Abstract
Discovery of genetic alterations that cause human birth defects provide key opportunities to improve the diagnosis, treatment, and family counseling. Frequently, however, these opportunities are limited by the lack of knowledge about the normal functions of the affected genes. In many cases, there is more information about the gene’s orthologs in model organisms, including Drosophila melanogaster. Despite almost a billion years of evolutionary divergence, over three-quarters of genes linked to human diseases have Drosophila homologs. With a short generation time, a twenty-fold smaller genome, and unique genetic tools, the conserved functions of genes are often more easily elucidated in Drosophila than in other organisms. Here we present how this applies to Cornelia de Lange syndrome, as a model for how Drosophila can be used to increase understanding of genetic syndromes caused by mutations with broad effects on gene transcription and exploited to develop novel therapies.
Keywords: NIPBL, Nipped-B, cohesin, AFF4, CHOP Syndrome
Introduction
Cornelia de Lange Syndrome displays multiple physical and intellectual birth defects, including distinctive facial features, poor growth, limb and organ abnormalities, learning and speech difficulties, and autism (Kline et al., 2007; Deardorff et al., 2016). The severity and spectrum of these birth defects vary from individual to individual. Cornelia de Lange Syndrome (CdLS) is caused by spontaneous dominant mutations in genes that alter the activity of the cohesin protein complex. Cohesin is a ring-like complex consisting of the SMC1A, SMC3, RAD21 and SA1 or SA2 proteins (Figure 1). Cohesin is highly conserved from yeast to human, and plays critical roles in sister chromatid cohesion, chromosome segregation, DNA repair, and regulation of gene transcription (Dorsett and Ström, 2012; Dorsett and Merkenschlager, 2013; Remeseiro et al., 2013). Cohesin encircles DNA and loaded topologically onto chromosomes by the kollerin complex consisting of the NIPBL (Nipped-B-Like) and MAU-2 proteins (Figure 1). It is removed by the releasin complex consisting of WAPAL and the PDS5B or PDS5A proteins. The SMC3 subunit is acetylated the ESCO1 and ESCO2 proteins to facilitate its function in sister chromatid cohesion, and is deacetylated by the HDAC8 enzyme.
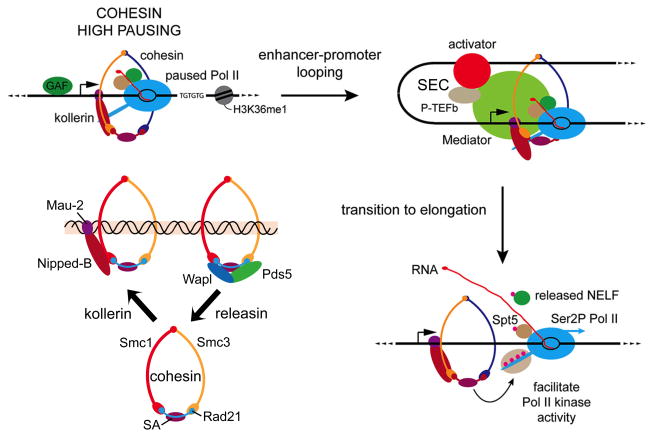
Figure 1. Nipped-B, cohesin, and potential role in transcriptional elongation and enhancer function.
Lower left: the kollerin complex, consisting of Nipped-B and the Mau-2 protein loads the cohesin complex topologically onto chromosomes, and the releasin complex, consisting of the Pds5 and Wapl proteins removes cohesin from chromosomes. Dominant loss-of-function mutations in the human NIPBL homolog of Nipped-B cause Cornelia de Lange syndrome (CdLS). Dominant mutations that maintain the reading frame of the human SMC1A and SMC3 cohesin subunits cause milder forms of CdLS. The diagram starting on the upper left illustrates a current model for one mechanisms by which cohesin promotes transition of promoter-proximal paused RNA polymerase into elongation by facilitating looping between enhancers and promoters, bringing transcriptional activator and the elongation factors they recruit into contact with the paused polymerase. These factors include the Super Elongation Complex (SEC) that contains P-TEFb, that phosphorylates (pink circles) the NELF and Spt5 pausing factors and the C terminal tail of polymerase to stimulate elongation. The SEC complex also contains AFF4, and gain-of-function AFF4 mutations cause CHOP syndrome, which has features overlapping those seen in CdLS. The Mediator complex helps connect activators with polymerase, and also interacts and functions together with cohesin to support enhancer-promoter communication (Kagey et al., 2010). Portions of this figure are modified from Schaaf et al., 2013a and Dorsett and Kassis, 2014.
The first CdLS-causing mutations were mapped to the NIPBL gene, and subsequently mutations in the SMC1A and SMC3 cohesin subunit genes, and the HDAC8 gene were discovered in individuals with mild to severe CdLS (Krantz et al., 2004; Tonkin et al., 2004, Musio et al., 2006; Deardorff et al., 2007; Deardorff et al., 2012a). Mutations in the RAD21 cohesin subunit gene were discovered to cause a distinct but overlapping set of birth defects (Deardorff et al., 2102b). Dominant loss-of-function NIPBL mutations are usually predicted to truncate the NIPBL protein and tend to cause more severe forms of CdLS.
