Abstract
Protamines are arginine-rich DNA-binding proteins that replace histones in elongating spermatids. This leads to hypercondensation of chromatin and ensures physiological sperm morphology, thereby protecting DNA integrity. In mice and humans, two protamines, protamine-1 (Prm1) and protamine-2 (Prm2) are expressed in a species-specific ratio. In humans, alterations of this PRM1/PRM2 ratio is associated with subfertility. By applying CRISPR/Cas9 mediated gene-editing in oocytes, we established Prm2-deficient mice. Surprisingly, heterozygous males remained fertile with sperm displaying normal head morphology and motility. In Prm2-deficient sperm, however, DNA-hypercondensation and acrosome formation was severely impaired. Further, the sperm displayed severe membrane defects resulting in immotility. Thus, lack of Prm2 leads not only to impaired histone to protamine exchange and disturbed DNA-hypercondensation, but also to severe membrane defects resulting in immotility. Interestingly, previous attempts using a regular gene-targeting approach failed to establish Prm2-deficient mice. This was due to the fact that already chimeric animals generated with Prm2+/− ES cells were sterile. However, the Prm2-deficient mouse lines established here clearly demonstrate that mice tolerate loss of one Prm2 allele. As such they present an ideal model for further studies on protamine function and chromatin organization in murine sperm.
The production of mature sperm starts with diploid spermatogonial stem cells, which undergo mitotic and meiotic division and finally differentiate into haploid spermatids. During spermiogenesis, round spermatids develop into elongated spermatids, with a species-specific head morphology. In developing male germ cells, DNA-binding histones are gradually replaced by testis-specific histone variants, transition proteins, and finally protamines, resulting in a hypercondensation of the paternal haploid genome1.
Protamines are rich in positively charged arginine residues, mediating strong binding to the minor groove of the negatively charged DNA2. In addition, cysteine residues form intra- and intermolecular disulfide bonds, thereby stabilizing the DNA-protamine complex2,3. The sperm-protamine complexes form toroids, leading to an at least 6-fold denser compaction compared to histone-bound DNA of mitotic chromosomes4. Each toroid comprises around 60 kb of DNA5. As a consequence, this hypercondensation in elongated spermatids causes a complete silencing of transcription and protects the genome from environmental insults6,7.
Although many mammals express only a single protamine gene, humans and rodents encode two protamine genes8,9,10. In mice and humans, the genes encoding protamine-1 (Prm1), protamine-2 (Prm2), and transition protein-2 (Tnp2) form a conserved and tightly regulated gene cluster located on chromosome 1611,12. PRM1 is translated as a mature protein, whereas PRM2 is translated as a precursor that matures by cleavage of its N-terminal part8,13,14. Interestingly, PRM1 and PRM2 are translated in a species-specific ratio. In humans, both isoforms are present in approximately equal amounts, whereas in mice, PRM2 is more abundant, accounting for approx. 65% of total protamine15. In some species, such as boar, bull, and rat, PRM2 is virtually absent due to mutations within the coding or promoter region16,17. In humans, aberrant protamine ratios are associated with subfertility and decreased DNA integrity18,19,20,21,22,23,24,25. Furthermore, altered protamine ratios are associated with decreased fertilization rates in men participating in assisted reproduction technology (ART) programs26,27,28. Using a standard gene-targeting approach, both murine protamine genes have been deleted29,30. However, chimeras generated by injecting ES cells heterozygous for one allele of either protamine gene resulted in infertility. Sperm displayed abnormal head morphology, disturbed DNA integrity, and decreased motility. As a consequence, Prm2-deficient mouse lines could not be established, hampering further experiments dealing with Protamine function.
In this study, we used the CRISPR/Cas9 technology to generate Prm2-deficient mice. Application of this genome editing tool in murine zygotes allows to precisely and efficiently edit genes without disturbing the overall exon-intron gene structure or deleting regulatory DNA sequences31. We demonstrate that mice heterozygous for Prm2 are fertile with normal sperm morphology and motility, whereas Prm2-null mice are infertile due to a complete loss of sperm motility and abnormal sperm head morphology.
Results
CRISPR/Cas9-mediated generation of Prm2-deficient mice
Previously, it was reported that chimeric mice generated by injecting Prm2+/− ES cells were sterile29. In general, ES cell lines are of male sex and mediate sex-reversal when injected into female host blastocysts. Thus, a mouse line harboring a mutation affecting the male germline, like the Prm2-heterozygosity, cannot be established by this technology, precluding detailed research of protamine biology. Since protamines are only required for male germ cell development, establishment of Prm2-deficient mouse lines should be possible from female founder animals. Therefore, we decided to apply CRISPR/Cas9-mediated gene-editing in murine zygotes to generate Prm2-deficient mice31.
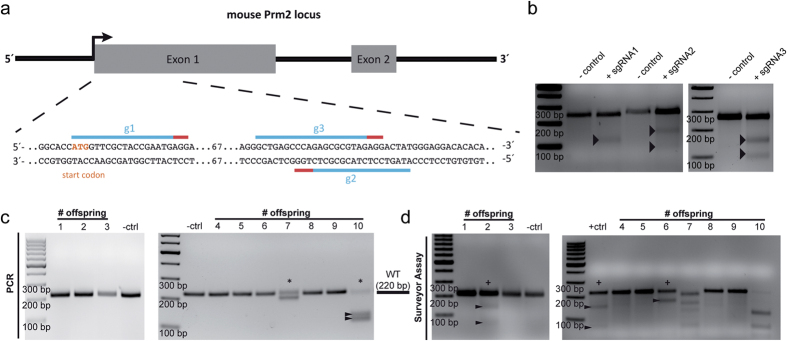
The murine Prm2 gene consists of two exons, encoding the 106 amino acids-long precursor protein (Fig. 1a). We derived three guide RNAs (gRNAs) targeting the first exon close to the translational start site (Fig. 1a). First, the gRNAs were validated in murine ES cells. CRISPR/Cas9 induces double-strand breaks, leading to error-prone non-homologous end-joining mediated repair32. This results in formation of small insertions or deletions (indels), which can be detected by the ‘Surveyor nuclease’, resulting in cleavage of PCR products. The gene-edited Prm2-coding region was amplified by PCR and subjected to the Surveyor assay33, which confirmed the functionality of derived gRNAs, as indicated by cleavage of the PCR products (Fig. 1b, cleavage products highlighted by arrowheads).
Figure 1. CRISPR/Cas9-mediated generation of Prm2-deficient mice.
(a) Schematic representation of the Prm2 gene locus and targeting sites of designed gRNAs. (b) Surveyor Assay. Intended target sites of gRNAs were amplified by PCR from genomic DNA of gene-edited E14TG2a ES cells. Wildtype and gene-edited PCR products were denaturated and re-annealed. Surveyor nuclease mediated cleavage of mismatched PCR products (highlighted by arrowheads) confirmed functionality of gRNAs. (c,d) Genotyping of offsprings by PCR and Surveyor assay identified 4/10 animals to be mutant as indicated by asterisks (PCR) and pluses (Surveyor).
To edit the Prm2 gene in murine zygotes, we microinjected in vitro transcribed single-gRNAs (sgRNAs) and Cas9 mRNA into the cytoplasm. A combination of sgRNAs 1 and 2 or sgRNAs 1 and 3 was applied to induce deletions of approximately 100 bp. From 336 injected zygotes, 245 (72.9%) survived (Table 1); 82 blastocysts were transferred and 10 viable pups were born (Table 1). Genotyping by PCR and Surveyor assay identified 4/10 pups carrying CRISPR/Cas9-induced mutations within the Prm2 locus (Fig. 1c,d). Of those, three animals (# 2, 6, 7) contained small indels, indicating that one sgRNA had catalyzed gene-editing. One female (#10) harbored the intended deletion of approximately 100 bp (Fig. 1c). Sequence analysis of genomic DNA from #10 revealed that the CRISPR/Cas9-mediated gene-editing resulted in four different alleles. Besides the wildtype allele, three alleles with deletions of 97 bp, 101 bp, and 103 bp were obtained. This suggests that gene-editing took place at the 2-cell-stage. Alleles with deletions of 97 bp and 101 bp were separated by back-crossing with C57BL/6 wildtype mice.
