Abstract
Upon pathogen infection, microbial killing pathways and cellular stress pathways are rapidly activated by the host innate immune system. These pathways must be tightly regulated because insufficient or excessive immune responses have deleterious consequences. Increasing evidence indicates that the nervous system regulates the immune system to confer coordinated protection to the host. However, the precise mechanisms of neural-immune communication remain unclear. Previously we have demonstrated that OCTR-1, a neuronal G protein-coupled receptor, functions in the sensory neurons ASH and ASI to suppress innate immune responses in non-neural tissues against Pseudomonas aeruginosa in Caenorhabditis elegans. In the current study, by using a mass spectrometry-based quantitative proteomics approach, we discovered that OCTR-1 regulates innate immunity by suppressing translation and the unfolded protein response (UPR) pathways at the protein level. Functional assays revealed that OCTR-1 inhibits specific protein synthesis factors such as ribosomal protein RPS-1 and translation initiation factor EIF-3.J to reduce infection-triggered protein synthesis and UPR. Translational inhibition by chemicals abolishes the OCTR-1-controlled innate immune responses, indicating that activation of the OCTR-1 pathway is dependent on translation upregulation such as that induced by pathogen infection. Because OCTR-1 downregulates protein translation activities, the OCTR-1 pathway could function to suppress excessive responses to infection or to restore protein homeostasis after infection.
The nematode Caenorhabditis elegans is a powerful model system for infection studies in the context of a whole animal. More than 40 microbes have been shown to be pathogenic to C. elegans, including bacteria, fungi and viruses, some of which are also human pathogens1. Unlike vertebrates or many other invertebrate species, C. elegans does not have an adaptive immune system; it relies on innate immunity and avoidance behavior to defend itself against microbial attacks. Upon pathogen infection, the nematode can mount protective responses by triggering evolutionarily conserved signaling cascades. These cascades include the mitogen-activated protein kinase (MAPK) pathways, the DAF-2/insulin-like receptor pathway, the DBL-1 pathway (homologous to the mammalian TGF-β cascade), the unfolded protein response (UPR), and programmed cell death2,3,4,5. Activation of these signaling pathways induces expression of defensive genes. The most common differentially expressed protein families in C. elegans during pathogenesis include C-type lectins, lysozymes, lipases and antimicrobial peptides1,2,6. These molecules are regarded as markers of immune responses because they are positively regulated in various organisms exposed to a broad range of pathogens. They are believed to be the immune effectors that act directly to fight off infection, although only a few markers have been demonstrated to have such activities7,8,9.
Upon pathogen infection, cellular stress pathways and microbial killing pathways are rapidly activated by the host innate immune system. These pathways must be tightly regulated as insufficient immune responses exacerbate infection, whereas excessive immune responses lead to prolonged inflammation, tissue damage and death. For example, in C. elegans, loss of the XBP-1-dependent UPR in the presence of pathogens leads to disruption of morphology of the endoplasmic reticulum (ER) and larval lethality10, while under physiological conditions results in constitutive ER stress11. Hyperstimulation of the p38 MAPK pathway is also toxic to C. elegans12. In humans, dysregulated innate immune responses have been linked to a myriad of human health concerns such as sepsis, autoimmune disease and chronic inflammatory disease. Increasing evidence indicates that the nervous system regulates the immune system to confer coordinated protection to the host13,14,15. Recent studies on neuronal G protein-coupled receptors (GPCRs) highlight the roles of specific neurons in the regulation of immune responses16,17,18,19. C. elegans deficient in NPR-1, a homologue of the neuropeptide Y receptor in mammals, shows decreased innate immune responses to infections by Pseudomonas aeruginosa, Salmonella enterica or Enterococcus faecalis but enhanced immune defense against Bacillus thuringiensis16,19. This suggests that the NPR-1-expressing neurons regulate innate immunity to certain pathogens. We have demonstrated that loss of expression of OCTR-1, a putative catecholamine GPCR, in the nematode’s sensory ASH and ASI neurons increases innate immune responses to P. aeruginosa, indicating that OCTR-1 functions in ASHs and ASIs to suppress innate immunity17,18. This regulation is partially achieved by down-regulating gene expression of the p38 MAPK pathway and the UPR pathways in pharyngeal and intestinal tissues that are the primary line of defense against microbial pathogens. It is currently unknown how the immuno-modulatory signals are relayed from ASH/ASI neurons to the non-neuronal tissues. The above studies investigated transcriptional regulatory mechanisms underlying neural regulation of innate immunity; however, it remains unclear how such regulation operates at the protein level.
Although one might expect a direct correspondence between mRNA transcripts and protein expression20, recent studies have shown that the mRNA-protein correlation can be low. Transcriptomic and proteomic studies under similar conditions often reported that only a small overlap exists between differentially regulated genes and proteins21,22,23,24,25,26. There are several reasons for this discordance. First, technological biases lead to a much lower proteome coverage than transcriptome coverage27,28. The most widely used technologies for transcriptomic profiling are DNA microarray and RNA sequencing. Mass spectrometry techniques are used for proteomic profiling. Technical resolution at the proteome level is much more constrained, resulting in identification of only those proteins with high difference in abundance. Second, physical properties of transcripts influence translation. For example, transcripts with weak Shine-Dalgarno (SD) sequences are translated less efficiently29,30. Third, codon bias affects the mRNA-protein correlation31,32. Fourth, ribosome-density, the number of ribosome in a transcriptional unit, has a major influence on efficiency of translation33,34,35. Finally, the correlation between half-lives of mRNAs and proteins is low36. For example, post-translational modifications significantly influence the stability of proteins. While RNA can serve as direct biological effectors, proteins are the effectors of most biological functions. Genome-wide analysis at the protein level is, therefore, a more direct reflection of gene expression.
Since the first use of the P. aeruginosa-C. elegans model for pathogenesis research in 199937,38, a number of transcriptomic studies have been carried out to investigate the nematode’s innate immunity39,40,41,42. In the current work, we examine how C. elegans responds to P. aeruginosa infection and how the nervous system regulates those responses at the protein level.
Results
P. aeruginosa infection induces proteomic changes in C. elegans
To investigate how C. elegans responds to P. aeruginosa infection at the protein level, we compared the proteomes of infected wild-type N2 worms to those of uninfected controls using a label-free quantitative proteomics approach. A schematic diagram of this approach is depicted in Fig. 1. Five biological replicates of each experimental group were individually analyzed with high resolution nano-HPLC tandem mass spectrometry. Protein identification and quantification were done using Thermo Scientific Proteome Discoverer software. In total, 4,413 proteins were identified and quantified, among which 1,312 proteins were identified with high confidence (1% false discovery rate, FDR). The levels of these 1,312 proteins were statistically compared between experimental groups using TIGR Multiexperiment Viewer (MeV). The proteomics data are available via ProteomeXchange with identifier PXD004173.
Figure 1. Scheme of label-free quantitative proteomics.
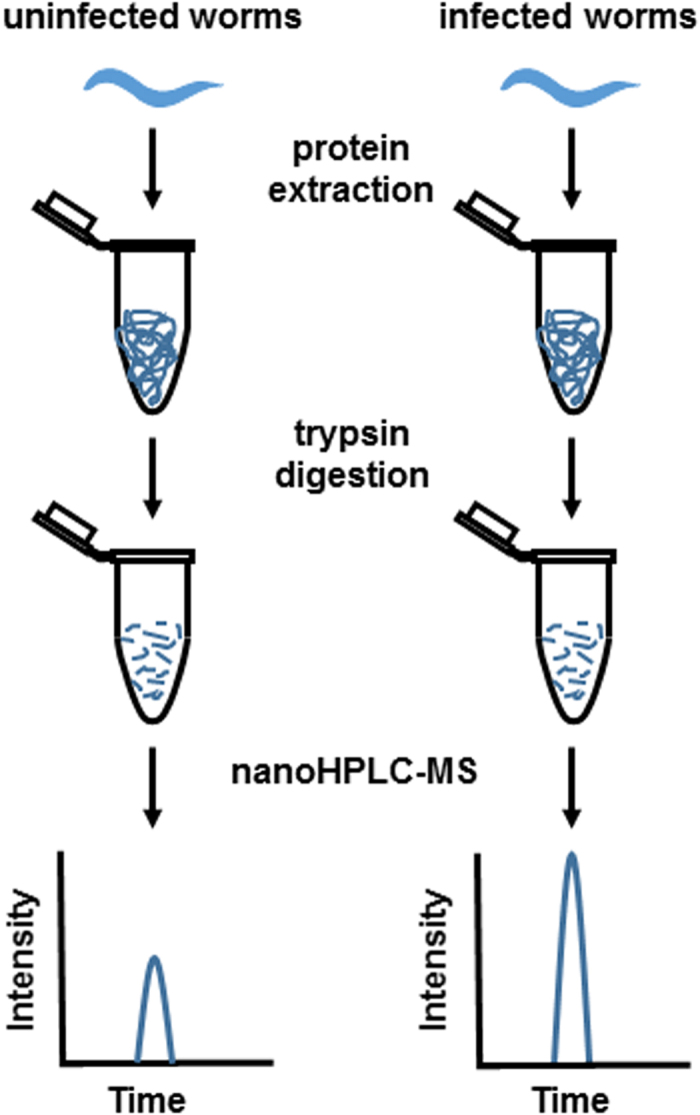
Synchronized animals at L4 stage were either fed E. Coli OP50 (uninfected controls) or P. aeruginosa PA14 (infected samples) for 4 hr, collected, and lysed. Total proteins were extracted and digested with trypsin. Peptides from control or infected samples were subject to nano-HPLC tandem MS analysis. Quantification is based on the comparison of peak intensity of same peptides in different samples.