CdLS caused by SMC1A and SMC3 mutations tends to display less severe structural abnormalities. All SMC1A and SMC3 mutations discovered to date preserve the open reading frame, and cause missense substitutions or deletion of a few amino acids. The SMC3 mutations are dominant. SMC1A is X-linked, and the mutations are dominant in females, with evidence that SMC1A escapes X-inactivation. The mutant allele is the only source of SMC1A in affected male individuals. Because SMC1A is an essential protein, it can be concluded that these individuals produce a protein functional for sister chromatid cohesion.
CdLS-causing mutations alter gene expression without overt effects on chromosome segregation
Because cohesin is crucial for sister chromatid cohesion, which ensures accurate chromosome segregation, it might be expected that individuals with CdLS would display chromatid cohesion and chromosome segregation abnormalities. However, examination of cell lines derived from individuals with CdLS have not revealed overt defects. Work in model organisms, including yeast and Drosophila, show that cohesin can by depleted by 80 to 90 percent without quantifiable effects on chromosome segregation (Schaaf et al., 2009; Heidinger-Pauli et al., 2010). Studies in CdLS cell lines revealed only slight defects in DNA repair (Vrouwe et al., 2007; Revenkova et al., 2009). Thus attention has focused on the potential effects of sister chromatid cohesion proteins on gene expression.
At the time that the first mutations in the NIPBL gene were discovered, mutations in the Drosophila ortholog, Nipped-B, had been shown to alter expression of genes with long-range transcriptional enhancers during development (Rollins et al., 1999). The known target genes included the cut homeobox gene, whose expression is driven by a remote wing margin enhancer during early wing development, and the Ultrabithorax HOX gene, whose expression is driven by distant enhancers in the segmentation pathway. Although heterozygous loss-of-function mutations alter expression of the wild-type alleles of these genes, the phenotypic consequences are minimal unless the enhancer-promoter communication in these genes is modestly compromised by insertion of a transposon containing an insulator sequence between the enhancer and promoter. Nonetheless, this provided the initial evidence that that partial reduction of Nipped-B activity could alter developmentally-regulated gene expression.
The early studies also revealed that the most severe Drosophila Nipped-B mutations, which are equivalent to null mutations by all genetic tests, only reduce Nipped-B mRNA levels by some 25% (Rollin et al., 2004). Combining strong Nipped-B alleles with in vivo RNAi could only reduce Nipped-B mRNA levels by a maximum of 50%, which is lethal, but without causing detectable defects in sister chromatid cohesion or chromosome segregation. It has thus been hypothesized that there is a feedback pathway that compensates for loss of one allele by increasing transcription of the remaining wild-type allele. Comparison of RNA-seq and genomic sequence confirms this idea (Wu et al. 2015). Strikingly, Nipbl (+/−) mice and CdLS cell lines with NIPBL mutations show a similar compensation, with maximal NIPBL mRNA reductions of 30%, indicating that this aspect of Nipped-B regulation is evolutionarily-conserved. The compensation mechanisms remain a mystery, but there is hope that if this natural compensation can be increased, then it may provide a treatment for individuals with NIPBL mutations by further increasing expression of the wild-type allele.
Stimulated by the findings in Drosophila, gene expression was measured in CdLS-derived cell lines, and in cells and tissues from mice with Nipbl(+/-) (Kawauchi et la., 2009; Liu et al., 2009a; Liu et al., 2009b). Many genes whose expression was altered relative to controls were discovered in cells with NIPBL and SMC1A mutations, and in cells from Nipbl(+/−) mice. The altered gene expression in human cells proved to be predictive for CdLS. Although many genes are affected, the magnitudes of the changes are typically modest, with most under 2-fold. These findings make it difficult except in rare cases to ascribe specific expression changes to developmental deficits. Also, the gene expression changes responsible for specific defects might occur at very critical times in development, which can only captured by following gene expression closely throughout development. This of course, is only feasible in model organisms such as Drosophila, zebrafish, and mice, where it also remains challenging. It is also likely that there are differences as to which developmental pathways are the most sensitive and relevant between organisms, given the developmental differences. Although Nipbl(+/−) mice display key overlapping phenotypes with CdLS, some features, such limb defects are much less severe, and some, such as heart defects, are more severe (Kawauchi et al., 2009).
The number and small magnitude of the effects on gene expression in CdLS also make it difficult to determine which effects on transcription are direct, and to delineate the mechanisms by which NIPBL and cohesin modulate transcription. As described in more detail in a later section, the Drosophila model has provided substantial information on how NIPBL and cohesin function in gene regulation, some of which have been validated in mammalian cells, and some of which have yet to be investigated. Understanding these mechanisms is likely to be crucial for inventing effective therapies, given the broad effects that NIPBL and cohesin have on gene expression.
