B., Singh R., Vohra R., Koul A., Meena L. S., Koduri H., Ghildiyal M., Deol P., Das T. K., Tyagi A. K., and Singh Y. (2003) Phosphoprotein phosphatase of Mycobacterium tuberculosis dephosphorylates serine-threonine kinases PknA and PknB. Biochem. Biophys. Res. Commun. 311, 112–120 [DOI] [PubMed] [Google Scholar]
- 31. Pullen K. E., Ng H. L., Sung P. Y., Good M. C., Smith S. M., and Alber T. (2004) An alternate conformation and a third metal in PstP/Ppp, the M. tuberculosis PP2C-family Ser/Thr protein phosphatase. Structure 12, 1947–1954 [DOI] [PubMed] [Google Scholar]
- 32. Wehenkel A., Bellinzoni M., Schaeffer F., Villarino A., and Alzari P. M. (2007) Structural and binding studies of the three-metal center in two mycobacterial PPM Ser/Thr protein phosphatases. J. Mol. Biol. 374, 890–898 [DOI] [PubMed] [Google Scholar]
- 33. Narayan A., Sachdeva P., Sharma K., Saini A. K., Tyagi A. K., and Singh Y. (2007) Serine threonine protein kinases of mycobacterial genus: phylogeny to function. Physiol. Genomics 29, 66–75 [DOI] [PubMed] [Google Scholar]
- 34. Kang C. M., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., and Husson R. N. (2005) The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19, 1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sajid A., Arora G., Gupta M., Upadhyay S., Nandicoori V. K., and Singh Y. (2011) Phosphorylation of Mycobacterium tuberculosis Ser/Thr phosphatase by PknA and PknB. PLoS One 6, e17871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pandey A. K., Raman S., Proff R., Joshi S., Kang C. M., Rubin E. J., Husson R. N., and Sassetti C. M. (2009) Nitrile-inducible gene expression in mycobacteria. Tuberculosis 89, 12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parikh A., Kumar D., Chawla Y., Kurthkoti K., Khan S., Varshney U., and Nandicoori V. K. (2013) Development of a new generation of vectors for gene expression, gene replacement, and protein-protein interaction studies in mycobacteria. Appl. Environ. Microbiol. 79, 1718–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Av-Gay Y., and Everett M. (2000) The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8, 238–244 [DOI] [PubMed] [Google Scholar]
- 39. Molle V., and Kremer L. (2010) Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol. Microbiol. 75, 1064–1077 [DOI] [PubMed] [Google Scholar]
- 40. Prisic S., and Husson R. N. (2014) Mycobacterium tuberculosis serine/threonine protein kinases. Microbiol. Spectr. 10.1128/microbiolspec.MGM2-0006-2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chakraborti P. K., Matange N., Nandicoori V. K., Singh Y., Tyagi J. S., and Visweswariah S. S. (2011) Signalling mechanisms in Mycobacteria. Tuberculosis 91, 432–440 [DOI] [PubMed] [Google Scholar]
- 42. Böhmer F., Szedlacsek S., Tabernero L., Ostman A., and den Hertog J. (2013) Protein tyrosine phosphatase structure-function relationships in regulation and pathogenesis. FEBS J. 280, 413–431 [DOI] [PubMed] [Google Scholar]
- 43. Pereira S. F., Goss L., and Dworkin J. (2011) Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol. Mol. Biol. Rev. 75, 192–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Osaki M., Arcondéguy T., Bastide A., Touriol C., Prats H., and Trombe M. C. (2009) The StkP/PhpP signaling couple in Streptococcus pneumoniae: cellular organization and physiological characterization. J. Bacteriol. 191, 4943–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kalinowski J., Bathe B., Bartels D., Bischoff N., Bott M., Burkovski A., Dusch N., Eggeling L., Eikmanns B. J., Gaigalat L., Goesmann A., Hartmann M., Huthmacher K., Krämer R., Linke B., et al. (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104, 5–25 [DOI] [PubMed] [Google Scholar]
- 46. Weidel W., and Pelzer H. (1964) Bagshaped macromolecules: a new outlook on bacterial cell walls. Adv. Enzymol. Relat. Areas Mol. Biol. 26, 193–232 [DOI] [PubMed] [Google Scholar]
- 47. Sieger B., Schubert K., Donovan C., and Bramkamp M. (2013) The lipid II flippase RodA determines morphology and growth in Corynebacterium glutamicum. Mol. Microbiol. 90, 966–982 [DOI] [PubMed] [Google Scholar]
- 48. Henriques A. O., Glaser P., Piggot P. J., and Moran C. P. Jr. (1998) Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28, 235–247 [DOI] [PubMed] [Google Scholar]
