Abstract
This chapter presents methods to conduct and analyze genome-wide chromatin immunoprecipitation of the cohesin complex and the Nipped-B cohesin loading factor in Drosophila cells using high-throughput DNA sequencing (ChIP-seq). Procedures for isolation of chromatin, immunoprecipitation, and construction of sequencing libraries for the Ion Torrent Proton high throughput sequencer are detailed, and computational methods to calculate occupancy as input-normalized fold-enrichment are described. The results obtained by ChIP-seq are compared to those obtained by ChIP-chip (genomic ChIP using tiling microarrays), and the effects of sequencing depth on the accuracy are analyzed. ChIP-seq provides similar sensitivity and reproducibility as ChIP-chip, and identifies the same broad regions of occupancy. The locations of enrichment peaks, however, can differ between ChIP-chip and ChIP-seq, and low sequencing depth can splinter broad regions of occupancy into distinct peaks.
Keywords: ChIP-chip, ChIP-seq, chromatin immunoprecipitation, cohesin, Drosophila, high throughput sequencing, Ion Torrent Proton, microarray, Nipped-B
1. Introduction
The cohesin complex and the proteins that regulate its binding to chromosomes, such as the Nipped-B loading factor, participate in multiple fundamental cellular processes, including sister chromatid cohesion and chromosome segregation, DNA repair, and control of gene expression (1–5). Knowing the chromosomal regions and genes they occupy in different cells and tissues can provide critical information on these basic processes and their roles in development and disease. However, cohesin and Nipped-B present unique challenges for determining their locations by chromatin immunoprecipitation (ChIP). Cohesin, consisting of the Smc1, Smc3, Rad21 and SA proteins, forms a ring-like structure that topologically entraps DNA, allowing it to slide along the chromosome (6). Thus cohesin makes less intimate contacts with DNA than proteins that recognize specific DNA sequences, and its exact position varies from cell to cell. Topological binding also predicts that cohesin crosslinks less efficiently to DNA with formaldehyde than most DNA binding proteins. Combined, the variable positioning and expected low crosslinking efficiency predict that the enrichment of cohesin-occupied sequences by ChIP sample will be low. These problems are further compounded when performing ChIP with tissues containing multiple cell types, where cohesin and Nipped-B are unlikely to occupy the same regions and genes in all cells. Many computational methods for detecting occupancy in genomic ChIP experiments presume that proteins bind in specific peaks, and thus are suboptimal for proteins such as Nipped-B and cohesin that occupy broad domains of several kilobases that are functionally significant.
Our laboratory previously addressed these challenges in Drosophila melanogaster cultured cells and tissues by using multiple antibodies to confirm regions of relatively low enrichment, and microarray-based genomic ChIP (ChIP-chip), which provides high sensitivity and reproducibility (7–9). We used computational methods, including the TiMAT (http://bdtnp.lbl.gov/TiMAT/) and MAT (10) programs that use windowing to reduce noise and quantify enrichment without relying on peak calling.
Here we describe methods for ChIP with high-throughput DNA sequencing (ChIP-seq) in Drosophila cultured cells that attain similar sensitivity for Nipped-B and cohesin occupancy as obtained by ChIP-chip. The methods used for isolating chromatin and immunoprecipitation are modified from those we used for performing ChIP-chip (7–9). For ChIP-seq data analysis we developed a windowing method to normalize the sequencing of ChIP samples to input chromatin sequencing to calculate enrichment. The results largely corroborate those obtained using ChIP-chip, but also reveal how details of enrichment profiles can depend on the methodology. The advantages over ChIP-chip include the ability to detect occupancy in regions with repetitive genes.
2. Materials
2.1 Antibodies
Antibody specificity and affinity are crucial variables for ChIP, particularly when low crosslinking efficiency and variable positioning are expected. We previously described several antibodies used for Drosophila cohesin and Nipped-B ChIP (7, 9). Although these are unlikely to be useful in other organisms, these reagents were made using general principles that can be applied to developing similar antibodies for other species.
Our strategy is to raise at least two independent polyclonal antiserum (rabbit and guinea pig) against 30 to 40 kD fragments of the target proteins, and antibodies against multiple cohesin subunits. Using polyclonal antibodies against larger proteins reduces the risk of epitope-masking or other selective recognition artifacts, which are concerns with monoclonal antibodies, or polyclonal antibodies against small peptide antigens, as is often the case with commercial antibodies. Chromosomal position-selective recognition can also occur with polyclonal antibodies against large proteins (9) and thus using multiple antibodies against the same target protein and antibodies against different subunits of the same complex can further reduce these concerns.