Table 1. Injection statistics.
| #ID | Zygotes injected | Intact after injection (% of injected) | Blastocysts transferred (% of intact) | Prm2 edited/pups born (%) |
|---|---|---|---|---|
| A | 150 | 108 (72.0) | 21 (19.4) | 1/3 (33.3) |
| B | 186 | 137 (73.7) | 61 (44.5) | 3/7 (42.9) |
| total | 336 | 245 (72.9) | 82 (33.5) | 4/10 (40.0) |
Efficiency of CRISPR/Cas9 microinjections displayed as survival rates of oocytes and number of modified alleles obtained to total pups born.
Haploinsufficiency of Prm2 does not affect male fertility
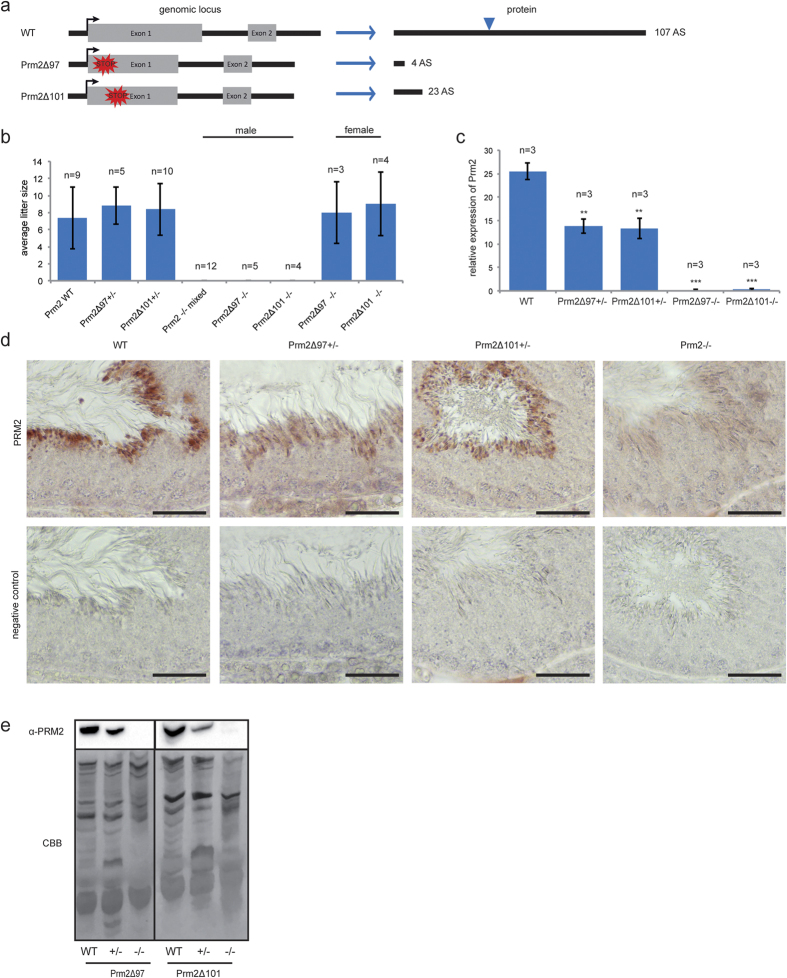
Two mouse lines, Prm2Δ97 (harboring a 97 bp deletion) and Prm2Δ101 (harboring a 101 bp deletion), were established. On both alleles, a frameshift was generated, resulting in nonsense transcripts with a premature stop codon and deletion of Prm2 (Fig. 2a). In contrast to earlier reports29, male mice heterozygous for either Prm2Δ97 or Prm2Δ101 were fertile and sired offspring. Matings of males heterozygous for either Prm2Δ97 or Prm2Δ101 produced an average litter size of 8.6 (Fig. 2b). There was no significant difference in the average litter size among the two mouse lines (8.8 vs. 8.4). This is comparable to published data for wildtype mice with a mean litter size of 6.2 (9.5 in the plateau period of reproduction)34,35 and our wildtype matings with an average litter size of 7.3. In contrast, male mice homozygous for either Prm2Δ97 or Prm2Δ101 were not able to reproduce (Fig. 2b), whereas female mice homozygous for either Prm2Δ97 or Prm2Δ101 were fertile and sired offspring with an average litter size of 8.5, when mated with heterozygous males (Fig. 2b). These results clearly indicate that loss of one allele of Prm2 can be tolerated, whereas complete lack of Prm2 results in male sterility.
Figure 2. Knockout validation and fertility analysis.
(a) Schematic representation of established Prm2-deficient mouse lines Prm2Δ97bp and Prm2Δ101bp. Blue triangle marks cleavage site within the premature PRM2 protein. (b) Mating statistics of Prm2-deficient males and females. Successful mating of Prm2−/− males with wildtype females was indicated by presence of a vaginal plug at 0.5 dpc. (c) Relative expression of Prm2 mRNA in murine testis of wildtype, Prm2+/−, and Prm2−/− mice measured by qRT-PCR. (d) Immunohistochemical staining of PRM2 on testicular sections of wildtype, Prm2+/− and Prm2−/− mice. Scale bar = 100 μm. (e) Immunoblot against PRM2 following acid urea gel electrophoresis of basic nuclear proteins isolated from epididymal sperm of Prm2Δ97bp or Prm2Δ101bp mice. Of note, the amount of protein loaded from Prm2-deficient samples were doubled compared to samples from wildtype and heterozygous animals. Coomassie-Brilliant-Blue staining (CBB) served as loading control. The antibody detects mature PRM2 as well as its precursor forms (pP2-a, pP2-b).
To validate that gene-editing generated a Prm2 loss-of-function allele, we analyzed full-length Prm2 RNA levels. Since Prm2 is expressed during spermiogenesis in step 7–15 spermatids, qRT-PCR was performed on RNA isolated from murine testis36. In heterozygous animals, relative Prm2 mRNA expression levels were reduced by approximately 50% compared to wildtype animals (Fig. 2c). Sperm from males homozygous for either mutation did not show any Prm2 signal, indicating that the gene-editing led to successful deletion of the Prm2 gene (Fig. 2c).
Immunohistochemical staining (IHC) using an anti-PRM2 antibody on testicular tissue sections detected the protein in spermatids at stage VIII of the seminiferous epithelial cycle of both, wildtype and Prm2+/− mice (Fig. 2d). The staining appeared less intense in heterozygous mice, suggesting a reduction in protein level due to decreased mRNA levels. In accordance with qRT-PCR data, sections from Prm2−/− testes were negative for PRM2 at all stages of the mouse seminiferous epithelial cycle, further confirming the deletion of the gene (Fig. 2d, right).
Western blot analysis of basic nuclear proteins isolated from epididymal sperm was performed to determine protein levels of PRM2. Males heterozygous for either Prm2Δ97 or Prm2Δ101 displayed an approximately 50% decrease of mature PRM2 compared to wildtype controls (Fig. 2e). In Prm2−/− mice, mature PRM2 protein was not detectable (Fig. 2e).
Prm2-deficient sperm display severe morphological defects
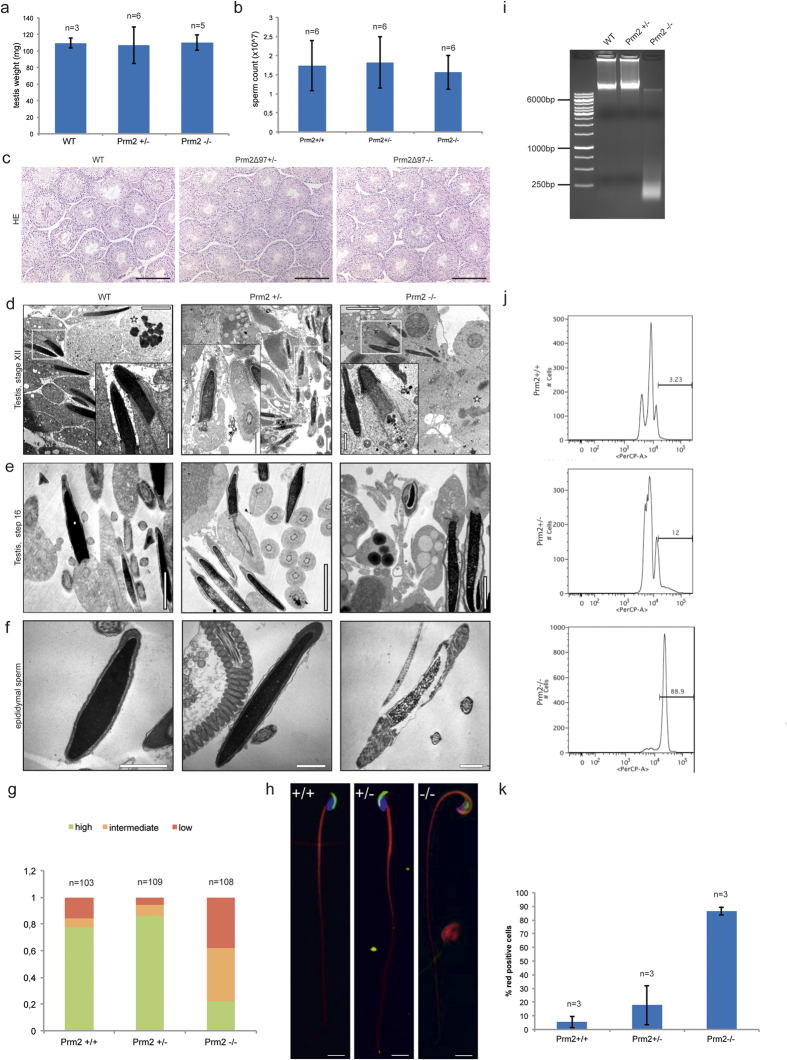
Because Prm2+/− males were fertile, we were able to breed Prm2−/− animals and analyze the effects of loss of Prm2 in more detail. Since testis weight (Fig. 3a), epididymal sperm count (Fig. 3b), and testis morphology was not changed in Prm2+/− and Prm2−/− animals compared to wildtype (Fig. 3c), we concluded that spermatogenesis is not affected. To investigate why Prm2-deficient male mice are infertile, we analyzed the morphology of testicular spermatids and epididymal sperm.
