Abstract
Perilipin-2 (PLIN2) is a constitutively associated cytoplasmic lipid droplet coat protein that has been implicated in fatty liver formation in non-alcoholic fatty liver disease. Mice with or without whole-body deletion of perilipin-2 (Plin2-null) were fed either Western or control diets for 30 weeks. Perilipin-2 deletion prevents obesity and insulin resistance in Western diet-fed mice and dramatically reduces hepatic triglyceride and cholesterol levels in mice fed Western or control diets. Gene and protein expression studies reveal that PLIN2 deletion suppressed SREBP-1 and SREBP-2 target genes involved in de novo lipogenesis and cholesterol biosynthetic pathways in livers of mice on either diet. GC-MS lipidomics demonstrate that this reduction correlated with profound alterations in the hepatic lipidome with significant reductions in both desaturation and elongation of hepatic neutral lipid species. To examine the possibility that lipidomic actions of PLIN2 deletion contribute to suppression of SREBP activation, we isolated endoplasmic reticulum membrane fractions from long-term Western diet-fed wild type (WT) and Plin2-null mice. Lipidomic analyses reveal that endoplasmic reticulum membranes from Plin2-null mice are markedly enriched in ω-3 and ω-6 long-chain polyunsaturated fatty acids, which others have shown inhibit SREBP activation and de novo lipogenesis. Our results identify PLIN2 as a determinant of global changes in the hepatic lipidome and suggest the hypothesis that these actions contribute to SREBP-regulated de novo lipogenesis involved in non-alcoholic fatty liver disease.
Keywords: endoplasmic reticulum stress (ER stress), gene knockout, lipid droplet, lipid metabolism, lipogenesis, liver metabolism, mass spectrometry (MS), membrane lipid, metabolic disease, perilipin-2
Introduction
Non-alcoholic fatty liver disease (NAFLD)2 is a major public health concern and the most prevalent liver disorder in developed nations (1). NAFLD caused by obesity has been linked to insulin resistance, metabolic syndrome, and liver cancer (2–4). NAFLD begins with subtle accumulation of neutral lipids in the liver, and if not reversed through dietary or lifestyle changes the condition can progress to non-alcoholic steatohepatitis and subsequent cirrhosis of the liver (5). Therefore, reducing or preventing hepatic neutral lipid accumulation is an area of significant therapeutic interest (6). Nonetheless, despite intense study over many decades, the specific mechanisms regulating hepatic lipid accumulation have remained confusing and elusive (7).
Recently, the lipid droplet coat proteins perilipins (PLINs) have emerged as physiologic regulators of lipid accumulation in many tissues, including liver (8). Thus far, five perilipin family members have been identified (PLIN1 through PLIN5), with family members exhibiting differential tissue expression under both physiologic and pathophysiologic states (9). Perilipin-2 (PLIN2) is a constitutively associated cytoplasmic lipid droplet (CLD) coat protein that is expressed in many organs, including the liver. Under high fat diet-induced NAFLD, PLIN2 is the predominant lipid droplet coat protein in hepatocytes in humans and rodents (10, 11). Therefore, PLIN2 has become a potentially valuable therapeutic target for the treatment or prevention of NAFLD development (12).
We previously generated mice with whole-body knock-out of PLIN2 (Plin2-null) (13). These animals are resistant to obesity, adipose tissue inflammation, and liver steatosis (NAFLD) when raised on a high fat diet. In this study, we sought to probe how loss of PLIN2 alters hepatic lipid accumulation and metabolism at the molecular level. Using Plin2-null mice on Western (WD) or control (CD) diets for 30 weeks, we show that loss of Plin2 prevents diet-induced steatosis largely by impairing activation of the SREBP-1 and SREBP-2 pathways through effects on membrane lipid composition. We also show that PLIN2 deletion impairs activation of SREBP-dependent lipogenic pathways in mice fed control diets low in fat, carbohydrate, and cholesterol. Although we had expected a reduction in total neutral lipid on WD, the alteration in the global hepatic lipidome was unexpected. The findings of this study further our understanding of the role of PLIN2 in diet-induced fatty liver formation and provide evidence of its function in coordinating content and composition of biologically active cellular lipids in the liver.
Results
Plin2-null Mice on Long-term Western Diet Are Protected from Obesity and Insulin Resistance
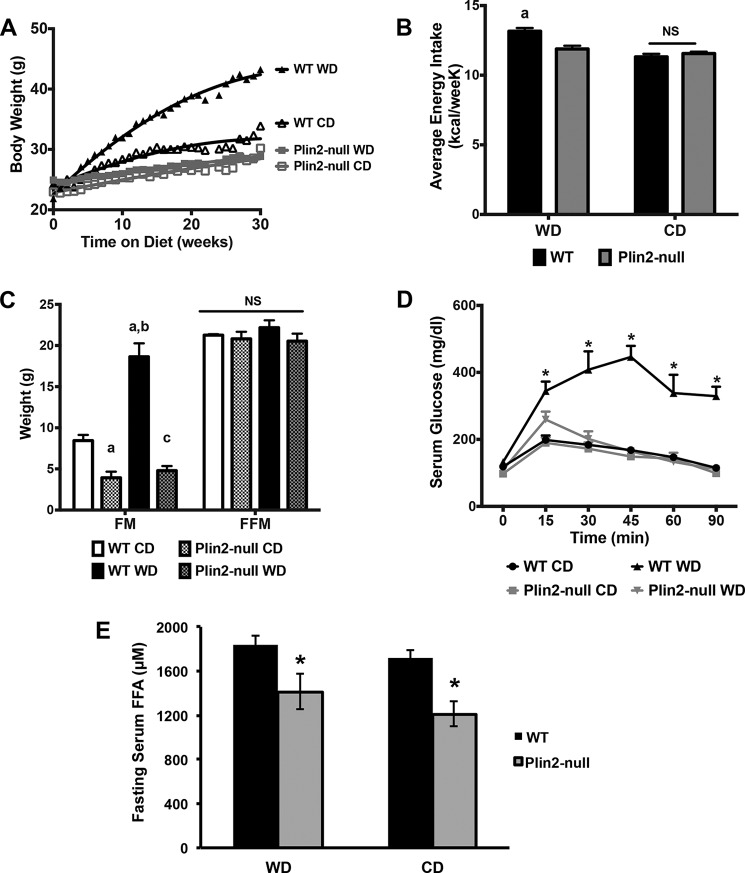
Wild type and Plin2-null mice were fed an obesity-promoting Western diet (WD), containing elevated levels of saturated fat, sucrose, and cholesterol, or a low fat CD, which had 75% less fat, 65% less sucrose, and no added cholesterol, ad libitum for 30 weeks. Consistent with previous studies of long-term high fat-fed mice (13), we found that WD-fed Plin2-null mice are protected from marked obesity produced by WD feeding of WT mice. The effects of feeding WT or Plin2-null mice WD or CD on weight gain, body composition, energy intake, and serum glucose regulation are shown in Fig. 1. Body weights of WD-fed WT mice doubled over 30 weeks, whereas body weights of CD-fed WT mice increased by 43% (Fig. 1A), and body weights of Western and control diet-fed Plin2-null animals increased by 16 and 31%, respectively. The rate of weight gain of WD-fed WT mice was significantly greater than that of CD-fed WT mice or of Western or control diet-fed Plin2-null mice (p < 0.001, two-way ANOVA). In addition, the rate of weight gain of CD-fed WT mice was significantly greater than that of Plin2-null mice fed WD or CD (p < 0.05, two-way ANOVA). Increased weight gains in WD-fed WT mice corresponded to modestly elevated energy intakes compared with the other groups (p < 0.001; one-way ANOVA). Differences in energy intake were not detected between WD- and CD-fed Plin2-null mice or between CD-fed WT mice and either WD- or CD-fed Plin2-null mice (Fig. 1B). Quantitative magnetic imaging analyses of mice after 30 weeks on WD or CD showed that body weight differences among WT and Plin2-null animals reflected differences in fat mass quantities (Fig. 1C). The average fat mass quantity of WD-fed WT mice was more than twice that of CD-fed animals and 4–5-fold greater than WD- and CD-fed Plin2-null mice, respectively (p < 0.001; two-way ANOVA). In addition, the average fat mass quantity of CD-fed WT mice was significantly greater than that of WD- or CD-fed Plin2-null mice (p < 0.05; two-way ANOVA). We did not detect significant differences in average fat-free (lean) mass quantities between WD- and CD-fed WT and Plin2-null mice (Fig. 1C).
FIGURE 1.
Plin2-null mice are protected from obesity and insulin resistance on Western diet. A, weight gain curves for WT and Plin2-null mice fed Western diet (WD) or control (CD) diets. B, average weekly energy consumption for WT and Plin2-null mice fed CD or WD for 30 weeks. a indicates significantly (p < 0.001, two-way ANOVA) increased average energy consumption by WD-fed WT mice compared with the other groups. C, body compositions of WT and Plin2-null mice fed WD or CD for 30 weeks. FM, fat mass; FFM, fat-free mass. a, indicates fat mass values are different from CD-fed WT mice (p < 0.05, two-way ANOVA); b, indicates fat mass values are different from CD-fed WT mice and CD- or WD-fed Plin2-null mice (p < 0.001, two-way ANOVA). D, glucose tolerance tests for WT and Plin2-null fed WD or CD at 30 weeks. Asterisks indicate significantly elevated serum glucose levels in WD-fed WT mice compared with the other groups, p < 0.05, two-way ANOVA. E, 18-h fasting serum free fatty acid (FFA) levels in WT and Plin2-null mice fed Western and control diets. Asterisks indicate significantly reduced serum FFA levels in Plin2-null mice, p < 0.05, Student's t test. NS, not statistically different.
WD-induced obesity is associated with glucose utilization abnormalities linked to insulin resistance (14). To determine whether protection from obesity by PLIN2 deletion in WD-fed mice also confers protection against abnormalities in glucose utilization, we determined serum glucose responses to intraperitoneal (i.p.) glucose injection in WT and Plin2-null mice fed WD or CD (Fig. 1D). Serum glucose levels in WD-fed WT mice remain significantly elevated 90 min after i.p. glucose injection but return to normal by 30 min in CD-fed WT mice. Serum glucose curves in WD- or CD-fed Plin2-null mice were similar to that of CD-fed WT mice, which demonstrates that protective effects of Plin2 deletion on WD-induced obesity are associated with protection against impaired glucose utilization, possibly through improved insulin sensitivity. Consistent with this possibility, we found that fasting serum-free fatty acid (FFA) levels, a measure of insulin inhibition of adipose lipolysis (15, 16), were 25% (p = 0.0403) lower in WD-fed Plin2-null mice compared with WT mice fed this diet (Fig. 1E). Collectively, these results agree with previous conclusions that PLIN2-deletion protects against insulin resistance associated with diet or genetically induced obesity (13, 17, 18). Interestingly, we also observed a 25% reduction (p = 0.0056) in fasting serum FFA for Plin2-null mice fed the control diet. As such, the reduction in fasting FFA associated with loss of PLIN2 appears to be independent of diet or effects on obesity, although direct effects on adipose properties remain possible (19).
