Abstract
O6-Methyl-2′-deoxyguanosine (O6-MeG) is a ubiquitous DNA lesion, formed not only by xenobiotic carcinogens but also by the endogenous methylating agent S-adenosylmethionine. It can introduce mutations during DNA replication, with different DNA polymerases displaying different ratios of correct or incorrect incorporation opposite this nucleoside. Of the “translesion” Y-family human DNA polymerases (hpols), hpol η is most efficient in incorporating equal numbers of correct and incorrect C and T bases. However, the mechanistic basis for this specific yet indiscriminate activity is not known. To explore this question, we report biochemical and structural analysis of the catalytic core of hpol η. Activity assays showed the truncated form displayed similar misincorporation properties as the full-length enzyme, incorporating C and T equally and extending from both. X-ray crystal structures of both dC and dT paired with O6-MeG were solved in both insertion and extension modes. The structures revealed a Watson-Crick-like pairing between O6-MeG and 2"-deoxythymidine-5"-[(α, β)-imido]triphosphate (approximating dT) at both the insertion and extension stages with formation of two H-bonds. Conversely, both the structures with O6- MeG opposite dCTP and dC display sheared configuration of base pairs but to different degrees, with formation of two bifurcated H-bonds and two single H-bonds in the structures trapped in the insertion and extension states, respectively. The structural data are consistent with the observed tendency of hpol η to insert both dC and dT opposite the O6-MeG lesion with similar efficiencies. Comparison of the hpol η active site configurations with either O6-MeG:dC or O6-MeG:dT bound compared with the corresponding situations in structures of complexes of Sulfolobus solfataricus Dpo4, a bypass pol that favors C relative to T by a factor of ∼4, helps rationalize the more error-prone synthesis opposite the lesion by hpol η.
Keywords: DNA damage, DNA polymerase, enzyme kinetics, mass spectrometry (MS), X-ray crystallography
Introduction
High fidelity replication of DNA is essential to all forms of life. However, DNA is extensively modified by both sources outside and inside mammalian bodies. One type of damage is alkylating agents, and the simplest and most common of these are methylating agents. Methylation of DNA by xenobiotic chemicals, including some used to treat cancer (1), was recognized at least as early as 1961 and then associated with tumors caused by simple alkyl N-nitrosamines (2). Although N7-MeG4 is the major DNA adduct formed by methylating agents (2), several lines of evidence indicate that O6-MeG is the most miscoding and mutagenic adduct (3). O6-MeG is formed from endogenous sources as well as exogenous (4), and all mammalian DNA samples show finite levels, generally attributed to extraneous methylation by S-adenosylmethionine (4).
Exactly how O6-MeG miscodes has been a topic of interest ever since it became possible to study miscoding. The general phenomenon with O6-MeG is the insertion of C, the correct base, or T (5). The purines (A, G) have not been incorporated to any major extent by any DNA polymerases studied to date.
Melting studies with oligonucleotides containing O6-MeG provide some (unexplained) results (6). Substitution of O6-MeG for dG lowered the Tm of a duplex of 12-mers by 19–26 °C. However, the most stable complex was with O6-MeG:C pairing, and the O6-MeG:T pair (favored in misincorporation studies) was the weakest (7, 8). Swann (6) proposed that polymerases either (a) mistake O6-MeG for A due to “physical similarities” and insert dTTP and/or (b) miscoding is a consequence of the alignment of the bases. Both of these explanations are vague, but Swann (6) proposed a wobble pair for O6-MeG:C and Watson-Crick pairing (both with only one H-bond) for O6-MeG:T pairing. These later proved to be the structures observed in the X-ray crystal structure with Sulfolobus solfataricus Dpo4 (with the O6-MeG:T pair being in a two H-bond pseudo-Watson-Crick pairing) (9).
NMR studies of oligonucleotides (in the absence of polymerases) provided evidence for the existence of a wobble base pair between O6-MeG and C (10). Crystallographic studies of such oligonucleotides (O6-MeG:C) (no polymerase) yielded split-hydrogen bond bifurcated pairing and Watson-Crick type pairing in Z-DNA (11, 12). NMR studies suggested that O6-MeG:T pairing retained Watson-Crick geometry, with a single hydrogen bond between the N2 exocyclic amine of O6-MeG and the O2 carbonyl of T (10), plus no bonding or possibly a “long” bond between the N1 atom of O6-MeG and the imino proton at the N3 atom of T. A crystal structure of oligonucleotides containing an O6-MeG:T pair (in a self-complementary system) showed Watson-Crick pairing (13).
Three sets of crystal structures of O6-MeG paired with C and T in DNA polymerases are available, and they differ dramatically. With DNA polymerase I of the thermophile Bacillus stearothermophilus, C was paired opposite O6-MeG in an isosteric Watson-Crick geometry, invoking a rare C tautomer (14). An O6-MeG:dTTP complex in the same enzyme involved an unusual electrostatic interaction between (O6-MeG) O-methyl protons and a dT carbonyl oxygen. S. solfataricus Dpo4 complexes show pairing predicted by NMR studies, i.e. wobble pairing between O6-MeG and C and pseudo-Watson-Crick pairing for O6-MeG and T (9). The only other reported DNA polymerase structure with O6-MeG (and the only one to date with a eukaryotic polymerase) is that of hpol ι, which shows Hoogsteen pairing (generally typical of this enzyme) for both the O6-MeG:C and O6-MeG:T complexes, with one H-bond in the T complex and two H-bonds in the C complex (15). Dpo4 favors C incorporation >T by a factor of ∼4 (9), but hpol ι and B. stearothermophilus pol I favor T > C incorporation by factors of ∼6 and 10, respectively (14–16). Thus, there appear to be multiple mechanisms by which DNA polymerases catalyze dCTP and dTTP incorporation opposite O6-MeG.
Of the human Y-Family DNA polymerases, hpol η (Uniprot entry Q9Y253, POLH) is by far the most efficient in catalyzing both dCTP and dTTP incorporation (16). This enzyme, like hpol δ, incorporated dCTP and dTTP with similar efficiencies (16). We solved insertion and extension structures with either dCTP (dC) or dTMP-NPP (dT) positioned opposite O6-MeG at the active site of hpol η. The structural data allow insight into the principles underlying the similar activities with both pyrimidine nucleotides displayed by this human TLS (translesion synthesis) polymerase.