Figure 3. Morphological testes and sperm analysis.
(a) Testes weight. (b) Epididymal sperm count of Prm2+/+, Prm2+/−, and Prm2−/− mice. (c) HE staining of testicular sections from wildtype, Prm2+/− and Prm2−/− mice. Scale bar = 200 μm. (d) Transmission electron micrographs (TEM) of stage XII seminiferous epithelium as indicated by meiotic spermatocytes (stars) showing step 12 elongating spermatids. Scale bar = 5 μm. Inset: enlargement of rectangle, scale bar = 1 μm. Note homogeneously condensed chromatin in wildtype in contrast to heterogeneously condensed fine-grained chromatin in the Prm2-deficient mutant, whereas formation of the acrosome and manchette as well as head shape did not differ. (e) TEMs of step 16 elongated spermatids prior to sperm release. Scale bar = 1 μm. (f) TEMs of epididymal sperm. Note nuclear matrix alteration, detachment of acrosome (arrow) and attachment of the flagellum to the head (arrowhead) in knockout sperm. Scale bar = 1 μm. (g) Quantification of epididymal sperm chromatin integrity. (h) Prm2−/− but not Prm2+/− sperm showed morphological defects in the acrosome and head-tail conjunction. The acrosome was labeled using PNA-FITC (green), DNA using DAPI (blue), and the flagellum using MitoTracker (red). Scale bar = 10 μm. (i) Agarose gel electrophoresis of sperm genomic DNA. (j,k) Sperm damage analysis referring to SCSA.
Electron microscopy of Prm2-deficient testicular spermatids revealed impaired nuclear matrix morphology, exhibiting a fine-grained, sometimes coarse-grained texture (Fig. 3d). Anomalies in chromatin condensation were obvious from step 12 spermatids onward. Until sperm release, the heterogeneity in chromatin packaging, including vesiculation, became more pronounced (Fig. 3e). However, sperm head elongation and shaping were not affected in testicular spermatids (Fig. 3d,e).
Next, epididymal sperm were co-stained with PNA-FITC to label the acrosome, MitoTracker to label mitochondria in the midpiece of the flagellum, and DAPI to label the DNA in the sperm head (Fig. 3h). Wildtype mice and mice heterozygous for Prm2Δ97 or Prm2Δ101 showed the typical sickle-shaped head with the acrosome lining the sperm head, whereas Prm2-deficient sperm lacked the acrosome and the midpiece of the flagellum was wrapped around the sperm head (Fig. 3h). The majority of Prm2-deficient sperm showed this phenotype, which was confirmed by electron microscopy (Fig. 3f). On the ultrastructural level, Prm2-deficient sperm displayed detachment of the acrosome at the acrosome-nuclear interface, giving rise to an abnormally shaped head with an undulating plasma membrane (Fig. 3f). In line with observations from testicular spermatids, chromatin integrity was highly disturbed in epididymal sperm (Fig. 3f). Quantification of chromatin integrity revealed that around 80% of Prm2-deficient sperm showed intermediate to low chromatin density. No difference was observed between wildtype and Prm2+/− animals with around 80% of sperm showing high chromatin integrity (Fig. 3g). Thus, loss of Prm2 not only affects DNA integrity in the sperm head, but also bending of the flagellum and anchorage of the acrosome. To analyze whether a loss of DNA integrity makes sperm more susceptible to DNA damage, we subjected genomic DNA from sperm to agarose gel electrophoresis. While a band indicative of high molecular weight DNA suggested little to no DNA damage in sperm of Prm2+/+ and Prm2+/− animals, the majority of sperm DNA from Prm2-deficient mice was strongly degraded into fragments of around 250 bp in size (Fig. 3i). Next, sperm were treated with a strong acid to induce denaturation of DNA, followed by staining with acridine orange and FACS analysis, referring to the sperm chromatin structure assay (SCSA). Intercalation of acridine orange into intact, double-stranded DNA results in emission of green fluorescence, whereas incorporation into damaged, single-stranded DNA results in red fluorescence. In accordance with the results from the agarose gels, red fluorescence intensity was strongly increased in Prm2-deficient sperm compared to Prm2+/+ and Prm2+/− animals (Fig. 3j). In total, more than 80% of Prm2-deficient sperm cells showed intense red fluorescence indicating a strong DNA damage. Although there is a slight increase in sperm damage in Prm2+/− animals compared to wildtype mice, more than 90% of Prm2+/− sperm presented intact (Fig. 3k).
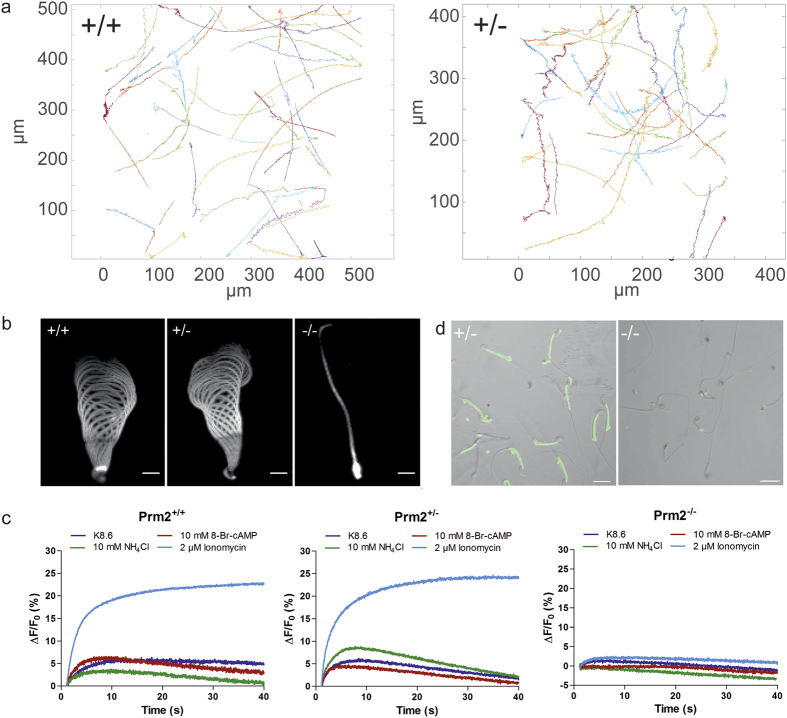
Prm2-deficient sperm are immotile
To reveal whether sperm motility in addition to sperm morphology was affected by loss of Prm2, we isolated wildtype, heterozygous, and Prm2-deficient sperm and analyzed the motility of freely swimming sperm using computer-assisted semen analysis (CASA) and the flagellar waveform of tethered sperm. While motility parameters of freely swimming sperm form wildtype and heterozygous Prm2Δ97 or Prm2Δ101 animals were not significantly different (Table 2, Fig. 4a), homozygously Prm2-deficient sperm were completely immotile (Fig. 4b). Wildtype and heterozygous mice displayed a symmetrical flagellar waveform, which was absent in Prm2-deficient sperm (Fig. 4b). Sperm function, in particular sperm motility, crucially relies on Ca2+ signaling37,38,39. Thus, we analyzed Ca2+ signaling in Prm2-deficient mice using the kinetic stopped-flow technique to measure changes in the intracellular Ca2+ concentration in sperm populations loaded with the Ca2+ dye Cal520-AM. We applied a number of different stimuli to evoke a Ca2+ influx in sperm. Stimulation with K8.6, a buffer that depolarizes and alkalizes the cell and thereby opens the principal Ca2+-channel in mammalian sperm, CatSper, evoked a Ca2+ response in wildtype and Prm2-heterozygous, but not in deficient mice (Fig. 4c). Similarly, alkalization of sperm using NH4Cl and direct opening of CatSper using 8-Br-cAMP evoked a Ca2+ response in wildtype and Prm2-heterozygous, but not in deficient sperm (Fig. 4c). As a control, we applied the Ca2+-ionophore ionomycin, which resulted in Ca2+ influx in controls and Prm2-heterozygous, but not in Prm2-deficient sperm (Fig. 4c). To test whether Prm2-deficient sperm were loaded with Cal520-AM, we compared Prm2-heterozygous and deficient sperm using fluorescence microscopy (Fig. 4d). Heterozygous sperm were loaded with the dye, showing prominent fluorescence in the midpiece, whereas loading of Prm2-deficient sperm was negative (Fig. 4d).