Compared with the proteins identified in the uninfected wild-type worms, 53 proteins were significantly upregulated at least 1.5-fold in the P. aeruginosa-infected wild-type worms (Table 1). Twenty-three of these proteins were only detected in the infected samples, indicating that their abundance was below the detection limit in the uninfected worms. We performed gene ontology (GO) enrichment analyses to identify significantly enriched biological processes using the web-based program Gorilla (http://cbl-gorilla.cs.technion.ac.il/)43. Analysis of the 53 upregulated proteins against a background of 1,312 proteins (the total number of proteins identified with high confidence) revealed 15 enriched biological processes, 14 of which involve the nematode’s responses to infection or other external stimuli (Table 2). For example, the most significantly enriched GO term is “innate immune response” (FDR = 8.7E-09, enriched 9.28-fold); and the highest enriched GO term is “defense response to Gram-negative bacterium” (FDR = 7.08E-05, enriched 12.85-fold) (Table 2). As P. aeruginosa is a Gram-negative bacterium and known to trigger innate immune responses in C. elegans39,40,41,44, these results validate the effectiveness of our proteomics approach for detecting differentially expressed proteins under infection conditions. We also found that 16 proteins were downregulated in P. aeruginosa-infected wild-type worms by more than 1.5-fold (Table S1). Gorilla analysis of these 16 proteins yielded no enriched GO terms.
Table 1. P. aeruginosa-induced proteins in wild-type N2 animals.
| Functional Group | Protein | Gene ID | Fold Change# | p-Value | Overlap with published microarray data* |
||
|---|---|---|---|---|---|---|---|
| Troemel et al. | Shapira et al. | Evan et al. | |||||
| CUB-like domain | DOD-24 | c32h11.12 | only in the infected | x | x | ||
| F55G11.2 | f55g11.2 | 10.1 | 7.58E-03 | x | x | x | |
| F55G11.4 | f55g11.4 | only in the infected | |||||
| C32H11.4 | c32h11.4 | only in the infected | x | x | x | ||
| DOD-17 | k10d11.1 | only in the infected | x | x | |||
| C17H12.8 | c17h12.8 | only in the infected | x | x | |||
| C-type lectin | CLEC-63 | f35c5.6 | 3.4 | 1.11E-04 | |||
| CLEC-66 | f35c5.9 | only in the infected | x | x | |||
| Lysozyme | LYS-2 | y22f5a.5 | only in the infected | x | |||
| LMP-1 | c03b1.12 | only in the infected | |||||
| ShK toxin domain | C14C6.5 | c14c6.5 | only in the infected | x | x | ||
| GST | GST-38 | f35e8.8 | only in the infected | x | x | ||
| Proteolysis/hydrolysis | ASP-14 | k10c2.3 | 3.4 | 5.97E-04 | x | x | |
| M60.2 | m60.2 | only in the infected | x | ||||
| BRE-1 | c53b4.7 | 2.3 | 1.18E-03 | ||||
| HEX-1 | t14f9.3 | only in the infected | |||||
| CAT-4 | f32g8.6 | only in the infected | |||||
| B0222.5 | b0222.5 | only in the infected | |||||
| oxidoreductases | F20D6.11 | f20d6.11 | only in the infected | x | |||
| VNA | r04b5.5 | 1.8 | 5.72E-03 | ||||
| C55A6.4 | c55a6.4 | only in the infected | |||||
| GLRX-5 | y49e10.2 | 3.3 | 4.56E-03 | ||||
| F53C11.3 | f53c11.3 | 5.1 | 2.63E-03 | x | x | ||
| DAF-22 | y57a10c.6 | 1.9 | 7.01E-03 | x | |||
| MAOC-1 | e04f6.3 | 2.0 | 2.93E-03 | x | |||
| Heat shock protein | Y55F3BR.6 | y55f3br.6 | 2.0 | 2.25E-03 | |||
| DNJ-19 | t05c3.5 | 3.1 | 2.28E-03 | ||||
| Development | NASP-2 | c50b6.2 | 2.5 | 6.75E-03 | |||
| Y71F9AL.9 | y71f9al.9 | 2.3 | 5.39E-03 | ||||
| CPG-2 | b0280.5 | 2.1 | 6.37E-03 | ||||
| MLC-4 | c56g7.1 | 2.2 | 2.14E-03 | ||||
| VPS-32.1 | c56c10.3 | 2.7 | 4.38E-03 | ||||
| T23D8.3 | t23d8.3 | only in the infected | |||||
| RMD-1 | t05g5.7 | only in the infected | |||||
| SEC-24.2 | zc518.2 | 2.1 | 6.44E-03 | ||||
| MRPL-34 | c25a1.13 | only in the infected | |||||
| Others or unknown | DSC-4 | k02d7.4 | 1.8 | 9.23E-03 | |||
| CEY-4 | y39a1c.3 | 1.7 | 4.01E-04 | ||||
| ZC247.1 | zc247.1 | 1.7 | 4.37E-03 | ||||
| PQN-59 | r119.4 | 1.7 | 6.29E-03 | ||||
| PUD-2.1 | f15e11.1 | 2.6 | 4.85E-05 | ||||
| Y44A6D.2 | y44a6d.2 | 1.8 | 1.59E-03 | ||||
| Y69A2AR.18 | y69a2ar.18 | 1.7 | 8.28E-03 | ||||
| VDAC-1 | r05g6.7 | only in the infected | |||||
| CLEC-209 | f56a4.2 | only in the infected | |||||
| C15C7.5 | c15c7.5 | 4.3 | 5.37E-04 | ||||
| W05H9.1 | w05h9.1 | 2.1 | 3.93E-03 | ||||
| GLB-1 | zk637.13 | 3.0 | 5.27E-03 | x | |||
| ZK418.9 | zk418.9 | 1.8 | 4.27E-03 | ||||
| LPD-8 | r10h10.1 | 2.1 | 1.34E-03 | ||||
| SNA-1 | w02f12.6 | only in the infected | |||||
| IRG-3 | f53e10.4 | only in the infected | x | x | |||
| LBP-4 | zk742.5 | 2.1 | 8.12E-03 | ||||
#Only in the infected: the protein was only detected in the infected animals, not in the uninfected animals.
*Troemel et al. 2006 PLoS Genet 2(11):183; Shapira et al. 2006 Proc Natl Acad Sci USA 103(38):14086–14091; Evans et al. 2008 PLoS Pahtog 4(10):e1000175.
Table 2. GO term (biological process) enrichment analysis of P. aeruginosa-induced proteins in wild-type N2 animals.
| GO term | Description | P-value# | FDR q-value* | Enrichment (N, B, n, b)§ |
|---|---|---|---|---|
| GO:0002376 | immune system process | 1.25E-11 | 2.63E-08 | 9.28 (1098, 36, 46, 14) |
| GO:0006955 | immune response | 1.25E-11 | 1.32E-08 | 9.28 (1098, 36, 46, 14) |
| GO:0045087 | innate immune response | 1.25E-11 | 8.77E-09 | 9.28 (1098, 36, 46, 14) |
| GO:0006952 | defense response | 7.98E-11 | 4.19E-08 | 7.46 (1098, 48, 46, 15) |
| GO:0006950 | response to stress | 7.15E-08 | 3.00E-05 | 3.91 (1098, 110, 46, 18) |
| GO:0050829 | defense response to Gram-negative bacterium | 2.03E-07 | 7.08E-05 | 12.85 (1098, 13, 46, 7) |
| GO:0050896 | response to stimulus | 2.27E-07 | 6.79E-05 | 3.64 (1098, 118, 46, 18) |
| GO:0043207 | response to external biotic stimulus | 1.23E-06 | 3.22E-04 | 10.44 (1098, 16, 46, 7) |
| GO:0098542 | defense response to other organism | 1.23E-06 | 2.86E-04 | 10.44 (1098, 16, 46, 7) |
| GO:0042742 | defense response to bacterium | 1.23E-06 | 2.57E-04 | 10.44 (1098, 16, 46, 7) |
| GO:0009617 | response to bacterium | 1.23E-06 | 2.34E-04 | 10.44 (1098, 16, 46, 7) |
| GO:0009607 | response to biotic stimulus | 1.23E-06 | 2.15E-04 | 10.44 (1098, 16, 46, 7) |
| GO:0051707 | response to other organism | 1.23E-06 | 1.98E-04 | 10.44 (1098, 16, 46, 7) |
| GO:0009605 | response to external stimulus | 3.87E-05 | 5.80E-03 | 6.68 (1098, 25, 46, 7) |
| GO:0051704 | multi-organism process | 4.65E-04 | 6.50E-02 | 3.64 (1098, 59, 46, 9) |
#P-value is the enrichment p-value computed according to the mHG model (Eden et al. 2007 PLos Comp Bio 3(3):e39).