Plin2-null Animals Are Resistant to Western Diet-induced Hepatic Steatosis
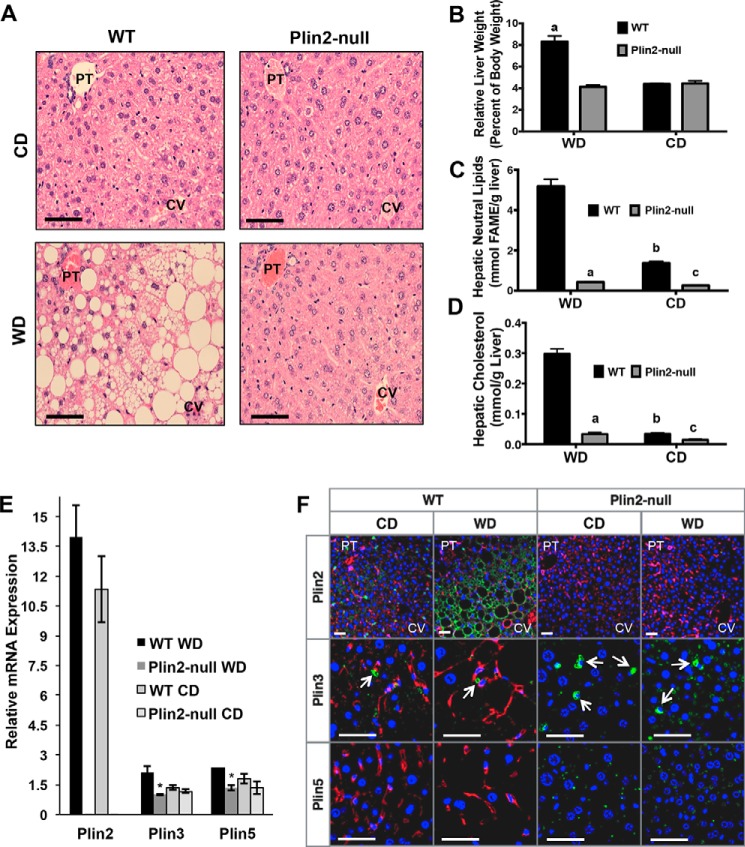
We next examined whether Plin2-null mice were protected from WD-induced hepatic steatosis. As expected, WD-fed WT livers show striking histologically detectable steatosis in all lobular zones characterized by a mixture of both micro- and macro-steatosis (Fig. 2A). In contrast, histologically detectable steatosis was not observed in livers of WD-fed Plin2-null mice or in livers of CD-fed WT or Plin2-null animals. Additionally, we detected an ∼2-fold increase in relative liver weights of WD-fed WT mice compared with CD-fed WT mice or Western or control diet-fed Plin2-null mice (Fig. 2B). To confirm the differences in lipid accumulation between WT and Plin2-null mice biochemically, we performed quantitative GC-MS of fatty acid methyl esters derived from neutral lipids in liver samples from WD and CD mice. In agreement with the histologic evidence, hepatic neutral lipid levels were greatly elevated in livers of WD-fed WT mice over those of WD Plin2-null animals or CD-fed WT or Plin2-null mice (p < 0.001, two-way ANOVA) (Fig. 2C). Similarly, the cholesterol content in the neutral lipid fraction of WD-fed WT mice was markedly elevated in WD-fed Plin2-null mice or in CD-fed WT or Plin2-null mice (p < 0.001, two-way ANOVA) (Fig. 2D). Surprisingly, we also observed that total hepatic neutral lipid and cholesterol levels in livers of CD-fed WT mice were significantly elevated over those of CD-fed Plin2-null mice (p < 0.001) (Fig. 2D).
FIGURE 2.
Plin2-null animals are resistant to Western diet-induced steatosis and experience reduced hepatic lipid accumulation on both diets. A, H&E staining of WT and Plin2-null animals following 30 weeks of feeding Western (WD) and control (CD) diets. Location of portal triad (PT) and central vein (CV) regions are indicated. Scale bars, 50 μm. B, liver weights at the time of harvest for WT and Plin2-null mice fed CD or WD for 30 weeks. C, hepatic neutral lipid-derived fatty acid methyl ester (FAME) concentrations in WT and Plin2-null mice after 30 weeks of feeding CD or WD. D, hepatic cholesterol content measured by GC-MS for WT and Plin2-null mice following 30 weeks of CD or WD feeding. E, relative RNA expression of Plin2, Plin3, and Plin5 in livers of WT and Plin2-null mice fed CD or WD for 30 weeks. *, p < 0.05. F, representative images of liver sections from WT and Plin2-null mice fed WD or CD for 30 weeks and immunostained for PLIN2, PLIN3, or PLIN5. Portal triad (PT) and central vein (CV) regions are indicated for PLIN2-immunostained sections. Stellate cells are indicated by arrows. Scale bars, 50 μm.
PLIN3 and PLIN5 Do Not Compensate for Loss of PLIN2 in WD and CD Livers
Because murine livers also express PLIN3 and PLIN5 (20, 21), we were interested in determining whether loss of PLIN2 altered their expression. We first examined Plin2, Plin3, and Plin5 gene expression in both WT and Plin2-null mice fed WD or CD for 30 weeks. As established previously (13), Plin2 gene expression is not detected in livers of Plin2-null mice (Fig. 2E). Interestingly, we found that Plin2 transcripts were up-regulated ∼1.3-fold in WT mice fed WD, although this difference was not significant. Importantly, we did not observe up-regulation of Plin3 gene expression in Plin2-null mice on either diet, nor did we observe up-regulation of Plin3 in WT mice fed WD compared with CD. On WD, we observed significantly lower Plin3 gene expression in Plin2-null mice. Although Plin3 gene expression is trending down in Plin2-null mice on CD, this difference was not significant. We observed a similar pattern of Plin5 expression. Plin5 is slightly up-regulated in WT livers on WD compared with CD, but we did not observe a significant difference in Plin5 gene expression on control diet. To confirm the gene expression studies, we conducted immunohistochemical analysis of PLIN2, PLIN3, and PLIN5 in livers of WD or CD-fed WT and Plin2-null mice (Fig. 2F). Again, PLIN2 was not detected by immunofluorescence in liver sections of Plin2-null mice on either diet, further validating its deletion. In livers of WD-fed WT mice, strong PLIN2 staining was detected primarily on large CLD present in hepatocytes, whereas in livers of CD-fed WT mice PLIN2 was detected on small hepatocyte CLD and on CLD in stellate cells (Fig. 2F). We did not detect PLIN5 staining in livers of WD-fed WT mice. In livers of WD- and CD-fed WT mice, PLIN3 but not PLIN5, immunostaining was observed on CLD in stellate cells as indicated by arrows (Fig. 2F). In livers of Plin2-null animals, PLIN3 and PLIN5 were detected on CLD in hepatocytes, and Plin3 was detected on CLD in stellate cells. These data demonstrate that both PLIN3 and PLIN5 are expressed by hepatocytes and localize to CLD in the absence of PLIN2. However, neither protein is capable of compensating for PLIN2 loss in mediating hepatocyte steatotic responses to WD feeding.
Loss of PLIN2 Suppresses de Novo Lipogenic Gene Expression in Liver in WD and CD-fed Mice
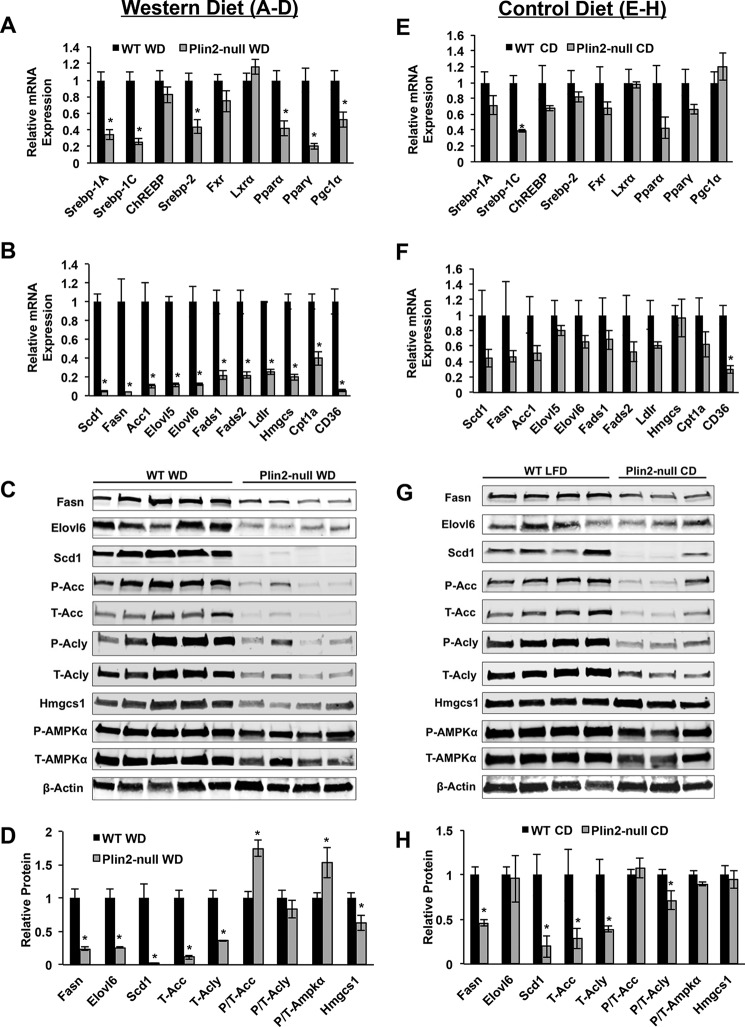
Because we observed improved glucose tolerance in Plin2-null mice on long-term WD, we suspected that induction of de novo lipogenic genes might be suppressed in Plin2-null mice. In the liver, insulin has been shown to induce SREBP1-c and activate lipogenic target gene expression. De novo lipogenesis is a major pathway in development of NAFLD both in humans and rodents fed long-term high fat, high carbohydrate diets. Additionally, reduction of hepatic Plin2 gene expression through the use of anti-Plin2 oligonucleotides in mouse models fed a long-term high fat diet has shown that de novo lipogenic gene expression is reduced (17). Given the improved glucose tolerance and previous gene expression studies in Plin2 antisense oligonucleotide mice, we suspected that SREBP-1 target genes would be reduced in Plin2-null animals. We first determined relative transcript levels of the Srebp transcription factors themselves. In livers of WD-fed mice, we found that PLIN2 deletion resulted in a 65% reduction in Srebp-1a (p = 0.0014), a 75% reduction in Srebp-1c (p = 0.0005), and a 50% reduction in Srebp-2 (p = 0.0048) mRNA (Fig. 3A). We then demonstrated drastic and statistically significant reductions in transcript levels of the entire suite of SREBP-1C target genes, including Scd1, Fasn, Acc1, Elovl5, Elovl6, Fads1, and Fads2. In most cases, PLIN2 deletion reduced SREBP-1c target gene mRNA levels by 80–90% compared with WT mice (Fig. 3B). Because Fasn and Acc1 are also targets of ChREBP, we determined the expression level of this transcription factor. We did not observe statistically significant differences in Chrebp mRNA levels in livers of WD-fed WT and Plin2-null mice (Fig. 3A). In addition to suppression of SREBP-1 target gene expression, we also determined expression levels of SREBP-2 target genes. Srebp-2 has been shown to be the primary physiologic regulator of cholesterol biosynthesis (22). Because we observed significant reduction of hepatic cholesterol in Plin2-null mice, we suspected that SREBP-2 might be altered as well. Gene expression of this transcription factor was reduced 50% in Plin2-null mice on WD compared with WT mice (Fig. 3A). Additionally, we observed 80% reductions in the transcript levels of SREBP-2 target genes, Hmgcs1 and Ldlr, in Plin2-null mice (p < 0.001) (Fig. 3B) (23). Because FXR and LXR are involved in Srebp gene transcription, we determined their gene expression responses as well. We did not observe a statistically significant difference in Fxr or Lxr gene expression. Finally, we checked both PPARγ and PPARα target gene expression. Pparγ itself is reduced 80% in Plin2-null mice on WD, and its target gene Cd36 is reduced 95%. Additionally, we observed Pparα and Cpt1a to be down 50% in the Plin2-null. This was surprising but is similar to what has been observed with anti-Plin2 antisense oligonucleotide treatment (17).
FIGURE 3.
PLIN2 deletion suppresses hepatic de novo lipogenic gene/protein expression in Western and control diet-fed mice. A–D, hepatic levels of transcripts and proteins in WD-fed mice. A, transcription factor genes. B, transcription factor target genes Scd1, Fasn, Acc1, Elovl5, Elovl6, Fads1, and Fads2 are targets of SREBP-1a and SREBP-1c. Ldlr and Hmgcs1 are target genes of SREBP-2. Cpt1α is a target of Pparα. Cd36 is a target gene of PPARγ. C, immunoblot analyses of SREBP-1- and SREBP-2-regulated lipid metabolism enzymes. D, immunoblot quantification of lipid metabolism enzymes and P-AMPKα to T-AMPKα ratios. E–H, hepatic levels of transcripts and proteins in CD-fed mice. E, transcription factor genes. F, transcription factor target genes. G, immunoblot analysis of SREBP-1- and SREBP-2-regulated lipid metabolism enzymes. H, immunoblot quantification of lipid metabolism enzymes and P-AMPKα to T-AMPKα ratios. *, p < 0.05.