Table 2. Motility parameters.
| genotype | VCL | VAP | VSL | LHD | STR |
|---|---|---|---|---|---|
| wildtype | 100 ± 43 | 49 ± 18 | 34 ± 22 | 2.2 ± 1.2 | 67 ± 30 |
| Prm2 Δ97+/− | 69 ± 25 | 45 ± 21 | 21 ± 14 | 2.1 ± 1.1 | 46 ± 29 |
| Prm2 Δ101+/− | 97 ± 49 | 62 ± 25 | 32 ± 22 | 2.4 ± 1.5 | 53 ± 32 |
Free-swimming sperm from wildtype or heterozygous Prm2Δ 97 or Prm2Δ 101 mice have been analyzed using dark-field microscopy. Data are presented as mean ± S.D.; VCL: curvilinear velocity μm/s), VAP: average path velocity (μm/s); VSL: straight-line velocity (μm/s), LHD: lateral head displacement (μm); STR: path straightness (VSL/VAP).
Figure 4. Functional sperm analysis.
(a) Tracks for freely swimming wildtype Prm2+/+ and heterozygous Prm2+/− sperm. (b) Flagellar waveform. Sperm were tethered with their heads to a glass surface and the flagellar waveform was analyzed. One beat cycle was projected. Scale bar: 10 μm. (c) Changes in the intracellular Ca2+ concentration in Prm2+/+, Prm2+/−, and Prm2−/− sperm. Sperm have been loaded with CAL520-AM and stimulated with K8.6 (blue), 10 mM 8-Br-cAMP (red), 10 mM NH4Cl (green), or 2 μM ionomycin (light blue). Experiments have been measured using the stopped-flow technique. (d) Loading of sperm with Cal520-AM. Loading of Prm2+/−, and Prm2−/− sperm was tested using fluorescence microscopy. Scale bar = 20 μm.
Of note, low levels of fluorescence could also indicate that the intracellular Ca2+ concentration in Prm2-deficient sperm is extremely low. However, this can be excluded since treatment with ionomycin did not evoke a Ca2+ response. Together with the results obtained by transmission electron microscopy (TEM), this suggests that Prm2-deficient sperm have severe plasma membrane defects, which might prevent dye loading.
Prm1 expression is maintained in Prm2-deficient mice
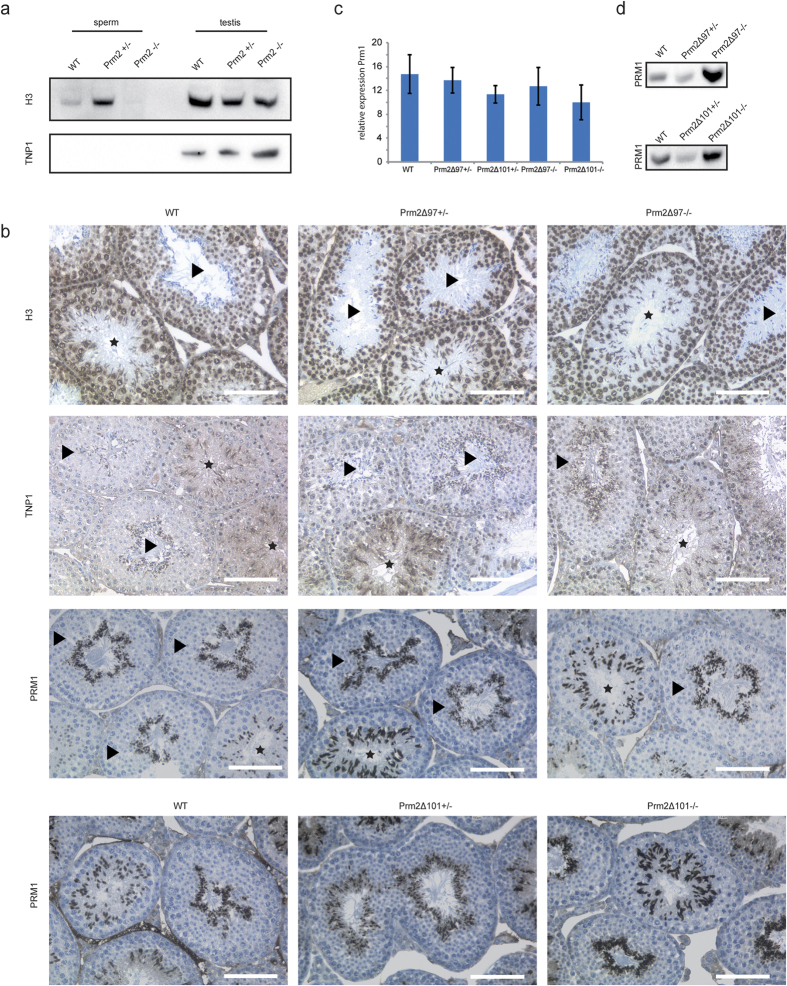
Next, we analyzed the effects of Prm2-deficiency on the overall histone-to-protamine exchange dynamics. Western blot analysis of basic nuclear proteins isolated from epididymal sperm revealed weak levels of histone H3 in wildtype sperm (Fig. 5a). This is in accordance with previous studies, which described that around 1% of histones remain bound to sperm chromatin after histone-to-protamine exchange40. Interestingly, an increased level of H3 was observed in sperm from Prm2+/− animals, whereas no H3 was detectable in Prm2-deficient sperm (Fig. 5a). No differences in H3 level were observed in protein extracts from testicular tissue (Fig. 5a). IHC stainings gave evidence that the histone replacement is functional. While spermatogonia, round spermatids, and elongating spermatids stained positive for H3, fully elongated spermatids displayed no or only very weak staining (Fig. 5b). Immunoblotting against TNP1 revealed a complete absence in epididymal sperm (Fig. 5a). As a control, presence of TNP1 in testicular tissue was shown. In accordance with this, IHC staining revealed expression of TNP1 in elongating spermatids. In elongated spermatids, where transition proteins are assumed to be completely exchanged by protamines, staining for TNP1 was only observed in residual bodies of Prm2+/+ and Prm2+/− spermatids (Fig. 5b). Interestingly, sections of Prm2-deficient animals showed an additional staining within elongated spermatids (Fig. 5b), suggesting a disturbed TNP1 exchange.
Figure 5. Effects of Prm2-deficiency on histone-to-protamine exchange dynamics.
(a) Immunoblot against H3 and TNP1 of basic nuclear proteins of epididymal sperm and testis from Prm2+/+, +/− and −/− mice (b) Immunohistochemical staining of total H3, TNP1 and PRM1 on testicular sections from Prm2Δ mice. Sections were counterstained with hematoxylin. Tubules with elongating spermatids were marked with a star, tubules with mature spermatids with arrowheads. Scale bar = 100 μm. (c) Relative expression of Prm1 mRNA in murine testis of wildtype, Prm2+/− and Prm2−/− mice measured by qRT-PCR (n = 3). (d) Immunoblot against PRM1 following acid urea gel electrophoresis of basic nuclear proteins isolated from epididymal sperm of Prm2+/+, Prm2+/−, and Prm2−/− mice. Reanalysis of blots shown in Fig. 2e.