Drosophila Nipped-B mutants display reduced growth
Although the role of Nipped-B (NIPBL) in developmental gene regulation was first recognized in Drosophila, heterozygous Nipped-B mutant flies do not display as overt morphological phenotypes as individuals with heterozygous NIPBL mutations. Until recently, most phenotypes associated with heterozygous Nipped-B mutations were apparent only in the presence of cryptic or visible mutations in other genes, such as wing defects in combination with weak cut mutations, or segmental transformations in the presence of weak Ultrabithorax mutations. Closer examination, however, revealed mild, low penetrance segmental transformations in heterozygous Nipped-B and Rad21 mutants, that become fully penetrant in flies with both Nipped-B and Rad21 mutations (Schaaf et al., 2013b).
In addition to the characteristic facial features, poor growth and intellectual deficits are the most consistent of the CdLS phenotypes. A recent study revealed that Drosophila Nipped-B mutants share these characteristics (Wu et al., 2015). The size of adult Nipped-B null mutants is reduced by some 8% relative to wild-type controls. Although this reduction appears modest, it is similar in magnitude to the size deficits caused by heterozygous null mutations affecting the highly conserved Myc and Tor growth regulators, and thus actually represents a substantial decrease.
Drosophila permits experimental manipulations to investigate the possible sources of the growth defect. Strikingly, these studies revealed an absence of developmental delays in Nipped-B mutants, with all hormonally-triggered developmental transitions occurring on time. Similarly, Nipped-B mutants showed the same developmental delays and dramatic reduction in size as wild-type in response to a severely restricted diet. Combined, these findings argue that the reduced size of Nipped-B mutants is not caused by changes in systemic hormonal signaling pathways that connect the brain and fat body (fat and liver equivalent) to control growth. Close examination of adult wings revealed that the reduced size of Nipped-B mutations stems from reductions in both cell number and size. Mutations in the Drosophila myc gene (diminutive) have a similar effect on both cell number and size, while Tor mutations reduced cell size, but not cell number.
These findings have implications for CdLS. Although the growth deficits are more severe in CdLS, with most individuals in the bottom 5th percentile, most enter puberty near the expected age. Thus it is possible that reduced cell number and size is also a key contributor to the small size of individuals with CdLS. If so, then it cannot be expected that correcting the common feeding problems observed CdLS will dramatically improve growth because the size deficit in Drosophila, relative to wild-type, was not substantially affected by nutrition. It is likely that the growth deficit reflects a change in gene expression in highly conserved pathways that reduce the frequency of cell division and cell size, and thus can only be corrected by restoring NIPBL expression to normal, or by simultaneously increasing expression of the multiple pathways that regulate cell growth and size. How much reduced cell number and size might contribute to the structural and intellectual deficits remains an open question.
The Drosophila studies indicate that the deficits in cell growth control in Nipped-B mutants cannot be precisely corrected by targeting only one key growth regulator. The myc gene in flies and zebrafish is under direct control of Nipped-B and cohesin (Rhodes et al., 2010), and there is reduced c-myc expression in CdLS-derived cells and mice with Nipbl mutations (Kawauchi et al., 2009; Liu et al., 2009a). Detailed analysis of genome-wide expression in developing Nipped-B mutant wing discs revealed modest decreases in myc expression, and similar decreases in many other genes that control growth.
Myc is a transcription factor that directly amplifies transcription of many genes that promote growth, so it was tested if increasing myc expression with a transgene would rescue the growth deficit in Nipped-B mutants. Although wing size was rescued, the rescue reflected primarily an increase in cell size, with little impact on cell number. Thus increasing myc expression alone does not provide a complete rescue of the mutant growth phenotype. Given that c-myc is an oncogene, and that excess expression can promote tumor growth in addition to apoptosis in other cells, increasing c-myc expression carries substantial risks as a potential therapy.
Key unexpected observations were made in the attempts to rescue Nipped-B mutants with a myc transgene. Additional myc expression reduced adult body weight in the wild-type controls, and although it increased wing size, it reduced the number of cells in the wing (Wu et al., 2015). A likely explanation is that excess Myc induces apoptosis, as observed in Drosophila and mammalian cells, and confirmed for wing discs in the Nipped-B mutant study. What these data also indicate, however, is that the reduction in Nipped-B activity prevents excess Myc from inducing apoptosis. This is crucial information because it indicates that the reduction in cell number in Nipped-B mutants is unlikely to result from an increase in cell death, and more likely reflects a reduced number of cell divisions.
Drosophila Nipped-B mutants display learning, memory, sleep deficits, and altered brain structure
Intellectual deficits are another consistent feature of CdLS, with the average IQ in the mid-50’s. Drosophila Nipped-B mutants also show learning and memory deficits. Learning is tested by training males with previously-mated females, who reject their courtship. A reduction in the number of mating attempts by a male over the course of an hour measures the male’s ability to learn. Short-term memory is measured by the frequency of trained male’s courtship behavior with a receptive female, relative to naïve males, after resting the trained male for an hour after training with a non-receptive female. Nipped-B mutant males show reduced learning and memory relative to wild-type males (Wu et al., 2015). Learning, which is necessary to test memory, is not completely ablated, but is significantly reduced. Thus Drosophila Nipped-B mutants also model CdLS in terms of reduced neurological function.