Results
Analysis of Misincorporation
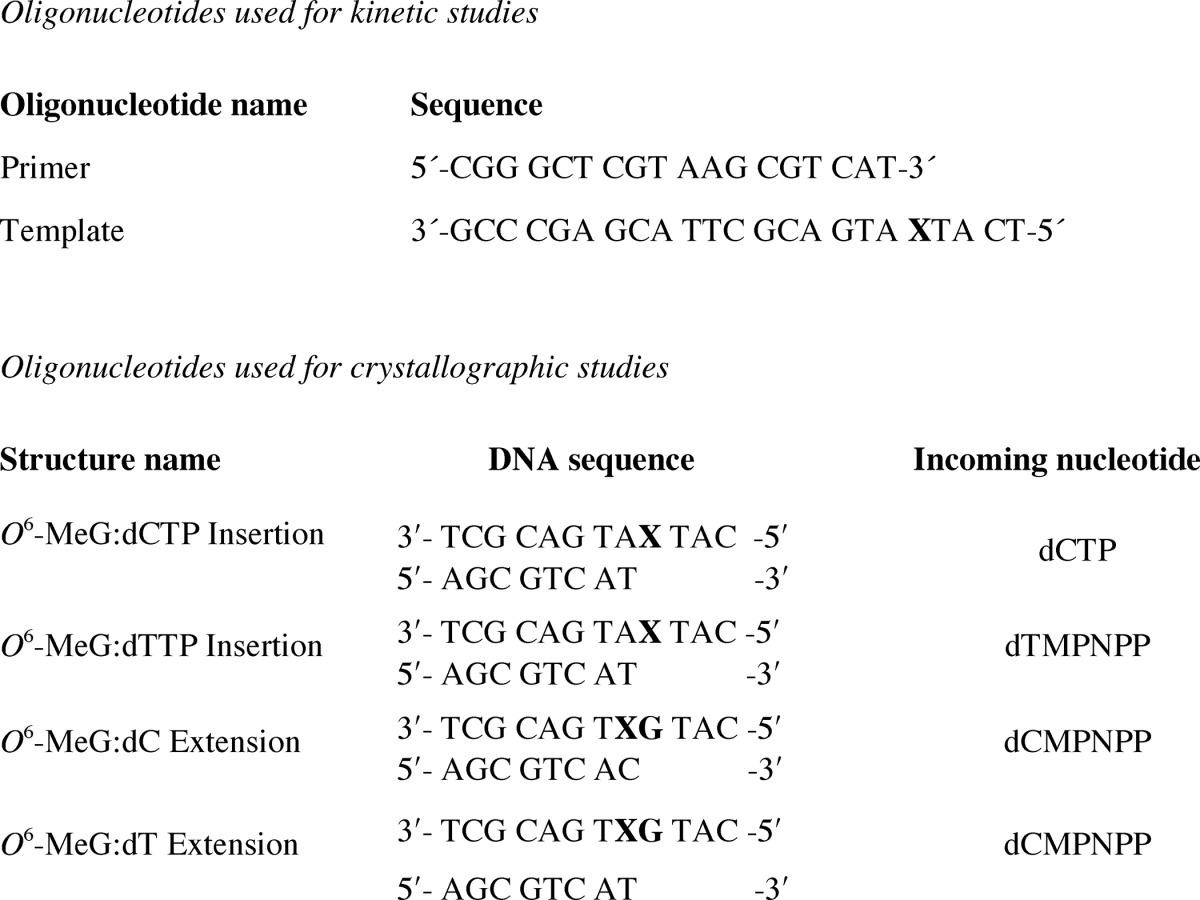
Studies with the catalytic core of hpol η and a 23-mer/18-mer template-primer DNA duplex construct with incorporated O6-MeG (Table 1) yielded results essentially identical to our findings with the full-length enzyme (16). The ratio of (kcat/Km)dCTP/(kcat/Km)dTTP was 1.1 (results not presented), identical to the value found earlier (16).
TABLE 1.
Oligonucleotides used in this study (X = O6-MeG)
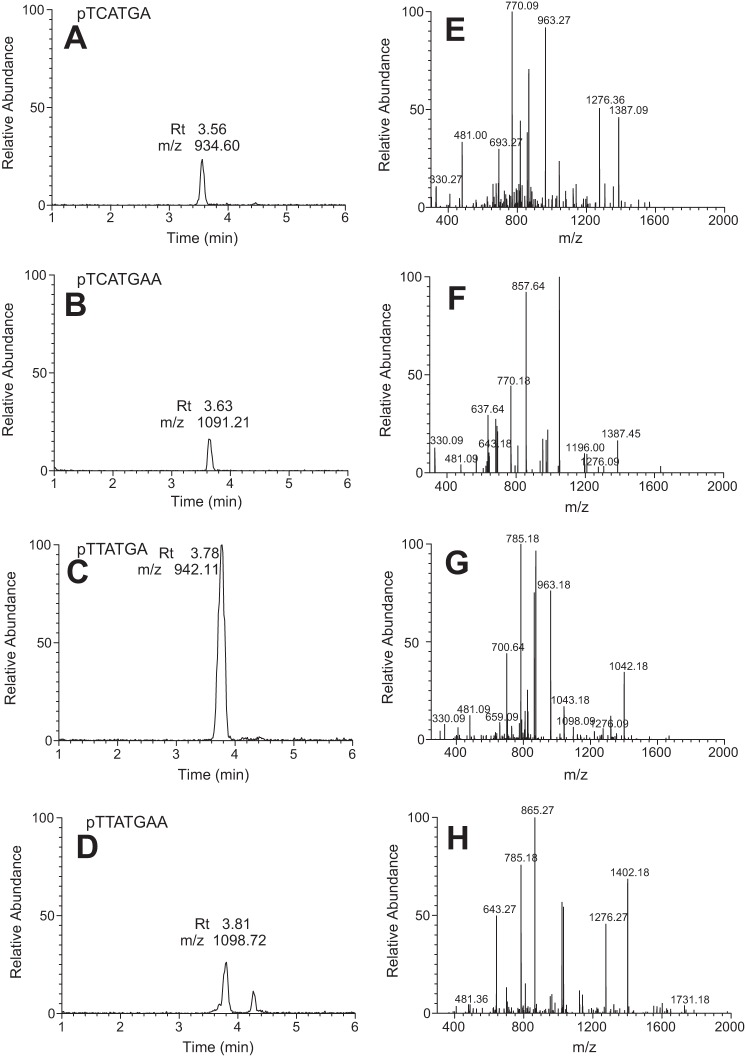
LC-MS analysis of the extended primer indicated the presence of only C and T incorporation opposite O6-MeG (47% C, 53% T) (Fig. 1, Table 2). The ratio is very similar to the results of assays done before with full-length hpol η (16) and in accord with the results of the steady-state kinetic analysis. Thus, C and T are equally preferred.
FIGURE 1.
LC-MS analysis of primer extension products. A–D, reconstructed chromatograms for the indicated ion. E–H, fragmentation patterns for each of the indicated primer extension products.
TABLE 2.