*FDR q-value is the correction of the above p-value for multiple testing using the Benjamini and Hochberg method (Benjamini and Hochberg 1995 J R Statist Soc B 57(1):289–300).
§Enrichment (N, B, n, b) is defined as follows:
N - is the total number of proteins.
B - is the total number of proteins associated with a specific GO term.
n - is the number of proteins in the target set.
b - is the number of proteins in the intersection.
Enrichment = (b/n)/(B/N).
The proteins upregulated by P. aeruginosa infection belong to a number of functional groups, as shown in Table 1. These include markers of immunity, such as CUB-like proteins, C-type lectins, lysozymes, proteins containing ShK toxin domain, and glutathione s-transferases (GSTs). Immune markers are a set of genes/proteins that are positively regulated in various organisms in response to a wide range of pathogens; they are believed to be the immune effectors that enhance the host’s ability to fight off pathogens1,2,6,45. Many immune markers were discovered by transcription studies. Our finding that marker genes are also induced at the protein level suggests that these genes are biologically functional during immune response. When compared with three whole-genome transcription studies in the literature39,40,41, which used microarray to detect differentially expressed genes in C. elegans upon P. aeruginosa infection, nine out of the 12 marker proteins that were upregulated in our proteomics study are also induced at the transcript level (Table 1). Table 1 also shows the upregulation of a group of 13 proteins that have proteolysis, hydrolysis or oxidoreductase activities, which might reflect increased metabolism in the nematode and the need for extra energy whilst fighting off an infection. About half of these proteins overlap with the induced transcripts revealed in the microarray studies39,40,41. Despite the fact that 70% of the 53 upregulated proteins have no match in the transcriptomic data (Table 1), the two types of studies showed good agreement on the enrichment of innate immune proteins and enzymes involved in macromolecule metabolism.
OCTR-1 regulates innate immune responses at the protein level
Previously we have shown that neuronally expressed OCTR-1 suppresses P. aeruginosa-triggered innate immune responses in C. elegans by down-regulating gene expression in the UPR pathways and the p38 MAPK pathway17,18. These studies were performed at the transcript level. To investigate how OCTR-1 regulates innate immunity at the level of protein expression, we examined the proteomic changes of octr-1(ok371) worms relative to wild-type worms exposed to P. aeruginosa. As shown in Table S2, infection upregulated 113 proteins in the mutant worms. Twenty-three of these are immunity marker proteins, which almost doubles the number of the immune proteins induced in the wild-type worms (Table 3 versus Table 1). The result is consistent with an inhibitory role of OCTR-1 in innate immunity17,18. Strikingly, all 12 immune proteins induced in wild-type worms are upregulated in octr-1(ok371) worms, illustrating the high-degree reproducibility of our proteomics method. Besides the immune proteins, we also observed significant induction of proteins in the UPR pathway in octr-1(ok371) worms (Table 3), demonstrating that OCTR-1 inhibits the UPR at the protein level. This correlates well with our previous genome microarray study that showed OCTR-1 suppresses the UPR at the transcript level17,18. Knockdown of a number of UPR genes individually (abu-1, -7, -8, -12, -13, xbp-1, or Y41C4A.11) by RNA interference (RNAi) partially or fully rescued the mutant phenotype of octr-1(ok371) worms, i.e. decreased the immunity of the mutants to the wild-type level17,18. Taken together, these results suggest that elevated UPR activity contributes to the enhanced immunity of octr-1 mutant worms.
Table 3. P. aeruginosa-induced proteins in octr-1(ok371) animals.
| Biological Functions | Protein | Gene ID | Description | Fold Change# | p-Value |
|---|---|---|---|---|---|
| Innate immune response | DOD-24 | c32h11.12 | CUB-like domain | infected only | |
| F55G11.2 | f55g11.2 | CUB-like domain | infected only | ||
| F55G11.4 | f55g11.4 | CUB-like domain | infected only | ||
| C32H11.4 | c32h11.4 | CUB-like domain | infected only | ||
| DOD-17 | k10d11.1 | CUB-like domain | infected only | ||
| CLEC-67 | f56d6.2 | C-type lectin | infected only | ||
| CLEC-66 | f35c5.9 | C-type lectin | infected only | ||
| CLEC-63 | f35c5.6 | C-type lectin | 3.5 | 6.05E-03 | |
| SKPO-1 | f49e12.1 | ShK toxin domain | infected only | ||
| C14C6.5 | c14c6.5 | ShK toxin domain | infected only | ||
| LYS-2 | y22f5a.5 | lysozyme | infected only | ||
| GST-38 | f35e8.8 | glutathione S-transferase | infected only | ||
| GST-5 | r03d7.6 | glutathione S-transferase | 9.5 | 6.98E-03 | |
| GST-4 | k08f4.7 | glutathione S-transferase | 2.8 | 9.81E-03 | |
| GST-7 | f11g11.2 | glutathione S-transferase | 3.9 | 9.31E-03 | |
| GCS-1 | f37b12.2 | glutathione biosynthesis | 2.7 | 8.72E-03 | |
| ASP-14 | k10c2.3 | aspartyl protease | 7.2 | 5.77E-03 | |
| M60.2 | m60.2 | ortholog of human endonuclease, polyU-specific | infected only | ||
| C17H12.8 | c17h12.8 | involved in innate immune response | infected only | ||
| F35E12.6 | f35e12.6 | involved in innate immune response | infected only | ||
| IRG-3 | f53e10.4 | infection response protein | infected only | ||
| SKR-3 | f44g3.6 | skp1 related (ubiquitin ligase complex component) | 2.3 | 6.32E-03 | |
| DJR-1.1 | b0432.2 | glutathione-independent glyoxalase DJR-1.1 | 1.8 | 2.25E-03 | |
| Sulfur amino acid biosynthesis | METR-1 | r03d7.1 | probable methionine synthase | infected only | |
| C01G10.9 | c01g10.9 | methylthioribose-1-phosphate isomerase | infected only | ||
| F58H1.3 | f58h1.3 | enolase-phosphatase E1 | 2.5 | 8.81E-05 | |
| SAMS-4 | c06e7.3 | probable S-adenosylmethionine synthase 4 | 1.8 | 9.82E-03 | |
| CTH-2 | zk1127.10 | putative cystathionine gamma-lyase 2 | 4.1 | 6.44E-03 | |
| CYSL-2 | k10h10.2 | bifunctional L-3-cyanoalanine synthase/cysteine synthase | 2.3 | 1.71E-04 | |
| HMG-11 | t05a7.4 | predicted to have DNA binding activity | infected only | ||
| Translation/protein synthesis | RPL-18 | y45f10d.12 | 60S ribosomal protein L18 | 2.1 | 5.23E-03 |
| RPS-10 | d1007.6 | ribosomal protein, small subunit | 2.1 | 5.22E-03 | |
| RPL-13 | c32e8.2 | 60S ribosomal protein L13 | 2.0 | 7.94E-03 | |
| RPS-11 | f40f11.1 | ribosomal protein, small subunit | 1.7 | 7.59E-03 | |
| RPS-1 | f56f3.5 | 40S ribosomal protein S3a | 1.6 | 8.48E-03 | |
| RPS-28 | y41d4b.5 | 40S ribosomal protein S28 | 1.4 | 9.56E-03 | |
| MRPL-12 | w09d10.3 | mitochondrial ribosomal protein, large | 2.5 | 5.09E-03 | |
| RUVB-1 | c27h6.2 | RuvB-like 1 | infected only | ||
