Abstract
Insulin-like growth factor-1 (IGF-1) and its receptor IGF-1R play a paramount role in tooth/bone formation while hsa-let-7c actively participates in the osteogenic differentiation of mesenchymal stem cells. However, the interaction between IGF-1/IGF-1R and hsa-let-7c on the committed differentiation of stem cells from apical papilla (SCAPs) remains unclear. In this study, human SCAPs were isolated and treated with IGF-1 and hsa-let-7c over/low-expression viruses. The odonto/osteogenic differentiation of these stem cells and the involvement of mitogen-activated protein kinase (MAPK) pathway were subsequently investigated. Alizarin red staining showed that hsa-let-7c low-expression can significantly promote the mineralization of IGF-1 treated SCAPs, while hsa-let-7c over-expression can decrease the calcium deposition of IGF-1 treated SCAPs. Western blot assay and real-time reverse transcription polymerase chain reaction further demonstrated that the expression of odonto/osteogenic markers (ALP, RUNX2/RUNX2, OSX/OSX, OCN/OCN, COL-I/COL-I, DSPP/DSP, and DMP-1/DMP-1) in IGF-1 treated SCAPs were significantly upregulated in Let-7c-low group. On the contrary, hsa-let-7c over-expression could downregulate the expression of these odonto/osteogenic markers. Moreover, western blot assay showed that the JNK and p38 MAPK signaling pathways were activated in Let-7c-low SCAPs but inhibited in Let-7c-over SCAPs. Together, the IGF-1/IGF-1R/hsa-let-7c axis can control the odonto/osteogenic differentiation of IGF-1-treated SCAPs via the regulation of JNK and p38 MAPK signaling pathways.
Mesenchymal stem cells (MSCs) are multipotent progenitor cells, originally isolated from the adult bone marrow or other issues, which can differentiate into many kinds of different cell types1. Various growth factors can mediate the osteo/dentinogenic differentiation of MSCs2,3,4,5, including stem cells from apical papilla (SCAPs)6. These SCAPs can differentiate into odontoblasts or osteoblasts in vitro, and form dentin-like tissues along with tooth root-like structures in vivo1,3,7.
Insulin-like growth factor (IGF) axis plays a paramount role in bone formation, maintenance of mineralized skeletons as well as the process of tooth formation. One of the polypeptide growth factors (i.e., IGF-1) is the most important member of this axis7. Previous studies have revealed that IGF-1 can significantly enhance the odonto/osteogenic differentiation of SCAPs, periodontal ligament stem cells (PDLSCs) and dental pulp stem cells (DPSCs)6,8,9. IGF-1 receptor (IGF-1R) is a cell membrane receptor that contains two transmembrane α subunits and two cytosolic β subunits covalently bonded through disulphide bridges7. It is commonly believed that IGF-1R can respond to IGF-1 to stimulate the osteoblast proliferation and bone matrix mineralization10,11. Moreover, IGF-1 can regulate the cell differentiation by activating the downstream signaling pathways through its receptor IGF-1R, and IGF-1/IGF-1R pathway is thereby known as IGF-1/IGF-1R axis.
MicroRNAs (miRNAs) are small RNA molecules and comprised of 21–25 nucleotides that modulate the inhibition of translation from mRNA to protein, promotion of mRNA degradation and control of gene transcription via binding to the 3′-untranslated region (UTR) of their target mRNAs12. The let-7 family of miRNAs is initially detected in caenorhabditis elegans and identified as a heterochronic gene, which is considered as a miRNA biomarker in osteoblastic and osteocytic differentiation of mesencymal stem cells13,14. Recent studies have demonstrated that the downregulation of let-7 can upregulate the expression of IGF1/IGF1R by mediating the IGF1 signaling cascade to promote the differentiation of spermatogonia into primary spermatocytes15. It has been reported that the overexpression of hsa-let-7c can downregulate the renewal capacity of stem cells subsets, and the upregulation of expression of hsa-let-7c is related with the mechanism of GH/IGF-1 axis signaling pathway16,17.
To date, the effects of IGF-1/IGF-1R/let-7c axis as a whole on the odonto/osteogenic differentiation of SCAPs and its underling mechanism remain unclear. In this study, we investigated the influence of IGF-1/IGF-1R/hsa-let-7c axis on the proliferation and differentiation of human SCAPs, and found that IGF-1/IGF-1R/hsa-let-7c axis can affect the odonto/osteogenic differentiation of SCAPs during which JNK and p38 MAPK signaling pathway were involved. The accumulating findings may implicate a novel approach to dental tissue regeneration as well as apexogenesis.
Results
Identification of SCAPs and screening of MOI values for lentiviral infection
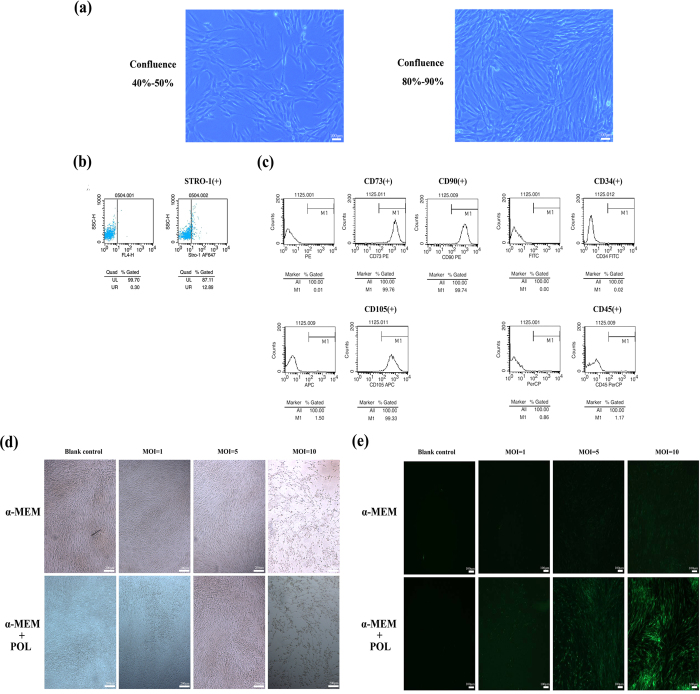
The isolated SCAPs presented a typical fibroblast- or spindle-like morphology under microscope at 40–50% and 80–90% confluence respectively (Fig. 1a). FCM results revealed that the isolated cells were immune-positive against STRO-1 (a mesenchymal stem cell marker, Fig. 1b). Moreover, CD73, CD90 and CD105 were highly expressed, while CD45 and CD34 were lowly expressed in these cells, indicating that these isolated cells were stem cells of mesenchymal origin (Fig. 1c) without contamination of hematopoietic precursors.
Figure 1. Identification of SCAPs and screening of MOI values for lentiviral infection.
(a) Isolated SCAPs with typical fibroblast- or spindle-like morphology. (b) Isolated SCAPs were immunopositive against STRO-1 by flow cytometry. (c) Isolated SCAPs were positive for CD73, CD90 and CD105, respectively, but negative for CD45 and CD34 by flow cytometry. (d) Cell morphology under microscope in α-MEM and α-MEM+POL (Polybrene) groups with different virus titers. (e) Cell fluorescence expression under an inverted microscope in α-MEM and α-MEM+POL groups with different virus titers.