In light of the reduction in hepatic neutral lipids and cholesterol associated with Plin2 loss in CD-fed mice, we hypothesized that Srebp target genes would be also reduced in livers of CD-fed Plin2-null mice. Both Srebp-1a and Srebp-1c mRNAs are reduced in Plin2-null livers, with the reduction in Srebp-1c being highly significant (p < 0.001) (Fig. 3E). Srebp-2 mRNA is trending downward in Plin2-null livers on CD, but this difference was not significant. Chrebp, Fxr, Lxr, Pparα, Pparγ, and Pgc1α were not statistically different between WT and Plin2-null livers on control CD. Among the entire suite of SREBP-1 target genes, we observe trending decreases of all genes, including Scd1, Fasn, Acc1, Elovl5, Elovl6, Fads1, and Fads2 (Fig. 3F). However, these decreases were not significant at the mRNA level. Among SREBP-2 targets, we observed a trending decrease for Ldlr but not for Hmgcs1. Interestingly, we did observe a significant 50% decrease in Cd36 mRNA. These data suggest that PLIN2 may contribute to the regulation of lipid metabolism genes even in the absence of obesity, when physiologic levels of lipid are present within the hepatocyte.
Protein Levels of Lipogenic Enzymes Are Significantly Reduced in Plin2-null Mice
Because Srebp-1 and Srebp-2 target proteins do not always correlate with mRNA expression, we performed extensive Western blotting analysis of SREBP target enzymes (Fig. 3, C and D). As expected, we observed remarkable decreases in SREBP-1 target protein quantities in liver extracts of WD-fed Plin2-null mice compared with levels in extracts of WT mice fed the WD. On WD, FASN levels were reduced in livers of Plin2-null mice by 75% (p = 0.0016) compared with WT levels, and ElOVL6 protein levels were reduced by a similar amount (p = 0.0019). Consistent with gene expression studies, we observed an astonishing 98% decrease (p = 0.0054) in levels of SCD1 in liver extracts of Plin2-null mice, with this enzyme being barely detectable by Western blotting. In Plin2-null mice on WD, total ACC levels are reduced 90% (p = 0.0003), whereas phospho/total ACC is increased by almost 1.5-fold (p = 0.0019). Because we observed this increase in phospho/total ACC, we also examined phospho/total AMPKα. Phospho/total AMPKα is increased by 70% in Plin2-null animals on WD (p = 0.0495). In addition, we quantified ATP citrate lyase (ACLY) protein levels because it is also a validated SREBP-1 target and an important link between carbohydrate metabolism and lipid biosynthesis (24, 25). Interestingly, we found total ACLY levels to be decreased 65% in Plin2-null mice on WD (p = 0.0024), although the phospho/total ACLY levels were not significantly different between the two genotypes. Finally, we checked protein expression levels of HMGCS1, a well validated target protein of SREBP-2 (23), and observed a 38% decrease in this enzyme in Plin2-null animals on WD compared with WD-fed WT mice (p = 0.0254).
Because we found such great differences in SREBP target gene expression on WD, we also checked both SREBP-1 and SREBP-2 target proteins via Western blotting for CD-fed animals (Fig. 3, G and H). As noted, we observed downward trends for most of these target genes, but the decreases were significant only for Srebp-1C itself and Cd36. To our surprise, protein levels of most SREBP-1 target proteins were significantly reduced in liver extracts of CD-fed Plin2-null mice compared with CD-fed WT mice. FASN was found to be reduced 50% (p = 0.0034), and SCD1 was decreased 75% (p = 0.0436). Although gene expression predicted we might observe a decrease in ELOVL6, this was not the case as there was no difference in the levels of this enzyme between the genotypes. A 65% reduction of total ACC was observed in Plin2-null animals on CD (p = 0.0425), whereas no difference was observed between the genotypes for phospho/total ACC. Accordingly, we did not observe any statistical difference in phospho/total AMPKα on this diet. Interestingly, we also observed a 50% reduction in total ACLY for Plin2-null on CD (p = 0.0369) as well as a slight decrease in phospho/total Acly (p = 0.0499) for animals lacking PLIN2. As expected, based on Srebp-2 gene expression on CD, we did not observe any difference in levels of HMGCS1 on this diet.
Loss of PLIN2 Results in Altered Hepatic Neutral Lipid Profiles in Plin2-null Mice Consistent with Reduction in SREBP Activity
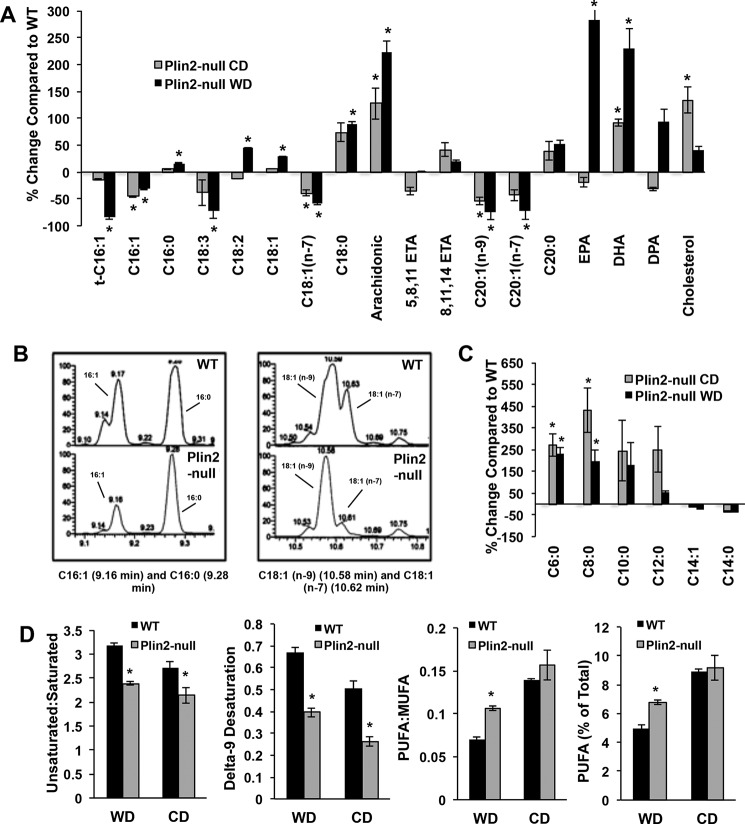
Because we observed significant changes in SREBP-1 and SREBP-2 target gene and protein expression, we suspected that loss of PLIN2 was preventing induction of de novo lipogenesis in Plin2-null mice on long-term WD as well as the CD. To better define the effects of PLIN2 deletion on this process at the physiologic level, we quantified lipid species in livers of WT and Plin2-null mice fed WD or CD for 30 weeks. Using a GC-MS lipidomics approach, we found that Plin2-null animals experience less desaturation and elongation of neutral lipid species on both diets. We first looked for evidence of steroyl-CoA desaturase 1 (SCD1) activity in neutral lipid profiles. We noticed a marked decrease in palmitoleic acid (C16:1) as a percentage of total neutral lipid in Plin2-null mice on both diets, with this particular species being reduced 31% on WD and 45% on CD compared with WT animals (Fig. 4A). Palmitoleic acid is one of the major products of SCD1 (and SCD2) activity and, like oleic acid, is formed by introducing n-9 desaturation in palmitic acid (C16:0) and stearic acid (C18:0) (26). SCD1 is the primary SCD isoform in the liver, with SCD2 expression reported as being extremely low (27). Because very little palmitoleic acid is found in either the WD or the CD (supplemental Table S1), we used the ratio of C16:1 to C16:0 as a measure for SCD1 activity. Differences in this ratio were particularly evident in the GC-MS chromatograms (Fig. 4B). WT animals on WD exhibit almost a 1:1 ratio of palmitoleic/palmitic acid in neutral lipids, whereas this ratio is close to 1:3 for Plin2-null animals on the same diet. We observed similar ratios for animals on CD (data not shown). We also calculated the δ-9 desaturation index for WT and Plin2-null animals on both diets (Fig. 4D). On WD, WT animals exhibit an index score of ∼0.7, whereas Plin2-null mice have an index score of 0.4 (p = 0.0003), indicating significantly less n-9 desaturase activity in knock-out animals. Likewise WT animals on the CD had a δ-9 desaturation score of 0.5, whereas the score for Plin2-null animals was 0.3 (p = 0.0017). Not unexpectedly, we noticed that δ-9 desaturation increased for both genotypes on WD compared with CD, indicating a dietary difference in SCD1 induction regardless of genotype. Furthermore, we calculated total unsaturated/saturated ratios for both WT and Plin2-null mice on each diet for hepatic neutral lipids. On WD, this ratio was found to be ∼3.25 for WT and ∼2.5 for Plin2-null (p < 0.0001), indicating more overall lipid desaturation in WT animals. Similarly, for CD-fed mice, the unsaturated/saturated species ratio was 2.75 for WT mice and 2.1 for Plin2-null animals (p = 0.0363), indicating the same trend for decreased desaturation in Plin2-null animals even on CD. Importantly, using this analytical approach, we observe a consistent reduction in δ-9 desaturase activity in Plin2-null animals on both diets, which is consistent with data from gene and protein analysis (Fig. 4D).
FIGURE 4.
PLIN2 deletion alters hepatic neutral lipid profiles in CD- and WD-fed mice. A, percent change compared with WT of neutrally derived long chain fatty acids and cholesterol. WT is set at zero. B, GC-MS chromatograms showing hepatic neutral lipid profiles for individual WT (top) and Plin2-null (bottom) mice fed the WD for 30 weeks. The left panel shows the peaks for palmitoleic acid (C16:1) at 9.16 min retention and palmitic acid (C16:0) at 9.28 min. The ratio of C16:1 to C16:0 is greatly reduced in the Plin2-null profile indicating less SCD1 activity. The right panel shows the peaks for oleic acid (C18:1 n-9) at 10.58 min of retention compared with vaccenic acid (C18:1 n-7) at 10.62 min. The ratio of neutral lipid-derived oleic to vaccenic acids is greatly increased in Plin2-null mice, signifying less elongation. C, percent change compared with WT of neutral lipid-derived SCFAs in Plin2-null mice on both WD and CD. WT is set at zero. D panels show ratio of neutral lipid-derived unsaturated to saturated fatty acids; δ-9 desaturation indices; ratio of PUFA to MUFA; and PUFA as a percentage of total neutral lipid in the neutral lipid fraction from livers of WT and Plin2-null mice fed WD or CD for 30 weeks. *, p < 0.05.
We also expected decreased elongase activity in Plin2-null animals on WD, given gene expression and Western blotting data. First, we looked at levels of short chain fatty acids (SCFAs) in neutral lipids of WT and Plin2-null mice on both diets. Because milk fat is the lipid source for both diets, the WD and the CD contain significant amounts of SCFAs (supplemental Table S1). Because hepatic FASN is elevated in livers of WT mice compared with Plin2-null mice on both Western and control diets, we expected that levels of SCFA would be reduced in WT livers due to increased FASN-dependent elongation activity. Accordingly, we observed decreases in C6:0 through C12:0 species as a percent of total neutral lipids in livers of WT mice compared with levels in livers of Plin2-null animals on both diets, with increases in both C6:0 and C8:0 being statistically significant (Fig. 4C and supplemental Tables S3 and S4).