We use several methods for validating antibodies, including western blots with control and RNAi-treated cells or mutant animals to confirm antigen size and identity, and to detect non-specific cross-reacting proteins. If there are significant cross-reacting proteins, we use western blots of nuclear extracts to determine if the cross-reacting proteins are in the nucleus, and also test if two independent antisera recognize the same cross-reacting proteins. Fluorescent immunostaining of control and RNAi-treated cells confirms the expected nuclear localization, and co-immunostaining of salivary gland polytene chromosomes can confirm co-occupancy detected by genomic ChIP.
2.2 Solutions and Reagents
All reagents are molecular biology grade, and all solutions are made using 18 MΩ H2O. Solutions are stored at room temperature except where noted.
Formaldehyde Solution, 36.5–38% (Sigma F-8775, stored at −20° in aliquots)
Phosphate-Buffered Saline pH 7.5 (PBS, cell culture grade)
2.5 M glycine in PBS pH 7.5
Wash Buffer A: 10 mM Hepes·KOH pH 7.9, 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton X-100
Wash Buffer B: 10 mM Hepes·KOH pH 7.9, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.01% Triton X-100
Sonication Buffer: 10 mM Hepes·KOH pH 7.9, 1 mM EDTA, 0.5 mM EGTA
10% (w/v) sodium lauroyl sarcosine (Sarkosyl, Sigma L-9150)
Adjustment Buffer: 10 mM Hepes·KOH pH 7.9, 0.23 M NaCl, 1 mM EDTA, 1.7% Triton X-100, 0.17% sodium deoxycholate
Ribonuclease A (4 mg per mL, Qiagen 158924), store at 4°.
Proteinase K (20 mg per mL, Roche) store at 4°.
100 mM phenylmethylsulfonyl fluoride (PMSF) in isopropanol (keep dry by adding molecular sieve pellets (Sigma) to storage bottle, store at 4°. CAUTION: PMSF is a toxin and should be handled using appropriate precautions. It is added to solutions just prior to their use.
cOmplete™ Mini EDTA-free Protease Inhibitor Cocktail Tablets (Roche 1836170001)
5 M NaCl
Phenol:Chloroform:Isoamyl Alcohol 25:24:1 Saturated with 10 mM Tris·HCl, pH 8.0, 1 mM EDTA (Sigma P3803), store at 4°. CAUTION: phenol-chloroform can cause significant skin and eye damage upon contact and should be handled accordingly.
Ethanol, 100%. CAUTION: ethanol is highly flammable and should be stored in a cabinet designed for flammable materials.
3 mM Tris.HCl, pH 8.0
Pierce™ Protein A Plus (22811) and/or Protein A/G Plus (20423) Agarose Beads (Life Technologies), store at 4°.
TE Buffer: 10 mM Tris·HCl pH 7.5, 1 mM EDTA
Modified RIPA Buffer: 140 mM NaCl, 10 mM Hepes·KOH pH 7.9, 1 mM EDTA, 1% (v/v) Triton X-100, 0.2% (w/v) sodium lauroyl sarcosine, 0.1% sodium deoxycholate
LiCl Buffer: 10 mM Tris.HCl pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate
Ion Plus Fragment Library Kit (Life Technologies, 4471252). Store components according to manufacturer’s recommendations.
Ion Xpress™ Barcode Adapters 1–16 Kit (Life Technologies, 4471250). Store at −20°.
Agencourt AMPure XP Magnetic Beads (Beckman-Coulter, A63881)
2.3 Special Equipment
Diagenode Bioruptor Bath Sonicator (or equivalent)
Qubit Fluorometer (Invitrogen/Life Technologies)
Agilent 2100 Bioanalyzer (Agilent Technologies)
2.4 Computer Software
3. Methods
3.1 Chromatin Isolation (see Note 1)
Grow Drosophila tissue culture cells at 25° to a density of 5 to 6 million cells per mL in the recommended media. We usually start with 500 to 600 million cells to make chromatin, but this can be adjusted as needed, considering that chromatin from 10 to 20 million cells is typically used for one immunoprecipitation.
Collect the cells in the growth medium by scraping (see Note 2), transfer to centrifuge tubes and fix with 1% formaldehyde (1:37 dilution of stock reagent, mix by gentle inversion) for 10 minutes at room temperature.
Quench fixation by addition of 2.5 M glycine in PBS pH 7.5 to give a final concentration of 0.125 M (1:20 dilution), mix gently, and incubate for 3 minutes at room temperature.
Collect the fixed cells by centrifugation at 1000g for 5 minutes at room temperature, and wash 3X by suspension in PBS and centrifugation.
Discard the supernatant and flash-freeze the cell pellet in liquid nitrogen and store at −80°. We typically freeze Drosophila cells at 180 to 200 million cells per tube. Fresh cell pellets can also be used to prepare chromatin without storage (see Note 3).