| EIF-3.J | y40b1b.5 | eukaryotic initiation factor | 1.7 | 4.35E-03 | |
| K07C5.4 | k07c5.4 | ortholog of human NOP56 ribonucleoprotein | 1.5 | 9.73E-03 | |
| SRP-6 | c03g6.19 | serpin (serine protease inhibitor) | infected only | ||
| proteolysis/hydrolysis | KLO-1 | c50f7.10 | KLOtho (mammalian aging-associated protein) homolog | 2.6 | 1.74E-03 |
| F13H6.3 | f13h6.3 | ortholog of human carboxylesterase 2 | 2.3 | 4.69E-03 | |
| TAX-6 | c02f4.2 | Serine/threonine-protein phosphatase | 2.0 | 8.34E-03 | |
| Y25C1A.13 | y25c1a.13 | ortholog of human enoyl CoA hydratase 1 | 2.0 | 9.29E-03 | |
| PAM-1 | f49e8.3 | puromycin-sensitive aminopeptidase | 1.8 | 2.09E-03 | |
| T22C1.6 | t22c1.6 | ortholog of human taxilin α, β, and γ | infected only | ||
| ER/UPR | Y41C4A.11 | y41c4a.11 | involved in ER UPR | 4.3 | 1.84E-03 |
| C14B9.2 | c14b9.2 | probable protein disulfide-isomerase A4 | 2.3 | 1.52E-03 | |
| HSP-4 | f43e2.8 | heat shock protein | 2.2 | 2.00E-03 | |
| PDI-1 | c14b1.1 | protein disulfide isomerase | 1.6 | 8.26E-03 | |
| SNA-1 | w02f12.6 | snRNP-binding protein | infected only | ||
| Transcription | HRP-1 | f42a6.7 | putative hnRNP | 5.5 | 6.47E-05 |
| T13F2.2 | t13f2.2 | predicted to have transcription coactivator activity | 3.3 | 1.74E-03 | |
| RPB-2 | c26e6.4 | RNA polymerase II (B) subunit | 3.1 | 3.22E-05 | |
| RPB-9 | y97e10ar.5 | RNA polymerase II (B) subunit | 2.5 | 6.86E-03 | |
| CEY-4 | y39a1c.3 | C. elegans Y-box | 2.0 | 1.05E-04 | |
| CEY-2 | f46f11.2 | C. elegans Y-box | 1.7 | 3.55E-03 | |
| CPG-2 | b0280.5 | chondroitin proteoglycan | infected only | ||
| Development/reproduction | C29E4.12 | c29e4.12 | ortholog of human C7orf55 | infected only | |
| Y18D10A.9 | y18d10a.9 | ortholog of human CIAO1 | infected only | ||
| T23D8.3 | t23d8.3 | ortholog of human and yeast LTV1 | infected only | ||
| K07H8.10 | k07h8.10 | predicted to have necleic acid and nucleotide binding activity | 8.2 | 1.56E-05 | |
| F55B11.2 | f55b11.2 | development | 4.4 | 9.81E-03 | |
| DNC-2 | c28h8.12 | dynactin complex component | 3.2 | 3.30E-03 | |
| F32A11.3 | f32a11.3 | involved in reproduction | 3.2 | 9.82E-03 | |
| Y71F9AL.9 | y71f9al.9 | ortholog of human SPATS2L and SPATS2 | 2.1 | 4.21E-04 | |
| DPY-30 | zk863.6 | dosage compensation protein | 2.0 | 4.34E-03 | |
| PRX-19 | f54f2.8 | Peroxisome assembly factor | 1.9 | 9.17E-03 | |
| MRPS-28 | y43f8c.8 | Mitochondrial ribosomal Protein | 1.8 | 1.63E-03 | |
| C44E4.4 | c44e4.4 | ortholog of human Sjogren syndrome atigen B | 1.6 | 7.28E-03 | |
| F20D6.11 | f20d6.11 | putative FAD-binding oxidoreductase | infected only | ||
| Cell redox homeostasis | F29C4.2 | f29c4.2 | ortholog of human cytochrome c oxidase subunit Vic | infected only | |
| Y47G6A.21 | y47g6a.21 | predicted to have oxidoreductase activity | 2.8 | 1.91E-03 | |
| F45H10.3 | f45h10.3 | predicted to have NADH dehydrogenase (ubiquinone) activity | 2.6 | 7.79E-03 | |
| PRDX-6 | y38c1aa.11 | preoxiredoxin 6 | 1.7 | 6.03E-03 | |
| KIN-10 | t01g9.6 | protein kinase | infected only | ||
| Protein Kinase | CAT-4 | f32g8.6 | GTP cyclohydrolase 1 | infected only | |
| MMCM-1 | zk1058.1 | methylmalonyl-CoA mutase homolog | 5.4 | 4.87E-03 | |
| compound metabolic process | C36A4.4 | c36a4.4 | probable UDP-N-acetylglucosamine pyrophosphorylase | 2.7 | 4.17E-03 |
| C39D10.7 | c39d10.7 | involved in chitin metabolic process | 2.0 | 6.91E-03 | |
| ZK1307.8 | zk1307.8 | ortholog of human protein kinase C substrate 80K-H | 1.8 | 8.37E-03 | |
| CTS-1 | t20g5.2 | citrate synthase | 1.7 | 4.81E-03 | |
| DPYD-1 | c25f6.3 | dihydropyrimidine dehydrogenase | 1.7 | 5.14E-03 |
#Infected only: the protein was only detected in the infected animals, not in the uninfected animals.
Interestingly, we observed significant induction of a number of proteins with functions in transcription, sulfur amino acid biosynthesis and translation in the mutant worms (Table 3). In contrast, abundance of these proteins in the wild-type worms did not change upon infection (Table 1). We attribute the induction of these proteins in the mutants to the lack of OCTR-1. Therefore, in addition to the UPR, OCTR-1 also potentially regulates transcription, sulfur amino acid biosynthesis and translation in response to P. aeruginosa infection. Surprisingly, the abundance of the transcripts of these proteins remained unchanged in octr-1(ok371) relative to wild-type worms in our previous whole-genome microarray study conducted under the same infection conditions17, suggesting that induction of these proteins occurs at the post-transcriptional level. Because enhanced transcription, sulfur amino acid biosynthesis and translation likely promote protein synthesis, it is reasonable to speculate that higher protein synthesis activity is responsible for the upregulation of many proteins in octr-1(ok371) worms, which in turn triggers strong UPR in the mutants.
OCTR-1 modulates protein synthesis in response to P. aeruginosa infection
Dunbar et al. showed that translation of green fluorescent protein (GFP) was blocked in the intestine of C. elegans during P. aeruginosa infection through the use of transgene hsp-16.2::GFP as a reporter for translation activity46. To investigate if OCTR-1 plays a direct role in modulating protein synthesis, we used a similar transgenic GFP reporter Phsp-4::GFP(zcls4). The C. elegans hsp-4 gene encodes a homologue of mammalian BiP/GRP-78; and intestinal expression of the transgene Phsp-4::GFP(zcIs4) has been used as an indicator of the XBP-1-dependent UPR18,47, which reflects protein accumulation and demand on protein folding in the endoplasmic reticulum. Previously we have demonstrated that upon P. aeruginosa infection, octr-1(ok371);Phsp-4::GFP(zcls4) animals exhibited significantly higher levels of GFP expression than Phsp-4::GFP(zcls4) animals (Fig. 2 in Sun et al.18). It is unclear whether the elevated GFP expression was due to upregulation of transcription or increased translation or both. To find out the cause of this phenomenon, we performed qRT-PCR to compare the levels of GFP mRNA in Phsp-4::GFP(zcls4) and octr-1(ok371);Phsp-4::GFP(zcls4) animals exposed to P. aeruginosa. As shown in Fig. 2, the levels of GFP mRNA were not significantly different between the two strains, indicating that the elevated GFP expression in the octr-1 mutants was due to upregulation of translation, not transcription. Hence, OCTR-1 modulates protein synthesis in response to P. aeruginosa infection.
Figure 2. qRT-PCR analysis of gfp gene expression in Phsp-4::GFP(zcls4) and octr-1(ok371);Phsp-4::GFP(zcls4) animals exposed to P. aeruginosa.
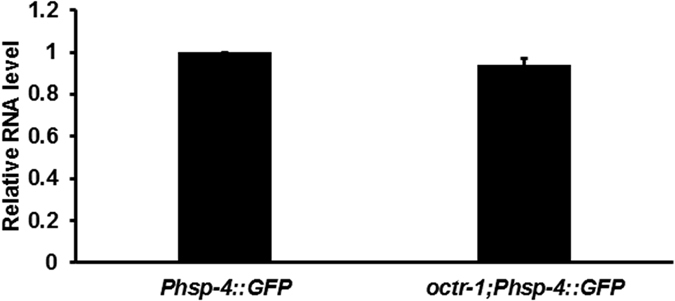
Relative fold-changes for gfp transcripts were normalized to pan-actin (act-1, -3, -4). Phsp-4::GFP(zcls4) versus octr-1(ok371);Phsp-4::GFP(zcls4), p = 0.07. Bars represent mean ± SEM. n = 3 independent experiments.