LC/MS analysis of full-length extension opposite O6-MeG
| Sequence | Charge/theoretical m/z | Measured m/z | Peak area (most abundant ion) | % Total |
|---|---|---|---|---|
| TCA TGA | −2/934.60 | 934.36 | 17,630 | 47 (C) |
| −3/622.73 | 622.91 | |||
| TCA TGAA | −2/1091.21 | 1090.82 | 3,497 | |
| −3/727.14 | 727.00 | |||
| TTA TGA | −2/942.11 | 941.82 | 19,807 | 53 (T) |
| −3/627.74 | 627.64 | |||
| TTA TGA A | −2/1098.72 | 1098.36 | 4,440 | |
| −3/732.14 | 732.27 |
Crystal Structures of Insertion-stage Ternary hpol η Complexes
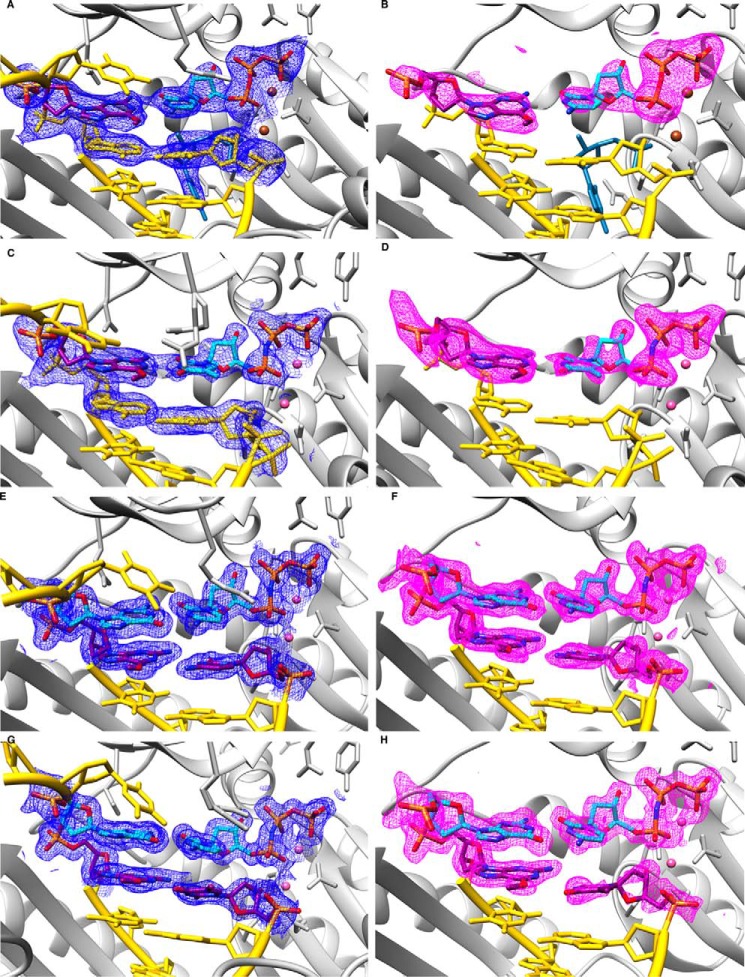
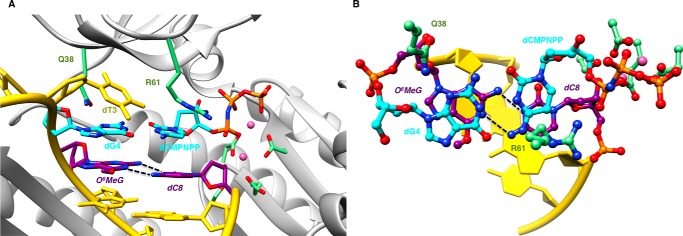
We determined two structures of complexes between hpol η and 12-mer template-8-mer primer duplexes (see Table 1 for sequences) containing O6-MeG, with nucleoside triphosphates paired opposite the adducted residue. The atomic coordinates and structure factors have been deposited in the Protein Data Bank under codes 5L1I (insertion O6-MeG opposite dCTP), 5L1J (insertion O6-MeG opposite dTMP-NPP), 5L1K (extension after O6-MeG opposite dC), and 5L1L (extension after O6-MeG opposite dT). In the first, determined to a resolution of 2.79 Å, the incoming nucleotide is dCTP, and in the second, determined to 1.94 Å resolution, the incoming nucleotide is dTMP-NPP. A summary of crystal data, data collection, and refinement parameters is provided in Table 3, and examples of the quality of the final electron density in the active site region are shown in Fig. 2, A and B. Crystals of the complex with the non-hydrolyzable dTTP analog dTMP-NPP were obtained readily, and electron density maps generated after molecular replacement and initial rigid body refinement revealed a fully occupied nucleoside triphosphate molecule. Conversely, dCMP-NPP in combination with Mg2+ in the crystal of the other insertion-stage complex was only partially ordered, with electron density visible for the triphosphate moiety and part of the sugar but not for the base. Subsequently, we resorted to dCTP at higher concentrations in the presence of Ca2+ (to prevent nucleotide insertion) to obtain crystals of the ternary complex in which the entire incoming nucleotide was surrounded by electron density (Fig. 2A). Somewhat surprisingly, the refined structure also revealed extension of the primer by dC (Fig. 3), indicating that hpol η possesses activity with Ca2+ as the metal ion cofactor. The primer strand makes a sharp turn at the active site to prevent a clash with the incoming dCTP, and the additional cytidine dC9 is located in the minor groove, where its base portion forms a H-bonding interaction with Arg-111 and stacks against the 2′-deoxyribose of template G7 (O6-MeG is residue 4; Fig. 3B).
TABLE 3.
Crystal data, data collection parameters, and structure refinement statistics
| Complex | O6-MeG:dCTP insertion | O6-MeG:dTMP -NPP insertion | O6-MeG:dC extension | O6-MeG:dT extension |
|---|---|---|---|---|
| Data Collection | ||||
| Wavelength (Å) | 0.97856 | 1.07810 | 0.97872 | 0.97872 |
| Space group | P61 | P61 | P61 | P61 |
| Resolution (Å) | 50-2.79 (2.79-2.84)a | 50-1.94 (1.94-1.97) | 50-1.82 (1.82-1.85) | 50-1.62 (1.62-1.65) |
| Unit cell a = b, c (Å) | 99.09, 81.89 | 99.00, 81.70 | 98.46, 82.11 | 98.36, 82.03 |
| Unique reflections | 11,577 (581) | 33,771 (1,684) | 40,671 (2,018) | 57,284 (2,831) |
| Completeness (%) | 99.9 (99.8) | 100 (100) | 100 (100) | 100 (100) |
| I/σ(I) | 10.65 (1.58) | 27.38 (1.78) | 14.55 (1.85) | 23.18 (2.04) |
| Wilson B-factor (Å2) | 35.0 | 27.0 | 15.4 | 15.5 |
| R-merge | 0.169 (0.966) | 0.091 (0.819) | 0.118 (0.809) | 0.098 (0.869) |
| Redundancy | 5.5 (5.2) | 7.6 (7.5) | 5.7 (5.6) | 7.6 (7.2) |
| Refinement | ||||
| R-work | 0.159 (0.226) | 0.159 (0.183) | 0.153 (0.203) | 0.156 (0.187) |
| R-free | 0.228 (0.266) | 0.201 (0.281) | 0.187 (0.232) | 0.193 (0.240) |
| Number of atoms protein/DNA | 3,372/409 | 3,450/395 | 3,412/390 | 3,433/391 |
| dNMP-NPP/Mg2+ | 28/2 (Ca2+) | 29/2 | 28/2 | 28/2 |
| Water/solute | 138/2 | 410/1 | 491/1 | 526/3 |
| Protein residues | 430 | 430 | 430 | 430 |
| B-factor (Å2) | ||||
| Average | 49.1 | 33.6 | 23.8 | 24.4 |
| Protein/DNA | 49.4/55.5 | 33.1/37.3 | 22.9/30.9 | 23.1/33.3 |
| dNMP-NPP/Mg2+ | 30.5/38.9 | 30.3/26.8 | 11.2/8.5 | 13.3/10.9 |
| Water/ glycerol | 37.2/50.7 | 41.1/25.9 | 32.0/15.0 | 34.6/ 33.3 |
| Root mean square deviations | ||||
| Bonds (Å) | 0.010 | 0.018 | 0.018 | 0.017 |
| Angles (degress) | 1.5 | 1.8 | 1.8 | 1.8 |
| Ramachandran | ||||
| Favored (%) | 95 | 97 | 97 | 98 |
| Allowed (%) | 4.8 | 2.5 | 2.8 | 1.8 |
| Outliers (%) | 0.2 | 0.5 | 0.2 | 0.2 |
| PDB ID code | 5L1I | 5L1J | 5L1K | 5L1L |
a Statistics for the highest resolution shell are shown in parentheses.