The importance of the species-specific ratio of protamines for male fertility suggests that Prm1 and 2 expression levels are tightly controlled. Therefore, we next analyzed the effects of Prm2-deficiency on Prm1 gene expression. qRT-PCR analysis revealed no significant differences in testicular Prm1 mRNA levels in males heterozygous or deficient for Prm2 compared to wildtype controls (Fig. 5c). IHC staining detected PRM1 protein in elongating spermatids of wildtype, heterozygous, and homozygous Prm2Δ97 and Prm2Δ101 animals (Fig. 5b). In the seminiferous epithelial cycle, cells of step 10 were the first cells expressing PRM1. Signal intensity was highest in step 12–15 elongated spermatids, marking the replacement of transition proteins by protamines. Western blot analysis of basic nuclear proteins isolated from epididymal sperm further demonstrated the successful incorporation of PRM1 into sperm chromatin (Fig. 5d). Interestingly, PRM1 protein level were slightly decreased in Prm2+/− animals, but increased in Prm2-deficient sperm compared to controls (Fig. 5d). Of note, for western blot analysis the basic proteins like the protamines were isolated from precipitated DNA. The fact that we could detect PRM1 in PRM2-deficient sperm suggests that PRM1 is incorporated into sperm chromatin independent of PRM2.
Discussion
In the present study, we used CRISPR/Cas9 gene-editing to generate Prm2-deficient mice to investigate the role of PRM2 in controlling chromatin hypercondensation and fertility. We demonstrate that Prm2-heterozygous mice remain fertile with sperm being morphologically and functionally indistinguishable from wildtype sperm. However, lack of Prm2 causes infertility with severe defects in sperm head morphology and sperm motility.
Although the role of protamines for sperm function has been described many times for different species41, our observation that loss of one Prm2 allele does not affect male fertility in mice is contradictory to previous studies. Cho et al. described that already chimeras produced by injecting Prm2+/− ES cells were sterile29. We hypothesize that these fundamental differences between our study and the observations made by Cho et al. might be due to the different experimental approaches used to generate Prm2-knockout alleles. Cho et al. applied a classical gene-targeting approach in ES cells to disrupt the Prm2 coding region by insertion of a Pgk-neo cassette29. This cassette might have affected the expression in the tightly regulated 11.7 kb spanning gene cluster encoding for Prm1-Prm2-Tnp212. Insertion of the Pgk-neo cassette into the Prm2 coding region might have resulted in silencing of the neighboring Prm1 gene. In fact, such a trans-silencing has been described for the knockout of the herculin (Myf6) gene. Here, the Pgk-neo cassette introduced by homologous recombination to disrupt the Myf6 locus silenced expression of the nearby myogenic factor 5 (Myf5) gene42,43. We propose that the trans-silencing of the Prm1 allele may be caused by the Pgk-neo cassette inserted into the Prm2 allele. This assumption is supported by the fact that testes of chimeric mice generated by injecting Prm2+/− ES cells displayed a strong reduction in PRM1 protein level29. In our model, qRT-PCR clearly demonstrates that expression of Prm1 is maintained in Prm2+/− males, while only a mild decrease in protein level is observed.
Further, the hypothesis that physiological PRM2 protein levels are required for proper incorporation of PRM1 into sperm chromatin has to be re-visited29, since sperm of Prm2+/− and Prm2−/− males displays certain degrees of DNA-hypercondensation indicative for DNA-protamine interaction.
Here, we utilized the CRISPR/Cas9 system to generate Prm2-deficient mice. This state-of-the-art technique allows to efficiently edit genes without introducing additional DNA elements like loxP sites and/or antibiotic resistance cassettes31. Off-target activity is a highly discussed issue in the field of CRISPR/Cas9-mediated genome editing. Nonetheless, different approaches analyzing potential off-target effects in genetically altered mice and ES cells, which closely resemble the situation in zygotes, showed either no or only a very low incidence for CRISPR/Cas9-mediated off-target activity44,45, when compared with analyses carried out (mainly) in cancer cell lines, which are hallmarked by genomic instability46,47,48. To reduce the risk for off-target events, we selected only gRNAs with very low predicted off-target activity. Furthermore, if possible we used 18-mer guide sequences instead of 20-mer as this was shown to further decrease the frequency of Cas9-mediated off-target events49. In addition, the Prm2Δ97 and Prm2Δ101 alleles were backcrossed with wildtype mice. Thus, potential off-target effects, which might have affected loci on other chromosomes, are lost during further breeding.
We established two mouse lines, Prm2Δ97 and Prm2Δ101, with N-terminal deletions of 97 and 101 bp, respectively. Both mutations did not interfere with or deleted regulatory regions and left the overall structure of the gene cluster intact. Male mice heterozygous for the respective mutation were fertile and sired offspring with an average litter size of 8.6, which is within the range of the reproductive performance of wildtype mice with a mean litter size of 6.2 and a maximum mean litter size of 9.5 in the plateau period of reproduction34,35. This clearly indicates that deletion of one Prm2 allele does not affect male reproductive performance in mice. The typical apical hook-like structure of sperm heads and intact sperm motility further suggests that sperm physiology and morphology are not affected.
This raises the question whether disturbed chromatin integrity or the loss of sperm motility and detachment of the acrosome is the main reason for infertility of knockout mice. The latter is supported by Takeda et al., who were able to generate offspring from Prm1+/− males by in vitro fertilization of zona-free oocytes, although chromatin integrity of those sperm was severely affected50. Nontheless, strong sperm DNA degradation observed in Prm2-deficient animals most likely prohibits successful reproduction.
Protamines are expressed in haploid spermatids, hence in Prm2+/− mice, only half of the spermatids express Prm2. However, IHC staining revealed PRM2 protein in all spermatids of seminiferous tubules. While transcript or protein sharing among syncytial spermatids has been discussed in general, the IHC stainings shown here clearly demonstrate that such sharing exists for Prm229,51,52,53.
Of note, knockout male mice heterozygous for the transition proteins TNP1 and TNP2 display preserved fertility as well, while complete deletion of both alleles resulted in subfertility54,55.
Prm2-heterozygous mice display reduced levels of PRM2 protein, compared to controls, and show maintained expression of PRM1. We demonstrate that these mice sire normal litter sizes indicative of full fertility. Western blot showed that in Prm2-heterozygous mice deletion of one allele of Prm2 not only led to a reduction of PRM2 protein but also caused a moderate decrease of PRM1 protein level. This is suggestive for a PRM1/2 coregulation in order to maintain the mouse specific PRM1/2 ratio. Recent analyses revealed that PRM1/PRM2 ratio range from 65% (M. castaneus and M. domesticus) to 99% (M. macedonicus and M. spicilegus) in different mouse subspecies56. This indicates that mice might be able to tolerate changes in the PRM1/2 ratio to some extent. This would be in contrast to the situation in humans, where changes of the PRM1/2 ratio are associated with reduced fertility. Further, the elevated levels of histones detected in such animals that remain bound to sperm chromatin might compensate for the lower total amount of protamines.
Prm2-deficient sperm displayed upregulation of PRM1. We hypothesize that the higher PRM1 levels are an attempt to compensate for the loss of PRM2. A similar compensatory effect has been described for Tnp2-deficient mice, which showed enhanced Tnp1-expression55. Sperm from Prm2−/− animals lost the characteristic hook-like structure: sperm exhibited round heads and were immotile. Interestingly, a recent study suggests that the cleaved N-terminal part of PRM2 rather than the chromatin-bound mature PRM2 is involved in controlling sperm head morphology57. However, the underlying molecular mechanism is not known. In TEM analysis, we were able to show a detachment of the acrosomal cap and a less-condensed chromatin. Similar observations were made for protamine-deficient human sperm58. Whereas the reason for the defect in chromatin packaging is obvious, the molecular mechanisms underlying the ultrastructural change in acrosomal cap formation and attachment to the nucleus need to be addressed in further studies.
But why are sperm from Prm2-deficient mice immotile? Once histones are replaced by protamines, transcription is silenced6. Thus, transcriptional regulation of other genes by Prm2 seems unlikely. Further, in Prm2-deficient males, development of testicular spermatids is not affected and sperm counts are comparable. Profound morphological aberrations in Prm2-deficient mice are first apparent in the epididymis with detachment of acrosome and bending of the flagellum towards the head. In parallel, treatment with Ca2+ ionophores indicates that epididymis derived Prm2-deficient sperm presents with defects in membrane function. It remains to be determined whether this is a defect in the self-assembly of sperm or a built-in quality control mechanism triggering a self-destructing program. Of note, impaired sperm motility is a common phenotype of Prm1, Prm2, Tnp1, and Tnp2 knockout mice29,50,54,55,59.