The Drosophila mushroom body is a brain structure required for consolidation of short and long-term memory. The mushroom body contains three types of neurons, and Drosophila Nipped-B mutants show a higher frequency of structural abnormalities, including missing and kinky lobes. It is too soon to say if these structural defects are responsible for the memory deficits, but these findings raise the clear possibility that the intellectual deficits in Drosophila, and possibly in CdLS, arise from morphological changes in the brain and central nervous system.
Individuals with CdLS often have abnormal sleep patterns, and altered sleep also occurs in Drosophila Nipped-B mutants (Wu et al., 2015). The changes varied with sex and mutant allele, but the overall pattern is of shorter sleep episodes, and reduced overall sleep. This may stem in part from disruption of circadian rhythm, observed in a large fraction of Drosophila Nipped-B mutants. As with sleep, the circadian defects are stronger and more frequent in females than in males.
Combined with the original findings on limb development and segmental identity changes, the growth defects, broad modest effects on gene expression and neurological dysfunction in Drosophila Nipped-B mutants strongly indicate that they provide a useful model for understanding CdLS. It thus likely worthwhile to use Drosophila to investigate the mechanisms by which Nipped-B and cohesin regulate gene expression, which could provide insights that aid efforts to improve therapeutic methods.
Nipped-B and cohesin modulate transcription by multiple mechanisms
As described below, strong depletion of Nipped-B or cohesin over a course of a few days in cultured Drosophila cells alters the expression of several hundred genes, with many large increases and decreases in expression. The findings with developing wing tissue in heterozygous Nipped-B mutants, however, are substantially different. Similar to findings in Nipbl(+/−) mice and CdLS cell lines, many genes were affected, but most changes in RNA levels were less than two-fold. Because the effects of the Nipped-B mutations are broad and modest, there is concern that standard methods for normalizing RNA-seq measurements might be inaccurate. Most RNA-seq data is normalized to total sequencing reads or total base pair coverage, assuming that most genes are unaffected by the treatment or mutation under study. This assumption, however, is questionable in the case of Nipped-B (Schaaf et al., 2013a). This kind of normalization forces the total gene expression to be equal between mutant and controls, when it might actually differ. For example, studies on the mammalian Myc protein, which directly amplifies transcription of most genes, standard normalization methods indicate that some genes increase in gene expression, simply because they decrease less in expression than average (Lovén et al., 2012). When Nipped-B mutant RNA-seq data is normalized to total histone RNA levels instead of total sequence coverage, it reveals that some 30% of active genes show significant but modest decreases in expression on the order of 25 to 30%, with very few genes increasing. The decreasing genes include many genes that promote protein synthesis, cell division and growth. It is difficult to pick out specific genes or pathways that correlate with adult phenotypes, given the broad modest overall decrease in gene expression. It is also difficult to dissect which effects on expression are direct, and which are indirect, given the clear indications of compensatory feedback mechanisms, including on Nipped-B expression itself. The overall picture that emerges is analogous to driving a car while applying the throttle and a little brake at the same time, impeding forward movement. This contrasts with the results in cultured cells with rapid strong depletion of Nipped-B and cohesin described below, which causes large changes in gene expression in both directions, and provides more direct insights into gene targets and the mechanisms by which Nipped-B and cohesin facilitate transcriptional regulation.
Genomic chromatin immunoprecipitation experiments in cultured cells and developing wing tissues revealed that Nipped-B and cohesin closely co-localize in Drosophila at active genes, transcriptional enhancers, and Polycomb Response Elements (PREs) that epigenetically silence developmental genes (Misulovin et al., 2008; Schaaf et al., 2013a; Schaaf et al., 2013b). Strong depletion of Nipped-B or cohesin by 80 – 90% in cultured cells increases transcription measured by precision run-on sequencing (PRO-seq) and steady-state RNA levels of several hundred genes, and decreases transcription and transcripts for a similar number (Schaaf et al., 2009; Schaaf et al., 2013a). There is a strong correlation between the effects of depleting Nipped-B and cohesin, both on transcription and the steady-state transcripts, indicating that Nipped-B and cohesin work together to control transcription, or that one of Nipped-B’s key roles in transcription is to load cohesin.