FIGURE 2.
Quality of the final models of ternary hpol η complexes with DNA template strands containing O6-MeG. Fourier (2Fo − Fc) sum electron density drawn at the 1σ threshold (blue meshwork; panels A, C, E, and G) and Fourier (Fo − Fc) omit electron density at the 3 σ level (magenta meshwork; panels B, D, F, and H) around the polymerase active site region. A and B, insertion stage with incoming dCTP opposite O6-MeG. C and D, insertion stage with incoming dTMP-NPP opposite O6-MeG. E and F, extension stage with dC opposite O6-MeG followed by dCMP-NPP opposite template dG. G and H, extension stage with dT opposite O6-MeG followed by dCMP-NPP opposite template dG. Selected active site residues are colored by atom with carbon atoms shown in purple (template O6-MeG and primer dC and dT in the extension complexes) or cyan (incoming nucleotide and template dG in the extension complexes). Base pairs at the −1 and −2 (panels A–D) or −2 (panels E–H) positions and template dT 5′-adjacent to the adducted residue are shown in yellow, with the DNA backbones highlighted by a ribbon, the protein is shown in light gray, and Mg2+ and Ca2+ ions are pink and brown spheres, respectively.
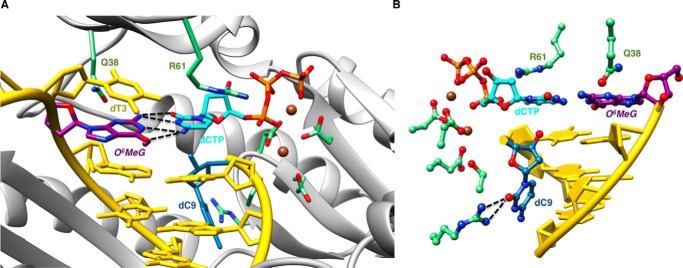
FIGURE 3.
Sheared configuration of incoming dCTP and O6-MeG in a hpol η insertion stage complex. A, the active site viewed into the DNA major groove. B, rotated by ∼180° around the vertical axis and viewed into the minor groove. Selected active site residues are colored by atom with carbon atoms shown in purple (O6-MeG), cyan (dCTP), or green (Arg-61 and Gln-38 from the finger domain as well as Asp/Glu coordinated to Ca2+ (brown spheres)). The remaining nucleotides are shown in yellow, except for dC9 (blue), which was added to the primer during crystallization. H-bonds involving the adducted residue are depicted as dashed lines.
In this insertion complex, O6-MeG and dCTP adopt a wobble-like, slightly sheared configuration whereby the O6-Me moiety is shifted into the major groove and O2 of C is shifted into the minor groove (Fig. 3A). This relative orientation of the two nucleobases allows for formation of two bifurcated H-bonds. In the first, N2(H)2 of O6-MeG donates to both the O2 and N3 acceptors of dCTP (distances of 2.9 Å and 3.2 Å, respectively). In the second, N4(H)2 of dCTP donates to both the N1 and O6 acceptors of O6-MeG (distances of 3.3 Å and 2.9 Å, respectively). The adduct base remains fully stacked between the 5′-adjacent T and the 3′-adjacent A in the template strand, but the cytosine of the incoming dCTP has shifted away from the penultimate T of the primer strand, probably as a result of the added dC (Fig. 3). Residues Gln-38 and Arg-61 from the finger domain of hpol η interact with the adducted residue and the incoming nucleotide, respectively. Gln-38 is engaged in a H-bond with O4′ of the sugar of O6-MeG, and Arg-61 forms a salt bridge to the α-phosphate group and an offset stacking interaction with the nucleobase of dCTP.
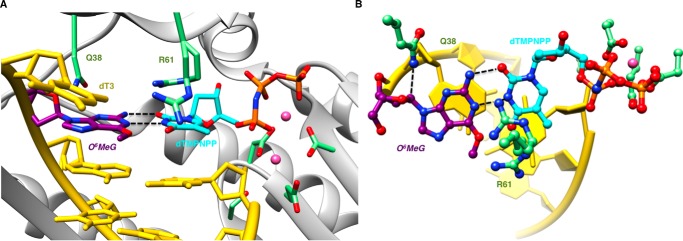
In the second insertion complex, the pairing configuration of the O6-MeG:dTMP-NPP pair and the interactions between the nascent pair and hpol η active site residues differ distinctly from the above dCTP insertion-stage complex. Thus, adducted base and thymine exhibit a Watson-Crick-like arrangement that permits formation of two H-bonds, one between N1 (O6-MeG) and N3(H) (dTMP-NPP) and the other between N2(H) (O6-MeG) and O2 (dTMP-NPP) (Fig. 4). On the template side, Gln-38 interacts with O4′ of the sugar of O6-MeG, and the adducted base remains stacked between adjacent residues (Fig. 4A). However, Arg-61 is rotated away from the phosphates of the incoming nucleotide compared with the dCTP complex and directed toward the base moieties of the nascent pair. There, the amino acid side chain adopts two conformations such that the guanidino moiety protrudes into the major groove in one orientation, whereas in the other it is above the Watson-Crick edge of the thymine base and also in close proximity of the O6 atom of the adducted base (Fig. 4B). Thus, the two insertion-stage complexes demonstrate that the active site of hpol η is capable of accommodating a range of chemistries and pairing configurations and that key residues such as Arg-61 exhibit considerable conformational freedom.
FIGURE 4.
Watson-Crick-like configuration of incoming dTMP-NPP and O6-MeG in a hpol η insertion stage complex. A, the active site viewed into the DNA major groove. B, rotated by ∼90° around the horizontal axis and viewed perpendicular to the base planes of dTMP-NPP and O6-MeG. Selected active site residues are colored by atom with carbon atoms shown in purple (O6-MeG), cyan (dTMP-NPP), or green (Arg-61 and Gln-38 from the finger domain as well as Asp/Glu coordinated to Mg2+ (pink spheres)). The remaining nucleotides are shown in yellow, and H-bonds involving the adducted residue are depicted as dashed lines.