Suspend fixed cell pellet (frozen or fresh) in Wash Buffer A at a ratio of 4 mL per 180 to 200 million cells, and gently mix at room temperature for 10 min, followed by centrifugation at 1000g for 5 min at room temperature.
Suspend the resulting pellet in the same volume of Wash Buffer B, gently mix for 10 min at room temperature, followed by centrifugation at 1300g for 5 min at room temperature.
Suspend the resulting pellet in Sonication Buffer at a ratio of 1 mL per 100 to 120 million cells, and divide into 0.3 mL aliquots in Diagenode 1.5 mL sonication tubes.
Sonicate the suspended cells in the Diagenode Bioruptor with a circulating ice water bath for 15 to 18 cycles of 30 sec, with 30 sec off between cycles, and groups of 5 to 6 sonication cycles separated by a few minutes (see Note 4).
Collect the sonicated chromatin into 2 mL microfuge tubes, add sodium lauroyl sarcosine to a final concentration of 0.5% and mix gently for 10 min at room temperature.
Remove insoluble material by centrifugation at top speed in a microfuge for 10 min at 4°. Divide the supernatant into 200 μL aliquots (~20 million cells per aliquot), flash freeze in liquid nitrogen, and store at −80°.
Determine the shearing efficiency by reverse crosslinking a 50 μL aliquot of chromatin using the procedure described below. Dissolve the purified DNA in 50 μL of 3 mM Tris.HCl pH 8.0, and quantify the amount using a Qubit Fluorometer (see Note 5), and measure the size of the purified DNA by agarose gel electrophoresis or with an Agilent Bioanalyzer. The expected amount of DNA in 50 μL of chromatin is ~2 μg for diploid Drosophila cells, and the mode of the DNA size distribution should be ~500 base pairs. Store the remaining DNA sample at −20° for preparing an input chromatin sequencing library.
3.2 Chromatin immunoprecipitation
We typically use 100 or 200 μL of chromatin (~10 or 20 million cells). For 100 μL of chromatin, add 100 μL of Sonication Buffer containing 0.5% sodium lauroyl sarcosine, and use the same buffer volumes as described below for 200 μL of chromatin.
Quickly thaw a 200 μL aliquot of chromatin and dilute with 300 μL of Adjustment Buffer containing a 2X concentration of protease inhibitors (2 mM PMSF and 2X cOmplete cocktail).
Pre-clear the diluted chromatin by incubation with 20 μL of Protein A agarose beads with gentle mixing for approximately 2 hours at 4°. Wash the Protein A beads with Modified RIPA Buffer several times before using them to pre-clear the diluted chromatin.
Pellet the Protein A beads by gentle centrifugation, transfer the supernatant to a new microfuge tube, and repeat the centrifugation and transfer to ensure complete bead removal.
Incubate the pre-cleared chromatin with the antibody of choice for 8 to 16 hours at 4° with gentle mixing. We typically use 10 to 15 μL of serum, but this should be optimized for each antibody (see Note 6).
Prepare 20 μL of Protein A or Protein A/G agarose beads per 500 μL of chromatin-antibody mix by washing three times in Modified RIPA Buffer, twice with PBS containing 0.5% BSA, and twice with a 2:3 mixture of Sonication Buffer and Adjustment Buffer.
Bind the antibody-chromatin complexes to the prepared Protein A beads by incubation for 3 to 4 hours at 4° with gentle mixing.
Collect the Protein A beads by gentle centrifugation and wash at room temperature sequentially with 1 mL aliquots of the following buffers (kept at 4°) in order, collecting by gentle centrifugation between washes: 5 times with Modified RIPA Buffer for 5 min, 1 time with LiCl solution for 30 sec, 1 time with LiCl solution for 5 min, and 3 times with TE.
Collect the washed beads by gentle centrifugation and suspend in 300 μL of TE.
Reverse the crosslinks by the following two steps: (a) Add 3 μL of DNase-free ribonuclease A (4 mg per mL), to give a final concentration of 40 μg per mL, mix and incubate for 30 min at 37°, (b) Add 15 μL of 10% SDS to give a final concentration of 0.5%, and 7.5 μL of Proteinase K (20 mg per mL) to give a final concentration of 0.5 mg per mL, mix and incubate for 6 to 16 hours at 65°.
After reversing the crosslinks, add 100 μL of TE containing 0.5 M NaCl and extract with 400 μL of phenol:chloroform:isoamyl alcohol. Shake vigorously for 30 sec and separate the phases by centrifugation at top speed in a microfuge for 10 min.