With the gradual increase of MOI values, the toxic effects of the virus on SCAPs increased too. SCAPs in conventional medium (α-MEM) groups and conventional medium plus polybrene (POL) groups became elongated as compared with the blank control under the microscope (Fig. 1d). The fluorescence expression of conventional medium groups was weaker than conventional medium plus POL groups as compared with the negative control under an inverted microscope. When the virus titer is 5 × 106 TU/mL, the cell morphology and fluorescence expression in conventional medium + POL group were better than other groups (Fig. 1e). The best MOI value equals to 5 via the calculation of fluorescence microscope images as well as the evaluation of cell vitality (Fig. 1d,e).
IGF-1R protein expression is negatively correlated with let-7c expression
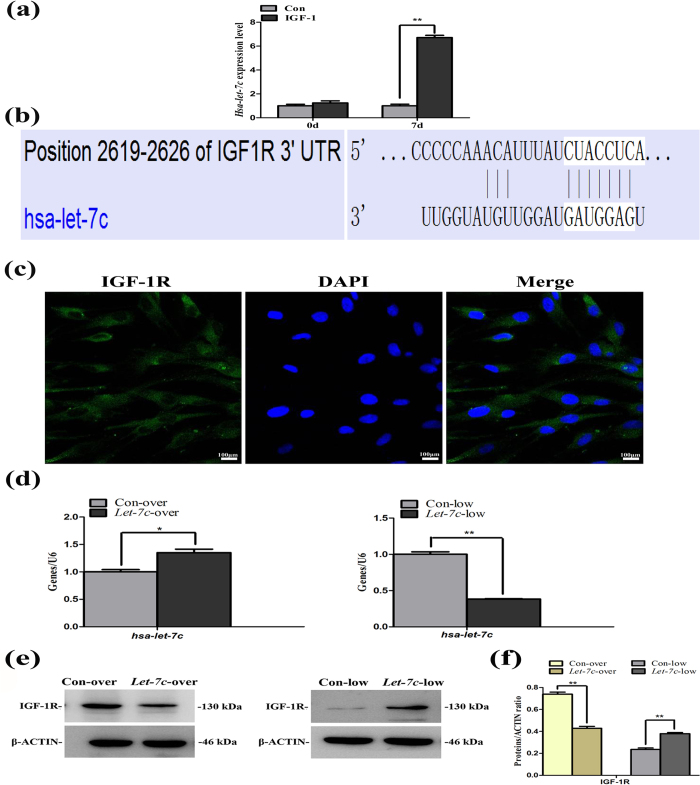
We utilized the miRDB database bioinformational analysis and found that IGF-1R was one member of the target genes of hsa-let-7c with a target score up to 91 points. Then we verified the target gene of hsa-let-7c through the gene and protein detection. We performed the miRNA microarray to detect the related miRNA levels in IGF1-treated SCAPs and found that hsa-let-7c expression in treated cells increased 6.68 folds as compared with control group (Fig. 2a). Meanwhile, we obtained the sequence of hsa-let-7c (UGAGGUAGUAGGUUGUAUGGUU) on the miRBase website. Then we found the predicted consequential pairing of target region (top) and miRNA (bottom) between IGF-1R and hsa-let-7c on the TargetScanHuman website (Fig. 2b), confirmed the specific sequence of hsa-let-7c (UGAGGUAGUAGGUUGUAUGGUU), and thus acquired the real-time sequence (Table 1).
Figure 2. Negative correlation between hsa-let-7c and IGF-1R.
(a) Hsa-let-7c detection in IGF-1-treated SCAPs by miRNA microarray. (b) The predicted consequential pairing of target region (top) and miRNA (bottom) between IGF-1R and hsa-let-7c on the TargetScanHuman website. (c) Immunofluorescent staining of IGF-1R in SCAPs. Nuclei are stained in blue and IGF-1 receptor was in green. (d) Real-time RT-PCR analysis for the expression of hsa-let-7c in Con-over group, Let-7c-over group, Con-low group and Let-7c-low group, respectively. Values were described as the means ± SD, n = 3. **2−∆∆Ct > 2, P < 0.01; *1 < 2−∆∆Ct < 2, P < 0.01. (e) Western blot assay for the expression of IGF-1R in Con-over group, Let-7c-over group, Con-low group and Let-7c-low group, respectively. (f) Quantitative analysis for western blot results. Values were described as the means ± SD, n = 3. **P < 0.01.
Table 1. Sense and antisense primers for real-time reverse transcription polymerase chain reaction.
| Genes | Sequences (5′-3′) |
|---|---|
| COL-I | F: GAGCTGGCTACTTCTCGC |
| R: TCTATCCGCATAGGACTGAC | |
| RUNX2 | F: AATGCCTCCGCTGTTATG |
| R: TTCTGTCTGTGCCTTCTTG | |
| OSX | F: GCCTACTTACCCGTCTGACTTT |
| R: GCCCACTATTGCCAACTGC | |
| OCN | F: AAGCCCAGCGACTCTGAGTCT |
| R: CCGGAGTCTATTCACCACCTTACT | |
| DSPP | F: CGTTCAGGGAGTCCTAGCGGGAACG |
| R: GTGACTCTCCCTTCCATCTCCTG | |
| DMP1 | F: TTTTAGGAAGTCTCGCATCT |
| R: TGGGACCATCTACGTTTT | |
| GAPDH | F: GCCTCGTCTCATAGACAAGATGGT |
| R: GAAGGCAGCCCTGGTAACC | |
| Hsa-let-7c | F: ACACTCCAGCTGGGTGAGGTAGTAGGTTGT |
| R: TGGTGTCGTGGAGTCG | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT |
The immunofluorescent results demonstrated that IGF-1R was mainly located in the cytoplasm of SCAPs (Fig. 2c). Real-time RT-PCR results confirmed that the expression of hsa-let-7c increased in Let-7c over-expression group (Let-7c-over), but decreased in Let-7c low-expression group (Let-7c-low, Fig. 2d, P < 0.01). Western blot revealed that the low expression of let-7c could upregulate the expression of IGF-1R, whereas the over-expression of let-7c downregulated the expression of IGF-1R (Fig. 2e,f).
Effects of IGF-1/IGF-1R/hsa-let-7c axis on proliferation of SCAPs
CCK8 assay and FCM were performed to investigate whether the IGF-1/IGF-1R/hsa-let-7c axis can affect the proliferation of SCAPs through the over/low-expression of let-7c. CCK8 assay on 11 consecutive days presented no significant difference (P > 0.05) between two groups (Fig. 3a,b). FCM analysis further showed no distinct difference (P > 0.05) in the proliferation index (PI = G2M+S) between Con-over group (17.42%) and Let-7c-over group (16.2%, Fig. 3c,d). Likewise, FCM analysis demonstrated no distinct difference (P > 0.05) in the proliferation index (PI) between Con-low group (17.19%) and Let-7c-low group (16.78%, Fig. 3e,f). Together, IGF-1/IGF-1R/hsa-let-7c axis had no significant effect on the proliferation of SCAPs.