Crystal Structures of Extension-stage Ternary hpol η Complexes
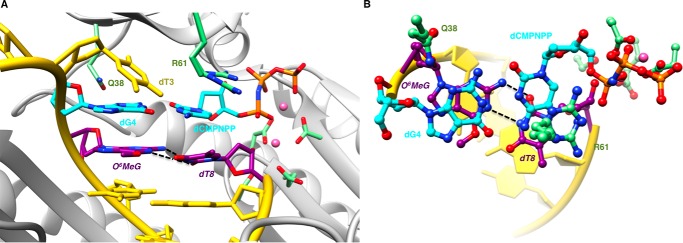
The crystal structures of two extension-stage complexes with O6-MeG opposite dC and dT and followed by a dG:dCMP-NPP pair were determined at resolutions of 1.82 Å and 1.62 Å, respectively. Selected crystallographic data are summarized in Table 3, and examples of the quality of the electron density around the final models are depicted in Fig. 2, C and D. In the complex with cytidine, O6-MeG and dC display more shearing than in the dCTP insertion complex. This precludes formation of bifurcated H-bonds, and the O6-MeG:dC pair is stabilized by two H-bonds that link N1(H) (O6-MeG) and N4 (dC) and N2(H) (O6-MeG) and N3 (dC) (Fig. 5). By comparison, the pairing mode between O6-MeG and dT in the other extension complex is virtually identical to that seen in the insertion-stage structure with the adduct paired opposite dTMP-NPP (Figs. 3 and 6). The base pair at the −1 position is stabilized by two H-bonds between N1 (O6-MeG) and N3(H) (dT) and between N2(H) (O6-MeG) and O2 (dT). In both complexes, Arg-61 assumes an orientation that is more commonly observed in complexes of hpol η, namely a curled conformation that allows an offset stacking interaction with the base of the incoming nucleotide and salt bridge formation with its α-phosphate group.
FIGURE 5.
Sheared configuration of primer dC and O6-MeG in a hpol η extension stage complex. A, the active site viewed into the DNA major groove. B, rotated by ∼90° around the horizontal axis and viewed perpendicular to the base planes of dCMP-NPP and dG. Selected active site residues are colored by atom with carbon atoms shown in purple (O6-MeG and dC at the −1 position), cyan (nascent dG:dCMP-NPP pair), or green (Arg-61 and Gln-38 from the finger domain as well as Asp/Glu coordinated to Mg2+ (pink spheres)). The remaining nucleotides are shown in yellow, and H-bonds involving the adducted residue are depicted as dashed lines.
FIGURE 6.
Sheared configuration of primer dT and O6-MeG in a hpol η extension stage complex. A, the active site viewed into the DNA major groove. B, rotated by ∼90° around the horizontal axis and viewed perpendicular to the base planes of dCMP-NPP and dG. Selected active site residues are colored by atom with carbon atoms shown in purple (O6-MeG and dT at the −1 position), cyan (nascent dG:dCMP-NPP pair), or green (Arg-61 and Gln-38 from the finger domain as well as Asp/Glu coordinated to Mg2+ (pink spheres)). The remaining nucleotides are shown in yellow, and H-bonds involving the adducted residue are depicted as dashed lines.
Discussion
Kinetic analysis of hpol η catalyzed polymerization past O6-MeG using either the catalytic core construct (present work) or full-length enzyme (16) shows efficient bypass synthesis and equal preference for either correct C or incorrect T. The roughly equal ratio of C to T incorporation sets hpol η apart from the Y-family pols Dpo4 from S. solfataricus and hpol ι that both display a clear preference for error-prone insertion of T over C opposite O6-MeG (4-fold (9) and 6-fold (15), respectively). The preference for T by hpol ι was rationalized by the Hoogsteen pairing mode of the nascent base pair at the active site of the enzyme. Thus, the O6-MeG:dTTP pair is stabilized by a H-bond between N7 of the adduct and N3-H of thymine (15). By comparison, N7 of O6-MeG is positioned opposite an acceptor atom (N3) in the pair with dCTP, although one can invoke a single H-bond upon protonation of C at lower values of pH.
The different preferences for C and T found for insertion opposite O6-MeG with S. solfataricus Dpo4 and hpol η are somewhat surprising given similarities between the two enzymes in the bypass of many adducts, including cyclic pyrimidine dimers (17, 18) and 8-oxoG (19, 20). However, as the analyses of the efficient and correct bypass of the 8-oxoG adduct by both pols demonstrated (19-fold (19) and 4-fold preference (19) for dCTP over dATP insertion opposite the adduct by Dpo4 and hpol η, respectively), the mechanisms underlying the mostly error-free bypass can differ significantly. Thus, Dpo4 relies on the Arg-332 residue from the little finger domain to maintain 8-oxoG in the template strand in the anti conformation to allow formation of a canonical Watson-Crick base pairing with incoming dCTP (19). Conversely, hpol η focuses less on the template residue but recruits Arg-61 from the finger domain to guide the position of the incoming nucleoside triphosphate by contacting either base moiety or α-phosphate, suggesting an inherent conformational flexibility of the side chain in response to a particular adduct (19) (Fig. 7).
FIGURE 7.

Conformational flexibility of finger residue Arg-61 at the active site of ternary hpol η-DNA-dNTP complexes. View into the major groove of the DNA template (left):primer (right) duplex with dNTP visible to the right of Arg-61 side chains that are drawn in stick mode. Two divalent metal ions (Mg2+ or Ca2+) between the 3′-terminal nucleotide of the primer strand and the triphosphate moiety of the incoming nucleotide are located at the center of crossing dashed lines that mark their coordination geometry. A total of 23 crystal structures with the following PDB ID codes were included in the overlay: 5L1I, 5L1J, 5K1K, and 5L1L (O6-MeG, this work), 5DLF, 5DLG, 5DQG, 5DQH, and 5DQI (O4-methyl- and O4-ethyl-T) (22), 4RNM, 4RNN, and 4RNO (abasic site) (21), 4RU9 (MeFapy-dG) (23), 4O3N (native G:dCMP-NPP Mg2+ complex), 4O3O, 4O3P, 4O3Q, 4O3R, and 4O3S (8-oxoG) (17), 5DG7, 5DG8, and 5DG9 (1,N6-etheno-dA) (24), and 3MR3 (cys-syn-cyclobutane thymine dimer) (18). Arg-61 adopts various orientations that we have marked with numbers 1 (curled conformation and interacting with the α- and β-phosphate groups of the incoming 2′-deoxynucleotide triphosphate, the most common position), 2 (curled conformation and stacked in an offset manner on the nucleobase of the incoming dNTP), 3 (extended conformation and interacting with the major groove edge of the template base at the 0 position, e.g. 8-oxoG (magenta carbons)), 4 (curled conformation and interacting with the template base at the +1 position, e.g. the second T of cys-syn-cyclobutane thymine dimer (orange carbons)), and 5 (curled conformation and fully stacked on the nucleobase of the incoming dNTP, e.g. O6-MeG and dTMP-NPP, this work (lilac carbons)).