With Prm2Δ97 and Prm2Δ101 mouse lines, we have established, for the first time, a robust and reliable model for functional studies on protamine-induced chromatin hypercondensation during spermiogenesis. These mice allow for a detailed investigation of basic regulatory mechanisms of haploid gene expression in sperm. Shedding light on these molecular mechanisms is an absolute requirement for a better understanding of aberrant protamine expression in subfertile men.
Material and Methods
Ethics statement
All animal experiments were conducted according to German law of animal protection and in agreement with the approval of the local institutional animal care committees (Landesamt für Natur, Umwelt und Verbraucherschutz, North Rhine-Westphalia, approval ID: AZ84-02.04.2013.A429).
Guide RNA design and cloning
Gene-specific guide sequences (gRNAs) (Supplementary Table 1) were designed using the CRISPR design tool (http://crispr.mit.edu)60 and inserted into the pX330 expression vector (pX330-U6-Chimeric_BB-CBh-hSpCas9 Addgene plasmid # 42230))61 as described previously62. Some gRNAs were truncated to 19- or 18-mers, to ensure initial guanine or adenine bases and to reduce potential off-target activity49.
Culture and transfection of mES cells
E14Tg2a mES cells (kind gift of Christof Niehrs, IMB Mainz, Germany) were maintained on gelatinized cell culture dishes with standard ES cell medium at 37 °C and 7.5% CO2. For transfection, 3 × 105 cells per well were seeded onto a gelatinized 12-well-plate in media without antibiotics. After 3 h cells were transfected with a 3:1 ratio of pX330: Lipofectamine2000, according to the manufacturers’ protocol (Thermo Fisher Scientific, Waltham, USA). 8 h later media was changed to ES media to remove DNA-Lipofectamine complexes.
Surveyor assay
Two days after transfection, genomic DNA was extracted from cells using the phenol-chloroform-isoamyl-alcohol (PCI) precipitation method as described previously63. Gene edited loci were amplified by PCR (see Supplementary Table 1 for primer sequences). Products were denaturated by heating to 95 °C for 10 min, re-annealed by stepwise cooling to room temperature (RT), and analyzed using the ‘SURVEYOR Mutation Detection Kit’ (Transgenomic, Manchester, GB).
Generation of Prm2-deficient mice
Female B6D2F1 mice were superovulated by intraperitoneal injection of pregnant mare’s serum (PMS, 5 i.u.) and human chorionic gonadotropin (hCG, 5 i.u.) and mated 2:1 with B6D2F1 males. Zygotes were isolated at 0.5 dpc from the oviducts and subjected to microinjection using an inverted microscope (Leica, Wetzlar, Germany) equipped with micromanipulators (Narishige, Japan) and piezo unit (Eppendorf, Hamburg, Germany). Injection pipettes (PIEZO 8-15-NS, Origio, Charlottesville, USA) were filled with Fluorinert (FC-770, Sigma-Aldrich, Taufkirchen, Germany) for appropriate piezo pulse propagation. In vitro transcribed single-guide RNAs (sgRNA) (50 ng/μl each) and Cas9 mRNA (100 ng/μl) (Sigma-Aldrich) were injected into the cytoplasm of zygotes, as described previously31. Surviving zygotes were cultured in KSOM medium for 3 days. Developing blastocysts were transferred into the uteri of pseudo-pregnant foster mice.
Quantitative reverse transcription–polymerase chain reaction (qRT-PCR)
RNA was isolated from testis tissue with TRIzol (Life Technologies, Carlsbad, USA). RNA concentrations and purity ratios were determined by NanoDrop measurement (Peqlab, Erlangen, Germany). qRT-PCR was performed as described previously64. First strand cDNA synthesis was carried out using RevertAid First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Rot, Germany) after DNAseI treatment of RNA. qRT-PCR was performed on ViiA 7 Real Time PCR System (Applied Biosystems, distributed by Life Technologies) using Maxima SYBR Green qPCR Master Mix (Life Technologies). At the end of each PCR run, a melting point analysis was performed. Actin-beta was used as reference gene for data normalization. See Supplementary Table 1 for primer sequences.
Antibodies
Mouse monoclonal antibodies anti-PRM1 (Hup1N) and anti-PRM2 (Hup2B) (Briar Patch Biosciences, Livermore, CA, USA, IB 1:2.000, IHC 1:100), rabbit anti-H3 (Abcam, Ab1791, IB 1:2.000, IHC 1:400) and rabbit anti-TNP1 (Abcam, Ab73135, IB 1:1.000, IHC 1:400) were used in this study. Secondary polyclonal antibodies were used as follows: rabbit anti-mouse HRP 1:1.000 (Dako, Hamburg, Germany), goat anti-rabbit HRP 1:2000 (Dako), goat anti-mouse BIOT 1:200 (Dako).
Isolation of epididymal sperm
Sperm were isolated by multiple incisions of the cauda followed by a swim-out in modified TYH medium65 or M2 medium. Sperm count was determined using a Neubauer hemocytometer. For all experiments, sexually mature males with an age of 2–5 months were used. Animals were derived from intercrosses of the F1 generation.
Isolation of sperm nuclear proteins
Nine million sperm were washed with phosphate-buffered saline (PBS, pH 7.4) containing protease inhibitors (Complete mini, Roche, Basel). Extraction of basic nuclear proteins was performed according to Deyebra and Oliva66.
Immunoblotting
Protein extracts were subjected to 15% polyacrylamide acid-urea gel electrophoresis67,68 and blotted onto a polyvinylidene fluoride (PVDF) membrane using the Trans-Blot Turbo system (BioRad, Munich, Germany). Equal protein loading was validated by staining with Coomassie Brillant Blue G-250. Membranes were blocked in 3% nonfat dry milk powder in Tris-buffered saline with Tween20 (TBST) for 2 h at RT and probed with primary (overnight at 4 °C) and secondary (1 h at RT) antibodies in blocking solution. Chemiluminescent signals were detected using ChemiDoc MP Imaging System (BioRad) after incubation of the membrane with PierceSuper Signal West Pico chemiluminescent substrate (Perbio, Bonn, Germany) or Westar Supernova substrate (7Bioscience, Hardtheim, Germany).
Immunohistochemistry
Bouin’s fixed testicular tissue was processed in paraffin wax. Immunohistochemistry (IHC) against PRM1 was performed as described previously64 using the Autostainer 480 S (Medac, Hamburg, Germany). For IHC of PRM2, sections were treated for 10 min at RT with decondensing-mix containing 25 mM dithiothreitol, 0.2% Triton X-100 and 200 IU heparin/ml in PBS to enhance antigen accessibility for the primary antibody69. Subsequently, sections were washed in 0.02 M PBS (pH 7.4), boiled for 20 min in sodium citrate buffer, treated for 20 min with 3% H2O2 in methanol and blocked for 20 min with 5% bovine serum albumin (BSA) in PBS. Incubation with the primary antibody was performed overnight at 4 °C, with biotinylated goat-anti-mouse secondary antibody (DAKO) and the Vectastain ABC Kit (Vector Laboratories, Burlingame, CA, USA) for 1 h at RT each. Immunoreaction was visualized using AEC (DAKO). Finally, sections were counterstained with hematoxylin and covered with glycerin gelatin.
Electron microscopy
Testis and epididymis perfused with PBS buffer (0.1 M) including 0.25% heparin followed by Yellow-Fix (2% paraformaldehyde, 0.1 M sodium cacodylate buffer pH 7.2, 0.02% picric acid mixed with 2% glutaraldehyde just before usage) via the heart were immersion-fixed in Yellow-Fix for 24 h at 4 °C. Smaller samples rinsed in PBS buffer (0.1 M) were then immersion-fixed in 1% osmium tetroxide at 4 °C for 2 h and rinsed in buffer again. Subsequently, the tissue was dehydrated and embedded in Epon. Ultrathin-sections were picked up on grids, stained with lead-citrate (0.2%) and examined with a Zeiss EM 109 (Zeiss, Oberkochen, Germany).
Assessment of sperm DNA damage
For isolation of sperm genomic DNA, ten million sperm were resuspended in 500 μl lysis buffer (10 mM Tris HCl, 25 mM EDTA, 1% SDS, 75 mM NaCl) and supplemented with 21 μl 1 M DTT, 2,5 μl Triton x-100 and 40 μl Proteinase K (10 mg/ml). The suspension was incubated at 55 °C for 2 h following PCI precipitation of DNA. SCSA-like analysis was performed referring to Evenson et al.70.