Figure 3. IGF-1/IGF-1R/hsa-let-7c axis had no significant influence on the proliferation of SCAPs.
(a) CCK8 assay of 13 consecutive days presented no significant difference (P > 0.05) between Con-over group and Let-7c-over group. (b) CCK8 assay of 13 consecutive days displayed no significant difference (P > 0.05) between Con-low group and Let-7c-low group. (c) Flow cytometry (FCM) analysis of Con-over group. (d) Flow cytometry analysis of Let-7c-over group. (e) Flow cytometry analysis of Con-low group. (f) Flow cytometry analysis of Let-7c-low group.
Effects of IGF-1/IGF-1R/hsa-let-7c axis on odonto/osteogenic differentiation of SCAPs
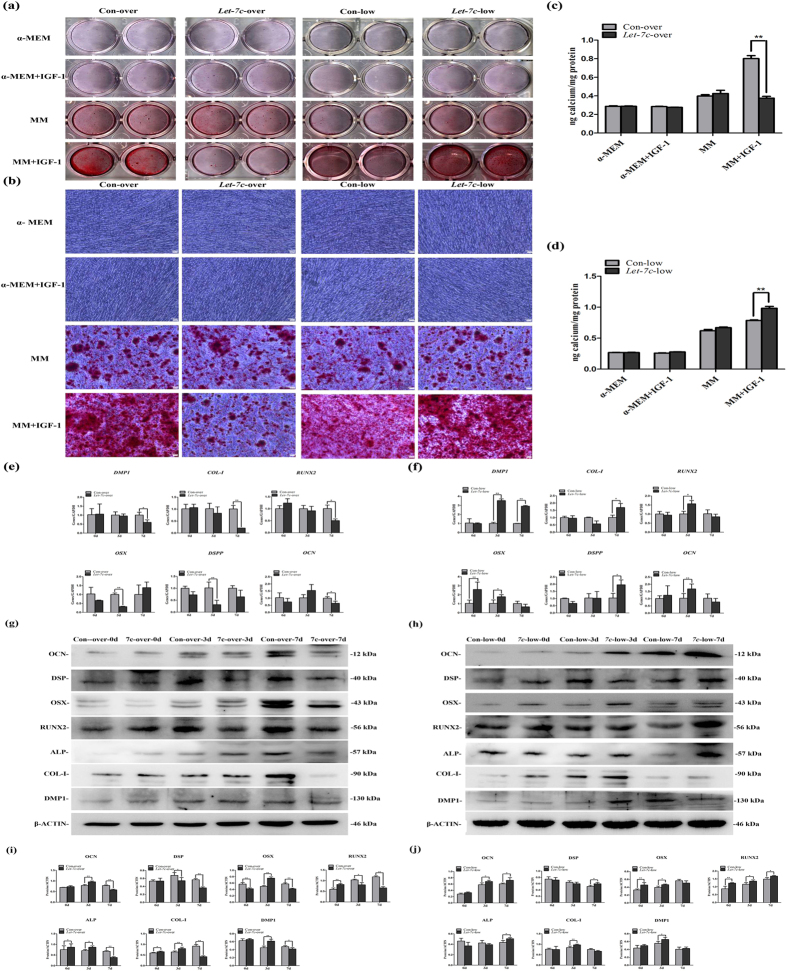
SCAPs were cultured in α-MEM or mineralization media (MM) containing IGF-1 after transfection with hsa-let-7c over/low-expression viruses. Alizarin red staining and quantitative calcium measurement demonstrated that SCAPs with the low-expression of hsa-let-7c produced more calcium nodules after 14 days of osteogenic induction in MM+IGF-1 group than the control group. However, SCAPs with the over-expression of hsa-let-7c generated less calcium nodules after 14 days of induction in MM+IGF-1 group than the control group (Fig. 4a–d, P < 0.01).
Figure 4. IGF-1/IGF-1R/hsa-let-7c axis can regulate the odonto/osteogenic differentiation of SCAPs.
(a) Alizarin red staining for control group, insulin-like growth factor-1 (IGF-1) group, mineralization-inducing media (MM) group and MM+IGF-1 group at day 14, respectively. (b) Calcium nodules in different groups under the inverted microscope. Scale bars = 100 μm. (c) Quantitative analysis for calcium contents in the Let-7c-over group. Values were the means ± SD, n = 3. **P < 0.01. (d) Quantitative analysis for calcium contents in the Let-7c-low group. Values were the means ± SD, n = 3. **P < 0.01. (e) Real-time RT-PCR analysis for the expression of DMP-1, COL-I, RUNX2, OSX, DSPP and OCN in Con-over group and Let-7c-over group respectively. (f) Real-time RT-PCR analysis for the expression of DMP-1, COL-I, RUNX2, OSX, DSPP and OCN in Con-low group and Let-7c-low group respectively. Values were described as the means ± SD, n = 3. **2−∆∆Ct > 2, P < 0.01; *1 < 2−∆∆Ct < 2, P < 0.01. (g) Western blot assay for the odonto/osteogenic proteins (OCN, DSP, OSX, RUNX2, ALP, COL-I and DMP1) in Con-over group and Let-7c-over group respectively. (h) Western blot assay for the odonto/osteogenic proteins (OCN, DSP, OSX, RUNX2, ALP, COL-I and DMP1) in Con-low group and Let-7c-low group respectively. (i) Quantitative analysis for western blot results in Con-over group and Let-7c-over group respectively. Values were described as the means ± SD, n = 3. *P < 0.05, **P < 0.01. (j) Quantitative analysis for western blot results in Con-low group and Let-7c-low group respectively. Values were described as the means ± SD, n = 3. *P < 0.05, **P < 0.01.
Real-time RT-PCR findings revealed that the odonto/osteogenic genes including DMP1, DSPP, RUNX2, OSX and OCN obviously decreased in IGF-1-treated SCAPs accompanying with the hsa-let-7c over-expression, in which the expression of DSPP and OSX reduced at day 3, while COL-I, RUNX2, OCN, DMP1 reduced at day 7 (Fig. 4e, P < 0.05). However, the mRNA levels of these markers were significantly enhanced in IGF-1-treated SCAPs accompanying with the low-expression of hsa-let-7c. In particular, the expression of RUNX2, OSX and OCN was remarkably increased at day 3, and the expression of COL-I was raised at day 7, while the expression of DMP1 was upregulated at day 3 and 7 (Fig. 4f, P < 0.05).
Western blot results demonstrated that the expression of the odonto/osteogenic proteins (DMP1, COL-I, ALP, RUNX2, OSX, DSP and OCN) was significantly decreased in IGF-1 treated SCAPs accompanying with the over-expression of hsa-let-7c, in which the expression of RUNX2 and DSP kept reducing from day 3 to day 7, and the expression of DMP1, COL-I, ALP, OSX and OCN obviously decreased at day 7 (Fig. 4g,i, P < 0.05). In contrast, these protein expressions were significantly upregulated in IGF-1-treated SCAPs accompanying with the low-expression of hsa-let-7c. In detail, the expression of DMP1, COL-I, and OSX increased at day 3, while the expression of ALP, RUNX2, and DSP was enhanced at day 7, and the expression of OCN was upregulated at both day 3 and 7 (Fig. 4h,j, P < 0.05).