Looking at the base pairing modes of the O6-MeG adduct with dCTP and dTMP-NPP as well as with dC and dT at the active site of hpol η (Figs. 3–6), we note that each pair is stabilized by two H-bonds, consistent with similar rates and extents of incorporation by this pol (16). The adduct pairing modes of this adduct (11) with dCTP and dC differ somewhat in that the former pair exhibits bifurcated H-bonds with less shearing and the latter two individual H-bonds with more shearing (similar to a wobble pair, e.g. G:U in RNA, but with the O2 atom of dT jutting into the minor groove rather than N2 of G; Fig. 5B). In fact, the two pairing modes of O6-MeG with dCTP and dC observed at the hpol η active site, with varying degrees of shearing, closely resemble those of O6-EtG:dC pairs in the structure of a DNA dodecamer with Hoechst 33258 bound in the minor groove (11) (PDB ID code 128D). In that duplex one of the pairs displays the two bifurcated H-bonds (dCTP insertion complex; Fig. 3), and the other one is stabilized by two individual H-bonds, with more shearing between O6-EtG and dC (dC extension complex; Fig. 5).
In the crystal structures of extension-stage Dpo4 complexes with a template strand containing O6-MeG and paired to either dC or dT, the former shows two H-bonds with shearing (9), just like the O6-MeG:dC pair at the active site of hPol η (Fig. 5). However, in the structure with dT the weak electron density around the thymine moiety indicated enhanced flexibility of the incoming nucleotide and perhaps reduced stability of the nascent base pair compared with O6-MeG:dC. This scenario is consistent with the preference of Dpo4 for dCTP relative to dTTP.
Although both Dpo4 complexes were trapped at the extension stage in the crystal structures, the ceiling of the active site in the pol from S. solfataricus lacks the two amino acids that protrude from the finger domain and contact the nascent base pair in the case of hpol η (Gln-38 and Arg-61, e.g. Fig. 3). Gln-38 interacts with sugar O4′ of the adduct in the insertion complexes (Figs. 3 and 4), and Arg-61 contacts the incoming nucleotide. However, the interaction modes observed for Arg-61 vis à vis dCTP and dTMP-NPP differ distinctly. In the former complex, the side chain is rotated toward the triphosphate moiety, and the guanidino group forms a salt bridge with the α-phosphate of dCTP (Fig. 3). In the complex with dTMP-NPP, the side chain adopts two orientations whereby the guanidino group in both is facing away from the triphosphate moiety (Fig. 4). In one orientation, the positive charge of the arginine side chain hovers directly above the space between the thymine and O6-methylguanine bases, in close vicinity of the O4 and O6 atoms (not H-bonded) and N3(H) and N1 atoms (H-bonded) (Fig. 4B). The particular position that the guanidino moiety assumes in the complex with dTMP-NPP is unique among structures of ternary hpol η complexes (Fig. 7, #5) and may help relieve a potential repulsion between the O6 and O4 acceptors from O6-MeG and dTMP-NPP, respectively, in the major groove.
Arg-61, which protrudes from the active site ceiling, can swivel around and adopt a range of orientations that allows it to interact with the α-phosphate of the incoming dNTP and partially stack on its base moiety (the most common orientation) but also stretch and interact with the major groove edges of template and incoming base (e.g. 8-oxoG and dGTP; Ref. 17), reach over to the template base of a cyclic thymine dimer (18), or even form a salt bridge with a phosphate from the template strand (e.g. abasic site; Ref. 21) (Fig. 7). However, even taking into account this conformational freedom of Arg-61, the orientation of the guanidino moiety in the complex with dTMP-NPP allows a face-to-face stacking interaction with thymine protrudes. It contrasts with the situation in the dCTP complex of hpol η and the corresponding complex of Dpo4 with dTTP and may account for the more favorable incorporation of dT by the human pol compared with that from S. solfataricus.
Although the dCTP and dTMP-NPP complexes of hPol η were crystallized under somewhat different conditions (Ca2+ versus Mg2+, respectively, and varying concentrations of the nucleoside triphosphate), the positions of Arg-61 in the respective complexes may reflect the particular electronic features of the nascent O6-MeG:dC and O6-MeG:dT pairs. In any case, the extension in situ of the primer in the presence of dCTP and Ca2+ demonstrates that the conditions used to crystallize the complex did not preclude activity (Fig. 3). Primer extension under conditions that employed Ca2+ in place of Mg2+ were also observed for hpol η crystallizations with a template containing O4-EtT (22) and had previously been seen in the structures of Dpo4 complexes with 8-oxoG (20, 25). Thus, both hpol η and Dpo4 can accommodate an additional primer nucleotide that is wrapped around into the minor groove in addition to an incoming nucleoside triphosphate. In both pols, the major groove side of the active site is wide open and more bulky O6 substituents such as benzyl are easily tolerated in bypass synthesis (26). Overall, it appears that hPol η is more capable of controlling and stabilizing incoming dTTP relative to dCTP compared with Dpo4 despite roomy active sites in both pols that allow a host of pairing modes and adducts, including the Watson-Crick type O6-MeG:dT and sheared wobble-like O6-MeG:dC pairs studied here.
In terms of the biological relevance of individual DNA polymerases in dealing with O6-MeG, the situation is not clear. In yeast, the methylating agent methyl methanesulfonate promoted the degradation of the Rad5-related human T-cell leukemia virus enhancer factor protein and stimulated the interaction of another Rad5-related protein, SHPRH, with Rad18 and (yeast) pol κ (27). The conclusion of that study was that yeast pol κ suppresses methyl methansulfonate-induced mutagenesis, presumably due to higher fidelity reading of O6-MeG lesions (27). hpol κ was also concluded to have a partial but significant role in protection of HeLa cells form cytotoxicity caused by another methylating agent (methyl nitrosourea), but hpol κ status did not affect mutagenicity (28). In another study involving glioblastoma cells overexpression of hpol κ conferred resistance to another methylating agent, temozoloomide (29). hpol κ inactivation facilitated temozolomide-induced Rad17 ubiquination and proteasomal degradation. As pointed out earlier in the current study, our own comparative studies with human DNA polymerases showed that hpol η was the most efficient in copying opposite O6-MeG (16). hpol η, hpol δ, and hpol κ all incorporated dCTP and dTTP opposite O6-MeG, and hpol ι was 10-fold more efficient in adding T > C (16). Although there is evidence that hpol κ may have roles with O6-MeG in cells (27–29), these studies do not address the issue of miscoding. Another confounding point is that hpol κ has a role as an “extender” of pairs/mispairs (30), and it may contribute in this way to overall biological properties. In our own work, hpol η was ∼50-fold more efficient in inserting both dCTP and dTTP opposite O6-MeG (16). The relative roles of the individual DNA polymerases still need to be discerned, but in summary, we have characterized modes of correct and incorrect incorporation opposite O6-MeG by a catalytically efficient human DNA polymerase.