Measurement of changes in sperm intracellular Ca2+
Sperm were isolated by incision of the cauda followed by a swim-out in modified TYH medium65. Sperm were loaded with the fluorescent Ca2+ indicator Cal-520, AM (AAT Bioquest, Sunnyvale, USA) (5 μM) in the presence of Pluronic F-127 (0.05% vol/vol) at 37 °C for 45 min. After incubation, excess dye was removed by three centrifugation steps (700 × g, 7 min, RT). The pellet was resuspended in TYH and equilibrated for 5 min at 37 °C. Pictures of loaded sperm were taken on a confocal microscope (FV1000; Olympus). Changes in [Ca2+]i were recorded in a rapid-mixing device in the stopped-flow mode (SFM400; Bio-Logic, Claix, France) as previously described65. Data acquisition was performed with a data acquisition pad (PCI-6221; National Instruments, Austin, USA) and Bio-Kine software v. 4.49 (Bio-Logic). Ca2+ signals are depicted as the percent change in fluorescence (ΔF) with respect to the mean of the first three data points recorded immediately after mixing (F0), that is, when a stable fluorescence signal was observed. The control (buffer) ΔF/F0 signal was subtracted from compound-induced signals.
Immunocytochemistry
Sperm were dried and fixed in 4% paraformaldehyde/PBS for 20 min. To block unspecific binding sites, sperm were incubated for 40 min with blocking buffer (0.5% Triton X-100 and 5% ChemiBLOCKER (Merck Millipore, Darmstadt, Germany) in 0.1 M phosphate buffer, pH 7.4. PSA-FITC (0.5 mg/ml, Sigma Aldrich) and MitoTracker Red CMXRos (1:2000, Thermo Fisher) were diluted in blocking buffer containing 0.5 mg/ml DAPI (Life Technologies) and incubated for 1 h. Pictures were taken on a confocal microscope (FV1000; Olympus).
Motility analyses
Sperm motility was studied in shallow observation chambers (depth 150 μm) using an inverted microscope (IX71; Olympus) equipped as described previously65.
To analyze the flagellar beat, cells were tethered to the glass surface by adjusting the BSA concentration in the buffer from 3 to 0.3 mg/ml. Cells that had their head attached to the glass surface and had a free beating flagellum were selected for imaging. Images were collected at 200 frames per second using a CMOS camera (Dimax; PCO, Kelheim, Germany). Quantification of the flagellar beat was performed using custom-made programs written in Java, which can be made available upon request. The program identified a point on the flagellum 25 μm apart from the center of the mouse head within every time frame. The flagellar beat parameters were determined within a time window of 1 s after each frame. The first point in this time window was chosen as reference point and the distance between the successive points found on the flagellum and the reference point was monitored. This distance varied in a sinusoid-like manner in time and the beat frequency was determined as the maxima in the Fourier spectrum.
For analyzing the swimming parameters of sperm cells, the BSA concentration in the buffer was 3 mg/ml, resulting in most of the cells swim freely at the glass surface. Images were collected at 50 frames per second (Dimax). Quantification of the swimming parameters was performed the Casa program71 adapted to MATLAB (MathWorks. Natick, USA).
Statistics
For mating experiments, sperm count analysis, testis weight, qRT-PCR, and FACS analysis mean values are shown. Error bars represent standard deviation (SD). Statistical significance was determined by two-tailed, unpaired Student’s t-test. Significance was assumed for p-values < 0.05 (*p < 0.05; **p < 0.005; ***p < 0.001).
Additional Information
How to cite this article: Schneider, S. et al. Re-visiting the Protamine-2 locus: deletion, but not haploinsufficiency, renders male mice infertile. Sci. Rep. 6, 36764; doi: 10.1038/srep36764 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG) to HS (SCH 503/15-1) and KS (STE 892/14-1), and DW (SFB645). DW was further supported by the Bonn Excellence Cluster for “ImmunoSensation”. We thank Jörg Bedorf, Gaby Beine, Mareike Buch-Heberling, Angela Egert, Barbara Fröhlich, Alexandra Hax, Andrea Jäger, Sigi Kettner, Blanca Randel, Barbara Reddemann, Susi Schubert-Porth and Susi Steiner for excellent technical assistance.
Footnotes
Author Contributions S.S., D.F., D.N., M.K., M.B., D.W., K.S. and H.S. conceived and designed the experiments. S.S. generated and analyzed gene-edited mice (qRT-PCR, western blot, mating statistics, HE). M.B. performed stopped-flow analyses of sperm. J.J. recorded and analyzed sperm motility parameters. S.J. performed qRT-PCR and FACS analyses, D.F. performed and analyzed IHC stainings, R.S. and M.K. analyzed sperm ultrastructure by TEM. S.S., D.W., K.S. and H.S. were major contributors in writing the manuscript. All authors read and approved the final manuscript.
References
- Rathke C., Baarends W. M., Awe S. & Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Chromatin and epigenetic regulation of animal development 1839, 155–168 (2014). [DOI] [PubMed] [Google Scholar]
- Balhorn R. A model for the structure of chromatin in mammalian sperm. J Cell Biol 93, 298–305 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelingh J. P., Rozijn T. H. & Monfoort C. H. Isolation and partial characterization of a basic protein from bovine sperm heads. Biochim Biophys Acta 188, 353–356 (1969). [DOI] [PubMed] [Google Scholar]
- Ward W. S. & Coffey D. S. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biology of Reproduction 44, 569–574 (1991). [DOI] [PubMed] [Google Scholar]
- Hud N. V., Allen M. J., Downing K. H., Lee J. & Balhorn R. Identification of the Elemental Packing Unit of DNA in Mammalian Sperm Cells by Atomic Force Microscopy. Biochemical and Biophysical Research Communications 193, 1347–1354 (1993). [DOI] [PubMed] [Google Scholar]
- Steger K. Transcriptional and translational regulation of gene expression in haploid spermatids. Anat Embryol (Berl) 199, 471–487 (1999). [DOI] [PubMed] [Google Scholar]
- Ward W. S. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod 16, 30–36 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammer H., Henschen A. & Lee C. H. Isolation and amino-acid sequence analysis of human sperm protamines P1 and P2. Occurrence of two forms of protamine P2. Biol Chem Hoppe Seyler 367, 515–522 (1986). [DOI] [PubMed] [Google Scholar]
- Bower P. A., Yelick P. C. & Hecht N. B. Both P1 and P2 protamine genes are expressed in mouse, hamster, and rat. Biology of Reproduction 37, 479–488 (1987). [DOI] [PubMed] [Google Scholar]
- Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 8, 227–227 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. H. et al. Mapping of PRM1 to human chromosome 16 and tight linkage of Prm-1 and Prm-2 on mouse chromosome 16. J Hered 80, 442–446 (1989). [DOI] [PubMed] [Google Scholar]
- Wykes S. M. & Krawetz S. A. Conservation of the PRM1 –> PRM2 –> TNP2 domain. DNA. Seq 14, 359–367 (2003). [DOI] [PubMed] [Google Scholar]
- McKay D. J., Renaux B. S. & Dixon G. H. Human sperm protamines. Amino-acid sequences of two forms of protamine P2. Eur J Biochem 156, 5–8 (1986). [DOI] [PubMed] [Google Scholar]
- Yelick P. C. et al. Mouse protamine 2 is synthesized as a precursor whereas mouse protamine 1 is not. Molecular and Cellular Biology 7, 2173–2179 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corzett M., Mazrimas J. & Balhorn R. Protamine 1: Protamine 2 stoichiometry in the sperm of eutherian mammals*. Mol. Reprod. Dev. 61, 519–527 (2002). [DOI] [PubMed] [Google Scholar]
- Maier W. M., Nussbaum G., Domenjoud L., Klemm U. & Engel W. The lack of protamine 2 (P2) in boar and bull spermatozoa is due to mutations within the P2 gene. Nucleic Acids Research 18, 1249–1254 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunick D., Balhorn R., Stanker L. H. & Hecht N. B. Expression of the rat protamine 2 gene is suppressed at the level of transcription and translation. Experimental Cell Research 188, 147–152 (1990). [DOI] [PubMed] [Google Scholar]
- Balhorn R., Reed S. & Tanphaichitr N. Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia 44, 52–55 (1988). [DOI] [PubMed] [Google Scholar]
- de Yebra L. et al. Detection of P2 precursors in the sperm cells of infertile patients who have reduced protamine P2 levels. Fertility and Sterility 69, 755–759 (1998). [DOI] [PubMed] [Google Scholar]
- Aoki V. W. DNA Integrity Is Compromised in Protamine-Deficient Human Sperm. Journal of Andrology 26, 741–748 (2005). [DOI] [PubMed] [Google Scholar]