ELOVL6 is the enzyme responsible for elongating fatty acids containing 12, 14, and 16 carbons, and this important SREBP-1 target has been shown to be a crucial element in development of dietary insulin resistance and development of NAFLD (28–31). To look for evidence of altered ELOVL6 activity in the neutral lipid profile, we first examined levels of vaccenic acid (C18:1 n-7). This product results from elongation of palmitoleic acid (C16:1 n-7) and is closely related to oleic acid (C18:1 n-9). Consistent with markedly reduced ELOVL6 expression in livers of WD-fed Plin2-null mice, we found that vaccenic acid levels in their neutral lipid fraction were significantly lower than those from WT mice (p < 0.01). This increase in vaccenic acid was prominent in the GC-MS chromatograms for WD animals (Fig. 4B, right panel). The ratio of oleic acid to vaccenic acid is 1:0.75 in WT animals, whereas this ratio is roughly 1:0.2 in Plin2-null animals on WD, which is consistent with greater hepatic elongase activity in WT compared with Plin2-null animals on WD. Importantly, we observed similar decreases of vaccenic acid as a percentage of total neutral lipids in Plin2-null compared with WT animals fed the CD. Vaccenic acid is reduced 39% in Plin2-null animals on CD (Fig. 4A). Although we observed trending reduced ELOVL6 gene expression in Plin2-null livers on CD, we were not able to detect a statistically significant difference at the protein level. ELOVL5 has also been shown to produce vaccenic acid from palmitoleic acid in hepatocytes (32, 33), so it is possible that this enzyme is significantly reduced in Plin2-null animals on CD. We observed striking decreases in Elovl5 gene expression for knock-out animals on WD. Finally, we also quantified levels of C20:1 n-9 and C20:1 n-7 as markers for altered elongase activity. C20:1 n-9 represents simple elongation of oleic acid, whereas C20:1 n-7 represents elongation by two carbons of vaccenic acid. As expected, we observed significantly reduced levels of these species in Plin2-null livers on both diets (Fig. 4A and supplemental Tables S3 and S4). On WD, C20:1 n-9 is reduced by 75% in knock-out animals compared with WT, but it is decreased by 54% on CD. Similarly, C20:1 n-7 is reduced by 73% in Plin2-null mice on WD with a decrease of 43% on CD. Given this analysis, we observed significant reduction in elongase activity in Plin2-null mice on both diets consistent with gene and protein expression of SREBP target genes.
Interestingly, we observed extremely significant increases in long chain polyunsaturated fatty acids (LC-PUFA) as a percentage of total neutral lipid in Plin2-null animals on WD. As a percentage of total lipid on WD, PUFAs account for ∼7% of total neutral lipid in Plin2-null mice, whereas these species account for 4.9% of total neutral lipid in WT mice (p = 0.001) (Fig. 4D). At the individual lipid species level, we observed increases in neutral lipid-derived linoleic acid (44% increase), arachidonic acid (224% increase), 8,11,14-eicosatrienoic acid (20% increase), eicosapentaenoic acid (283% increase), docosahexaenoic acid (230% increase), and docosapentaenoic acid (95% increase) (Fig. 4A and supplemental Table S2). Additionally, the polyunsaturated/monounsaturated (PUFA/MUFA) ratio in Plin2-null mice on WD is 0.11, whereas this ratio is 0.07 in WT mice (p = 0.0004) (Fig. 4D). This is counter to what we had expected from gene expression and protein analysis. Plin2-null animals exhibit statistically significant reduction of Fads1 and Fads2 mRNA levels (Fig. 3B), and they also showed decreased Elovl2 and Elovl3 gene expression (data not shown). FADS1, FADS2, ELOVL2, and ELOVL3 are important enzymes for synthesizing very long chain PUFA from linolenic and linoleic acid (30, 34, 35). On CD, we observed a slight trending increase in PUFA/MUFA for Plin2-null mice, but we did not observe a statistically significant increase in PUFA as a percentage of total lipid (Fig. 4D). However, at the individual species level, we did observe significant increases in neutral lipid-derived arachidonic acid (128% increase), 8,11,14-eicosatrienoic acid (42% increase), and docosahexanoic acid (92% increase) (Fig. 4A). Therefore, the loss of PLIN2 drastically alters the neutral lipid-derived LC-PUFA profile on WD and to a lesser extent on CD at the individual species level.
Loss of PLIN2 Alters ER Membrane Lipid Composition and Likely Suppresses SREBP-1 and SREBP-2 by This Mechanism
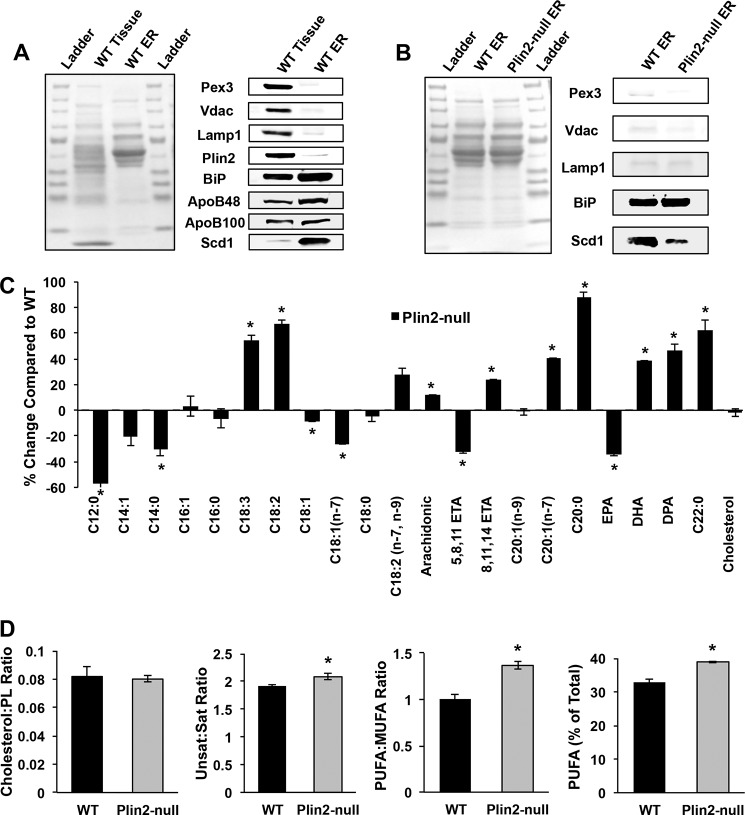
Because we observed evidence of decreased SREBP-1 and SREBP-2 activity in Plin2-null mice on both diets at the gene expression, protein, and neutral lipid profile levels, we reasoned that PLIN2 deletion may lead to altered membrane lipid properties known to regulate SREBP activation (36, 37). To test this hypothesis, we determined the lipid composition in isolated microsomal membrane fractions from livers of WD-fed WT and Plin2-null mice. We confirmed the purity of microsome preparation by assessing both whole tissue and microsomal proteins in quantitative Western blotting assays. Almost no PEX3 protein was observed in isolated microsomes, indicating that our fractions did not contain significant peroxisomal contamination (Fig. 5A). We similarly observed negligible VDAC and LAMP1 proteins (markers for mitochondria and lysosomes, respectively) in isolated microsomal membrane fractions. Importantly, there was negligible Plin2 protein in microsomal fractions, indicating that lipid droplet-derived phospholipids would not interfere with microsomal membrane phospholipid analysis. We observed a slight enrichment of GRP78 (BiP), a well known ER chaperone in our microsomes compared with whole tissue. The limited extent of enrichment of this ER chaperone is likely because it is a soluble species, which is probably lost due breakage of the microsomes during their isolation. However, it is possible that some of the microsomal membrane GRP78 was associated with proteins by virtue of their translation at the ER membrane. Similarly, we observed only a very slight enrichment of ApoB48 and ApoB100 in our microsomal membrane fractions. Murine livers secrete VLDL with both ApoB isoforms (38). Similar to BiP, it is likely that most of the soluble ApoB48/100 (in the form of VLDL) was lost upon rupture of the microsomes during centrifugation. In contrast, protein levels of the ER membrane enzyme SCD1 exhibited a substantial (∼70-fold) enrichment (Fig. 5A) in our microsomal membrane fractions. Based on these data, we concluded that our isolated microsomal membranes were highly enriched and predominantly composed of ER membranes rather than luminal ER contents.
FIGURE 5.
PLIN2 deletion alters hepatic microsomal membrane lipid composition in WD-fed mice. A, microsomal membrane properties. Protein (left panel) and immunoblot staining (right panel) profiles of starting liver extracts and isolated microsomal fractions containing 37.5 μg of protein from WD-fed WT mice. Antibodies to markers of peroxisomes (PEX3), mitochondria (VDAC), lysosomes (LAMP1), cytoplasmic lipid droplet (PLIN2), endoplasmic reticulum lumen (BiP/Grp78), membrane (SCD1), and VLDL vesicles (ApoB48/ApoB100) were used to estimate microsomal membrane enrichment and contamination by other cellular fractions. B, comparison of microsomal membrane preparations from WD-fed WT and Plin2-null mice. Representative protein (left panel) and immunoblot staining profiles (right panel) for 37.5 μg of protein from microsomal membrane fractions isolated from livers of WD-fed WT and Plin2-null mice. Immunoblots show staining profiles for PEX3, VDAC, LAMP1, GRP78, and SCD1. C, percent change of microsomal membrane phospholipid species and cholesterol of Plin2-null mice compared with WT. WT is set at zero. Asterisks indicate values that are significantly different from those of WT mice (p < 0.01, two-way ANOVA). D, panels show cholesterol to phospholipid ratio; ratio of unsaturated fatty to saturated fatty acids; ratio of PUFA to MUFA fatty acids; and polyunsaturated fatty acids as a percent of total phospholipid species in microsomal fractions from livers of WD-fed WT and Plin2-null mice. Asterisks indicate values that are significant different from those of WT mice (p < 0.05, two-way ANOVA).
The microsomal membrane proteins derived from WT and Plin2-null mice were then compared. As with the WT microsomes, we observed no significant contamination of peroxisomes, lysosomes, or mitochondria in Plin2-null microsomal membrane fractions (Fig. 5B). We also observed similar levels of GRP78 in microsome membranes from the two genotypes. However, we observed significantly more SCD1 in microsomal membranes from WT mice on long-term WD compared with Plin2-null mice. To assess membrane lipid composition, we performed GC-MS lipidomics on isolated microsomal membranes. We did not observe any difference in total membrane lipids per mg of protein. We first suspected we might observe increased ER membrane cholesterol in Plin2-null mice because loss of PLIN2 might direct more of this lipid species into cellular membranes. Increased cholesterol could explain SREBP suppression, as this has been shown to be a classic regulation mechanism of the SREBP family (24, 39). However, we observed no difference in the cholesterol/phospholipid ratio between the two genotypes, thus obviating the simple explanation that increased cholesterol might lead to SREBP suppression (Fig. 5D). We then looked at relative lipid species contained in the microsomal membrane phospholipids (PL). To our surprise, we observed striking alterations in ER membrane's PL composition in microsomes isolated from Plin2-null mice compared with WT (Fig. 5C and supplemental Table S5), with LC-PUFAs being significantly increased in the Plin2-null membranes. The PL profile of Plin2-null ER membranes is very different from that of the neutral lipid pool with substantial increases in PUFA observed. Compared with WT species as a percentage of total lipid, we found significant increases in linolenic acid (55% increase), linoleic acid (68% increase), conjugated linoleic acid (28% increase), arachidonic acid (12% increase), 8,11,14-eicosaenoic acid (24% increase), docosahexaenoic acid (38% increase), and docosapentaenoic acid (47% increase) in microsomal membranes from Plin2-null mice. Additionally, we observe a slight but statistically significant (p = 0.0413) increase in the unsaturated/saturated lipid species in Plin2-null microsomal membranes compared with WT mice (Fig. 5D). Importantly, we observed significant increases in PUFA/MUFA as well as PUFA as a percent of total phospholipid in Plin2-null microsomal membranes relative to membranes from WT mice (Fig. 5D). Because we observe striking changes in ER membrane PUFA in Plin2-null mice, we hypothesize that this is partly responsible for SREBP suppression in these animals. Abundant data from multiple laboratories have shown that increased PUFA content inhibits SREBP induction both at the membrane level by preventing proteolytic processing of SREBP as well transcription of the SREBP family by inhibiting binding of the LXR/retinoic X receptor dimer to the SREBP promoter (26, 36, 40–43).