Protein synthesis factors are involved in the OCTR-1-dependent immunity
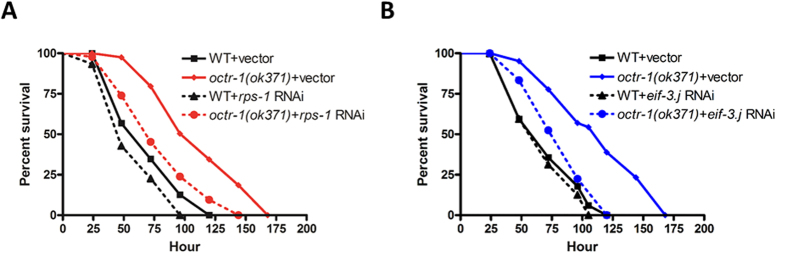
Eight ribosomal proteins (RPs) were upregulated by P. aeruginosa in octr-1(ok371) animals relative to wild-type animals, including RPS-1, RPS-11, MRPL-12, RPL-13, RPL-18, RPS-10, RPS-28, and K07C5.4 (Table 3). Recent studies revealed the multifunctional roles of RPs ranging from protein synthesis to apoptosis48,49,50. Identification of these upregulated RPs raises an important question: do RPs play any roles in innate immunity? To answer this question, we knocked down the expression of the above RPs individually by RNAi in both wild-type and octr-1(ok371) animals, then measured the nematode’s survival against P. aeruginosa infection, as well as their lifespan on a standard food source E. coli OP50. This approach allows us to determine gene contribution to immunity independently of possible functions important for development or lifespan. The RNAi experiment demonstrated that silencing the RPs in many cases led to delay in development or even to maternal sterility (Table S3). The results are in agreement with the previous studies in zebrafish that showed knockdown of many individual RPs causes cell death or defective development51,52. As components of ribosome, the expression of RPs is tightly regulated to provide the appropriate ratio between RPs and rRNAs as well as among RPs. Thus, perturbation of ribosome biogenesis results in ribosomal stress, which can trigger p53-dependent cell cycle arrest and apoptosis53,54,55. Nonetheless, we observed that RNAi of rps-1 did not alter the development (Table S3) or lifespan of C. elegans (Figure S1), but strongly enhanced susceptibility to P. aeruginosa in octr-1 mutant animals and also exerted a subtle effect on the susceptibility of wild-type animals (Fig. 3A). The result indicates that this RP plays a role in innate immunity of C. elegans, which adds value to the extraribosomal functions of RPs.
Figure 3. Protein synthesis factors are involved in the OCTR-1-depedent immunity.
(A) Wild-type and octr-1(ok371) animals grown on double-stranded RNA (dsRNA) for vector control or dsRNA for rps-1 were exposed to P. aeruginosa PA14 and scored for survival over time. WT+ vector versus WT+rps-1 RNAi: p = 0.0386; octr-1(ok371)+vector versus octr-1(ok371)+rps-1 RNAi: p < 0.0001. Shown is a representative assay of three independent experiments. n = 45 young adult animals per strain. Significant knockdown of rps-1 expression by RNAi was confirmed by qRT-PCR (p < 0.001, Figure S2A). (B) Wild-type and octr-1(ok371) animals grown on dsRNA for vector control or dsRNA for eif-3.j were exposed to P. aeruginosa PA14 and scored for survival over time. WT+vector versus WT+eif-3.j RNAi: p = 0.4873; octr-1(ok371)+vector versus octr-1(ok371)+ eif-3.j RNAi: p = 0.0001. Shown is a representative assay of three independent experiments. n = 45 young adult animals per strain. p values < 0.05 are considered significant. Significant knockdown of eif-3.j expression by RNAi was confirmed by qRT-PCR (p < 0.001, Figure S2B).
Besides the above RPs, two more proteins (RUVB-1 and EIF-3.J) involved in protein synthesis were upregulated in octr-1(ok371) animals compared to wild-type animals upon P. aeruginosa infection (Table 3 versus Table 1). RUVB-1 is an AAA+ ATPase orthologous to the RUVBL1 family of ATPases and functions as a component of the Target of Rapamycin (TOR) signaling pathway that enables robust protein synthesis56. EIF-3.J is an orthologue of the j-subunit of human translation initiation factor EIF3 that participates in nearly all steps of translation initiation57. While RNAi of ruvb-1 caused developmental delay (Table S3), knockdown of eif-3.j increased pathogen susceptibility in octr-1 mutants, whereas the same knockdown had no effects in wild-type animals (Fig. 3B). This result indicates that EIF-3.J is important for the OCTR-1-controlled immunity.
Protein synthesis factors RPS-1 and EIF-3.J contribute to the elevated UPR in infected octr-1(ok371) animals
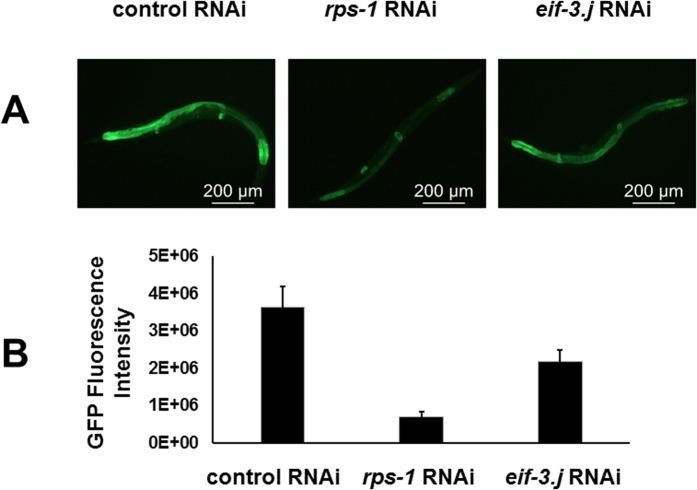
Because both RPS-1 and EIF-3.J have important roles in protein translation, lack of either protein is expected to reduce protein synthesis. This would also lower the elevated UPR in octr-1(ok371) animals exposed to P. aeruginosa. To test this prediction, we took advantage of the established correlation between intestinal expression of Phsp-4::GFP(zcls4) and XBP-1-dependent UPR18,47, and used Phsp-4::GFP(zcls4) as a reporter to examine if knockdown of rps-1 or eif-3.j by RNAi could lower the elevated UPR in P. aeruginosa-infected octr-1(ok371);Phsp-4::GFP(zcls4) animals18. As shown in Fig. 4, RNAi of rps-1 or eif-3.j in infected octr-1(ok371);Phsp-4::GFP(zcls4) animals significantly reduced Phsp-4::GFP expression, as compared to the control RNAi with an empty vector. The above result demonstrated that both RPS-1 and EIF-3.J contribute to the elevated UPR in P. aeruginosa-infected octr-1(ok371) animals, which in turn enhances their immunity. Because expression of the same transgene Phsp-4::GFP(zcls4) also reflects OCTR-1-dependent protein synthesis (Section 3 in Results), the contribution of RPS-1 and EIF-3.J in UPR can be attributed to their protein synthesis activity.
Figure 4. Knockdown of rps-1 or eif-3.j by RNAi reduces the XBP-1-dependent UPR in octr-1 mutant animals.
(A) Images of octr-1(ok371);Phsp-4::GFP(zcls4) animals. Animals were grown on double-stranded RNA for vector control or dsRNA for rps-1 or eif-3.j, and young adult animals were exposed to P. aeruginosa PA14. Animals that best represent the fluorescence level of the population were shown. (B) GFP quantification of octr-1(ok371);Phsp-4::GFP(zcls4) animals exposed to P. aeruginosa PA14. Binary mean intensity of the region of interest (ROI) that corresponds to an entire animal was measured by Image J software. n = 10–20 animals, error bars represent SEM. A two-sample t test was performed to compare the fluorescence intensity between populations. rps-1 RNAi versus control RNAi: p = 6.1 × 10−10; eif-3.j RNAi versus control RNAi: p = 4.8 × 10−7. p values < 0.05 are considered significant. Significant knockdown of rps-1 or eif-3.j expression by RNAi was confirmed by qRT-PCR (p < 0.001, Figure S3).