Transfer the aqueous phase to a new tube and precipitate the DNA by adding 12 μL of 5 M NaCl, 2 μL of glycogen (20 mg per mL) as carrier, and 900 μL of ethanol. Mix and incubate at −20° for 15 min.
Collect the precipitated DNA by centrifugation at top speed in a microfuge for 20 min at 4°. Wash the pellet with 70% ethanol, air dry, and dissolve in 25 μL of 3 mM Tris.HCl pH 8.0. Measure the concentration using a Qubit Fluorometer and store at −20° until use. From Nipped-B and cohesin ChIP, we typically obtain 2.5 to 15 ng of total DNA, starting from 100 to 200 μL of chromatin.
3.3 Library construction
Shear the DNA isolated from input chromatin and from the ChIP samples by diluting 2 to 3 ng with 40 μL of 3 mM Tris.HCl pH 8.0 and sonicating with the Diagenode Bioruptor for 18 cycles with a circulating ice water bath (see Note 7). Each cycle is 30 sec with 30 sec off between cycles.
Check the size distribution of the sonicated DNA using an Agilent Bioanalyzer. The mode of the distribution should be ~200 bp (see Note 8).
-
Construct the sequencing libraries from the sheared DNA using the Ion Plus Fragment Library Kit. The steps below are adapted from those described in the manufacturer’s protocol (Life Technologies Publication Number 4473623, revision B). The reagent names refer to those in the kit. Because the amounts of DNA are small, it is important to use coated low-binding microfuge tubes for all steps.
Repair the ends of the sheared DNA by incubating the following reaction mix for 30 min at room temperature: 20 μL of sheared DNA (1 to 1.5 ng), 5 μL of 5X End Repair Buffer, 0.5 μL of End Repair Enzyme.
Bind the repaired DNA to 45 μL (1.8X vol) of AMPure magnetic beads by mixing with the beads and incubating for 5 min at room temperature.
Collect the beads on a magnetic rack for 3 min, discard the supernatant and wash the beads twice for 30 sec with 70% ethanol. Remove all residual 70% ethanol.
Air dry the beads for 1 to 2 min, and elute the DNA by suspending the beads in 12 μL of Low TE. Collect the beads on a magnetic rack and transfer the supernatant with the purified DNA to a new tube.
Ligate adaptors for amplification and sequencing to the purified repaired DNA by incubating the following reaction mix for 30 min at room temperature: 12 μL of repaired DNA, 9 μL of H2O, 2.5 μL of 10X Ligase Buffer, 0.5 μL of In P1 Adapter diluted 1:1 with H2O, 0.5 μL of Ion Xpress™ Barcode Adapter (chosen from 1 – 96) diluted 1:1, 0.5 μL of DNA Ligase.
Bind the adapter-ligated DNA to 37.5 μL (1.5X vol) of AMPure beads by mixing with the beads and incubating for 5 min at room temperature.
Collect the beads in a magnetic rack for 3 min, discard the supernatant and wash the beads twice with 70% ethanol for 30 sec each wash. Remove all residual 70% ethanol.
Air dry the beads for 1 to 2 min, and elute the DNA by suspending the beads in 10 μL of Low TE. Pellet the beads on a magnetic rack and transfer the supernatant with the adapter-ligated DNA to a new tube.
Amplify the adapter-ligated DNA in thermal cycler by making the following reaction mix in a 0.2 mL PCR tube, and using the indicated cycler program: (i) Amplification Mix: 10 μL of adapter-ligated DNA, 60 μL of Platinum® PCR SuperMix High Fidelity, 1.25 μL of Library Amplification Primer Mix; (ii) Cycler Program: 72° for 20 min, 95° for 5 min, then 17 cycles of 97° for 15 sec, 60° for 15 sec, 70° for 1 min. Set the cycler to finish at 4° until purification (< 1 hour).
Bind the amplified DNA to 105 μL (1.5X vol) of AMPure beads by mixing with the beads and incubating for 5 min at room temperature.
Collect the beads on a magnetic rack for 3 min, discard the supernatant and wash twice with 70% ethanol. Remove all residual 70% ethanol.
Air dry the beads and elute the DNA by suspending the beads in 12 μL of H2O. Collect the beads on a magnetic rack and transfer the supernatant with the amplified DNA to a new tube.
Measure the DNA concentration using a Qubit Fluorometer. The total yield is typically 140 to 280 ng.