Loss of PLIN2 Prevents Diet-induced ER Stress
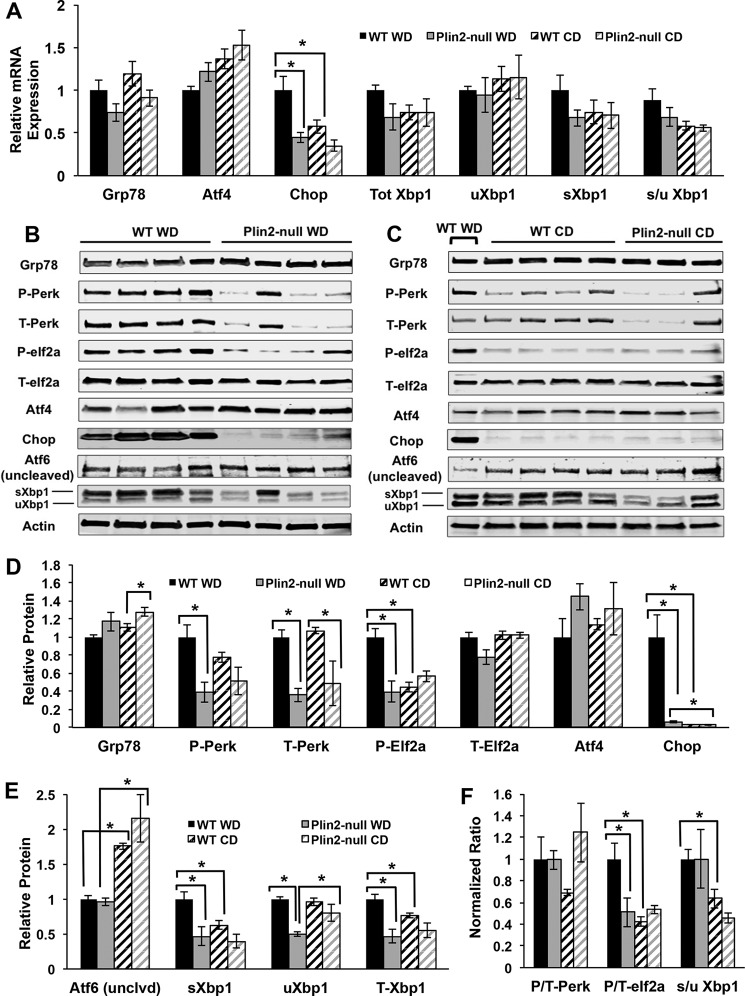
Increased hepatic ER stress has been observed in mice with diet-induced and genetic predisposition to obesity (44, 45). Because we observed changes in ER membrane lipid composition, we were interested in whether we might observe differences in ER stress activation between the two genotypes. Increases in saturation of ER membrane lipids, such as those observed in our WT animals on WD, have been shown to cause an ER stress response that can induce XBP1 and further enhance lipogenesis (46–48). Furthermore, changes in membrane lipid saturation have been shown to influence IRE1 and PERK activities via effects on their transmembrane domains (46), with increased membrane saturation inducing an ER stress response independent of their luminal stress-sensing domains. Because we demonstrated significant increases in PUFA as well as an overall increase in the unsaturated/saturated ratio in Plin2-null ER membrane lipids, we hypothesized that we might observe evidence of reduced ER stress in Plin2-null animals compared with WT. We tested this hypothesis by first quantifying transcript levels of ER stress markers Grp78, Atf4, Chop, total Xbp1, unspliced Xbp1 (uXbp1), spliced Xbp1 (sXbp1), and spliced/unspliced Xbp1 (s/u Xbp1) for WT and Plin2-null animals on WD and CD (Fig. 6A). We did not observe statistically significant differences in Grp78 or Atf4 transcript levels. However, consistent with induction of ER stress in WD-fed WT mice, we found a significant increase in Chop mRNA compared with CD-fed mice (p = 0.0481). Chop mRNA levels in Plin2-null animals on WD were significantly lower those in WD-fed WT mice (p = 0.0165), indicating reduced ER stress in these animals. WD-fed WT animals also exhibited a trending increase in total Xbp1 and spliced Xbp1 compared with their CD-fed counterparts and to WD- or CD-fed Plin2-null mice, but this did not reach significance. Additionally, the spliced/unspliced Xbp1 ratio was trending higher in WT animals on WD compared with Plin2-null animals on WD.
FIGURE 6.
Loss of PLIN2 protects mice from WD-induced ER stress. A, gene expression analysis of ER stress markers Grp78, Atf4, Chop, total Xbp1, uXbp1, sXbp1, and the ratio of spliced/unspliced Xbp1 (s/u Xbp1) for WT and Plin2-null mice on Western and control diets. Asterisks indicate significant changes (p < 0.05, Student's t test). B, Western blotting analyses of GRP78, phospho-PERK, total PERK, phospho-EIF2a, total EIF2a, ATF4, CHOP, and uncleaved (full-length) ATF6 for WT and Plin2-null mice on WD. C, Western blotting analyses of GRP78, phospho-PERK, total PERK, phospho-EIF2a, total EIF2a, ATF4, CHOP, and uncleaved (full-length) ATF6 for WT and Plin2-null mice on control diet. The 1st lane contains a representative WT animal on WD. D, quantification of GRP78, phospho-PERK, total PERK, phospho-EIF2a, total EIF2a, ATF4, and CHOP protein in WT and Plin2-null animals on Western and control diets. Asterisks indicate significant differences (p < 0.05). E, quantification of ATF6 (full-length), spliced XBP1 (sXBP1), unspliced XBP1 (uXBP1), and total XBP1 (T-XBP1) protein in WT and Plin2-null animals on Western and control diets. Asterisks indicate significant differences (p < 0.05). F, normalized ratios of phospho/total PERK (P/T-PERK), phospho/total EIF2a (P/T-EIF2a), and spliced/unspliced XBP1 (s/u XBP1) in WT and Plin2-null animals on Western and control diets. WT animals on WD are set to 1. Asterisks indicate significant differences (p < 0.05).
To further validate differences in ER stress responses between WT and Plin2-null mice, we performed Western blotting analysis of ER stress proteins GRP78, phosphorylated PERK (P-PERK), total PERK (T-PERK), phosphorylated EIf2a (P-EIF2a), total eIf2a (T-EIF2a), ATF4, CHOP, uncleaved ATF6, and XBP1 (spliced and unspliced) in WT and Plin2-null animals on Western and control diets (Fig. 6, B–F). We did not detect significant differences in GRP78 levels between WD- and CD-fed WT or between WD-fed WT or Plin2-null mice. However, Plin2-null animals on CD had slightly elevated GRP78 compared with WT animals on CD (p = 0.0472). In contrast, we found significant, 60%, reductions in the levels of total (p = 0.0009) and P-PERK (p = 0.0113) in WD-fed Plin2-null animals compared with WD-fed WT animals. Interestingly, we also found that T-PERK levels were reduced by 50% (p = 0.0418) in Plin2-null compared with WT animals fed the control diet, but we did not detect significant differences in P-PERK levels between WT and Plin2-null mice on this diet. Overall, the P-PERK to T-PERK ratio for WT animals did not differ from that of Plin2-null mice on the WD and was marginally (but not statistically) elevated over WT mice on the CD. Consistent with increased PERK levels in WD-fed WT mice, phospho-eIf2a (the product of PERK activity) was elevated 2.5-fold (p = 0.0021) over that of CD-fed WT mice and WD-fed Plin2-null animals (p = 0.0065). Total EIF2a levels were not affected by diet or PLIN2 deletion (Fig. 6D), which resulted in a selective increase in the P-EIF2a/T-EIF2a in WD-fed WT mice compared with WD-fed Plin2-null mice (p = 0.0491) (Fig. 6F). Additionally, we observed a 2-fold increase in the P/T-EIF2a ratio in WD-fed WT mice compared with CD-fed WT mice (p = 0.01). We did not detect differences in the effects of PLIN2-deletion or diet on ATF4, whose levels are regulated by EIF2a activity. However, we did find a striking 17-fold increase in the levels of CHOP, a target of ATF4 activity, in WD-fed WT mice compared with Plin2-null animals on the same diet (p = 0.0081) and a 30-fold increase (p = 0.0071) compared with WT animals on control diet. We also found that in livers of WD-fed Plin2-null mice that CHOP levels were approximately twice that of those fed the control diet (p = 0.0284). Collectively, these data are consistent with PLIN2 deletion interfering with activation of the PERK ER stress pathway caused by WD feeding.
Next, because we observed differences in the PERK-CHOP pathway, we sought to probe whether changes in the IRE1-XBP1 and ATF6 pathways would also be observed. At the protein level, we nevertheless observed a statistically significant increase in spliced XBP1 (sXBP1) in WD-fed WT mice compared with WT mice fed CD (p = 0.0219) (Fig. 6, B, C and E). Importantly, we also observed a 2-fold increase in sXBP1 in WD-fed WT mice compared with WD-fed Plin2-null mice (p = 0.0218) (Fig. 6E). For unspliced XBP1 (uXBP1), there was a 2-fold increase in WT animals fed WD compared with Plin2-null animals fed that diet (p < 0.0001). Plin2-null animals on CD experience a marginal increase in uXBP1 compared with Plin2-null animals on WD (p = 0.0411). Interestingly, we observed differences in total XBP1 (T-XBP1) between the two genotypes, with T-XBP1 increased 2-fold in WT mice fed WD compared with Plin2-null animals fed WD (p = 0.0044). WT mice on WD also experience a slight but significant increase in T-XBP1 compared with WT mice on CD (p = 0.0273). Because of total XBP1 being different between the genotypes, there was no difference in the spliced/unspliced XBP1 ratio (s/u XBP1) on WD (Fig. 6F). However, there was a 1.7-fold increase in the s/u XBP1 ratio in WD-fed WT mice compared with CD-fed WT mice (p = 0.0276). In summary, we show definitive changes in the PERK-CHOP and XBP1 ER stress pathways, with loss of PLIN2 preventing activation of these pathways on WD by an unconventional mechanism, reduced levels of total PERK and reduced total XBP1.
We were unable to definitively establish effects of diet or PLIN2 deletion on ATF6 activation, because our antibodies only detected its uncleaved form. However, we found a 40% reduction in the inactive uncleaved form of ATF6 (p < 0.0001) in livers of WD-fed compared with CD-fed WT animals. In addition, we found a 60% reduction in uncleaved ATF6 in WD compared with CD-fed Plin2-null mice (Fig. 6, B–D). Although these data suggest that the ATF6 pathway may be activated for both genotypes on WD compared with CD, additional work is needed to define the specific effects of diet and PLIN2 deletion on ATF6 activation.
Discussion
Despite intense study, the specific molecular mechanisms of how PLIN2 regulates hepatic lipid accumulation under both physiologic as well as pathophysiologic conditions have remained elusive. Previous studies using antisense oligonucleotides targeting PLIN2 in ob/ob mice and in mice fed high fat diets suggested that SREBP target genes might be altered upon loss of PLIN2 (11, 12, 17). In this study, we show that both SREBP-1 and SREBP-2 target genes do indeed experience extreme alterations with loss of PLIN2. These changes are evident at the gene transcript level, protein levels of the SREBP target enzymes themselves, and in the hepatic neutral lipid profiles of Plin2-null mice. Although we expected reductions in total neutral lipids in Plin2-null animals, we had not anticipated the global alterations in neutral lipid profiles. Similarly, the observation that loss of PLIN2 leads to a reduction in hepatic neutral lipid levels and profiles even on control diet was unexpected. The change in neutral lipid profiles in the Plin2-null animals on both diets is most likely due to the observed changes in SREBP-1 gene and protein expression. In addition to documenting these changes in SREBP target expression, we also sought to probe the mechanisms by which SREBPs are altered in these mice. Our data suggest that alterations in ER membrane LC-PUFA content are largely responsible for SREBP suppression in Plin2-null mice. Previous studies examining the loss of PLIN2 in relation to NAFLD development have largely ignored the effects on animals fed control diets. Although it was shown that whole-body knock-out of Plin2 by targeting exons 2/3 led to a reduction in hepatic TG even on a chow diet (49), changes in the neutral lipid profile were not observed in this model. This could possibly be due to incomplete knock-out of the PLIN2 protein (50) and/or differences in dietary sucrose. Additionally, the majority of studies examining the effect on hepatic TG accumulation with the loss of Plin2 have been performed in the context of leptin deficiency (11, 18) and high fat diet-only feeding studies (12, 17). Thus, complete characterization of the gene, protein, and lipid profile changes on control diet represents a novel area of study with regard to the role of PLIN2 in normal hepatic lipid metabolism.