- Torregrosa N. et al. Protamine 2 precursors, protamine 1/protamine 2 ratio, DNA integrity and other sperm parameters in infertile patients. Hum. Reprod. 21, 2084–2089 (2006). [DOI] [PubMed] [Google Scholar]
- García-Peiró A. et al. Protamine 1 to protamine 2 ratio correlates with dynamic aspects of DNA fragmentation in human sperm. Fertility and Sterility 95, 105–109 (2011). [DOI] [PubMed] [Google Scholar]
- Steger K. et al. Both protamine-1 to protamine-2 mRNA ratio and Bcl2 mRNA content in testicular spermatids and ejaculated spermatozoa discriminate between fertile and infertile men. Hum. Reprod. 23, 11–16 (2008). [DOI] [PubMed] [Google Scholar]
- Steger K. et al. Decreased protamine-1 transcript levels in testes from infertile men. Mol Hum Reprod 9, 331–336 (2003). [DOI] [PubMed] [Google Scholar]
- Ni K., Spiess A.-N., Schuppe H.-C. & Steger K. The impact of sperm protamine deficiency and sperm DNA damage on human male fertility: a systematic review and meta-analysis. Andrology, doi: 10.1111/andr.12216 (2016). [DOI] [PubMed] [Google Scholar]
- Rogenhofer N. et al. The sperm protamine mRNA ratio as a clinical parameter to estimate the fertilizing potential of men taking part in an ART programme. Hum. Reprod. 28, 969–978 (2013). [DOI] [PubMed] [Google Scholar]
- Aoki V. W. et al. Sperm protamine 1/protamine 2 ratios are related to in vitro fertilization pregnancy rates and predictive of fertilization ability. Fertility and Sterility 86, 1408–1415 (2006). [DOI] [PubMed] [Google Scholar]
- Nasr-Esfahani M. H. et al. Effect of protamine-2 deficiency on ICSI outcome. Reproductive BioMedicine Online 9, 652–658 (2004). [DOI] [PubMed] [Google Scholar]
- Cho C. et al. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet 28, 82–86 (2001). [DOI] [PubMed] [Google Scholar]
- Cho C. et al. Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biology of Reproduction 69, 211–217 (2003). [DOI] [PubMed] [Google Scholar]
- Yang H., Wang H. & Jaenisch R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 9, 1956–1968 (2014). [DOI] [PubMed] [Google Scholar]
- Kim H. & Kim J.-S. A guide to genome engineering with programmable nucleases. Nat Rev Genet 15, 321–334 (2014). [DOI] [PubMed] [Google Scholar]
- Qiu P. et al. Mutation detection using Surveyor nuclease. Biotechniques 36, 702–707 (2004). [DOI] [PubMed] [Google Scholar]
- Verley F. A., Grahn D., Leslie W. P. & Hamilton K. F. Sex ratio of mice as possible indicator of mutation rate for sex-linked lethals. J Hered 58, 285–290 (1967). [DOI] [PubMed] [Google Scholar]
- Biggers J. D., Finn C. A. & McLaren A. Long-term reproductive performance of female mice. J Reprod Fertil 3, 313–330 (1962). [DOI] [PubMed] [Google Scholar]
- Mali P. et al. Stage-specific expression of nucleoprotein mRNAs during rat and mouse spermiogenesis. Reprod Fertil Dev 1, 369–382 (1989). [DOI] [PubMed] [Google Scholar]
- Ren D. et al. A sperm ion channel required for sperm motility and male fertility. Nature 413, 603–609 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennemuth G., Carlson A. E., Harper A. J. & Babcock D. F. Bicarbonate actions on flagellar and Ca2+-channel responses: initial events in sperm activation. Development 130, 1317–1326 (2003). [DOI] [PubMed] [Google Scholar]
- Carlson A. E., Hille B. & Babcock D. F. External Ca2+ acts upstream of adenylyl cyclase SACY in the bicarbonate signaled activation of sperm motility. Developmental Biology 312, 183–192 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner A. M., Nanni P. & Mansuy I. M. Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics & Chromatin 7, 2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R. Protamines and male infertility. Human Reproduction Update 12, 417–435 (2006). [DOI] [PubMed] [Google Scholar]
- Yoon J. K., Olson E. N., Arnold H.-H. & Wold B. J. DifferentMRF4Knockout Alleles Differentially Disrupt Myf-5 Expression:cis-Regulatory Interactions at theMRF4/Myf-5Locus. Developmental Biology 188, 349–362 (1997). [DOI] [PubMed] [Google Scholar]
- Olson E. N., Arnold H. H., Rigby P. W. J. & Wold B. J. Know Your Neighbors: Three Phenotypes in Null Mutants of the Myogenic bHLH Gene MRF4. Cell 85, 1–4 (1996). [DOI] [PubMed] [Google Scholar]
- Wu X. et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 1–9, doi: 10.1038/nbt.2889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. et al. One-Step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-Mediated Genome Engineering. Cell 154, 1370–1379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33, 187–197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31, 839–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J. D., Reyon D., Cascio V. M. & Joung J. K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32, 279–284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N. et al. Viable offspring obtained from Prm1-deficient sperm in mice. Sci. Rep. 6, 27409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell K. A. & Handel M. A. Protamine transcript sharing among postmeiotic spermatids. Proceedings of the National Academy of Sciences 88, 2407–2411 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun R. E., Behringer R. R., Peschon J. J., Brinster R. L. & Palmiter R. D. Genetically haploid spermatids are phenotypically diploid. Nature 337, 373–376 (1989). [DOI] [PubMed] [Google Scholar]
- Dadoune J.-P., Siffroi J.-P. & Alfonsi M.-F. In International Review of Cytology Volume 237, 1–56 T2 – (Academic Press, 2004). [DOI] [PubMed] [Google Scholar]
- Yu Y. E. et al. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proceedings of the National Academy of Sciences of the United States of America 97, 4683–4688 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adham I. M. et al. Teratozoospermia in mice lacking the transition protein 2 (Tnp2). Mol Hum Reprod 7, 513–520 (2001). [DOI] [PubMed] [Google Scholar]
- Luke L., Campbell P., Varea Sanchez M., Nachman M. W. & Roldan E. R. S. Sexual selection on protamine and transition nuclear protein expression in mouse species. Proc Biol Sci 281, 20133359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke L., Tourmente M., Dopazo H., Serra F. & Roldan E. R. S. Selective constraints on protamine 2 in primates and rodents. BMC Evol Biol 16, 21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranpour F. G. The effects of protamine deficiency on ultrastructure of human sperm nucleus. Adv Biomed Res 3, 24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiko D. et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 3, 1–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L. et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettersheim D. et al. NANOG promoter methylation and expression correlation during normal and malignant human germ cell development. Epigenetics 6, 114–122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettersheim D. et al. Analysis of TET expression/activity and 5mC oxidation during normal and malignant germ cell development. PLoS ONE 8, e82881 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen V. et al. Controlling fertilization and cAMP signaling in sperm by optogenetics. Elife 4, 326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyebra L. & Oliva R. Rapid Analysis of Mammalian Sperm Nuclear Proteins. Analytical Biochemistry 209, 201–203 (1993). [DOI] [PubMed] [Google Scholar]
- Panyim S. & Chalkley R. High resolution acrylamide gel electrophoresis of histones. Archives of Biochemistry and Biophysics 130 IS, 337–346 (1969). [DOI] [PubMed] [Google Scholar]
- Shechter D., Dormann H. L., Allis C. D. & Hake S. B. Extraction, purification and analysis of histones. Nat Protoc 2, 1445–1457 (2007). [DOI] [PubMed] [Google Scholar]
- van der Heijden G. W. et al. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Developmental Biology 298, 458–469 (2006). [DOI] [PubMed] [Google Scholar]
- Evenson D. P., Larson K. L. & Jost L. K. Sperm chromatin structure assay: its clinical use for detecting sperm DNA fragmentation in male infertility and comparisons with other techniques. Journal of Andrology 23, 25–43 (2002). [DOI] [PubMed] [Google Scholar]
- Wilson-Leedy J. G. & Ingermann R. L. Development of a novel CASA system based on open source software for characterization of zebrafish sperm motility parameters. Theriogenology 67, 661–672 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.