Select the appropriate DNA size for sequencing (160–340 bp) using AMPure magnetic beads in the following steps: (i) Dilute 8 μL of amplified DNA with 17 μL of H2O and bind to 18 μL of AMPure beads by mixing with the beads and incubating for 5 min at room temperature. (ii) Collect the beads on a magnetic rack and transfer the supernatant to a new tube. Discard the beads, which retain long DNA. (iii). Bind the DNA in the supernatant (~43 μL) to 20 μL of AMPure beads by mixing with the beads and incubating for 5 min at room temperature. (iv). Collect the beads on a magnetic rack, discard the supernatant and wash the beads twice with 70% ethanol. Remove all residual 70% ethanol. (v). Air dry the beads and elute the DNA by mixing the beads with 12 μL of H2O. Collect the bead on a magnetic rack and transfer the supernatant with the size-selected DNA to a new tube. (vi). Measure the size-selected DNA concentration using a Qubit Fluorometer. Typically 50 to 70% of the amplified DNA is recovered.
Determine the size range of the size-selected DNA using an Agilent Bioanalyzer and to confirm the concentration. The majority of the DNA should be between 160 to 340 bp in length.
3.4 Sequencing
The size-selected DNA libraries are sequenced on a Ion Torrent Proton with the P1 sequencing chip, using the Ion Chef Hi-Q and Ion Torrent Hi-Q sequencing reagents according to the manufacturer’s (Life Technologies) protocols.
The barcoded libraries are diluted to a 50 pM concentration and pooled together for sequencing. This typically provides 60 to 75 million reads averaging 125 to 150 bases in length from one P1 chip. By default, reads shorter than 25 bp are discarded during generation of the sequence files, and the Torrent Suite software also allows PCR duplicates to be marked and filtered out. The sequencing depth is sufficient to permit pooling of three Drosophila ChIP-seq libraries on one P1 chip for the depth needed to obtain the most accurate results (see below). If the same chromatin preparation is used for several ChIP experiments, one input sequencing library is used to normalize all.
3.5 Data Processing and Analysis
To simplify comparisons to existing ChIP-chip data, we align the input and ChIP sample fastq sequence files to the Drosophila melanogaster release 5 (April 2006) genome sequence. We use a repeat masked fasta file, modified by deleting the chrU and Uextra contigs, which are primarily repetitive sequence and mostly redundant.
Examples of the commands and annotated R scripts used to calculate ChIP enrichment are provided in Supplemental File 1. The total sequencing coverage (the number of times each individual base pair in the genome is present in a sequencing read) is measured for the input chromatin and ChIP samples, and the ChIP sample coverage at many intervals across the genome is then normalized to the input coverage for the same interval. Normalization to input adjusts for differences in chromatin isolation, amplification and sequencing efficiency, although as noted below, this is likely insufficient for regions with very low coverage. The intervals are overlapping sliding windows of a selected length, with a selected step size between the windows. The use of sliding windows reduces noise and increases sensitivity, but decreases spatial resolution. The resolution loss is insignificant, however, for proteins that spread out over several kilobases. For Nipped-B and cohesin we use windows of 250 bp and a step size of 50 bp. The total base pair coverage in each window in the ChIP and input samples are internally normalized to the total sequencing coverage for each sample to adjust for differences in sequencing depth, and window values are also adjusted to account for the distorted coverage distribution caused by enrichment of specific genomic regions by ChIP. The adjusted ChIP coverage in each window is then is divided by the adjusted coverage in the corresponding input window to calculate the ChIP enrichment for each window.
The computational methods are optimized for Ion Torrent sequencing, which generates reads that vary in length. By using the total base pair coverage more information is available than if only the number of reads that start in a given region were used, as is typically done when all sequence reads are equal in length.
The TMAP sequence aligner is used because its algorithms are optimized for reads of varying length (11). To ensure high quality alignments we prevent soft-clipping of reads to accommodate alignment, and allow reads that align to multiple positions in the genome to be assigned only once to the position with the highest alignment score, or randomly to one of multiple positions with the highest score. This prevents loss of information, and as described below can allow ChIP enrichment calculations in regions with repetitive genes, such as the histone gene cluster. We typically find that 75 to 95% of reads align, with efficiencies over 90% in most cases.
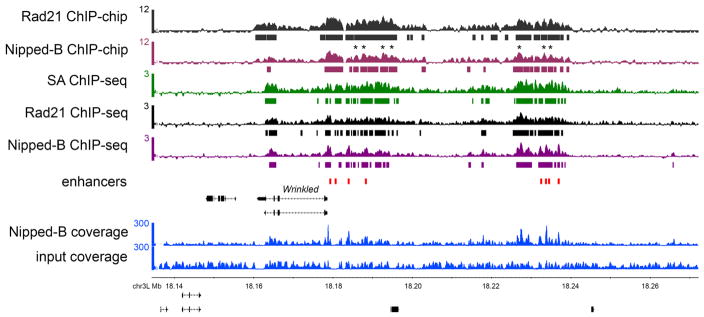
The ChIP-seq method described here give results similar to those obtained by ChIP-chip. Figure 1 shows a genome browser view of a region containing the Wrinkled gene and its transcriptional enhancers occupied by Nipped-B and cohesin in the ML-DmBG3-c2 cell line derived from larval central nervous system. The tracks show ChIP-chip data for Nipped-B and Rad21, and ChIP-seq for Nipped-B, Rad21 and SA. Two tracks show the raw sequence coverage files used to calculate ChIP-seq enrichment for Nipped-B.