The genes that bind Nipped-B and cohesin are more likely to show altered transcription than genes that do not, indicating that many changes reflect their direct roles in transcription (Schaaf et al., 2009; Schaaf et al., 2013a). There are, however, also many indirect effects, and it is not possible to determine for any one gene how much of the effect is direct and how much is indirect. Nonetheless, genome-wide trends clearly point to specific roles of cohesin. One of the significant findings is that depletion of Nipped-B or cohesin does not substantially alter the amount of transcriptionally-engaged paused RNA polymerase at gene promoters, indicating that it plays little role in transcription initiation (Schaaf et al., 2013a). A majority of cohesin-binding genes, however, show a change in how much elongating Pol II is in the gene body. At most active genes, there is a decrease in gene body transcription, but there are also increases at many genes, including genes that are repressed, but not fully silenced by Polycomb Group complexes (Fay et al., 2011; Schaaf et al., 2013a, Schaaf et al., 2013b). Thus Nipped-B and cohesin modulate the transition of paused Pol II into active elongation.
The roles of Nipped-B and cohesin in transcriptional elongation is particularly interesting in light of the facts that CHOP and CHARGE syndromes, which show overlapping phenotypes with CdLS, are caused by mutations affecting the elongation factors AFF4 (Izumi et al., 2015) and CHD7 (Vissers et al., 2004) respectively. AFF4 is a critical subunit in the Super Elongation Complex (SEC) in humans and Drosophila (Smith et al., 2011). The Drosophila ortholog of CHD7, Kismet, like Nipped-B and cohesin, promotes an early step in transcriptional elongation (Srinivasan et al., 2005).
One mechanism by which Nipped-B and cohesin aid transition to elongation is by facilitating enhancer–promoter contact (Figure 1). Cohesin is present at all active enhancers and roughly a third of active promoters (Schaaf et al., 2013a). Transition to elongation requires phosphorylation of the serine 2 residues in the heptad repeats in the C terminal domain (CTD) of the Rpb1 Pol II subunit and the NELF and DSIF pausing factors (Figure 1; Li and Gilmour, 2011). The phosphorylation is mediated by the Cdk9 kinase in the P-TEFb subcomplex of the Super Elongation Complex (SEC). Genomic ChIP detects significant levels of phosphorylated Pol II at active transcriptional enhancers that show little or no transcription by run-on sequencing. If these enhancers directly contact the promoter regions by looping, it could allow polymerase transcribing the gene to crosslink to the non-transcribed enhancer. At many genes, depletion of cohesin reduces the amount of phosphorylated Pol II at enhancers, consistent with the idea that enhancer-promoter looping is reduced (Schaaf et al., 2013a). This is consistent with evidence in mouse and human cells showing a role for cohesin and NIPBL in enhancer-promoter looping (Kagey et al., 2010; Chien et al., 2011) and with the original isolation of Nipped-B mutations in a genetic screen for factors that facilitate long-range gene activation (Rollins et al., 1999).
Nipped-B and cohesin also impact the transcription of many genes that they do not directly occupy, and this has been interpreted as indicating their role in facilitating expression of other proteins that regulate transcription, including myc (Rhodes et al., 2010; Schaaf et al. 2013a) which amplifies expression of a majority of active genes (Lin et al., 2012; Nie et al., 2012).
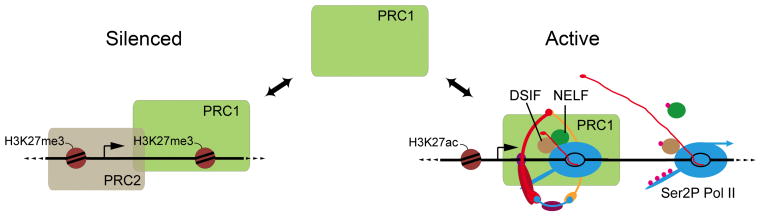
Drosophila experiments also revealed functional interactions between cohesin and the Polycomb Group protein complexes that epigenetically maintain gene silencing. The PRC2 complex makes the histone H3K27me3 modification that marks silenced genes, and facilitates binding of the PRC1 complex. The first cohesin mutation (verthandi = Rad21) in Drosophila was isolated in a screen for dominant suppressors of the haploinsufficient segmental transformation phenotypes caused by mutation of the Polycomb (Pc) gene encoding a subunit of the PRC1 complex (Kennison and Tamkun, 1988). It was later discovered that Nipped-B mutations also dominantly reverse Polycomb mutant phenotypes, and that combining heterozygous Nipped-B and Rad21 cohesin mutations cause the opposite segmental identity transformations as Polycomb mutations (Hallson et al., 2008; Schaaf et al., 2013b). While these findings were interpreted as indicating that cohesin promotes expression of genes that Polycomb silences, later discoveries provided an alternative explanation. It was discovered that the PRC1 complex directly interacts with cohesin, and is recruited by cohesin to active genes in the absence of PRC2 (Strübbe et al., 2011; Schaaf et al., 2013b). When cohesin is depleted, PRC1 levels decrease at active genes, and increase at silenced genes marked by H3K27me3. In other words, cohesin sequesters PRC1 at active genes, and reducing cohesin releases PRC1, which then compensates for a global deficiency in PRC1 (Figure 2).
Figure 2. Functional interactions between Polycomb Group silencing and cohesin.