Effects of IGF-1/IGF-1R/hsa-let-7c axis on MAPK pathway in SCAPs
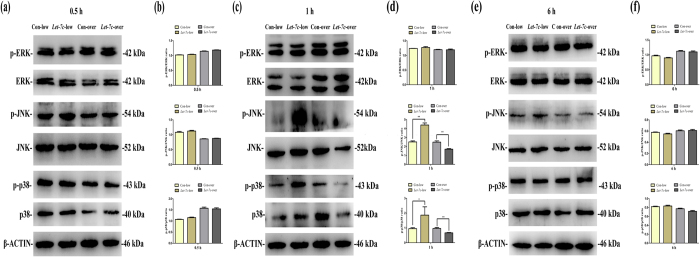
To elucidate the potential involvement of MAPK signaling pathway in IGF-1/hsa-let-7c mediated differentiation of SCAPs, we respectively collected the cytoplasmic proteins in Con-over group, Let-7c-over group, Con-low group and Let-7c-low group and investigated the expression of MAPK related proteins by western blot at 0.5 hour (Fig. 5a,b), 1 hour (Fig. 5c,d) and 6 hour (Fig. 5e,f) respectively. The levels of phosphorylated ERK, phosphorylated JNK and p38 almost did not change at both 0.5 hour and 6 hour (Fig. 5a,e, respectively). Quantitatively, the ratio of p-ERK/ERK, p-JNK/JNK and p-p38/p38 were not affected in both Let-7c-low group and Let-7c-over group (Fig. 5b,f, respectively). As shown in Fig. 5c (1 hour), the level of phosphorylated ERK almost did not change. However, the levels of phosphorylated JNK and p38 were significantly enhanced in Let-7c-low group, as compared with Let-7c-over group (Fig. 5c). Quantitatively, the ratio of p-ERK/ERK was not affected in both Let-7c-low group and Let-7c-over group (Fig. 5d). However, the ratios of p-JNK/JNK and p-p38/p38 noticeably increased at 1 hour in Let-7c-low group, while the ratios of p-JNK/JNK and p-p38/p38 remarkably decreased at 1 hour in Let-7c-over group (Fig. 5d, P < 0.05 or P < 0.01).
Figure 5. IGF-1/IGF-1R/hsa-let-7c axis can regulate MAPK signaling pathway in SCAPs.
(a) Western blot assay for the expression of MAPK related proteins at 0.5 hour (ERK, phosphorylated ERK, JNK, phosphorylated JNK, p38 and phosphorylated p38, respectively). (b) The ratio changes of p-ERK/ERK, p-JNK/JNK and p-p38/p38 at 0.5 hour in different groups. Values were described as the means ± SD, n = 3. *P < 0.05, **P < 0.01. (c) Western blot assay for the expression of MAPK related proteins at 1 hour. (d) The ratio changes of p-ERK/ERK, p-JNK/JNK and p-p38/p38 at 1 hour in different groups. Values were described as the means ± SD, n = 3. *P < 0.05, **P < 0.01. (e) Western blot assay for the expression of MAPK related proteins at 6 hour. (f) The ratio changes of p-ERK/ERK, p-JNK/JNK and p-p38/p38 at 6 hour in different groups. Values were described as the means ± SD, n = 3. *P < 0.05, **P < 0.01.
Discussion
The let-7 family which consists of several members (let-7a/b/c/d/e/f/g/i) plays a crucial role in various cellular activities including the committed differentiation of multiple cell types14,18,19. All different let-7 family members have the common seed motif thereby likely sharing the same target genes15, in which IGF-1R is identified as a target gene of let-7 miRNAs15. Magnucki et al. have detected that IGF-1R is expressed in both DPSCs and impacted third molars20. Other studies have reported that IGF-1R is deployed within the cytoplasm of human unbilical cord blood-derived neural stem cells and localized mainly in the cytoplasm of large luteal cells21,22. These findings have promoted us to speculate that the over/low-expression of let-7 family may modulate the expression of IGF-1R in dental stem cells. Thus, we selected the let-7c miRNA, one of the let-7 family members, as the target to observe its interaction with IGF-1R in SCAPs. In the present study, IGF-1/IGF-1R/hsa-let-7c axis plays an important role during the committed differentiation of stem cells in which hsa-let-7c acts as a major modulator in IGF-1 treated SCAPs via the regulation of IGF-1R.
Meanwhile, we identified IGF-1R was localized within cytoplasm in SCAPs as the target gene of hsa-let-7c, and both were negatively correlated. Transfection with hsa-let-7c low-expression virus brought about the upregulation of IGF-1R and the enhanced odonto/osteogenic differentiation in IGF-1-treated SCAPs, as indicated by the upregulation of several odonto/osteogenic markers (DMP1/DMP1, COL-I/COL-I, ALP, RUNX2/RUNX2, OSX/OSX, DSPP/DSP and OCN/OCN) in vitro. On the contrary, the hsa-let-7c over-expression downregulated the level of IGF-1R and decreased the odonto/osteogenic differentiation in IGF-1-treated SCAPs. It is commonly believed that IGF-1 takes actions mostly via IGF-1R23,24. These findings indicate that IGF-1, IGF-1R and hsa-let-7c act as a whole especially in the process of cell differentiation.
Dentin-sialoprotein (DSP) is one member of the major non-collagenous proteins, which is derived from a single dentin-sialophosphoprotein (DSPP) gene. Both DSP protein and DSPP gene are highly expressed in odontoblast lineages, and considered as the typical odontogenic markers for the differentiation of human MSCs25,26. Dentin matrix protein-1 (DMP1) is expressed in odontoblasts that secrete the matrix proteins in the process of dentinogenesis and mineralization during postnatal tooth development. Some studies have suggested a potential functional and synergistical relationship between DSPP and DMP1, in which DSPP acts as the downstream effector molecule of DMP1 and can be controlled by DMP1 during the dentinogenesis and osteogenesis27,28,29.
Some osteogenic markers (e.g., ALP, RUNX2, OSX and COL-I) are secreted during the early stage of osteo/odontoblastic differentiation, whereas OCN is involved in the late-stage differentiation process30,31,32,33. ALP is expressed and secreted from a proliferation phase to the deposition of mature extracellular matrix including calcium phosphate during the cell differentiation34. RUNX2 and OSX are expressed in osteogenic mesenchyme in all stages of craniofacial bone development, and RUNX2 can modulate bone and tooth development directly or through a RUNX2 signaling pathway related to OSX32. COL-I comprises almost 90% of organic material during the formation of inorganic bone matrix to support the structure of osteogenesis33,35,36. OCN is a small c-carboxyglutamate protein, which is synthesized only by mature cementoblasts, osteoblasts and odontoblasts, and its function is controlled by RUNX2 in the later-stage of mineralized tissues35,36,37. Our previous studies have proved that IGF-1 can stimulate the odonto/osteogenic differentiation of SCAPs and PDLSCs in vitro6,8. In this study, all early and late phase markers of odonto/osteogenic differentiation were highly expressed in IGF-1-treated SCAPs with the low-expression of hsa-let-7c, but significantly downregulated in Let-7c overexpression group, indicating that hsa-let-7c was paramount to the IGF-1/IGF-1R/hsa-let-7c axis.