Figure 1.
ChIP-chip and ChIP-seq of cohesin and Nipped-B. The browser view show the Wrinkled gene and its predicted transcriptional enhancers (red boxes) (8). The ChIP-chip track scales are MAT scores (10) and the bars underneath indicate where enrichment is significant at a p value ≤ 0.001. The ChIP-seq track scales are log2 enrichment (ChIP/input), and the bars underneath indicate where enrichment is ≥ 2σ for ≥ 300 bp (six windows), which gives a similar pattern as the statistical threshold used for ChIP-chip. The ChIP sample and input coverage tracks used to calculate the Nipped-B enrichments are shown. The ChIP-chip and ChIP-seq tracks show very similar global enrichment patterns, but for both Rad21 and Nipped-B there are cases where the ChIP-chip peaks are valleys in the ChIP-seq track. Some of these are marked with asterisks (*) above the Nipped-B ChIP-chip track. These are usually where input and ChIP sequence coverage are lower than average, suggesting that normalization of ChIP to input incompletely compensates for regions that amplify or sequence poorly.
The ChIP-seq data agrees well with the ChIP-chip data regarding the larger domains occupied by Nipped-B and cohesin, although close examination shows that some ChIP-chip summits occur in ChIP-seq valleys (Figure 1). Although it is possible that this reflects higher resolution of ChIP-seq compared to ChIP-chip, many of these valleys occur where the sequence coverage is low, suggesting that poor amplification and/or sequencing efficiency contributes to the reduced enrichment. Thus it is feasible that a broad region of cohesin occupancy has been artificially split into two by a region that sequences poorly. Although normalization to input should compensate for low coverage, it may be insufficient where sequencing coverage is particularly low. As described below, simply reducing sequencing depth can cause broad regions of enrichment to be fractured into multiple peaks. Comparison of the ChIP-chip to ChIP-seq data thus cautions against using peak-calling for genomic cohesin ChIP data, and against drawing conclusions based on the precise positions of peak summits.
The ChIP-seq procedure is reproducible. The Pearson correlation coefficient between two independent Nipped-B ChIP-seq experiments was 0.87. Similarly, we find high correlations between Nipped-B and cohesin (Nipped-B: Rad21, r = 0.76), and between cohesin subunits (Rad21: SA, r = 0.80). These are similar to genome-wide correlations obtained by ChIP-chip (7). At least two independent biological replicate ChIP-seq experiments for each antibody are recommended, and the data can be combined if the correlation between the two experiments is high (r ≥ 0.8).
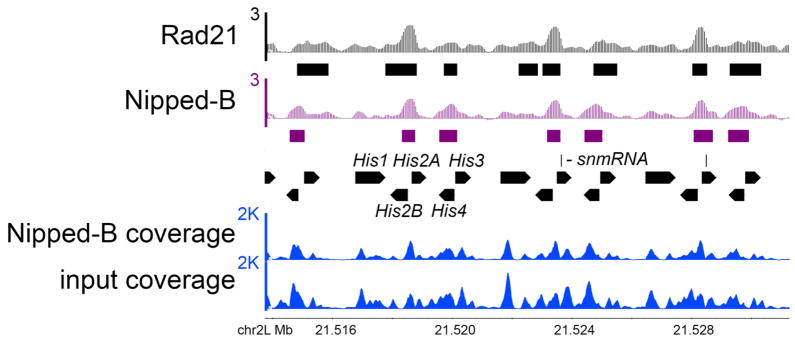
Figure 2 shows a region of the histone gene cluster, revealing that assigning reads of repetitive sequences to only one position allows detection of ChIP enrichment in repetitive gene clusters. We see Nipped-B and Rad21 occupancy in the divergent promoter regions of the histone H2A and H2B genes, and the promoter regions of the H3 and H4 genes, but not at the H1 genes. Note that this method cannot identify specifically which histone genes are occupied, and a more accurate picture might be obtained by making a composite of the repeats. Nonetheless, this region is inaccessible by ChIP-chip, and thus ChIP-seq provides the first evidence that Nipped-B and cohesin bind to these genes.
Figure 2.