Experimental Procedures
Reagents
HPLC purified oligonucleotides were purchased from Midland Certified Reagent Co. (Midland, TX), Integrated DNA Technologies (Coralville, IA), or TriLink Biotechnologies (San Diego, CA). The oligonucleotides containing O6-MeG were from TriLink. Oligonucleotide sequences are listed in Table 1. The catalytic core (amino acids 1–432) of hpol η was expressed and purified as described previously (18). Unlabeled dNTPs, T4 polynucleotide kinase, and uracil DNA glycosylase (UDG) were obtained from New England BioLabs (Ipswich, MA). A mixture of four dNTPs was purchased from Invitrogen. All non-hydrolyzable dCMP-NPP and dTMP-NPP nucleotides were obtained from Jena Bioscience (Jena, Germany). [γ-32P] ATP (specific activity 3000 Ci/mmol) was purchased from PerkinElmer Life Sciences. Biospin columns were from Bio-Rad.
Assays
Steady-state kinetic assays were done using varying concentrations of dCTP and dTTP as described in detail elsewhere (31, 32) For the estimation of kinetic parameters (kcat/Km), data points were fit to hyperbolic plots using non-linear regression in Prism (GraphPad, La Jolla, CA). The LC-MS analysis of extended primers was performed as described in detail elsewhere (33–35).
Crystallization
The hanging drop vapor diffusion technique was used to grow all crystals. Two dodecamer DNA sequences modified with the O6-MeG lesion either at the fourth or fifth position of the template and complementary primer strands were used in the crystallization experiments and are listed in Table 1. DNA template and primer oligonucleotide solutions were mixed in a 1:1 molar ratio in the presence of 10 mm sodium HEPES buffer (pH 8.0), 0.1 mm EDTA, and 50 mm NaCl and annealed at 85 °C for 10 min followed by slow cooling to room temperature. The DNA solution was then mixed with hpol η protein in a 1.2:1 molar ratio in the presence of 50 mm Tris-HCl (pH 7.5) containing 450 mm KCl and 3 mm DTT followed by the addition of either 5 μl of 100 mm MgCl2 or 5 μl of 100 mm CaCl2. The protein-DNA mixture solution was placed in an Amicon filter (Millipore, Billerica, MA), and the solution was concentrated to a final concentration of ∼2 mg of protein/ml. Either dCTP or one of the non-hydrolyzable nucleoside triphosphate analogs was added to the concentrated mixtures containing Ca2+ or Mg2+. The ternary complex solution was mixed with an equal volume of reservoir solution containing 0.10 m sodium MES (pH 5.5), 5 mm CaCl2 (or 5 mm MgCl2), and 14–18% (w/v) PEG 2000 monomethyl ether and equilibrated against 500 μl of reservoir solutions. Crystals commonly appeared after overnight incubation at 18 °C and were allowed to grow for 1 week to several weeks. They were transferred to cryoprotectant solution containing reservoir solution along with 25% glycerol (v/v) and then frozen in liquid nitrogen for data collection.
Data Collection and Structure Refinement
X-ray diffraction data were collected at 100 K on the 21-ID-D, 21-ID-F, or the 21-ID-G beam lines at the Advanced Photon Source (APS), Argonne National Laboratory (Argonne, IL). Data were processed using HKL2000 (36), and all data collection statistics are summarized in Table 3. Structures were determined by molecular replacement in MOLREP (37, 38) using the coordinates of the complex between hpol η and native DNA (PDB ID code 4O3N) as the search model. Structures were refined either with PHENIX (39) or REFMAC (40), and model building was carried out in COOT (41). Model statistics and geometric parameters are summarized in Table 3. All structural figures were generated with the program UCSF Chimera (42).
Author Contributions
A. P. crystallized the complexes and solved the structures, Q. Z. performed the kinetic and LC-MS analyses, and F. G. and M. E. conceived the studies and wrote the manuscript along with A. P.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 ES010375 and R01 ES010546 (to F. G.) and P01 CA160032 (to M. E.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 5L1I, 5L1J, 5L1K, and 5L1L) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MeG
- methylguanine
- hpol
- human DNA polymerase
- dCMP-NPP
- 2′-deoxycytidine-5′-[(α,β)-imido]triphosphate
- dTMP-NPP
- 2′-deoxythymidine-5′-[(α,β)-imido]triphosphate.
References
- 1. Lawley P. D. (1995) Alkylation of DNA and its aftermath. Bioessays 17, 561–568 [DOI] [PubMed] [Google Scholar]
- 2. Brookes P., and Lawley P. D. (1961) The alkylation of guanosine and guanylic acid. J. Chem. Soc. 10.1039/JR9610003923 [DOI] [Google Scholar]
- 3. Loveless A. (1969) Possible relevance of O6-alkylation of deoxyguanosine to the mutagenicity and carcinogenicity of ntrosamines and nitrosamides. Nature 223, 206–207 [DOI] [PubMed] [Google Scholar]
- 4. Barrows L. R., and Shank R. C. (1981) Aberrant methylation of liver DNA in rats during hepatotoxicity. Toxicol. Appl. Pharmacol. 60, 334–345 [DOI] [PubMed] [Google Scholar]
- 5. Loechler E. L., Green C. L., and Essigmann J. M. (1984) In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc. Natl. Acad. Sci. U.S.A. 81, 6271–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Swann P. F. (1990) Why do O6-alkylguanine and O4-alkylthymine miscode? The relationship between the structure of DNA containing O6-alkylguanine and O4-alkylthymine and the mutagenic properties of these bases. Mutat. Res. 233, 81–94 [DOI] [PubMed] [Google Scholar]
- 7. Gaffney B. L., Marky L. A., and Jones R. A. (1984) Synthesis and characterization of a set of four dodecadeoxyribonucleoside undecaphosphates containing O6-methylguanine opposite adenine, cytosine, guanine, and thymine. Biochemistry 23, 5686–5691 [DOI] [PubMed] [Google Scholar]
- 8. Gaffney B. L., and Jones R. A. (1989) Thermodynamic comparison of the base pairs fromed by the carcinogenic lesion O6-methylguanine with reference both to Watson-Crick pairs and to mismatch pairs. Biochemistry 28, 5881–5889 [DOI] [PubMed] [Google Scholar]
- 9. Eoff R. L., Irimia A., Egli M., and Guengerich F. P. (2007) Sulfolobus solfataricus DNA polymerase Dpo4 is partially inhibited by “wobble” pairing between O6-methylguanine and cytosine, but accurate bypass is preferred. J. Biol. Chem. 282, 1456–1467 [DOI] [PubMed] [Google Scholar]
- 10. Patel D. J., Shapiro L., Kozlowski S. A., Gaffney B. L., and Jones R. A. (1986) Structural studies of the O6-MeG.T interaction in the d(C-G-T-G-A-A-T-T-C-O6MeG-C-G) duplex. Biochemistry 25, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 11. Sriram M., van der Marel G. A., Roelen H. L., van Boom J. H., and Wang A. H. (1992) Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342. EMBO J. 11, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ginell S. L., Kuzmich S., Jones R. A., and Berman H. M. (1990) Crystal and molecular structure of a DNA duplex containing the carcinogenic lesion O6-methylguanine. Biochemistry 29, 10461–10465 [DOI] [PubMed] [Google Scholar]