The differentiation potential of stem cells is subject to a variety of internal and external factors regulated by signaling pathways. Many external factors, which play a physiological role through diverse signaling pathways, can affect the biological properties of SCAPs38,39,40,41,42. In this study, JNK and p38 MAPK pathways were activated through hsa-let-7c low-expression and IGF-1R over-expression, suggesting that the activation of this pathway is associated with IGF-1/IGF-1R/hsa-let-7c axis (Fig. 6) during the odonto/osteogenic differentiation of SCAPs.
Figure 6. Schematic diagram for IGF-1/IGF-1R/hsa-let-7c MAPK axis.
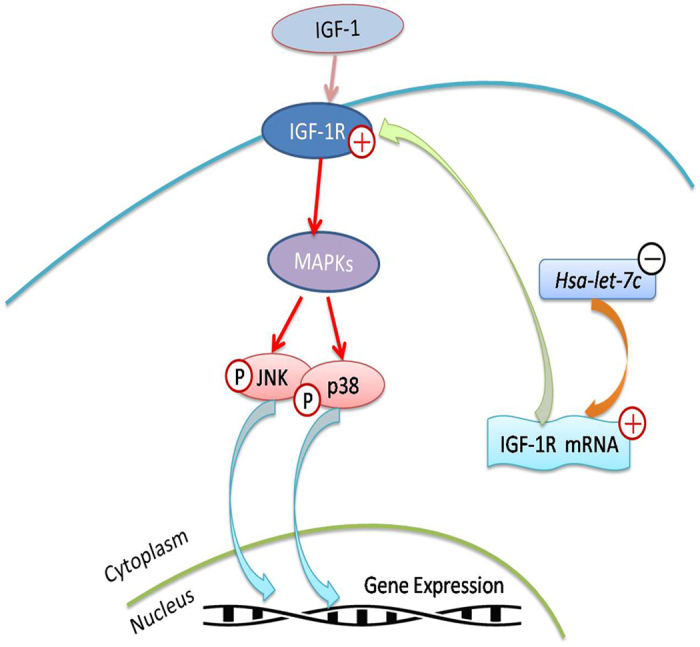
ERK, JNK and p38 MAPKs are members of MAPK family which can be activated by a variety of environmental factors. JNK and p38, which are activated by hsa-let-7c low-expression and IGF-1R over-expression, can translocate to the nucleus where they phosphorylate the transcription factors and subsequently regulate the downstream gene expression.
Mitogen-activated protein kinases (MAPKs) belong to the serine-threonine kinase family and are important component in the signal transduction networks. MAPK pathway, which is a non-classical pathway, plays a crucial role in cell growth, proliferation, differentiation and apoptosis. It mainly consists of three parallel mechanisms, i.e., extracellular signal regulated kinases (ERKs), c-Jun N-terminal kinases (JNKs) and p38 MAPK43. The three parallel pathways play crucial roles in regulating the odonto/osteogenic differentiation of many cell types, such as DPSCs, PDLSCs, SCAPs and MSCs8,9,38,44.
Different stimuli can activate different MAPK pathway members, and have different downstream targets. Let-7 can mediate cell growth and differentiation via the activation of MAPK signaling pathway45. In detail, let-7 can regulate the ERK MAPK and Akt/PI3K signaling pathways by targeting the estrogen receptor (ER)-α3617. In this study, our findings demonstrated that hsa-let-7c can control the activity of JNK and p38 MAPK pathways by targeting the IGF-1R. As a target of let-7 family, IGF-1R can promote the cell proliferation through the crosstalk between IRS-2/Akt and MAPK pathways46,47. The activated IGF-1R can combine corresponding molecules to trigger the downstream signaling cascades including MAPK pathway, thereby regulating the cell transformation, growth and survival48,49. In particular, IGF-1R and p38 MAPK pathway can control the quiescence of hDPSCs and will be inactivated when DPSCs mitosis occurs50. As the upstream regulators of MAPK pathway, IGF-1R and let-7 work closely as a whole in cell differentiation. Furthermore, ERK MAPK pathway can negatively regulate the expression of let-7 via LIN28 expression induced by Myc transcription51. ERK MAPK signaling pathway can modulate the phosphorylation of miRNA to generate the complex, thereby affects the regulation of cell mitosis signals.
Taken together, we have a general knowledge of the relationship among three members in the IGF-1/IGF-1R/hsa-let-7c axis (Fig. 6). As shown in our study, IGF-1, IGF-1R and hsa-let-7c were tightly associated with the odonto/osteogenic differentiation of SCAPs, while IGF-1R and hsa-let-7c were negatively correlated. IGF-1 takes actions via binding to IGF-1R, and the latter itself can also regulate the stem cell differentiation. Moreover, hsa-let-7c can control cell differentiation induced by IGF-1 and IGF-1R through modulating IGF-1R expression.
IGF-1/IGF-1R/hsa-let-7c axis has a key influence on the odonto/osteogenic differentiation of IGF-1-treated SCAPs as well as MAPK signaling pathway. These findings indicate that we can take advantage of the modulation IGF-1/IGF-1R/hsa-let-7c axis on the odonto/osteogenic differentiation for tooth regeneration in the future. Further study needs to be performed to explore the downstream signals of MAPK pathway, especially changes in the level of transcription factors.
Methods
Hsa-let-7c target genes prediction
Prediction and analysis of target genes of hsa-let-7c were performed using bioinformatics methods. The miRDB is an online database for miRNA target prediction. And the whole targets in miRDB are predicted by a bioinformatics tool. The miRDB can predict miRNA targets in five species, including human. In this study, we utilized the miRDB database to perform the bioinformational analysis of target genes of hsa-let-7c.
Cell isolation and identification
Impacted third molars (n = 24) were collected from young patients (17–20 years old) in Oral Surgery Department of Jiangsu Provincial Stomatological Hospital after the informed consent from patients was obtained. This study was approved by the Ethical Committee of the Stomatological School of Nanjing Medical University (Reference #200900128). All procedures were carried out according to the Human Care Guidelines of the Ethical Committee of Nanjing Medical University. The apical papillae were gently detached from the immature roots, minced and digested in a solution containing 3 mg/mL collagenase type I (Sigma, St. Louis, MO) and 4 mg/mL dispase (Sigma, St. Louis, MO) for 30 min at 37 °C. Then, these cells were purified by using rabbit anti-STRO-1 antibody (Santa Cruz, Delaware, CA) and sheep anti-rabbit IgG Dynabeads (Dynal Biotech, Oslo, Norway) according to the standard procedures for magnetic activated cell sorting (MACS). Purified stem cells from apical papilla (SCAPs) were seeded at 1 × 104 cells/mL into 10 cm culture dishes and cultured in alpha minimum essential medium (α-MEM, Gibco, Life Technologies, Grand Island, NY) supplemented with 6% fetal bovine serum (Gibco, USA), 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C in 5% CO2. The fresh medium was changed every 2 days. Cells were subcultured at the ratio of 1:3 when they reached 75–85% confluence. SCAPs at 1–3 passages were used for subsequent experiments. The expression of IGF-1R in purified SCAPs was detected by the confocal laser scanning microscopy (CLSM) method (IGF-1R antibody was purchased from Boster Technology, China). Then, these stem cells were cultured in α-MEM containing 100 ng/mL IGF-1 (Peprotech, USA)6,8. Stem cells were routinely examined under the phase-contrast inverted microscope (Olympus). Generally, SCAPs at the same passage were used in each experiment.