Nipped-B and cohesin occupy the histone gene cluster. The tracks show the calculated log2 enrichment of Rad21 and Nipped-B for three of the histone gene repeats and the raw Nipped-B ChIP and input coverage used to calculate enrichment. The raw coverage of the histone gene repeats is significantly higher than average, even though each read was only assigned once, indicating that the genome fasta file does not contain all the repeats present in the genome. Nonetheless, normalization of the ChIP coverage to the input coverage reveals occupancy of the promoter regions of the His2A, His2B, His3 and His4 genes, and no significant occupancy over the His1 genes. Because reads were assigned once randomly to only one of the best matches, we cannot know which histone genes are actually occupied.
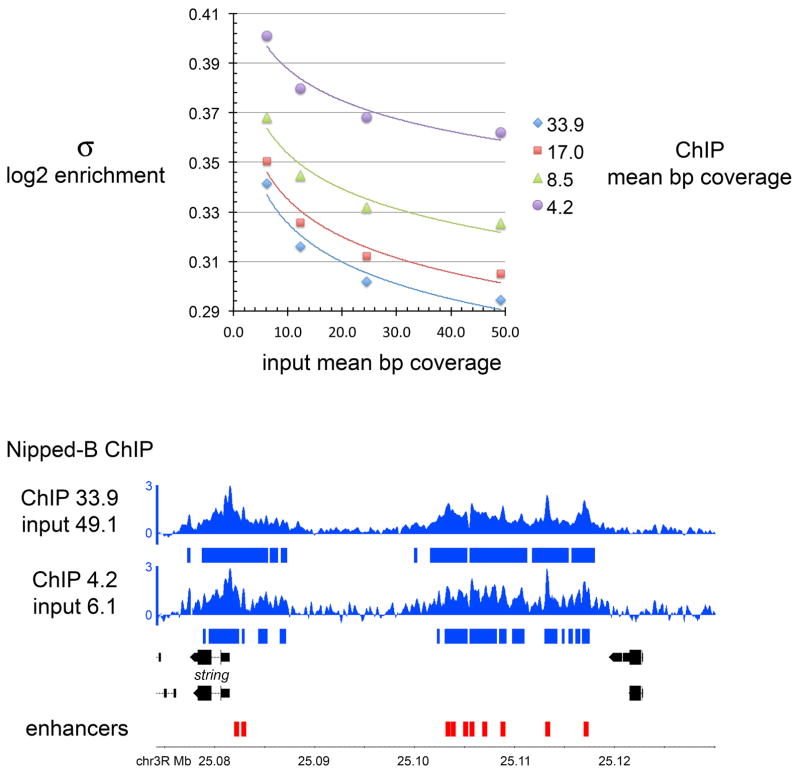
Figure 3 shows how sequencing depth affects the sensitivity and accuracy of the calculated enrichment. Nipped-B ChIP and input bam files from one experiment were randomly down-sampled using SAMtools (12), and used to calculate enrichment with different sequencing depths. The Nipped-B ChIP sample was used at a mean base pair coverage from 4.2 to 33.9, and the input was used from a depth of 6.1 to 49.1. As expected, the standard deviation (σ) of the log2 enrichment values decrease with both increased ChIP and input coverage (Figure 3, top panel). This affects the threshold used to call significant enrichment, as shown for the string gene and its enhancers in the genome browser view in the bottom panel of Figure 3. This compares the enrichment calculated using the highest coverage of both the ChIP and input samples to the enrichment calculated with the lowest coverage for both. For both, enrichment was called significant when ≥ 2σ above the baseline for 300 bp (six windows) or longer, as denoted by the bars underneath the ChIP tracks.
Figure 3.
Influence of sequencing depth on the accuracy of the enrichment calculation. Nipped-B ChIP and input chromatin bam files were randomly sampled at the different depths indicated (mean coverage per base pair), and used to calculate Nipped-B ChIP enrichment. The top panel shows the genome-wide standard deviation (σ) of the log2 enrichment values obtained with the various combinations of ChIP and input coverage. The bottom panel shows the string gene and its enhancers (red boxes), with the top track showing the enrichment calculated at the highest coverage for both ChIP and input, and the bottom track showing the enrichment calculated with the lowest coverage for both. The bars underneath show where the calculated enrichment is ≥ 2σ for ≥ 300 bp. The lower coverage levels fracture many extended regions of enrichment into peaks, and the unoccupied regions are noisier, showing more fluctuation around the baseline.
Inspection reveals that the although the overall broad pattern and maximum enrichment levels are not significantly affected, reduced coverage causes some broad regions of enrichment to be fractured into multiple peaks. Thus, in some cases, peak-calling algorithms might artificially call more peaks with lower coverage than with higher coverage. It can also be seen that unoccupied regions are noisier with lower coverage, showing more fluctuation around the baseline. Based on this analysis, it can be recommended that for cohesin and Nipped-B, that both the ChIP and input samples are sequenced to at least 15-fold coverage (~20 million aligned reads) for an accurate picture, and that conclusions are not drawn from precise peak locations. This comparison also reveals, however, that even low coverage levels on the order of 5-fold (~7 million reads) can provide a reasonably accurate broad view of cohesin and Nipped-B occupancy.