Effects of Insulin Activity on SREBP Regulation
Insulin has been shown to exert effects on transcription and activation of SREBP-1c (12, 51). Indeed, we observed impaired glucose tolerance for WT mice on WD. Therefore, it is likely that the hyperinsulinemia and insulin resistance in WT mice on WD is partly responsible for the drastic difference in SREBP target gene expression between our two genotypes on this diet. This is consistent with previous work showing that reduction of PLIN2 by the use of antisense oligonucleotides improves liver insulin sensitivity, insulin resistance, and reduction of SREBP-1 target genes (17). Importantly, however, we also observe suppression of SREBP-1 target genes even on control diet. On this diet, we did not observe differences in glucose tolerance despite a reduction in SREBP targets at the mRNA, protein, and lipid profile levels. It is therefore unlikely that impaired insulin activity accounts for SREBP suppression in Plin2-null mice on the control diet. Moreover, it has also been shown that increased activation of AMPK can directly phosphorylate and inhibit SREBP (52). Indeed, we observe increased phospho/total AMPKα in Plin2-null mice on WD. So, on this particular diet, increased AMPKα activity likely explains some of the suppression of SREBP activity in knock-out mice. Again, however, we did not observe differences in AMPKα activation between our two genotypes on the CD.
Contributions of LC-PUFA in SREBP Regulation
It has been hypothesized that altered ER membrane phospholipid composition and increases in LC-PUFA can regulate SREBP family members both at the ER membrane level as well as transcription of the SREBP family members themselves (26, 40, 41, 53). Because we observe alterations in LC-PUFAs in the neutral lipid fractions on both WD and CD, we hypothesize that altered ER membrane PUFA content is regulating SREBP activity in Plin2-null animals. Accordingly, we demonstrated drastic increases in PUFA content in microsomal membrane fractions of Plin2-null mice on WD compared with WT mice. We also observe significantly decreased SREBP-1 and SREBP-2 mRNA expression. Membrane-derived PUFA can be transported to the nucleus where they interfere with LXR/RXR dimer binding to the promoter region of the SREBP transcription factors (42). PUFAs have also been shown to inhibit maturation of SREBP at the ER membrane level (36, 37). Specifically, it has been shown that arachidonic acid inhibits proteasomal degradation of INSIG-1 by leading to membrane retention of the ubiquitinated protein. Retention of INSIG-1 in the ER membrane enhances sterol-mediated activation of SREBP and subsequent translocation of these transcription factors to the Golgi apparatus. ω-3 PUFAs have also been shown to exert effects on transcription of SREBP via activation of FXR (54). ω-3 FFAs can activate FXR, which leads to induction of SHP (a well known inhibitor of LXR-mediated transcription of the SREBP family), although we did not observe changes in the levels of LXR itself. Importantly, docosahexaenoic acid (DHA) specifically has been shown to suppress de novo lipogenesis, inhibit PUFA biosynthesis, decrease both SREBP-1 and ChREBP nuclear abundance, and decrease AKT phosphorylation (53). Additionally, DHA has been shown to accelerate the rate of SREBP-1 degradation via an ERK and 26S proteasome-mediated mechanism (55). Finally, it has been suggested that changes in membrane saturation can activate the unfolded protein response (UPR) and contribute to ER stress (46). Activation of the UPR can enhance lipogenesis by increasing levels of XBP1, which has been shown to be an important component regulating phospholipid biosynthesis and ER remodeling (47, 48). It is possible that the increased microsomal PUFA content in Plin2-null mice suppresses the ER stress response and abrogates induction of lipogenic pathways. Because we observed increased PUFAs, including ω-3 species, in Plin2-null animals, it is likely that SREBP is being inhibited through these species.
Further study is needed to elucidate the cause for the increased LC-PUFA in Plin2-null animals. One possibility is that Plin2-null animals could be synthesizing more LC-PUFA such as arachidonic acid and/or DHA compared with WT animals. However, because Fads1, Fads2, Elovl2 (data not shown), and Elovl3 (data not shown) mRNA expressions are all decreased in Plin2-null animals compared with WT on both diets, it is unlikely that loss of PLIN2 leads to increased hepatic synthesis of LC-PUFA from linoleic or linolenic acid. It has also been demonstrated that a high fat diet can increase reactive oxygen species (ROS) production in rodents (56, 57). PUFAs are extremely sensitive to oxidative damage, with each additional double bond increasing the probability of destruction exponentially (56). It is possible that Plin2-null animals experience reduced mitochondrial ROS production compared with their WT counterparts. We observed reduced Pgc1a and Cpt1a expression in Plin2-null mice, so it is possible they could be experiencing less β-oxidation with a concomitant reduction in ROS. It is also possible that Plin2-null animals are producing less oxidized eicosanoids compared with WT mice. For example, Plin2-null animals might be producing less pro-inflammatory eicosanoids such as hydroxyeicosatetraenoic acids thereby leading to an accumulation of LC-PUFA species that would otherwise be used for this purpose.
We also considered the possibility that the increased LC-PUFA in Plin2-null mice could be due to increased lipolysis in the adipose tissue and/or enhanced delivery of adipose-derived PUFA to the liver. However, fasting serum FFA concentrations are significantly reduced in Plin2-null mice on WD. This is likely due to increased insulin sensitivity on this diet, and it suggests that lack of whole-body PLIN2 might actually decrease adipose lipolytic activity. Indeed, we observed a similar magnitude reduction of fasting FFA in Plin2-null animals even on a standard chow diet (data not shown). It is also unlikely that livers of Plin2-null mice exhibit increased PUFA uptake. We observe a significant and substantial reduction in hepatic Cd36 (FAT) mRNA expression in livers of Plin2-null mice on both diets, suggesting that enhanced delivery of periphery-derived LC-PUFA to the livers of these animals is unlikely. However, loss of Plin2 in adipose tissue could be altering the lipid profile in that tissue (and hence the profile of the exported FFAs to other organs), and we plan to explore this possibility in future studies. Differences in VLDL secretion rates might also explain differences in microsomal membrane PUFA content. Animals treated with anti-Plin2 antisense oligonucleotides show reduced rates of VLDL secretion (17), which may lead to an accumulation of PUFA that would otherwise be exported as part of VLDL phospholipids in WT animals. Finally, more work is needed to elucidate whether the enrichment in phospholipid PUFA is limited to certain species of phospholipid or whether it is an effect across all PL species. We are currently undertaking a liquid chromatography mass spectrometry (LC-MS) approach to help clarify this.
Effects of ER Membrane Lipid Saturation on ER Stress
It has been shown previously that ER membrane lipid saturation is an important determinant of ER stress (58–60). Studies have suggested that the mechanism by which membrane lipids activate the stress response might be independent of the traditional GRP78-mediated unfolded protein-sensing mechanism (46), and it may involve lipid-sensing transmembrane domains of PERK and IRE1. Here, we show that loss of PLIN2 protects mice from development of WD-induced ER stress activation. The effects of Plin2 loss on ER stress are associated with altered PUFA content of ER membranes and appear to be independent of effects on GRP-78. Interestingly, our data suggest Plin2 loss reduces ER stress responses by reducing total PERK and XBP1 levels, without altering their relative activities. The cause for reduced PERK and XBP1 levels in WD-fed Plin2-null mice is unknown and warrants further investigation. It has been shown that UPR activation alone can up-regulate key ER stress-sensing components such as Ire1, Perk, Atf4, and ATF6 (61). Thus, it is possible that WT and Plin2-null livers experience differences in basal regulation of ER stress components such as PERK and XBP1. Interestingly, links between reduced SCD1 activity and increased ER stress have been proposed to be mediated by increased membrane lipid saturation (62–64). Surprisingly, we found that ER stress resistance of Plin2-null animals is associated with both striking reductions in SCD1 and increased unsaturated/saturated ER membrane lipid ratio compared with WT animals, which appears to be driven primarily by substantial increases in long chain PUFA in Plin2-null mice. Thus, Plin2-null animals are protected from development of ER stress on WD likely by a combination of increased PUFA content in their membranes and by a reduction in key ER stress components.
Other Possible Pathways That Might Be Affected by Loss of PLIN2
It is also worth noting that we observed changes in other lipid-signaling pathways in addition to SREBP. We had originally hypothesized that Plin2-null livers might exhibit increased CPT1a expression that would result in increased β-oxidation and reduced hepatic triglycerides in these animals. However, both Pparα and Cpt1a mRNA are reduced in Plin2-null mice, contrary to our expectations but consistent with previous findings (17). This reduction in both Pparα and Cpt1α gene expression suggests that Plin2-null livers perhaps do not experience a massive increase in β-oxidation. In fact, the data suggest they may experience less oxidation than WT animals. More study is needed to elucidate the specific effects that loss of hepatic PLIN2 has on lipid oxidation rates, and we plan to explore this in further detail.
A second potential mechanism of action that could lead to resistance of hepatic steatosis was suggested by the data from our studies. We observed a striking reduction in mRNA for both Pparγ and its target gene Cd36 (fatty acid translocase) in Plin2-null animals on both diets. It has previously been shown that PPARγ and CD36 are up-regulated in the liver during obesity, insulin resistance, and NAFLD (65, 66). It is possible that Plin2-null animals experience decreased uptake of free fatty acids from the periphery. However, given that the WT animals were obese on WD, these animals likely experienced adipose insulin resistance (as evidenced by impaired glucose tolerance testing) and persistent increases in serum non-esterified fatty acid compared with the lean Plin2-null cohort. Therefore, reduction in hepatic CD36 in Plin2-null animals was probably counterbalanced by the fact that they experience reduced circulating non-esterified fatty acid on WD. On the control diet, it is possible that suppressed CD36 contributes significantly to the reduction in hepatic neutral lipids. Therefore, we cannot rule out that reduction of PPARγ and its target genes is contributing to the reduction in hepatic steatosis in livers lacking Plin2.
In conclusion, we show that whole-body loss of PLIN2 exerts a protective effect in animals exposed to long-term Western diet in part by suppressing hepatic SREBP-1 and SREBP-2 activity. Furthermore, in addition to its traditional function of promoting lipid storage, PLIN2 also functions in lipid-partitioning pathways and membrane composition. Future work will focus on whether loss of PLIN2 exerts similar effects on the SREBP family in other tissues. Additionally, more work is needed to examine how loss of PLIN2 affects hepatic lipid oxidation rates and VLDL secretion.
Experimental Procedures
Animal Housing and Feeding
All animal protocols adhered to the University of Colorado Denver Institutional Animal Care and Use Committee (IACUC) guidelines. Plin2-null mice were generated as described previously (13). Mice were housed at the University of Colorado Anschutz Medical Campus animal facility at ambient temperatures (22–24 °C) on a 14:10-h light/dark cycle. Feeding was ab libitum with free access to food and water. Mice were fed standard chow from weaning until 8 weeks upon which they were fed either control diet (Teklad TD.08485) or a Western-style diet containing elevated fat and cholesterol. (Teklad TD.88137) for 30 weeks. Lipid composition of both diets as analyzed by GC-MS can be found in supplemental Table S1. Body weight and food intake were monitored weekly. Upon completion of the feeding experiments, the body composition of each animal was determined by quantitative magnetic resonance imaging (EchoMRI-900 Whole-body Composition Analyzer; Echo Medical Systems, Houston, TX), and mice were euthanized by CO2 exposure and cardiac puncture. Intraperitoneal glucose tolerance testing was performed as described previously (13). 18-Hour fasted serum FFA analysis was performed using a standard FFA calorimetric detection kit (ZenBio Inc.) with 5 μl of serum per reaction.
Histology and Immunohistochemistry
Samples from freshly excised livers were fixed for 24–36 h in 4% paraformaldehyde, subsequently embedded in paraffin, and sectioned as described previously. H&E staining and sectioning for histologic samples were performed by the Pathology Core at the University of Colorado Denver Anschutz Medical Campus. Immunohistochemical staining of PLIN2, PLIN3, and PLIN5 was performed as described previously (13).