Selection of the best MOI value
Multiplicity of infection (MOI) refers to the average number of viral particles in a cell with an active viral infection. The best MOI value means that viral particles exert the least impact on the growth morphology of the cells, but can obtain the highest transfection efficiency. In the present study, SCAPs after transfection were plated in triplicate in 8 wells of 96-well plates at 5 × 104 cells per well, and divided into eight groups as described in Table 2. 10 μL titer virus was added to the respective wells except control group. 12 hours later, virus was replaced with the conventional medium and cells were continuously cultured for 4 days to observe the growth features and fluorescence expression under an inverted microscope. Finally, MOI was calculated according to the photography records.
Table 2. Virus titers for multiplicity of infection (MOI) in eight groups.
| Virus titers | +Conventional medium | +Conventional medium+ POL |
|---|---|---|
| 1 × 107 TU/mL virus | 1 | 2 |
| 5 × 106 TU/mL virus | 3 | 4 |
| 1 × 106 TU/mL virus | 5 | 6 |
| Blank control group | 7 | 8 |
Virus transfection
IGF-1 treated SCAPs were transfected with hsa-let-7c over/low-expression lentiviruses (Shanghai Genechem Co., LTD, Shanghai, China) and thus four groups were applied in this study, i.e., Con-over group (cells transfected with over-expression control vectors), Let-7c-over group (cells transfected with hsa-let-7c over-expression lentivirus), Con-low group (cells transfected with scramble control vectors) and Let-7c-low group (cells transfected with hsa-let-7c low-expression lentivirus). In the preliminary experiment, the best parameter of MOI was screened and its value is equal to 5. SCAPs were seeded in 25 cm2 culture dishes, and the culture medium was replaced after 24 hours. At the confluence of 60–70%, cells were transfected with the hsa-let-7c over/low-expression lentiviral vectors in 2 mL α-MEM medium containing 6% fetal bovine serum and 8 μg/mL polybrene (POL). 10 hours later, virus media were replaced with 3 mL conventional media for further culture and screening.
Cell counting kit 8 (CCK8) assay
Transfected SCAPs were plated in triplicate in a 96-well plate at 2 × 103 cells per well and incubated in the absence or presence of IGF-1. The proliferation features were determined by using the cell counting kit-8 (CCK8, Dojindo, Japan) at different time points (days 0, 1, 3, 5, 7, 9, 11, 13, respectively). After SCAPs were treated with CCK8 at 37 °C for 2 hours, the optical density was measured by microplate reader scanning at 490 nm according to the manufacturer’s instruction52.
Flow cytometry
For cell cycle analysis, 5 × 105 cells were harvested by trypsinization (Beyotime, China), washed twice with PBS, and then fixed with 75% ice-cold ethanol for 24 hours at 4 °C. The fixed cells were then stained with propidium iodide (PI) for 30 min at 37 °C. Finally, all samples were analyzed by flow cytometry. Cell cycle analysis was measured by FACScan flow cytometer (BD Biosciences, San Jose, CA, USA). The experiment was performed in triplicate.
Alizarin red staining
Transfected SCAPs in Con-over group, Let-7c-over group, Con-low group and Let-7c-low group were respectively seeded into 24-well plates (Nunc, USA) at a density of 1 × 104 cells/well and cultured in routine media or mineralization-inducing media (MM) (containing α-MEM, 6% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, 100 μM ascorbic acid, 2 mM 2-glycerophosphate and 10 nM dexamethasone). At day 14, alizarin red staining was carried out as described before38,53,54,55 and images were acquired using a scanner. Then, nodule staining was destained by 10% cetylpyridinium chloride (CPC) in 10 mM sodium phosphate for 30 minutes at room temperature. The calcium concentration was determined by measuring the absorbance at 526 nm with a universal microplate reader (BioTek Instruments, USA). This experiment was performed in triplicate and the results were presented as the means ± SD.
Real-time reverse transcription polymerase chain reaction
Total cell RNA was extracted by using TRIzol reagent (Invitrogen, New York, NY, USA) according to the manufacturer’s protocols. The concentration and purity of RNA samples were determined by the absorbance of RNA at 230, 260, and 280 nm, respectively. The mRNA was reverse-transcribed into cDNA by using a PrimeScript RT Master Mix kit (TaKaRa Biotechnology, China). Real-time RT-PCR was performed using SYBR Green Master (Roche, Indianapolis, IN, USA) and ABI 7300 real-time PCR system. Real-time RT-PCR reaction conditions for hsa-let-7c were: 95 °C for 10 minutes; followed by 40 cycles of 95 °C for 15 seconds, 60 °C for 60 seconds; then 72 °C for 45 seconds, 72 °C for 7 minutes, 4 °C for 5 minutes. U6 was used as an internal control and the expression of hsa-let-7c was measured by the method of 2−∆∆Ct as previously reported38,56. Data were described as the means ± SD of three independent experiments. Real-time RT-PCR reaction conditions for dentinsialophosphoprotein (DSPP), dentin matrix protein 1 (DMP1), Runt-related transcription factor 2 (RUNX2), osterix (OSX), type I collagen (COL-I) and osteocalcin (OCN) were: 95 °C for 10 minutes; followed by 40 cycles of 95 °C for 15 seconds, 60 °C for 60 seconds. Primers used in this experiment were listed in Table 1. GAPDH was used as an internal control and the expression of osteo/odontoblast-related genes (DSPP, DMP1, RUNX2, OSX, COL-I and OCN) was measured by the method of 2−∆∆Ct as previously reported38,56. Data were described as the means ± SD of three independent experiments.
Western blot analysis
To investigate the effects of the over-expression/low-expression of let-7c on the odonto/osteogenic differentiation of IGF-1 treated SCAPs, IGF-1 treated SCAPs in Con-over group, Let-7c-over group, Con-low group and Let-7c-low group were respectively cultured for 0, 3 and 7 days and then collected. For the evaluation of MAPK pathway-related proteins, IGF-1 treated SCAPs at 80–90% confluence were collected after let-7c transfection for 0.5 hour, 1 hour and 6 hours. Cells in different groups were washed twice with cold PBS and lysed in RIPA lysis buffer (Beyotime, China) containing 1 mM PMSF. Cell debris was eliminated by centrifugation at 12,000 rpm for 10 minutes. The cytoplasm protein and nucleoprotein were obtained with an NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo, USA). Protein concentration was measured by Bradford protein assay. Thirty microgram protein per lane was loaded onto a 10% SDS-PAGE gel for electrophoresis, and then transferred to 0.22 μm PVDF membranes (Millipore, Bedford, MA) at 300 mA for 1 hour. Membranes were blocked in blocking solution (5% Bovine Serum Albumin, 0.01 M TBS, 0.1% Tween-20) at room temperature for 2 hours, and incubated with primary antibodies (DMP1, 1:1000, Abcam; DSP, 1:1000, Santa Cruz; RUNX2, 1:1000, Abcam; OSX, 1:1000, Abcam; OCN, 1:1000, Millipore; ERK1/2, 1:1000, Bioworld; phosphor-ERK1/2, 1:1000, Bioworld; JNK1/2/3, 1:1000, Bioworld; phosphor-JNK1/2/3, 1:1000, Bioworld; p38, 1:1000, Bioworld; phosphor-p38, 1:1000, Bioworld; IGF-1Rα, 1:1000, Abcam; β-ACTIN, 1:1000, Bioworld) overnight at 4 °C. β-ACTIN was used as the internal control. Finally, the membranes were washed with TBST for 10 minutes × 3 followed by incubation in secondary antibodies (1:10,000, Boster) for 1 hour at 37 °C, visualized with ImageQuant LAS4000 system (GE Healthcare, USA). The results were quantified with Image J software (National Institutes of Health, USA). The experiment was repeated three times.