Translational inhibition by chemicals abolishes the OCTR-1-controlled innate immune responses
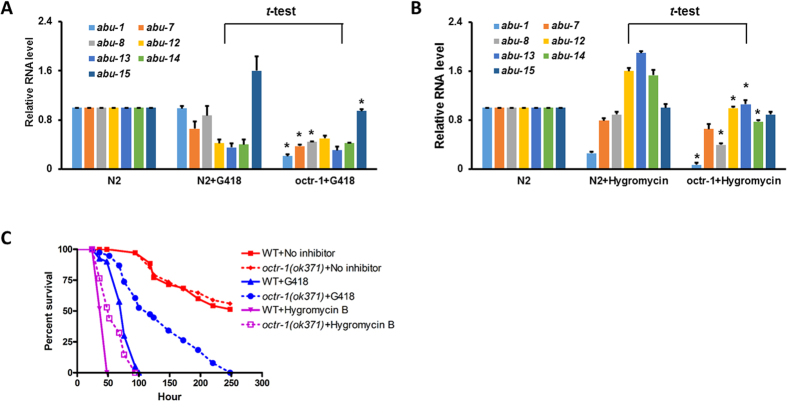
Translational inhibition is a very common pathogenic attack strategy; consequently, response to such inhibition is a conserved form of host defense46,58,59,60,61. However, how this response is regulated in the host remains largely unknown. Because OCTR-1 downregulates protein synthesis activities, the OCTR-1 pathway could function to suppress excessive responses to translational inhibition or to restore protein homeostasis after infection. To test this possibility, we treated wild-type and octr-1(ok371) animals with translation inhibitors G418 and Hygromycin B. These chemicals have been used to mimic P. aeruginosa-mediated host translational suppression in C. elegans46,58. We evaluated the effects of such inhibition by assessing: 1) expression levels of the OCTR-1-dependent immune genes; and 2) animal survival against the inhibition. Previously, we have shown that OCTR-1 suppresses innate immunity by downregulating the expression of non-canonical UPR abu (activated in blocked unfolded protein response) genes17. Here we examined the expression of seven abu genes by qRT-PCR, including abu-1, abu-7, abu-8, abu-12, abu-13, abu-14 and abu-15. As shown in Fig. 5A,B, G418 or Hygromycin B itself had various effects on the expression of abu genes in wild-type animals. Specifically, G418 downregulates abu-7, abu-12, abu-13 and abu-14 and upregulates abu-15, while Hygromycin B downregulates abu-1 and upregulates abu-12, abu-13 and abu-14. It is unclear why these translational inhibitors differentially regulate the abu genes. Nonetheless, a comparison of gene expression levels between G418-treated wild-type and octr-1(ok371) animals showed that abu-1, abu-7, abu-8, and abu-15 were significantly downregulated in the octr-1 mutants (Fig. 5A). Similarly, a comparison of gene expression levels between Hygromycin B-treated wild-type and octr-1(ok371) animals showed that abu-1, abu-8, abu-12, abu-13, and abu-14 were significantly downregulated in the octr-1 mutants (Fig. 5B). Interestingly, none of the abu genes increased expression in the inhibitor-treated octr-1(ok371) animals compared to wild-type animals with or without treatments (Fig. 5A,B). These results indicate that activation of the OCTR-1-controlled innate immune responses (i.e. upregulation of abu genes in octr-1 mutants relative to wild-type animals) is dependent on active translation. Despite lack of the OCTR-1-controlled immune responses, octr-1(ok371) animals are more resistant than wild-type animals to G418 or Hygromycin B treatment, although the chemical treatments shortened the survival time of both strains (Fig. 5C). It is likely that the higher protein translation activities in the mutants offset some of the inhibitory effects of the chemicals, thus allow their longer survival than the wild-type strains. These results suggest that protein translation is important for the OCTR-1-controlled innate immunity.
Figure 5. Translational inhibition abolishes the OCTR-1-controlled innate immune responses.
(A) qRT-PCR of abu-1, abu-7, abu-8, abu-12, abu-13, abu-14, and abu-15 expression in wild-type animals with or without exposure to G418 and in octr-1(ok371) animals exposed to G418. n = 3; bar graphs correspond to mean ± SEM. t-tests were performed between N2 + G418 and octr-1 + G418. *indicates significant difference. (B) qRT-PCR of abu-1, abu-7, abu-8, abu-12, abu-13, abu-14, and abu-15 expression in wild-type animals with or without exposure to Hygrocymin B and in octr-1(ok371) animals exposed to Hygromycin B. n = 3; bar graphs correspond to mean ± SEM. t-tests were performed between N2 + G418 and octr-1 + G418. *indicates significant difference. (C) Wild-type and octr-1(ok371) animals were exposed to NGM/E. coli OP50 (control) or NGM/E. coli OP50 containing G418 or Hygromycin B, and scored for survival over time. WT + No inhibitor versus octr-1(ok371)+No inhibitor: p = 0.7521; WT + G418 versus octr-1(ok371)+G418: p < 0.0001; WT + Hygromycin B versus octr-1(ok371)+Hygromycin B: p < 0.0001. Shown is a representative assay of two independent experiments. n = 45 young adult animals per strain. p values < 0.05 are considered significant.
Discussion
We have examined the proteomic changes in C. elegans upon P. aeruginosa infection using a label free quantitative proteomics approach. Fifty-three proteins were significantly upregulated in the infected wild-type worms relative to the uninfected controls. The data are in good agreement with the previously published transcriptomic studies39,40,41 on the enrichment of innate immune proteins and enzymes involved in macromolecule metabolism (Table 1). However, about 70% of the 53 proteins have no match in the transcriptomic data, including a set of nine proteins with known functions in development and a number of proteins with unknown functions (Table 1). Whether these proteins play any roles in immunity or pathogenesis warrant further investigation.
Previously we have demonstrated that OCTR-1 functions in ASH and ASI neurons to suppress transcription of non-canonical UPR genes of the pqn/abu family17,18. In the current proteomics study, we have identified ten PQN proteins, including PQN-22, PQN-24, PQN-27, PQN-32, PQN-41, PQN-51, PQN-52, PQN59, PQN-74 and PQN87. Surprisingly, none of these proteins have known functions in the non-canonical UPR pathway17,62. Many abu and pqn genes that were found significantly upregulated in our previous genome microarray study17 have not been identified at the protein level in the current proteomics study. This discrepancy is most likely caused by the different technologies used in the two studies. The C. elegans GeneChip (Affymetrix, Santa Clara, CA) was used in the genome microarray study, which examines the expression of 22,500 transcripts, while label-free quantitative nano-HPLC tandem mass spectrometry was used in the proteomics study, which only identified 4,413 proteins with 1,312 proteins identified with high confidence. Technical resolution at the proteome level is much more constrained than that at the transcriptome level, leading to a much lower proteome coverage than transcriptome coverage and identification of only those proteins with high difference in abundance63.
Consistent with our genome microarray study17, here we show that OCTR-1 inhibits translation and the UPR at the protein level. Our RNAi experiments and functional assays support the following conclusions: 1) OCTR-1 inhibits expression of specific protein synthesis factors, such as ribosomal protein RPS-1 and translation initiation factor EIF-3.J, which reduces infection-triggered protein synthesis and UPR; 2) upregulation of UPR proteins and elevated UPR in octr-1(ok371) animals contribute to their enhanced immunity; 3) RPS-1 and EIF-3.J contribute to the elevated UPR in infected octr-1(ok371) animals; and 4) activation of the OCTR-controlled immune response is dependent on active translation. The contributions of the upregulated protein synthesis activities to the enhanced immunity of infected octr-1(ok371) animals are two-fold: 1) higher protein synthesis activities lead to production of more immune proteins, which enhance the nematode’s ability to fight off invading pathogens; and 2) increased protein production results in elevated UPR, which in turn improves the animal immunity. As translational inhibition is a very common pathogenic attack strategy and OCTR-1 has homology in various species including humans, the OCTR-1-dependent pathway may be a conserved signaling pathway that the nervous system uses to control protein homeostasis during host immune defense.
Materials and Methods
C. elegans and bacterial strains
The following C. elegans strains were cultured under standard conditions and fed E. coli OP5064. Wild-type worms were C. elegans Bristol N2. Octr-1(ok371) and Phsp-4::gfp (zcIs4) strains were obtained from the Caenorhabditis elegans Genetics Center (University of Minnesota, Minneapolis, MN). The mutant octr-1(ok371);Phsp-4::GFP(zcIs4) was constructed using standard genetic techniques18. E. coli strain OP50 and P. aeruginosa strain PA1444 were grown in Luria-Bertani (LB) broth at 37 °C.
C. elegans survival assay
C. elegans wild-type worms and mutants were maintained as hermaphrodites at 20 °C and fed with E. coli OP50 on modified nematode growth medium (NGM) agar plates (0.35% instead of 0.25% peptone) as described64. The bacterial lawn used for C. elegans killing assays were prepared by placing a 25 μl drop of an overnight culture of the bacterial strains on modified NGM agar plates (3.5 cm diameter Petri plates). Plates were incubated at 37 °C for 16 hr. Plates were cooled down at room temperature for at least 1 hr before seeding with synchronized worms. The survival assays were performed at 25 °C and live worms were transferred daily to fresh plates. Worms were scored at the times indicated and were considered dead when they failed to respond to touch.
C. elegans infection and collection
Synchronized wild-type and octr-1(ok371) worms grown to L4 larval stage were infected with P. aeruginosa at 25 °C for 4 hr, as we described previously17. Infected worms and uninfected controls (on E. coli OP50) were collected and washed 5 times with M9 buffer in the presence of protease inhibitors (ProteaseArrest, G-Biosciences, St. Louis, MO) to remove surface bound bacteria. In the last wash, after centrifugation and removal of the supernatant, each worm pellet was immediately frozen in liquid nitrogen. Five biological replicates of infected and uninfected worms were collected. The frozen worm samples were submitted to the Tissue Imaging and Proteomics Laboratory at Washington State University (Pullman, WA) for mass spectrometry-based quantitative proteomics analyses.
Protein sample preparation
The worm pellet was ground into fine powder using a single 2.8 mm i.d. steel ball with a TissueLyser II (Qiagen, Valencia, CA) at a frequency of 30 Hz for 30 sec, followed by the addition of 100 μl extraction solvent of PBS buffer, pH 7.5, with protease inhibitor and vortexing. Supernatants were collected after centrifugation at 16,000 × g (10 min, 4 °C). Protein was then quantified with a Qubit Protein Assay Kit (Life Technologies, Carlsbad, CA) in compliance with the manufacturer’s protocol. Disulfide bonds were reduced using 100 mM dithiothreitol (DTT) at a ratio of 1:10 DTT/sample volume and incubated at 50 °C for 45 min. Cysteine bonds were then alkylated with 200 mM iodoacetamide at the same volume ratio for 20 min at room temperature. Finally, protein was digested with trypsin (G-Biosciences, St. Louis, MO) at a 1:50 ratio of trypsin/protein, and incubated at 37 °C for 12 hr.