Gene Expression Analysis
RNA was extracted from liver tissue using the RNeasy Plus mini kit (Qiagen). cDNA was synthesized from 1 μg of RNA using the iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was performed on a Bio-Rad CFX96 instrument using SYBR green probes. qRT-PCR MasterMix was either iTaq Universal SYBR Green Supermix (Bio-Rad) or 2× SYBR Green qPCR Mastermix (BioTools). All gene expression was normalized to 18S rRNA, and relative expression levels were calculated using the ΔΔCt method. All primer sequences used for qRT-PCR can be found in the supplemental material (see supplemental Table S2).
Western Blotting Analysis
Proteins were extracted from frozen liver by bead homogenization in 0.1% Triton X-100, 150 mm NaCl, and 50 mm Tris-HCl, pH 7.4, with standard protease and phosphatase inhibitors (Thermo Pierce). The homogenate was extracted twice with 2:1 chloroform/methanol to clear triglycerides and precipitate protein. Precipitated protein pellets were washed twice with 100% methanol, dried briefly, and resuspended in 10% SDS. For Western blotting analysis, 37.5 μg of protein were separated on 4–20% Tris-glycine gels (Bio-Rad), transferred to nitrocellulose, and probed with indicated primary antibodies at 1:1000 dilution. Immunoreactive bands were detected with corresponding fluorescently labeled secondary antibodies and quantified by imaging on a Li-COR CLx instrument. Antibodies to SCD1, VDAC, LAMP1, FASN, phospho-ACC (Ser79), total ACC, phospho-AMPKα (Thr-172), total AMPKα, phospho-ACLY (Ser-455), total ACLY, ATF4, phospho-PERK (Thr-980), total PERK, phospho-EIF2a (Ser-51), total EIF2a, CHOP, and XBP1 (spliced and unspliced) were purchased from Cell Signaling Technologies. Antibodies to ELOVL6, ApoB (pan), and HMGCS1 were purchased from Abcam. The antibody to ATF6 was obtained from Novus Biologicals. The antibody to PEX3 was purchased from Aviva Systems Biology. The antibody to BiP (GRP78) was obtained from Thermo Scientific. The antibody to Plin2 was obtained from Fitzgerald Industries. β-Actin antibodies for normalization were purchased from Abcam and Sigma.
Hepatic Lipid Analysis
Neutral lipid was analyzed as described previously (67). Briefly, frozen liver tissue was homogenized in (v/v) Folch reagent (2:1 CHCl3/MeOH) containing 300 μg of tritridecanoin reference standard (Nu-Check Prep Inc., Elysian, MN) by bead homogenization for two cycles of 2 min at 30 Hz. Homogenates were diluted further with Folch reagent, treated with 800 μl of 0.9% sodium chloride solution, vortexed, and centrifuged at 4000 rpm for 5 min. The organic phase was removed and dried under N2 gas. Total lipids were resuspended in 330 μl of 100% chloroform and applied to HyperSep SI SPE columns (Thermo Scientific, Waltham, MA) pre-equilibrated with 15 column volumes chloroform. Neutral lipids were eluted with a total of 3 ml of chloroform, dried under N2, and resuspended in 1 ml of methanol containing 2.5% H2SO4. Fatty acid methyl ester (FAME) production was initiated by heating at 80 °C for 1.5 h. 1 ml of HPLC-grade water was added to quench the reactions, and FAMEs/cholesterol was extracted with 200 μl of hexane. A Trace 1310 GC with a TG-5MS column (Thermo Scientific, Waltham, MA) was used to separate lipids chromatographically, and lipids were analyzed with an ISQ single quadrupole mass spectrometer. Xcalibur software (Thermo Scientific) was used to calculate peak areas. Areas were normalized to the tritridecanoin reference standard and then to tissue weight.
Microsomal Membrane Isolation and Lipidomics
Microsomal membranes were isolated as described previously (68). For isolation and subsequent lipidomic analysis, livers of 4-h fasted WT and Plin2-null mice (n = 5 for each group) were excised, washed with ice-cold PBS, minced in 0.25 m sucrose containing 0.1 m potassium phosphate, pH 7.2, 1 mm EDTA, and protease inhibitors (Thermo Pierce, Inc.), and homogenized using 10 strokes of a Dounce homogenizer. Homogenates were centrifuged at 8500 × g for 5 min at 4 °C to obtain the post-nuclear supernatant. After removing the top layer of fat, homogenates were re-centrifuged; the supernatant below the remaining fat was diluted into the isolation buffer described above and centrifuged at 15,000 × g for 20 min at 4 °C to obtain the post-mitochondrial supernatant. Post-mitochondrial homogenates were overlaid with PBS and centrifuged at 15,000 × g. The PBS layer containing any remaining lipid was carefully removed from the underlying homogenate containing microsomes. The microsomal fraction was then centrifuged at 125,000 × g to pellet microsomal membranes. The pellets were washed twice with 12 ml of isolation buffer, resuspended in 200 μl of PBS, and frozen at −80 °C until analysis. Frozen microsomal membranes were extracted in Folch reagent as described above. Lipid fractions were dried under N2, resuspended in chloroform, and loaded onto HyperSep SPE columns (Thermo Scientific, Inc.). Neutral lipid was eluted with 3 ml of chloroform followed by 5 ml of acetone/methanol (9:1) to elute glycolipids. Phospholipids were then eluted with 3 ml of 100% methanol, dried, and subjected to FAME production and GC-MS analysis as described above. The insoluble protein pellets were washed twice with methanol, solubilized in 10% SDS, and stored at −80 °C.
Statistics
Two-tailed unpaired Student's t tests and ANOVA were used to derive p values. All variance is presented as S.E. unless otherwise indicated. p < 0.05 was considered to be statistically significant. All statistical tests were performed with Prism Graphpad software.
Author Contributions
A. E. L. performed gene expression, serum FFA analysis, Western blotting analysis, neutral lipid lipidomics, microsomal membrane isolation/characterization, phospholipid lipidomics, and manuscript preparation. E. B. performed histologic staining, body weight measurements, feeding, and body composition analysis. D. J. O. performed histologic analysis and assisted with preparation of the manuscript. J. L. M. advised on and helped plan experiments and assisted with manuscript preparation.
Supplementary Material
Acknowledgments
We thank Miguel Lanaspa Garcia and Isabel Schlaepfer for generous donation of antibodies. We have enormous gratitude to Tânia Reis for use of the GC-MS instrument, use of facilities, and helpful discussions on lipidomics. We also thank Robert Eckel, Hong Wang, Tian Yu, and Kimberly Bruce for generous donation of materials. We are grateful to Andy Bradford and Jen Monks for helpful comments during preparation of this manuscript.
This work was supported by National Institutes of Health Grant 2RO1-HD045965 (to J. L. M.) and University of Colorado CCTSI/National Institutes of Health Grant TL1-TR001081 (to A. E. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1–S5.
- NAFLD
- non-alcoholic fatty liver disease
- ER
- endoplasmic reticulum
- WD
- Western diet
- CD
- control diet
- ANOVA
- analysis of variance
- UPR
- unfolded protein response
- MUFA
- monounsaturated fatty acid
- CLD
- cytoplasmic lipid droplet
- s/u
- spliced/unspliced
- qRT
- quantitative RT
- SCFA
- short chain fatty acid
- PL
- phospholipid
- Ppar
- peroxisome proliferator-activated receptor
- LC
- long chain
- P/T
- phospho/total
- sXBP1
- spliced Xbp1
- uXBP1
- unspliced Xbp1
- DHA
- docosahexaenoic acid
- ROS
- reactive oxygen species
- FXR
- farnesoid X receptor
- LXR
- liver X receptor
- FAME
- fatty acid methyl ester
- SREBP
- sterol regulatory element-binding protein
- ChREBP
- carbohydrate-response element-binding protein
- ACC
- acetyl-CoA carboxylase
- AMPK
- 5′-AMP-activated protein kinase
- TG
- triglyceride.
References
- 1. Catenacci V. A., Hill J. O., and Wyatt H. R. (2009) The obesity epidemic. Clin. Chest Med. 30, 415–444 [DOI] [PubMed] [Google Scholar]
- 2. Vanni E., Bugianesi E., Kotronen A., De Minicis S., Yki-Järvinen H., and Svegliati-Baroni G. (2010) From the metabolic syndrome to NAFLD or vice versa? Dig. Liver Dis. 42, 320–330 [DOI] [PubMed] [Google Scholar]
- 3. Bugianesi E., Gastaldelli A., Vanni E., Gambino R., Cassader M., Baldi S., Ponti V., Pagano G., Ferrannini E., and Rizzetto M. (2005) Insulin resistance in non-diabetic patients with non-alcoholic fatty liver disease: sites and mechanisms. Diabetologia 48, 634–642 [DOI] [PubMed] [Google Scholar]
- 4. Torres D. M., and Harrison S. A. (2012) Nonalcoholic steatohepatitis and noncirrhotic hepatocellular carcinoma: fertile soil. Semin. Liver Dis. 32, 30–38 [DOI] [PubMed] [Google Scholar]
- 5. Anstee Q. M., Targher G., and Day C. P. (2013) Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat. Rev. Gastroenterol. Hepatol. 10, 330–344 [DOI] [PubMed] [Google Scholar]
- 6. Musso G., Cassader M., Rosina F., and Gambino R. (2012) Impact of current treatments on liver disease, glucose metabolism and cardiovascular risk in non-alcoholic fatty liver disease (NAFLD): a systematic review and meta-analysis of randomised trials. Diabetologia 55, 885–904 [DOI] [PubMed] [Google Scholar]
- 7. Choi S. S., and Diehl A. M. (2008) Hepatic triglyceride synthesis and nonalcoholic fatty liver disease. Curr. Opin. Lipidol. 19, 295–300 [DOI] [PubMed] [Google Scholar]
- 8. Ducharme N. A., and Bickel P. E. (2008) Lipid droplets in lipogenesis and lipolysis. Endocrinology 149, 942–949 [DOI] [PubMed] [Google Scholar]
- 9. Sztalryd C., and Kimmel A. R. (2014) Perilipins: lipid droplet coat proteins adapted for tissue-specific energy storage and utilization, and lipid cytoprotection. Biochimie 96, 96–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu X., Xue R., Ji L., Zhang X., Wu J., Gu J., Zhou M., and Chen S. (2014) Activation of farnesoid X receptor (FXR) protects against fructose-induced liver steatosis via inflammatory inhibition and ADRP reduction. Biochem. Biophys. Res. Commun. 450, 117–123 [DOI] [PubMed] [Google Scholar]
- 11. Imai Y., Varela G. M., Jackson M. B., Graham M. J., Crooke R. M., and Ahima R. S. (2007) Reduction of hepatosteatosis and lipid levels by an adipose differentiation-related protein antisense oligonucleotide. Gastroenterology 132, 1947–1954 [DOI] [PubMed] [Google Scholar]
- 12. Imai Y., Boyle S., Varela G. M., Caron E., Yin X., Dhir R., Dhir R., Graham M. J., and Ahima R. S. (2012) Effects of perilipin 2 antisense oligonucleotide treatment on hepatic lipid metabolism and gene expression. Physiol. Genomics 44, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McManaman J. L., Bales E. S., Orlicky D. J., Jackman M., MacLean P. S., Cain S., Crunk A. E., Mansur A., Graham C. E., Bowman T. A., and Greenberg A. S. (2013) Perilipin-2-null mice are protected against diet-induced obesity, adipose inflammation, and fatty liver disease. J. Lipid Res. 54, 1346–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Manrique C., DeMarco V. G., Aroor A. R., Mugerfeld I., Garro M., Habibi J., Hayden M. R., and Sowers J. R. (2013) Obesity and insulin resistance induce early development of diastolic dysfunction in young female mice fed a Western diet. Endocrinology 154, 3632–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guilherme A., Virbasius J. V., Puri V., and Czech M. P. (2008) Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9, 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ruan H., and Lodish H. F. (2003) Insulin resistance in adipose tissue: direct and indirect effects of tumor necrosis factor-α. Cytokine Growth Factor Rev. 14, 447–455 [DOI] [PubMed] [Google Scholar]