Cohesin recruits the PRC1 Polycomb complex to active genes via direct interactions, and thereby controls how much PRC1 is available for silencing. This model is consistent with the in vivo antagonism between Polycomb silencing and cohesin, described in the text. The PRC1 at active genes influences RNA polymerase function, and may serve to govern cohesin’s ability to promote transition of paused polymerase into elongation (Dorsett and Kassis, 2014). This diagram is modified from Schaaf et al., 2013b.
The PRC1 recruited by cohesin to active genes influences Pol II function. Depletion of PRC1 increases the amount of Pol II in gene bodies, but it is underphosphorylated, indicating that it may be deficient in elongation or its ability to interact with RNA processing factors (Schaaf et al., 2013b). This is reflected in reduced mRNA levels at many genes. While cohesin-PRC1 interactions have yet to be observed in mammals, it seems likely that they occur, given the conservation of Polycomb complexes. If similar functional interacts do occur in human cells, it expands the potential explanations for the gene expression alterations that occur in CdLS.
Do opposing transcription factors offer potential therapeutic targets for CdLS?
Although it will not be possible to treat the structural birth defects associated with CdLS by postnatal drug therapy, it is likely that growth and neurological development, which occur over several years after birth, can be improved. Thus there is an impetus to develop therapeutic methods. The opposing in vivo effects of cohesin and PRC1 described above raise the potential for ameliorating the cohesin functional deficits by targeting PRC1 function. One of the key PRC1 subunits (Sce in Drosophila, Ring1b in mammals) is a histone ubiquitin ligase, and this enzymatic activity provides a potential drug target. The NIPBL deficit in CdLS, even in severe cases, is modest, with only a 30% reduction in NIPBL mRNA (Liu et al., 2009a). This suggests that even a modest up-regulation of the remaining functional allele would be beneficial.
PRC1 is not the only transcription factor with an activity that opposes cohesin function. Mi-2 is an ATP-dependent chromatin-remodeling enzyme that acts on nucleosomes, and a subunit of the NuRD repressor complex. Mi-2 co-localizes with cohesin along Drosophila chromosomes, and modest overexpression of Mi-2 destabilizes cohesin binding, decreasing cohesin levels on chromosomes (Fasulo et al., 2012). Strikingly, a heterozygous Mi-2 gene mutation suppresses wing defects caused by a heterozygous Nipped-B mutation. Thus a developmental deficit caused a modest deficit in loading cohesin onto chromosomes can be directly counteracted by a modest decrease in a factor that destabilizes cohesin binding. Mi-2 is conserved in humans thus may also provide a potential drug target for treatment for CdLS, if it proves to be true that human Mi-2 opposes cohesin function in the same manner as in Drosophila.
The releasin complex consisting of the Pds5 and Wapl proteins also counteracts Nipped-B by removing cohesin from chromosomes (Figure 1), and a heterozygous pds5 null mutation in Drosophila increases cohesin chromosome binding (Gause et al., 2010). Although it remains to be tested, it is possible that a mild pds5 mutation could counteract some of the developmental deficits in Nipped-B mutants. If this proves to be the case, Pds5 provides another potential therapeutic target.
Using Drosophila for drug screens
In addition to targeting factors that oppose cohesin binding or function, it is also possible to use Drosophila to screen for drugs that may ameliorate CdLS symptoms. Such a screen using a library of some 1,500 FDA-approved drugs was performed by selecting compounds that improve the growth of yeast cohesion factor mutants, and then selecting the top hits to test their ability to reverse Drosophila wing and eye defects that are Nipped-B dependent (Maria Gause, Ziva Misulovin, Adam Coster, Justin Fay, DD, unpublished). This screen identified indomethacin and one of its derivatives, acemetacin, as compounds that reverse Drosophila mutant phenotypes, and increase steady-state Nipped-B mRNA levels in cultured Drosophila cells. Indomethacin also normalizes NIPBL mRNA levels in CdLS fibroblasts (Ziva Misulovin, Ian D Krantz, DD, unpublished), and in cultured C2C12 mouse cells and embryonic fibroblasts from Nipbl(+/−) mice (Ziva Misulovin, DD, Arthur D Lander, Anne Calof, unpublished). Thus, indomethacin may treat the diverse developmental deficits observed in CdLS at their source, which is a modest deficiency in NIPBL function. A patent for the Drosophila screening method and the use of indomethacin for treating CdLS has been granted, giving hope that this prospective therapeutic lead will move forward, and that other potentially useful compounds can be identified. It is important to note that indomethacin is used to treat the Bartter’s syndrome, a kidney function deficiency, on a daily basis from a few weeks of age, and even prenatally (Fichman et al., 1976; Konrad et al., 1999) indicating that it likely to be safe to use in CdLS, if it proves effective.