High resolution nano-HPLC tandem mass spectrometry analysis
The peptide samples were subjected to Thermo Scientific Orbitrap Fusion Tribrid with an Easy-nLC 1000 ultra-high pressure LC on a Thermo Scientific PepMap 100 C18 column (2 μm, 50 μm × 15 cm). The peptides were separated over 115 min gradient eluted at 400 nL/min with 0.1% formic acid (FA) in water (solvent A) and 0.1% FA in acetonitrile (solvent B) (5–30% B in 85 min, followed by 30–50% B over 10 min and 50–97% B over 10 min). The run was completed by holding a 97% B for 10 min. MS1 data was acquired on an Orbitrap Fusion mass spectrometry using a full scan method according to the following parameters: scan range 400–1500 m/z, Orbitrap resolution 120,000; AGC target 400,000; and maximum injection time of 50 ms. MS2 data were collected using the following parameters: rapid scan rate, HCD collision energy 35%, 1.6 m/z isolation window, AGC 2,000 and maximum injection time of 50 ms. MS2 precursors were selected for a 3 s cycle. The precursors with an assigned monoisotopic m/z and a charge state of 2–7 were interrogated. The precursors were filtered using a 60 s dynamic exclusion window. MS/MS spectra were searched using Thermo Scientific Proteome Discoverer software version 2.0 with SEQUEST® against uniprot Caenorhabditis elegans database (TaxID = 6239). Precursor and fragment mass tolerances were set to 10 ppm and 0.8 Da respectively and allowing up to two missed cleavages. The static modification used was carbamidomethylation (C). The high confidence level filter with false discovery rate (FDR) of 1% was applied to the peptides. Protein relative quantitation was achieved by extracting peptide areas with the Proteome Discoverer 2.0 (Thermo Scientific, San Jose, CA) and 3 unique peptides per protein were used for the protein quantitation analysis. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE65 partner repository with the dataset identifier PXD004173.
RNA interference
RNA interference was conducted by feeding C. elegans with E. coli strain HT115(DE3) expressing double-stranded RNA (dsRNA) that is homologous to the target gene of interest66,67. Briefly, E. coli with the appropriate vectors was grown in LB broth containing ampicillin (100 μg/ml) at 37 °C overnight, and plated onto NGM plates containing 100 μg/ml ampicillin and 3 mM isopropyl β-D-thiogalactoside (IPTG). RNAi-expressing bacteria were allowed to grow overnight at 37 °C. L2 or L3 larval worms were placed on RNAi or vector control plates for 2 days at 20 °C until nematodes became gravid. Gravid adults were then transferred to fresh RNAi-expressing bacterial lawns and allowed to lay eggs for 1 hr at 25 °C to synchronize a second-generation RNAi population. The gravid adults were removed and eggs were allowed to develop at 20 °C to reach young adult stage for subsequent assays. Clone identity was confirmed by sequencing. unc-22 RNAi was included as a positive control in all experiments to account for RNAi efficiency.
Fluorescence imaging
Gravid adult Phsp-4::GFP(zcIs4) or octr-1(ok371);Phsp-4::GFP(zcIs4) were transferred to NGM plates seeded with E. coli OP50 for 1 hr at 25 °C to lay eggs. Gravid adults were removed from NGM plates and the eggs were allowed to hatch at 20 °C. Young adults were then exposed to P. aeruginosa PA14 or E. coli OP50 at 20 °C for 24 hr. Worms were observed under Leica MZ10F microscope and fluorescence images were taken using LAS V4.5 software. Binary mean intensity was measured by ImageJ software.
Translation inhibitor treatment
Three thousand one hundred twenty five μl of 50 mg/ml G418 (Roche, New York, NY) or 750 μl of 50 mg/ml Hygromycin B (Mediatech, Inc. Manassas, VA) was added to 250 ml NGM medium, respectively, to make corresponding G418 or Hygromycin B plates using 6 cm Petri dishes. 100 ml overnight E. coli OP50 cell culture was centrifuged at 4000 rpm for 10 min. Concentrated E. coli OP50 was seeded on the above translational inhibitor plates. Synchronized wild-type and octr-1(ok371) worms at young adult stage were transferred to NGM plates containing E. coli OP50 with or without translation inhibitor for survival assays and scored over time. The survival assays were performed at 25 °C and live worms were transferred daily to fresh plates. Worms were scored at the times indicated and were considered dead when they failed to respond to touch.
RNA isolation
Gravid adult wild-type and octr-1(ok371) worms were transferred to NGM plates seeded with E. coli OP50 for 1 hr at 25 °C to lay eggs. Gravid adults were removed from NGM plates and the eggs were allowed to hatch at 20 °C for 3 days. Young adult worms were then exposed to NGM plates containing E. coli OP50 with or without translation inhibitor at 25 °C for 4 hr. After 4 hr, worms were collected and washed with M9 buffer, and RNA was extracted using QIAzol lysis reagent and purified with RNeasy Plus Universal Kit (Qiagen, Valencia, CA).
Quantitative real-time PCR (qRT-PCR)
Total RNA was obtained as described above and subjected to reverse transcription as suggested by High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Quantitative PCR was conducted on StepOnePlus Real-Time PCR system using Power SYBR Green PCR Master Mix in a 96-well plate format (Applied Biosystems). Fifty nanograms of cDNA were used for real-time PCR. Twenty-five microliter reactions were set-up and performed as outlined by the manufacturer (Applied Biosystems). Relative fold-changes for transcripts were calculated using the comparative CT (2−ΔΔCT) method and normalized to pan-actin (act-1, -3, -4). Cycle thresholds of amplification were determined by StepOne Software v2.3 (Applied Biosystems). All samples were run in triplicate. Primer sequences are available on request.
Statistical Analysis
For the proteomics study, protein levels between experimental groups were statistically compared using TIGR Multiexperiment Viewer v4.9 (MeV, http://www.tm4.org/mev.html). The relative quantities of the proteins were compiled as tab-delimited text files and input into MeV v4.9. t-tests were performed between experimental groups with 5 replicates in each group. The program generates 2 text files with significantly different proteins and non-significantly different proteins in separate files along with P-values of each comparison. P-values < 0.05 are considered significant. For the C. elegans survival assays, animal survival was plotted as a non-linear regression curve using the PRISM (version 6, GraphPad Software, Inc. La Jolla, CA) computer program. Survival curves were considered different than the appropriate control indicated in the main text when P-values were < 0.05. Prism uses the product limit or Kaplan-Meier method to calculate survival fractions and the logrank test (equivalent to the Mantel-Heanszel test) to compare survival curves. A two-sample t test for independent samples was used to analyze qRT-PCR results; P-values < 0.05 are considered significant. All the experiments were repeated at least 3 times, unless otherwise indicated.
Availability of data and material
The mass spectrometry proteomics data of “N2 versus octr-1 shotgun proteomics nano MS/MS” have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD004173.
Additional Information
How to cite this article: Liu, Y. et al. Neuronal GPCR OCTR-1 regulates innate immunity by controlling protein synthesis in Caenorhabditis elegans. Sci. Rep. 6, 36832; doi: 10.1038/srep36832 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Jennifer Watts, Alejandro Aballay, Robert Schoborg, and Eva Szentirmai for critical reading of the manuscript. We thank the staff at the Tissue Imaging and Proteomics Lab, WSU for assistance with proteomics data analysis and thank Jeong Jin Park for submitting the proteomics data to PRIDE. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the Department of Biomedical Sciences, Elson S. Floyd College of Medicine, WSU-Spokane and the WSU New Faculty Seed Grant to J. S.
Footnotes
Author Contributions Y.L., D.S., and J.S. designed and performed the experiments. Y.L., D.S., and J.S. analyzed the data. Y.L. and J.S. wrote the manuscript. All authors read and approved the final manuscript.