- 49. Botta G. A., and Buffa D. (1981) Murein synthesis and β-lactam antibiotic susceptibility during rod-to-sphere transition in a pbpA(Ts) mutant of Escherichia coli. Antimicrob. Agents Chemother. 19, 891–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chawla Y., Upadhyay S., Khan S., Nagarajan S. N., Forti F., and Nandicoori V. K. (2014) Protein kinase B (PknB) of Mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J. Biol. Chem. 289, 13858–13875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hatzios S. K., Baer C. E., Rustad T. R., Siegrist M. S., Pang J. M., Ortega C., Alber T., Grundner C., Sherman D. R., and Bertozzi C. R. (2013) Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proc. Natl. Acad. Sci. U.S.A. 110, E5069–E5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mir M., Asong J., Li X., Cardot J., Boons G. J., and Husson R. N. (2011) The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 7, e1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nagarajan S. N., Upadhyay S., Chawla Y., Khan S., Naz S., Subramanian J., Gandotra S., and Nandicoori V. K. (2015) Protein kinase A (PknA) of Mycobacterium tuberculosis is independently activated and is critical for growth in vitro and survival of the pathogen in the host. J. Biol. Chem. 290, 9626–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prisic S., Dankwa S., Schwartz D., Chou M. F., Locasale J. W., Kang C. M., Bemis G., Church G. M., Steen H., and Husson R. N. (2010) Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. U.S.A. 107, 7521–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nakedi K. C., Nel A. J., Garnett S., Blackburn J. M., and Soares N. C. (2015) Comparative Ser/Thr/Tyr phosphoproteomics between two mycobacterial species: the fast growing Mycobacterium smegmatis and the slow growing Mycobacterium bovis BCG. Front. Microbiol. 6, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fortuin S., Tomazella G. G., Nagaraj N., Sampson S. L., Gey van Pittius N. C., Soares N. C., Wiker H. G., de Souza G. A., and Warren R. M. (2015) Phosphoproteomics analysis of a clinical Mycobacterium tuberculosis Beijing isolate: expanding the mycobacterial phosphoproteome catalog. Front. Microbiol. 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zheng J., Liu L., Liu B., and Jin Q. (2015) Phosphoproteomic analysis of bacillus Calmette-Guerin using gel-based and gel-free approaches. J. Proteomics 126, 189–199 [DOI] [PubMed] [Google Scholar]
- 58. Thakur M., and Chakraborti P. K. (2006) GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J. Biol. Chem. 281, 40107–40113 [DOI] [PubMed] [Google Scholar]
- 59. Sureka K., Hossain T., Mukherjee P., Chatterjee P., Datta P., Kundu M., and Basu J. (2010) Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS One 5, e8590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60. Chiaradia L. D., Martins P. G., Cordeiro M. N., Guido R. V., Ecco G., Andricopulo A. D., Yunes R. A., Vernal J., Nunes R. J., and Terenzi H. (2012) Synthesis, biological evaluation, and molecular modeling of chalcone derivatives as potent inhibitors of Mycobacterium tuberculosis protein tyrosine phosphatases (PtpA and PtpB). J. Med. Chem. 55, 390–402 [DOI] [PubMed] [Google Scholar]
- 61. Mascarello A., Mori M., Chiaradia-Delatorre L. D., Menegatti A. C., Delle Monache F., Ferrari F., Yunes R. A., Nunes R. J., Terenzi H., Botta B., and Botta M. (2013) Discovery of Mycobacterium tuberculosis protein tyrosine phosphatase B (PtpB) inhibitors from natural products. PLoS One 8, e77081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van Kessel J. C., and Hatfull G. F. (2007) Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 [DOI] [PubMed] [Google Scholar]
- 63. Wei J. R., Krishnamoorthy V., Murphy K., Kim J. H., Schnappinger D., Alber T., Sassetti C. M., Rhee K. Y., and Rubin E. J. (2011) Depletion of antibiotic targets has widely varying effects on growth. Proc. Natl. Acad. Sci. U.S.A. 108, 4176–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Soni V., Upadhayay S., Suryadevara P., Samla G., Singh A., Yogeeswari P., Sriram D., and Nandicoori V. K. (2015) Depletion of M. tuberculosis GlmU from infected murine lungs effects the clearance of the pathogen. PLoS Pathog. 11, e1005235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jain P., Hsu T., Arai M., Biermann K., Thaler D. S., Nguyen A., González P. A., Tufariello J. M., Kriakov J., Chen B., Larsen M. H., and Jacobs W. R. Jr. (2014) Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. MBio 5, e01245–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Singh L. K., Dhasmana N., Sajid A., Kumar P., Bhaduri A., Bharadwaj M., Gandotra S., Kalia V. C., Das T. K., Goel A. K., Pomerantsev A. P., Misra R., Gerth U., Leppla S. H., and Singh Y. (2015) clpC operon regulates cell architecture and sporulation in Bacillus anthracis. Environ. Microbiol. 17, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fay A., and Glickman M. S. (2014) An essential nonredundant role for mycobacterial DnaK in native protein folding. PLoS Genet. 10, e1004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. de Jong I. G., Beilharz K., Kuipers O. P., and Veening J. W. (2011) Live cell imaging of Bacillus subtilis and Streptococcus pneumoniae using automated time-lapse microscopy. J. Vis. Exp. 10.3791/3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.