Although indomethacin is a cyclooxygenase (COX) inhibitor, COX enzymes do not appear to be the critical target for increasing NIPBL mRNA levels. Drosophila has only one cyclooxygenase expressed only in the female germline. Indomethacin also inhibits the myogenesis and promotes adipogenesis pathway in C2C12 cells in a COX-independent manner (Styner et al., 2010). These facts indicate that there is an unknown evolutionarily-conserved indomethacin target, and identification of this target in future work may facilitate development of new therapeutics. The variety of molecular and genetic tools available in Drosophila should be helpful in this hunt.
Acknowledgments
The author thanks the continuing efforts of the members of his laboratory on investigating the functions of Nipped-B and cohesin in gene regulation, and Ian Krantz, Anne Calof and Arthur Lander for their continuing collaboration in the efforts to understand CdLS. The author also thanks Laird Jackson for his continued support of our laboratory’s efforts, and of the CdLS Foundation and their families. Work in the author’s laboratory is supported by grants from the US National Institutes of Health (R01 GM108872, R01 GM108714, P01 HD052860).
Footnotes
Conflict of Interest Disclosure
The author is an inventor on a patent (US9138424B2) entitled “Methods and compositions for treating Cornelia de Lange syndrome”.
References
- Chien R, Zeng W, Kawauchi S, Bender MA, Santos R, Gregson HC, Schmiesing JA, Newkirk DA, Kong X, Ball AR, Jr, Calof AL, Lander AD, Groudine MT, Yokomori K. Cohesin mediates chromatin interactions that regulate mammalian β-globin expression. J Biol Chem. 2011;286:17870–8. doi: 10.1074/jbc.M110.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, Saitoh K, Komata M, Katou Y, Clark D, Cole KE, De Baere E, Decroos C, Di Donato N, Ernst S, Francey LJ, Gyftodimou Y, Hirashima K, Hullings M, Ishikawa Y, Jaulin C, Kaur M, Kiyono T, Lombardi PM, Magnaghi-Jaulin L, Mortier GR, Nozaki N, Petersen MB, Seimiya H, Siu VM, Suzuki Y, Takagaki K, Wilde JJ, Willems PJ, Prigent C, Gillessen-Kaesbach G, Christianson DW, Kaiser FJ, Jackson LG, Hirota T, Krantz ID, Shirahige K. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–7. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodríguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–94. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Noon SE, Krantz ID. Cornelia de Lange Syndrome. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews® [Internet] Seattle (WA): University of Washington, Seattle; 2005. 1993–2016. [updated 2016 Jan 28] [Google Scholar]
- Deardorff MA, Wilde JJ, Albrecht M, Dickinson E, Tennstedt S, Braunholz D, Mönnich M, Yan Y, Xu W, Gil-Rodríguez MC, Clark D, Hakonarson H, Halbach S, Michelis LD, Rampuria A, Rossier E, Spranger S, Van Maldergem L, Lynch SA, Gillessen-Kaesbach G, Lüdecke HJ, Ramsay RG, McKay MJ, Krantz ID, Xu H, Horsfield JA, Kaiser FJ. RAD21 mutations cause a human cohesinopathy. Am J Hum Genet. 2012;90:1014–27. doi: 10.1016/j.ajhg.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Kassis JA. Checks and balances between cohesin and polycomb in gene silencing and transcription. Curr Biol. 2014;24:R535–9. doi: 10.1016/j.cub.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol. 2013;25:327–33. doi: 10.1016/j.ceb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Ström L. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol. 2012;22:R240–50. doi: 10.1016/j.cub.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasulo B, Deuring R, Murawska M, Gause M, Dorighi KM, Schaaf CA, Dorsett D, Brehm A, Tamkun JW. The Drosophila MI-2 chromatin-remodeling factor regulates higher-order chromatin structure and cohesin dynamics in vivo. PLoS Genet. 2012;8:e1002878. doi: 10.1371/journal.pgen.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, Gilmour DS, Dorsett D. Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr Biol. 2011;21:1624–34. doi: 10.1016/j.cub.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichman MP, Telfer N, Zia P, Speckart P, Golub M, Rude R. Role of prostaglandins in the pathogenesis of Bartter’s syndrome. Am J Med. 1976;60:785–97. doi: 10.1016/0002-9343(76)90892-5. [DOI] [PubMed] [Google Scholar]
- Gause M, Misulovin Z, Bilyeu A, Dorsett D. Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol Cell Biol. 2010;30:4940–51. doi: 10.1128/MCB.00642-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallson G, Syrzycka M, Beck SA, Kennison JA, Dorsett D, Page SL, Hunter SM, Keall R, Warren WD, Brock HW, Sinclair DA, Honda BM. The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc Natl Acad Sci U S A. 2008;105:12405–10. doi: 10.1073/pnas.0801698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol. 2010;20:957–63. doi: 10.1016/j.cub.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi K, Nakato R, Zhang Z, Edmondson AC, Noon S, Dulik MC, Rajagopalan R, Venditti CP, Gripp K, Samanich J, Zackai EH, Deardorff MA, Clark D, Allen JL, Dorsett D, Misulovin Z, Komata M, Bando M, Kaur M, Katou Y, Shirahige K, Krantz ID. Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat Genet. 2015;47:338–44. doi: 10.1038/ng.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, Hoang MP, Chua A, Lao T, Lechner MS, Daniel JA, Nussenzweig A, Kitzes L, Yokomori K, Hallgrimsson B, Lander AD. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A. 1988;85:8136–40. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline AD, Krantz ID, Sommer A, Kliewer M, Jackson LG, FitzPatrick DR, Levin AV, Selicorni A. Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet A. 2007;143A:1287–96. doi: 10.1002/ajmg.a.31757. [DOI] [PubMed] [Google Scholar]