- 13. Leonard G. A., Thomson J., Watson W. P., and Brown T. (1990) High-resolution structure of a mutagenic lesion in DNA. Proc. Natl. Acad. Sci. U.S.A. 87, 9573–9576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Warren J. J., Forsberg L. J., and Beese L. S. (2006) The structural basis for the mutagenicity of O6-methyl-guanine lesions. Proc. Natl. Acad. Sci. U.S.A. 103, 19701–19706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pence M. G., Choi J.-Y., Egli M., and Guengerich F. P. (2010) Structural basis for proficient incorporation of dTTP opposite O6-methylguanine by human DNA polymerase ι. J. Biol. Chem. 285, 40666–40672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choi J.-Y., Chowdhury G., Zang H., Angel K. C., Vu C. C., Peterson L. A., and Guengerich F. P. (2006) Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 281, 38244–38256 [DOI] [PubMed] [Google Scholar]
- 17. Ling H., Boudsocq F., Plosky B. S., Woodgate R., and Yang W. (2003) Replication of a cis-syn thymine dimer at atomic resolution. Nature 424, 1083–1087 [DOI] [PubMed] [Google Scholar]
- 18. Biertümpfel C., Zhao Y., Kondo Y., Ramón-Maiques S., Gregory M., Lee J. Y., Masutani C., Lehmann A. R., Hanaoka F., and Yang W. (2010) Structure and mechanism of human DNA polymerase eta. Nature 465, 1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patra A., Nagy L. D., Zhang Q., Su Y., Müller L., Guengerich F. P., and Egli M. (2014) Kinetics, structure, and mechanism of 8-oxo-7,8-dihydro-2′-deoxyguanosine bypass by human DNA polymerase η. J. Biol. Chem. 289, 16867–16882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zang H., Irimia A., Choi J.-Y., Angel K. C., Loukachevitch L. V., Egli M., and Guengerich F. P. (2006) Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 281, 2358–2372 [DOI] [PubMed] [Google Scholar]
- 21. Patra A., Zhang Q., Lei L., Su Y., Egli M., and Guengerich F. P. (2015) Structural and kinetic analysis of nucleoside triphosphate incorporation opposite an abasic site by human translesion DNA polymerase η. J. Biol. Chem. 290, 8028–8038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Flaherty D. K., Patra A., Su Y., Guengerich F. P., Egli M., and Wilds C. J. (2016) Lesion orientation of O4-alkylthymidine influences replication by human DNA polymerase η. Chem. Sci. 7, 4896–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patra A., Banerjee S., Johnson Salyard T. L., Malik C. K., Christov P. P., Rizzo C. J., Stone M. P., and Egli M. (2015) Structural basis for error-free bypass of the N5-methylformamidopyrimidine-dG lesion by human DNA polymerase η and Sulfolobus solfataricus P2 polymerase IV (Dpo4). J. Am. Chem. Soc. 137, 7011–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patra A., Su Y., Zhang Q., Johnson K. M., Guengerich F. P., and Egli M. (2016) Structural and kinetic analysis of miscoding opposite the DNA adduct 1,N6-ethenodeoxyadenosine by human translesion DNA polymerase η. J. Biol. Chem. 291, 14134–14145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Irimia A., Loukachevitch L. V., Eoff R. L., Guengerich F. P., and Egli M. (2010) Metal-ion dependence of the active-site conformation of the translesion DNA polymerase Dpo4 from Sulfolobus solfataricus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eoff R. L., Angel K. C., Egli M., and Guengerich F. P. (2007) Molecular basis of selectivity of nucleoside triphosphate incorporation opposite O6-benzylguanine by Sulfolobus solfataricus DNA polymerase Dpo4: steady-state and pre-steady-state kinetics and X-ray crystallography of correct and incorrect pairing. J. Biol. Chem. 282, 13573–13584 [DOI] [PubMed] [Google Scholar]
- 27. Lin J.-R., Zeman M. K., Chen J.-Y., Yee M.-C., and Cimprich K. A. (2011) SHPRH and HLTF act in a damage-specific manner to coordinate different forms of postreplication repair and prevent mutagenesis. Mol. Cell 42, 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lupari E., Ventura I., Marcon F., Aquilina G., Dogliotti E., and Fortini P. (2012) Pol κ partially rescues MMR-dependent cytotoxicity of O6-methylguanine. DNA Repair 11, 579–586 [DOI] [PubMed] [Google Scholar]
- 29. Peng C., Chen Z., Wang S., Wang H. W., Qiu W., Zhao L., Xu R., Luo H., Chen Y., Chen D., You Y., Liu N., and Wang H. (2016) The error-prone DNA polymerase κ promotes temozolomide resistance in glioblastoma through Rad17-dependent activation of ATR-Chk1 signaling. Cancer Res. 76, 2340–2353 [DOI] [PubMed] [Google Scholar]
- 30. Haracska L., Prakash L., and Prakash S. (2002) Role of human DNA polymerase κ as an extender in translesion synthesis. Proc. Natl. Acad. Sci. U.S.A. 99, 16000–16005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boosalis M. S., Petruska J., and Goodman M. F. (1987) DNA polymerase insertion fidelity: gel assay for site-specific kinetics. J. Biol. Chem. 262, 14689–14696 [PubMed] [Google Scholar]
- 32. O'Flaherty D. K., and Guengerich F. P. (2014) Steady-state kinetic analysis of DNA polymerase single-nucleotide incorporation products. Curr. Protoc. Nucleic Acid Chem. 59, 7.21.21–27.21.13 ( 10.1002/0471142700.nc0721s5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zang H., Goodenough A. K., Choi J.-Y., Irimia A., Loukachevitch L. V., Kozekov I. D., Angel K. C., Rizzo C. J., Egli M., and Guengerich F. P. (2005) DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: Analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J. Biol. Chem. 280, 29750–29764 [DOI] [PubMed] [Google Scholar]
- 34. Christov P. P., Angel K. C., Guengerich F. P., and Rizzo C. J. (2009) Replication past the N5-methylformamidopyrimidine lesion of deoxyguanosine by DNA polymerases and an improved procedure for sequence analysis of in vitro bypass products by mass spectrometry. Chem. Res. Toxicol. 22, 1086–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chowdhury G., and Guengerich F. P. (2011) Liquid chromatography-mass spectrometry analysis of DNA polymerase reaction products. Curr. Protoc. Nucleic Acid Chem. Chapter 7, Unit 7.16.1–7.16.11 ( 10.1002/0471142700.nc0716s47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 37. Vagin A., and Teplyakov A. (2010) Molecular replacement withMOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 38. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 39. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., and Murshudov G. N. (2004) REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 41. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 42. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]