The output enrichment graph files normalized to the same input chromatin sequencing all have the same data points across the genome, allowing one to be easily subtracted from the other. Subtraction of the log2 enrichment values measures the fold-change in the enrichment in a window. This can be used to quantify differences in ChIP enrichment for a protein between control and treated cells, using methods similar to those used with ChIP-chip data for measuring changes in RNA polymerase, cohesin, and PRC1 polycomb complex occupancy (8, 9). It is recommended that at least two biological replicates are used for these comparisons, which provide insights into how different proteins influence each other’s association with chromosomes.
Supplementary Material
Acknowledgments
This work was supported by an NIH research grant (R01 GM108872). The authors thank Seth Peterson and Brandon Blakey of Life Technologies for advice in the use of sequencing library reagents and Ion Torrent sequencing.
Footnotes
This chromatin isolation method uses relatively mild detergent conditions, in contrast to some published procedures that use up to 1% SDS for lysis. We have not evaluated how high SDS levels might affect ChIP of Nipped-B and cohesin. Thus the lysis conditions may need to be evaluated for how they could affect epitope recognition if a different chromatin isolation method is used.
Scraping of adherent cells can also be done in PBS after washing cells with PBS a few times, but in this case the PBS should contain some fetal bovine serum (3%) to avoid losing cells.
With some cells, freezing is beneficial for chromatin isolation because the freeze-thaw increases the efficiency of lysis. Where possible, it is useful to examine aliquots of cells by microscopy to visually confirm that lysis is complete.
The number of sonication cycles for a given cell type should be determined empirically, based on the size distribution of the DNA fragments observed after reversing the crosslinks. We typically try to achieve a distribution mode of ~500 bp. It is helpful to rest the sonicator for a few minutes after 5 or 6 cycles before continuing to ensure that 4° is maintained, and that the sonicator still functions at maximal energy output. Some epitopes may be sensitive to sonication, and thus using alternative shearing methods such as limited micrococcal nuclease digestion might need to be considered.
We find that measuring DNA concentration at this stage by a UV spectrophotometer, including a Nanodrop, can overestimate the amount of DNA as much as 5 to 8-fold, presumably because other UV-absorbing molecules are present in the preparation. The DNA yield measured with a Qubit Fluorometer is close to the expected 0.4 pg of DNA per diploid Drosophila cell. This number needs to be adjusted for cell lines that are aneuploid, such as the commonly used Schneider Line 2 (S2) cell line.
All the Nipped-B and cohesin antibodies we use do not need to be affinity-purified for use in ChIP experiments. For purified antibodies, the amount will need to be optimized, but is usually in the range of 2 to 5 μg for a 200 μL aliquot of chromatin.
It is important that there is no EDTA in the buffer used for sonication because EDTA can interfere with some steps in library construction.
Once the optimal sonication conditions have been determined, they remain consistent and it is not necessary to check the size distribution every time.
References
- 1.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 2.Dorsett D, Ström L. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol. 2012;22:R240–R250. doi: 10.1016/j.cub.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorsett D, Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol. 2013;25:327–333. doi: 10.1016/j.ceb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Remeseiro S, Cuadrado A, Losada A. Cohesin in development and disease. Development. 2013;140:3715–3718. doi: 10.1242/dev.090605. [DOI] [PubMed] [Google Scholar]
- 5.Ball AR, Jr, Chen YY, Yokomori K. Mechanisms of cohesin-mediated gene regulation and lessons learned from cohesinopathies. Biochim Biophys Acta. 2014;1839:191–202. doi: 10.1016/j.bbagrm.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ocampo-Hafalla MT, Uhlmann F. Cohesin loading and sliding. J Cell Sci. 2011;124:685–691. doi: 10.1242/jcs.073866. [DOI] [PubMed] [Google Scholar]
- 7.Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;11:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, Dorsett D. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 2013;9:e1003382. doi: 10.1371/journal.pgen.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaaf CA, Misulovin Z, Gause M, Koenig A, Gohara DW, Watson A, Dorsett D. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 2013;9:e1003560. doi: 10.1371/journal.pgen.1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homer N. TMAP: the torrent mapping program. 2011 https://github.com/iontorrent/TMAP/blob/master/doc/tmap-book.pdf.
- 12.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. http://www.R-project.org. [Google Scholar]
- 15.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.