- Konrad M, Leonhardt A, Hensen P, Seyberth HW, Köckerling A. Prenatal and postnatal management of hyperprostaglandin E syndrome after genetic diagnosis from amniocytes. Pediatrics. 1999;103:678–83. doi: 10.1542/peds.103.3.678. [DOI] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–5. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gilmour DS. Promoter proximal pausing and the control of gene expression. Curr Opin Genet Dev. 2011;21:231–5. doi: 10.1016/j.gde.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Feldman R, Zhang Z, Deardorff MA, Haverfield EV, Kaur M, Li JR, Clark D, Kline AD, Waggoner DJ, Das S, Jackson LG, Krantz ID. SMC1A expression and mechanism of pathogenicity in probands with X-Linked Cornelia de Lange syndrome. Hum Mutat. 2009b;30:1535–42. doi: 10.1002/humu.21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang Z, Bando M, Itoh T, Deardorff MA, Clark D, Kaur M, Tandy S, Kondoh T, Rappaport E, Spinner NB, Vega H, Jackson LG, Shirahige K, Krantz ID. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009a;7:e1000119. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. Revisiting global gene expression analysis. Cell. 2012;151:476–82. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–30. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, Zhao K, Levens D. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S, Cuadrado A, Losada A. Cohesin in development and disease. Development. 2013;140:3715–8. doi: 10.1242/dev.090605. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Focarelli ML, Susani L, Paulis M, Bassi MT, Mannini L, Frattini A, Delia D, Krantz I, Vezzoni P, Jessberger R, Musio A. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum Mol Genet. 2009;18:418–27. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JM, Bentley FK, Print CG, Dorsett D, Misulovin Z, Dickinson EJ, Crosier KE, Crosier PS, Horsfield JA. Positive regulation of c-Myc by cohesin is direct, and evolutionarily conserved. Dev Biol. 2010;344:637–49. doi: 10.1016/j.ydbio.2010.05.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–11. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–93. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Kwak H, Koenig A, Misulovin Z, Gohara DW, Watson A, Zhou Y, Lis JT, Dorsett D. Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 2013a;9:e1003382. doi: 10.1371/journal.pgen.1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Gause M, Koenig A, Gohara DW, Watson A, Dorsett D. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 2013;9:e1003560. doi: 10.1371/journal.pgen.1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CA, Misulovin Z, Sahota G, Siddiqui AM, Schwartz YB, Kahn TG, Pirrotta V, Gause M, Dorsett D. Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS One. 2009;4:e6202. doi: 10.1371/journal.pone.0006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–72. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, Tamkun JW. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–35. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- Strübbe G, Popp C, Schmidt A, Pauli A, Ringrose L, Beisel C, Paro R. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc Natl Acad Sci U S A. 2011;108:5572–7. doi: 10.1073/pnas.1007916108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styner M, Sen B, Xie Z, Case N, Rubin J. Indomethacin promotes adipogenesis of mesenchymal stem cells through a cyclooxygenase independent mechanism. J Cell Biochem. 2010;111:1042–50. doi: 10.1002/jcb.22793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–41. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- Vissers LELM, van Ravenswaaij CMA, Admiraal R, Hurst JA, de Vries BBA, Janssen IM, van der Vliet WA, Huys EHLPG, de Jong PJ, Hamel BCJ, Schoenmakers EFPM, Brunner HG, Veltman JA, Geurts van Kessel A. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nature Genet. 2004;36:955–7. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- Vrouwe MG, Elghalbzouri-Maghrani E, Meijers M, Schouten P, Godthelp BC, Bhuiyan ZA, Redeker EJ, Mannens MM, Mullenders LH, Pastink A, Darroudi F. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: evidence for impaired recombinational repair. Hum Mol Genet. 2007;16:1478–87. doi: 10.1093/hmg/ddm098. [DOI] [PubMed] [Google Scholar]
- Wu Y, Gause M, Xu D, Misulovin Z, Schaaf CA, Mosarla RC, Mannino E, Shannon M, Jones E, Shi M, Chen WF, Katz OL, Sehgal A, Jongens TA, Krantz ID, Dorsett D. Drosophila Nipped-B mutants model Cornelia de Lange syndrome in growth and behavior. PLoS Genet. 2015;11:e1005655. doi: 10.1371/journal.pgen.1005655. [DOI] [PMC free article] [PubMed] [Google Scholar]