References
- Sun J., Aballay A. & Singh V. Cellular responses to infections in Caenorhabditis elegans. Encyclopedia of Cell Biology 2, 845–852 (2015). [Google Scholar]
- Engelmann I. & Pujol N. Innate immunity in C. elegans. Advances in experimental medicine and biology 708, 105–121 (2010). [DOI] [PubMed] [Google Scholar]
- Ermolaeva M. A. & Schumacher B. Insights from the worm: the C. elegans model for innate immunity. Seminars in immunology 26, 303–309, doi: 10.1016/j.smim.2014.04.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R. & Ausubel F. M. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Current opinion in immunology 24, 3–9, doi: 10.1016/j.coi.2011.10.004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewbank J. J. Signaling in the immune response. WormBook: the online review of C. elegans biology, 1–12, doi: 10.1895/wormbook.1.83.1 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen K. T., Gallego S. F., Faergeman N. J. & Kallipolitis B. H. Strength in numbers: “Omics” studies of C. elegans innate immunity. Virulence 3, 477–484, doi: 10.4161/viru.21906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y. et al. abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. The Biochemical journal 361, 221–230 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyai L. & Patthy L. Amoebapore homologs of Caenorhabditis elegans. Biochimica et biophysica acta 1429, 259–264 (1998). [DOI] [PubMed] [Google Scholar]
- Ideo H. et al. A Caenorhabditis elegans glycolipid-binding galectin functions in host defense against bacterial infection. The Journal of biological chemistry 284, 26493–26501, doi: 10.1074/jbc.M109.038257 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. E., Kooistra T. & Kim D. H. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463, 1092–1095, doi: 10.1038/nature08762 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. E., Kinkel S. & Kim D. H. Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS genetics 7, e1002391, doi: 10.1371/journal.pgen.1002391 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman H. K. et al. Aberrant Activation of p38 MAP Kinase-Dependent Innate Immune Responses Is Toxic to Caenorhabditis elegans. G3 (Bethesda) 6, 541–549, doi: 10.1534/g3.115.025650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman L. Elaborate interactions between the immune and nervous systems. Nature immunology 5, 575–581, doi: 10.1038/ni1078 (2004). [DOI] [PubMed] [Google Scholar]
- Sternberg E. M. Neural regulation of innate immunity: a coordinated nonspecific host response to pathogens. Nature reviews. Immunology 6, 318–328, doi: 10.1038/nri1810 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. & Zhang Y. Neural-immune communication in Caenorhabditis elegans. Cell host & microbe 5, 425–429, doi: 10.1016/j.chom.2009.05.003 (2009). [DOI] [PubMed] [Google Scholar]
- Styer K. L. et al. Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322, 460–464, doi: 10.1126/science.1163673 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Singh V., Kajino-Sakamoto R. & Aballay A. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332, 729–732, doi: 10.1126/science.1203411 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Liu Y. & Aballay A. Organismal regulation of XBP-1-mediated unfolded protein response during development and immune activation. EMBO reports 13, 855–860, doi: 10.1038/embor.2012.100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakad R. et al. Contrasting invertebrate immune defense behaviors caused by a single gene, the Caenorhabditis elegans neuropeptide receptor gene npr-1. BMC genomics 17, 280, doi: 10.1186/s12864-016-2603-8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. Central dogma of molecular biology. Nature 227, 561–563 (1970). [DOI] [PubMed] [Google Scholar]
- Irazoqui J. E. et al. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS pathogens 6, e1000982, doi: 10.1371/journal.ppat.1000982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaerts A., Beets I., Temmerman L., Schoofs L. & Verleyen P. Proteome changes of Caenorhabditis elegans upon a Staphylococcus aureus infection. Biol Direct 5, 11, doi: 10.1186/1745-6150-5-11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A. et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS genetics 7, e1001393, doi: 10.1371/journal.pgen.1001393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C. & Marcotte E. M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature reviews. Genetics 13, 227–232, doi: 10.1038/nrg3185 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi S. P., Rochon Y., Franza B. R. & Aebersold R. Correlation between protein and mRNA abundance in yeast. Molecular and cellular biology 19, 1720–1730 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics 1, 304–313 (2002). [DOI] [PubMed] [Google Scholar]
- Yang W. et al. Overlapping and unique signatures in the proteomic and transcriptomic responses of the nematode Caenorhabditis elegans toward pathogenic Bacillus thuringiensis. Developmental and comparative immunology 51, 1–9, doi: 10.1016/j.dci.2015.02.010 (2015). [DOI] [PubMed] [Google Scholar]
- Lovric J. Introducing Proteomics: From Concepts to Sample Separation, Mass Spectrometry and Data Analysis. (Wiley, 2011). [Google Scholar]
- Shine J. & Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature 254, 34–38 (1975). [DOI] [PubMed] [Google Scholar]
- Shine J. & Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proceedings of the National Academy of Sciences of the United States of America 71, 1342–1346 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C., Govindarajan S. & Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol 22, 346–353, doi: 10.1016/j.tibtech.2004.04.006 (2004). [DOI] [PubMed] [Google Scholar]
- Lithwick G. & Margalit H. Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome research 13, 2665–2673, doi: 10.1101/gr.1485203 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldad N. & Arava Y. A ribosomal density-mapping procedure to explore ribosome positions along translating mRNAs. Methods in molecular biology 419, 231–242, doi: 10.1007/978-1-59745-033-1_16 (2008). [DOI] [PubMed] [Google Scholar]
- Ingolia N. T., Ghaemmaghami S., Newman J. R. & Weissman J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223, doi: 10.1126/science.1168978 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D., Colangelo C., Williams K. & Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome biology 4, 117, doi: 10.1186/gb-2003-4-9-117 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342, doi: 10.1038/nature10098 (2011). [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Tan M. W., Rahme L. G. & Ausubel F. M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96, 47–56 (1999). [DOI] [PubMed] [Google Scholar]
- Tan M. W., Rahme L. G., Sternberg J. A., Tompkins R. G. & Ausubel F. M. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proceedings of the National Academy of Sciences of the United States of America 96, 2408–2413 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E. R. et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS genetics 2, e183, doi: 10.1371/journal.pgen.0020183 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M. et al. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proceedings of the National Academy of Sciences of the United States of America 103, 14086–14091, doi: 10.1073/pnas.0603424103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. A., Kawli T. & Tan M. W. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS pathogens 4, e1000175, doi: 10.1371/journal.ppat.1000175 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block D. H. et al. The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult C. elegans. PLoS genetics 11, e1005265, doi: 10.1371/journal.pgen.1005265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D. & Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC bioinformatics 10, 48, doi: 10.1186/1471-2105-10-48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M. W., Mahajan-Miklos S. & Ausubel F. M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America 96, 715–720 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H., Hoeppner M. P., Weiner J. 3rd & Bornberg-Bauer E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology 213, 237–250, doi: 10.1016/j.imbio.2007.12.004 (2008). [DOI] [PubMed] [Google Scholar]
- Dunbar T. L., Yan Z., Balla K. M., Smelkinson M. G. & Troemel E. R. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell host & microbe 11, 375–386, doi: 10.1016/j.chom.2012.02.008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M. et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96, doi: 10.1038/415092a (2002). [DOI] [PubMed] [Google Scholar]
- Wang W. et al. Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med Res Rev 35, 225–285, doi: 10.1002/med.21327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Leslie P. & Zhang Y. Ribosomal proteins as unrevealed caretakers for cellular stress and genomic instability. Oncotarget 5, 860–871, doi: 10.18632/oncotarget.1784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. R. & McIntosh K. B. How common are extraribosomal functions of ribosomal proteins? Mol Cell 34, 3–11, doi: 10.1016/j.molcel.2009.03.006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uechi T. et al. Ribosomal protein gene knockdown causes developmental defects in zebrafish. PloS one 1, e37, doi: 10.1371/journal.pone.0000037 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N., Sakamoto K. M. & Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood 112, 5228–5237, doi: 10.1182/blood-2008-01-132290 (2008). [DOI] [PubMed] [Google Scholar]
- Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M. & Vousden K. H. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3, 577–587 (2003). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Molecular and cellular biology 23, 8902–8912 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M. S. et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Molecular and cellular biology 24, 7654–7668, doi: 10.1128/MCB.24.17.7654-7668.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D. L. & Mango S. E. Genetic suppressors of Caenorhabditis elegans pha-4/FoxA identify the predicted AAA helicase ruvb-1/RuvB. Genetics 177, 819–833, doi: 10.1534/genetics.107.076653 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Hellen C. U. & Pestova T. V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature reviews. Molecular cell biology 11, 113–127, doi: 10.1038/nrm2838 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D. L., Kirienko N. V. & Ausubel F. M. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell host & microbe 11, 364–374, doi: 10.1016/j.chom.2012.02.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J. A. & Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149, 452–466, doi: 10.1016/j.cell.2012.02.050 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngner J. S., Stinebring W. R. & Taube S. E. Influence of inhibitors of protein synthesis on interferon formation in mice. Virology 27, 541–550 (1965). [DOI] [PubMed] [Google Scholar]
- Fontana M. F. et al. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS pathogens 7, e1001289, doi: 10.1371/journal.ppat.1001289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F. et al. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. The Journal of cell biology 158, 639–646, doi: 10.1083/jcb.200203086 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider S. & Pal R. Integrated analysis of transcriptomic and proteomic data. Curr Genomics 14, 91–110, doi: 10.2174/1389202911314020003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino J. A. et al. 2016 update of the PRIDE database and its related tools. Nucleic acids research 44, D447–D456, doi: 10.1093/nar/gkv1145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A. G. et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330, doi: 10.1038/35042517 (2000). [DOI] [PubMed] [Google Scholar]
- Timmons L. & Fire A. Specific interference by ingested dsRNA. Nature 395, 854, doi: 10.1038/27579 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.