Statistics
Two-sample t test and Chi-square test were respectively performed to compare the means and constituent ratios between two groups. For multiple comparisons between experimental groups and control groups, Dunnett’s test was used to check the significant differences. Two-tailed P-values less than 0.05 were considered statistically significant. All statistical analysis was performed with SPSS 17.0 software (SPSS Inc., Chicago, IL, USA).
Additional Information
How to cite this article: Ma, S. et al. IGF-1/IGF-1R/hsa-let-7c axis regulates the committed differentiation of stem cells from apical papilla. Sci. Rep. 6, 36922; doi: 10.1038/srep36922 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81371144), Qing Lan Project (2014–23), Zhoushan 5313 Project, Natural Science Foundation of Jiangsu Province (No. BK20131392), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, No. 2014–37).
Footnotes
Author Contributions S.M. made substantial contributions to the conception and design of the study, carried out the study, performed the experiments and drafted the manuscript. G.X.L. participated in the design of the study, collected all data, performed data analysis and helped to draft the manuscript. L.J. performed the selection of the best MOI value and some of the realtime RT-PCR experiments. X.Y.P. contributed to the virus transfection and some of the western blot experiments. Y.Q.W. was responsible for cell culture and manuscript drafting. Z.L.W. and Y.Y. contributed to the isolation and expansion of mesenchymal stem cells and helped to draft the manuscript. J.H.Y. conceived the study, and contributed to the study design and coordination, data analysis and interpretation, and manuscript revision. All authors read and approved the final manuscript.
References
- Estrela C., Alencar A. H., Kitten G. T., Vencio E. F. & Gava E. Mesenchymal stem cells in the dental tissues: perspectives for tissue regeneration. Braz Dent J 22, 91–98 (2011). [DOI] [PubMed] [Google Scholar]
- Ye L. et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 11, 50–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoyama W. et al. Mesenchymal stem cell-mediated functional tooth regeneration in swine. PLoS One 1, e79 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standards for the assessment of estrogen receptors in human breast cancer. Report of a workshop on September 29, 1972, at the Antoni van Leeuwenhoek-Huis, Amsterdam. Eur J Cancer 9, 379–381 (1973). [DOI] [PubMed] [Google Scholar]
- Liu D. et al. Demethylation of IGFBP5 by Histone Demethylase KDM6B Promotes Mesenchymal Stem Cell-Mediated Periodontal Tissue Regeneration by Enhancing Osteogenic Differentiation and Anti-Inflammation Potentials. Stem Cells 33, 2523–2536 (2015). [DOI] [PubMed] [Google Scholar]
- Wang S. et al. Insulin-like growth factor 1 can promote the osteogenic differentiation and osteogenesis of stem cells from apical papilla. Stem Cell Res 8, 346–356 (2012). [DOI] [PubMed] [Google Scholar]
- Al-Kharobi H., El-Gendy R., Devine D. A. & Beattie J. The role of the insulinlike growth factor (IGF) axis in osteogenic and odontogenic differentiation. Cell Mol Life Sci 71, 1469–1476 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. et al. Insulin-like growth factor 1 enhances the proliferation and osteogenic differentiation of human periodontal ligament stem cells via ERK and JNK MAPK pathways. Histochem Cell Biol 137, 513–525 (2012). [DOI] [PubMed] [Google Scholar]
- Lv T. et al. Insulin-like growth factor 1 promotes the proliferation and committed differentiation of human dental pulp stem cells through MAPK pathways. Arch Oral Biol 72, 116–123 (2016). [DOI] [PubMed] [Google Scholar]
- Zhao G. et al. Targeted overexpression of insulin-like growth factor I to osteoblasts of transgenic mice: increased trabecular bone volume without increased osteoblast proliferation. Endocrinology 141, 2674–2682 (2000). [DOI] [PubMed] [Google Scholar]
- Zhang M. et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem 277, 44005–44012 (2002). [DOI] [PubMed] [Google Scholar]
- Stratford E. W. et al. The tankyrase-specific inhibitor JW74 affects cell cycle progression and induces apoptosis and differentiation in osteosarcoma cell lines. Cancer Med 3, 36–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B. J. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 (2000). [DOI] [PubMed] [Google Scholar]
- Eguchi T. et al. OstemiR: a novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS One 8, e58796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G. et al. Upstream and downstream mechanisms for the promoting effects of IGF-1 on differentiation of spermatogonia to primary spermatocytes. Life Sci 101, 49–55 (2014). [DOI] [PubMed] [Google Scholar]
- Huang C. W. et al. Differential expression patterns of growth-related microRNAs in the skeletal muscle of Nile tilapia (Oreochromis niloticus). J Anim Sci 90, 4266–4279 (2012). [DOI] [PubMed] [Google Scholar]
- Zhao Y. et al. let-7 microRNAs induce tamoxifen sensitivity by downregulation of estrogen receptor alpha signaling in breast cancer. Mol Med 17, 1233–1241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyerinas B., Park S. M., Hau A., Murmann A. E. & Peter M. E. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer 17, F19–F36 (2010). [DOI] [PubMed] [Google Scholar]
- Wulczyn F. G. et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J 21, 415–426 (2007). [DOI] [PubMed] [Google Scholar]
- Magnucki G. et al. Expression of the IGF-1, IGFBP-3 and IGF-1 receptors in dental pulp stem cells and impacted third molars. J Oral Sci 55, 319–327 (2013). [DOI] [PubMed] [Google Scholar]
- Janowski M., Lukomska B. & Domanska-Janik K. Migratory capabilities of human umbilical cord blood-derived neural stem cells (HUCB-NSC) in vitro. Acta Neurobiol Exp (Wars) 71, 24–35 (2011). [DOI] [PubMed] [Google Scholar]
- Uniyal S. et al. Expression and localization of insulin-like growth factor system in corpus luteum during different stages of estrous cycle in water buffaloes (Bubalus bubalis) and the effect of insulin-like growth factor I on production of vascular endothelial growth factor and progesterone in luteal cells cultured in vitro. Theriogenology 83, 58–77 (2015). [DOI] [PubMed] [Google Scholar]
- Romand S. et al. Improving expression of recombinant human IGF-1 using IGF-1R knockout CHO cell lines. Biotechnol Bioeng 113, 1094–1101 (2016). [DOI] [PubMed] [Google Scholar]
- Jiang G. et al. Insulin growth factor-1 (IGF-1) enhances hippocampal excitatory and seizure activity through IGF-1 receptor-mediated mechanisms in the epileptic brain. Clin Sci (Lond) 129, 1047–1060 (2015). [DOI] [PubMed] [Google Scholar]
- Yamazaki H., Kunisada T., Miyamoto A., Tagaya H. & Hayashi S. Tooth-specific expression conferred by the regulatory sequences of rat dentin sialoprotein gene in transgenic mice. Biochem Biophys Res Commun 260, 433–440 (1999). [DOI] [PubMed] [Google Scholar]