- 17. Varela G. M., Antwi D. A., Dhir R., Yin X., Singhal N. S., Graham M. J., Crooke R. M., and Ahima R. S. (2008) Inhibition of ADRP prevents diet-induced insulin resistance. Am. J. Physiol. Gastrointest. Liver Physiol. 295, G621–G628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang B. H., Li L., Saha P., and Chan L. (2010) Absence of adipose differentiation related protein upregulates hepatic VLDL secretion, relieves hepatosteatosis, and improves whole body insulin resistance in leptin-deficient mice. J. Lipid Res. 51, 2132–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takahashi Y., Shinoda A., Kamada H., Shimizu M., Inoue J., and Sato R. (2016) Perilipin2 plays a positive role in adipocytes during lipolysis by escaping proteasomal degradation. Sci. Rep. 6, 20975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carr R. M., and Ahima R. S. (2016) Pathophysiology of lipid droplet proteins in liver diseases. Exp. Cell Res. 340, 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Langhi C., Marquart T. J., Allen R. M., and Baldán A. (2014) Perilipin-5 is regulated by statins and controls triglyceride contents in the hepatocyte. J. Hepatol. 61, 358–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Madison B. B. (2016) SREBP2: A master regulator of sterol and fatty acid synthesis. J. Lipid Res. 57, 333–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tao R., Xiong X., DePinho R. A., Deng C. X., and Dong X. C. (2013) Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J. Lipid Res. 54, 2745–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horton J. D., Goldstein J. L., and Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chypre M., Zaidi N., and Smans K. (2012) ATP-citrate lyase: a mini-review. Biochem. Biophys. Res. Commun. 422, 1–4 [DOI] [PubMed] [Google Scholar]
- 26. Ntambi J. M. (1999) Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J. Lipid Res. 40, 1549–1558 [PubMed] [Google Scholar]
- 27. Zhang S., Yang Y., and Shi Y. (2005) Characterization of human SCD2, an oligomeric desaturase with improved stability and enzyme activity by cross-linking in intact cells. Biochem. J. 388, 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matsuzaka T., Atsumi A., Matsumori R., Nie T., Shinozaki H., Suzuki-Kemuriyama N., Kuba M., Nakagawa Y., Ishii K., Shimada M., Kobayashi K., Yatoh S., Takahashi A., Takekoshi K., Sone H., et al. (2012) Elovl6 promotes nonalcoholic steatohepatitis. Hepatology 56, 2199–2208 [DOI] [PubMed] [Google Scholar]
- 29. Matsuzaka T., and Shimano H. (2009) Elovl6: a new player in fatty acid metabolism and insulin sensitivity. J. Mol. Med. 87, 379–384 [DOI] [PubMed] [Google Scholar]
- 30. Matsuzaka T., Shimano H., Yahagi N., Kato T., Atsumi A., Yamamoto T., Inoue N., Ishikawa M., Okada S., Ishigaki N., Iwasaki H., Iwasaki Y., Karasawa T., Kumadaki S., Matsui T., et al. (2007) Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat. Med. 13, 1193–1202 [DOI] [PubMed] [Google Scholar]
- 31. Yamada K., Mizukoshi E., Sunagozaka H., Arai K., Yamashita T., Takeshita Y., Misu H., Takamura T., Kitamura S., Zen Y., Nakanuma Y., Honda M., and Kaneko S. (2015) Characteristics of hepatic fatty acid compositions in patients with nonalcoholic steatohepatitis. Liver Int. 35, 582–590 [DOI] [PubMed] [Google Scholar]
- 32. Tripathy S., and Jump D. B. (2013) Elovl5 regulates the mTORC2-Akt-FOXO1 pathway by controlling hepatic cis-vaccenic acid synthesis in diet-induced obese mice. J. Lipid Res. 54, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tripathy S., Lytle K. A., Stevens R. D., Bain J. R., Newgard C. B., Greenberg A. S., Huang L. S., and Jump D. B. (2014) Fatty acid elongase-5 (Elovl5) regulates hepatic triglyceride catabolism in obese C57BL/6J mice. J. Lipid Res. 55, 1448–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lattka E., Illig T., Koletzko B., and Heinrich J. (2010) Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr. Opin. Lipidol. 21, 64–69 [DOI] [PubMed] [Google Scholar]
- 35. Wang Y., Botolin D., Christian B., Busik J., Xu J., and Jump D. B. (2005) Tissue-specific, nutritional, and developmental regulation of rat fatty acid elongases. J. Lipid Res. 46, 706–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hannah V. C., Ou J., Luong A., Goldstein J. L., and Brown M. S. (2001) Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 276, 4365–4372 [DOI] [PubMed] [Google Scholar]
- 37. Lee J. N., Zhang X., Feramisco J. D., Gong Y., and Ye J. (2008) Unsaturated fatty acids inhibit proteasomal degradation of Insig-1 at a postubiquitination step. J. Biol. Chem. 283, 33772–33783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chirieac D. V., Davidson N. O., Sparks C. E., and Sparks J. D. (2006) PI3-kinase activity modulates apo B available for hepatic VLDL production in apobec-1−/− mice. Am. J. Physiol. Gastrointest. Liver Physiol. 291, G382–G388 [DOI] [PubMed] [Google Scholar]
- 39. Shimano H., Yahagi N., Amemiya-Kudo M., Hasty A. H., Osuga J., Tamura Y., Shionoiri F., Iizuka Y., Ohashi K., Harada K., Gotoda T., Ishibashi S., and Yamada N. (1999) Sterol regulatory element-binding protein-1 as a key transcription factor for nutritional induction of lipogenic enzyme genes. J. Biol. Chem. 274, 35832–35839 [DOI] [PubMed] [Google Scholar]
- 40. Sekiya M., Yahagi N., Matsuzaka T., Najima Y., Nakakuki M., Nagai R., Ishibashi S., Osuga J., Yamada N., and Shimano H. (2003) Polyunsaturated fatty acids ameliorate hepatic steatosis in obese mice by SREBP-1 suppression. Hepatology 38, 1529–1539 [DOI] [PubMed] [Google Scholar]
- 41. Espenshade P. J. (2006) SREBPs: sterol-regulated transcription factors. J. Cell Sci. 119, 973–976 [DOI] [PubMed] [Google Scholar]
- 42. Yoshikawa T., Shimano H., Yahagi N., Ide T., Amemiya-Kudo M., Matsuzaka T., Nakakuki M., Tomita S., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Takahashi A., Sone H., Osuga Ji J., et al. (2002) Polyunsaturated fatty acids suppress sterol regulatory element-binding protein 1c promoter activity by inhibition of liver X receptor (LXR) binding to LXR response elements. J. Biol. Chem. 277, 1705–1711 [DOI] [PubMed] [Google Scholar]
- 43. Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., and Mangelsdorf D. J. (2000) Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 14, 2819–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Winnay J. N., Boucher J., Mori M. A., Ueki K., and Kahn C. R. (2010) A regulatory subunit of phosphoinositide 3-kinase increases the nuclear accumulation of X-box-binding protein-1 to modulate the unfolded protein response. Nat. Med. 16, 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ozcan U., Cao Q., Yilmaz E., Lee A. H., Iwakoshi N. N., Ozdelen E., Tuncman G., Görgün C., Glimcher L. H., and Hotamisligil G. S. (2004) Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306, 457–461 [DOI] [PubMed] [Google Scholar]
- 46. Volmer R., van der Ploeg K., and Ron D. (2013) Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. U.S.A. 110, 4628–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee A. H., Scapa E. F., Cohen D. E., and Glimcher L. H. (2008) Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320, 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sriburi R., Jackowski S., Mori K., and Brewer J. W. (2004) XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 167, 35–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chang B. H., Li L., Paul A., Taniguchi S., Nannegari V., Heird W. C., and Chan L. (2006) Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol. Cell. Biol. 26, 1063–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Russell T. D., Palmer C. A., Orlicky D. J., Bales E. S., Chang B. H., Chan L., and McManaman J. L. (2008) Mammary glands of adipophilin-null mice produce an amino-terminally truncated form of adipophilin that mediates milk lipid droplet formation and secretion. J. Lipid Res. 49, 206–216 [DOI] [PubMed] [Google Scholar]
- 51. Li S., Brown M. S., and Goldstein J. L. (2010) Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 3441–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li Y., Xu S., Mihaylova M. M., Zheng B., Hou X., Jiang B., Park O., Luo Z., Lefai E., Shyy J. Y., Gao B., Wierzbicki M., Verbeuren T. J., Shaw R. J., Cohen R. A., and Zang M. (2011) AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 13, 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jump D. B., Botolin D., Wang Y., Xu J., Demeure O., and Christian B. (2008) Docosahexaenoic acid (DHA) and hepatic gene transcription. Chem. Phys. Lipids 153, 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Scorletti E., and Byrne C. D. (2013) ω-3 fatty acids, hepatic lipid metabolism, and nonalcoholic fatty liver disease. Annu. Rev. Nutr. 33, 231–248 [DOI] [PubMed] [Google Scholar]
- 55. Botolin D., Wang Y., Christian B., and Jump D. B. (2006) Docosahexaenoic acid (22:6,n-3) regulates rat hepatocyte SREBP-1 nuclear abundance by Erk- and 26S proteasome-dependent pathways. J. Lipid Res. 47, 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Browning J. D., and Horton J. D. (2004) Molecular mediators of hepatic steatosis and liver injury. J. Clin. Invest. 114, 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vial G., Dubouchaud H., Couturier K., Cottet-Rousselle C., Taleux N., Athias A., Galinier A., Casteilla L., and Leverve X. M. (2011) Effects of a high-fat diet on energy metabolism and ROS production in rat liver. J. Hepatol. 54, 348–356 [DOI] [PubMed] [Google Scholar]
- 58. Kitai Y., Ariyama H., Kono N., Oikawa D., Iwawaki T., and Arai H. (2013) Membrane lipid saturation activates IRE1α without inducing clustering. Genes Cells 18, 798–809 [DOI] [PubMed] [Google Scholar]
- 59. Volmer R., and Ron D. (2015) Lipid-dependent regulation of the unfolded protein response. Curr. Opin. Cell Biol. 33, 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ariyama H., Kono N., Matsuda S., Inoue T., and Arai H. (2010) Decrease in membrane phospholipid unsaturation induces unfolded protein response. J. Biol. Chem. 285, 22027–22035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rutkowski D. T., and Hegde R. S. (2010) Regulation of basal cellular physiology by the homeostatic unfolded protein response. J. Cell Biol. 189, 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Minville-Walz M., Pierre A. S., Pichon L., Bellenger S., Fèvre C., Bellenger J., Tessier C., Narce M., and Rialland M. (2010) Inhibition of stearoyl-CoA desaturase 1 expression Induces CHOP-dependent cell death in human cancer cells. PLoS One 5, e14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu X., Strable M. S., and Ntambi J. M. (2011) Stearoyl CoA desaturase 1: role in cellular inflammation and stress. Adv. Nutr. 2, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Peter A., Weigert C., Staiger H., Machicao F., Schick F., Machann J., Stefan N., Thamer C., Häring H. U., and Schleicher E. (2009) Individual stearoyl-CoA desaturase 1 expression modulates endoplasmic reticulum stress and inflammation in human myotubes and is associated with skeletal muscle lipid storage and insulin sensitivity in vivo. Diabetes 58, 1757–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Miquilena-Colina M. E., Lima-Cabello E., Sánchez-Campos S., García-Mediavilla M. V., Fernández-Bermejo M., Lozano-Rodríguez T., Vargas-Castrillón J., Buqué X., Ochoa B., Aspichueta P., González-Gallego J., and García-Monzón C. (2011) Hepatic fatty acid translocase CD36 upregulation is associated with insulin resistance, hyperinsulinaemia and increased steatosis in non-alcoholic steatohepatitis and chronic hepatitis C. Gut 60, 1394–1402 [DOI] [PubMed] [Google Scholar]
- 66. Tailleux A., Wouters K., and Staels B. (2012) Roles of PPARs in NAFLD: potential therapeutic targets. Biochim. Biophys. Acta 1821, 809–818 [DOI] [PubMed] [Google Scholar]
- 67. Perez C. L., and Van Gilst M. R. (2008) A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 8, 266–274 [DOI] [PubMed] [Google Scholar]
- 68. Garg M. L., Thomson A. B., and Clandinin M. T. (1988) Effect of dietary-cholesterol and or ω-3 fatty-acids on lipid-composition and Δ-5-desaturase activity of rat-liver microsomes. J. Nutr. 118, 661–668 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.