- Iejima D., Sumita Y., Kagami H., Ando Y. & Ueda M. Odontoblast marker gene expression is enhanced by a CC-chemokine family protein MIP-3alpha in human mesenchymal stem cells. Arch Oral Biol 52, 924–931 (2007). [DOI] [PubMed] [Google Scholar]
- Ye L. et al. Deletion of dentin matrix protein-1 leads to a partial failure of maturation of predentin into dentin, hypomineralization, and expanded cavities of pulp and root canal during postnatal tooth development. J Biol Chem 279, 19141–19148 (2004). [DOI] [PubMed] [Google Scholar]
- Gibson M. P. et al. The rescue of dentin matrix protein 1 (DMP1)-deficient tooth defects by the transgenic expression of dentin sialophosphoprotein (DSPP) indicates that DSPP is a downstream effector molecule of DMP1 in dentinogenesis. J Biol Chem 288, 7204–7214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani P. H. et al. Transgenic expression of Dspp partially rescued the long bone defects of Dmp1-null mice. Matrix Biol 52-54, 95–112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. W. et al. In vitro osteogenic differentiation of cultured human dental papilla-derived cells. J Oral Maxillofac Surg 67, 507–514 (2009). [DOI] [PubMed] [Google Scholar]
- Komori T. Regulation of osteoblast differentiation by Runx2. Adv Exp Med Biol 658, 43–49 (2010). [DOI] [PubMed] [Google Scholar]
- Chen S. et al. Runx2, osx, and dspp in tooth development. J Dent Res 88, 904–909 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni P. et al. Preparation of poly(ethylene glycol)/polylactide hybrid fibrous scaffolds for bone tissue engineering. Int J Nanomedicine 6, 3065–3075 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroleva A. et al. Osteogenic differentiation of human mesenchymal stem cells in 3-D Zr-Si organic-inorganic scaffolds produced by two-photon polymerization technique. PLoS One 10, e0118164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farea M. et al. Synergistic effects of chitosan scaffold and TGFbeta1 on the proliferation and osteogenic differentiation of dental pulp stem cells derived from human exfoliated deciduous teeth. Arch Oral Biol 59, 1400–1411 (2014). [DOI] [PubMed] [Google Scholar]
- Niu L. N. et al. Intrafibrillar-silicified collagen scaffolds enhance the osteogenic capacity of human dental pulp stem cells. J Dent 42, 839–849 (2014). [DOI] [PubMed] [Google Scholar]
- Feng X. et al. Insulin-like growth factor 1 can promote proliferation and osteogenic differentiation of human dental pulp stem cells via mTOR pathway. Dev Growth Differ 56, 615–624 (2014). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. 17beta-estradiol promotes the odonto/osteogenic differentiation of stem cells from apical papilla via mitogen-activated protein kinase pathway. Stem Cell Res Ther 5, 125 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Effects of canonical NF-kappaB signaling pathway on the proliferation and odonto/osteogenic differentiation of human stem cells from apical papilla. Biomed Res Int 2014, 319651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M. et al. Mineral trioxide aggregate promotes the odonto/osteogenic differentiation and dentinogenesis of stem cells from apical papilla via nuclear factor kappa B signaling pathway. J Endod 40, 640–647 (2014). [DOI] [PubMed] [Google Scholar]
- Mu C. et al. Mechanical stress stimulates the osteo/odontoblastic differentiation of human stem cells from apical papilla via erk 1/2 and JNK MAPK pathways. Biomed Res Int 2014, 494378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liu B., Gu S. & Liang J. Effects of Wnt/beta-catenin signalling on proliferation and differentiation of apical papilla stem cells. Cell Prolif 45, 121–131 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. et al. Mineral trioxide aggregate promotes odontoblastic differentiation via mitogen-activated protein kinase pathway in human dental pulp stem cells. Mol Biol Rep 39, 215–220 (2012). [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J., Song W. & Yu J. Mineral trioxide aggregate upregulates odonto/osteogenic capacity of bone marrow stromal cells from craniofacial bones via JNK and ERK MAPK signalling pathways. Cell Prolif 47, 241–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricarte-Filho J. C. et al. Effects of let-7 microRNA on Cell Growth and Differentiation of Papillary Thyroid Cancer. Transl Oncol 2, 236–241 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L. et al. IGF-1R, a target of let-7b, mediates crosstalk between IRS-2/Akt and MAPK pathways to promote proliferation of oral squamous cell carcinoma. Oncotarget 5, 2562–2574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. S., Cho H. Y., Hoyt K. R., Naegele J. R. & Obrietan K. IGF-1 receptor-mediated ERK/MAPK signaling couples status epilepticus to progenitor cell proliferation in the subgranular layer of the dentate gyrus. Glia 56, 791–800 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis M. M., Yuen J. S., Protheroe A. S., Pollak M. & Macaulay V. M. The type 1 insulin-like growth factor receptor pathway. Clin Cancer Res 14, 6364–6370 (2008). [DOI] [PubMed] [Google Scholar]
- Johnson G. L. & Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298, 1911–1912 (2002). [DOI] [PubMed] [Google Scholar]
- Vandomme J. et al. Insulin-like growth factor 1 receptor and p38 mitogen-activated protein kinase signals inversely regulate signal transducer and activator of transcription 3 activity to control human dental pulp stem cell quiescence, propagation, and differentiation. Stem Cells Dev 23, 839–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi-Garimella S. et al. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J 28, 347–358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying X. et al. Effect of boron on osteogenic differentiation of human bone marrow stromal cells. Biol Trace Elem Res 144, 306–315 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang Q. B., Cao W., Liu Y. R., Cui S. M. & Yan Y. Y. Effects of Sirtuin 1 on the proliferation and osteoblastic differentiation of periodontal ligament stem cells and stem cells from apical papilla. Genet Mol Res 15 (2016). [DOI] [PubMed] [Google Scholar]
- Diao S. et al. Enriched trimethylation of lysine 4 of histone H3 of WDR63 enhanced osteogenic differentiation potentials of stem cells from apical papilla. J Endod 41, 205–211 (2015). [DOI] [PubMed] [Google Scholar]
- Yu J. et al. Differentiation potential of STRO-1+ dental pulp stem cells changes during cell passaging. BMC Cell Biol 11, 32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Estrogen deficiency inhibits the odonto/osteogenic differentiation of dental pulp stem cells via activation of the NF-kappaB pathway. Cell Tissue Res 352, 551–559 (2013). [DOI] [PubMed] [Google Scholar]