Abstract
ATM (ataxia-telangiectasia, mutated) is an important cancer susceptibility gene that encodes a key apical kinase in the DNA damage response pathway. ATM mutations in the germ line result in ataxia-telangiectasia (A-T), a rare genetic syndrome associated with hypersensitivity to double-strand DNA breaks and predisposition to lymphoid malignancies. ATM expression is limited by a tightly regulated nonsense-mediated RNA decay (NMD) switch exon (termed NSE) located in intron 28. In this study, we identify antisense oligonucleotides that modulate NSE inclusion in mature transcripts by systematically targeting the entire 3.1-kb-long intron. Their identification was assisted by a segmental deletion analysis of transposed elements, revealing NSE repression upon removal of a distant antisense Alu and NSE activation upon elimination of a long terminal repeat transposon MER51A. Efficient NSE repression was achieved by delivering optimized splice-switching oligonucleotides to embryonic and lymphoblastoid cells using chitosan-based nanoparticles. Together, these results provide a basis for possible sequence-specific radiosensitization of cancer cells, highlight the power of intronic antisense oligonucleotides to modify gene expression, and demonstrate transposon-mediated regulation of NSEs.
Keywords: : alternative splicing, transposon, nonsense-mediated RNA decay, antisense oligonucleotides, ATM, lymphoid cancer, nanoparticles
Introduction
Eukaryotic genes contain intervening sequences or introns that must be removed by a large and highly dynamic RNA protein complex termed the spliceosome to ensure accurate protein synthesis [1]. The cell requires excessive energy and time to complete transcription of intron-containing precursor messenger RNAs (pre-mRNAs) from at least a quarter of the human genome and also needs to synthesize noncoding RNAs and >200 different spliceosomal proteins to achieve this task [1]. Although once regarded a selfish or junk DNA, introns are now recognized as critical functional components of eukaryotic genes that enhance gene expression and regulate alternative RNA processing, mRNA export, and RNA surveillance [2,3]. They are also an important source of new gene-coding and regulatory sequences [1,4,5] and noncoding RNAs, including microRNAs and circular RNAs [6,7]. Their removal process is tightly coupled with transcription, mRNA export, and translation, with most human introns eliminated from pre-mRNA cotranscriptionally [8]. However, their potential as targets for nucleic acid therapy is only beginning to be unleashed.
Spliceosomes assemble ad hoc on each intron in an ordered manner, starting with recognition of the 5′ splice site (5′ss) by the U1 small nuclear ribonucleoprotein or the 3′ss by the U2 pathway [1,9]. In addition to traditional splice site recognition sequences (5′ss, branch point, polypyrimidine tract, and 3′ss), accurate splicing requires auxiliary sequences or structures that activate or repress splice sites, known as intronic or exonic splicing enhancers or silencers. These elements allow genuine splice sites to be recognized among a vast excess of cryptic or pseudosites in vertebrate genomes that have similar sequences, but outnumber authentic sites by an order of magnitude [10]. Activation of cryptic splice sites can introduce premature termination codons (PTCs) in translational reading frames and may lead to genetic disease [11]. Such transcripts are usually recognized by a nonsense-mediated RNA decay (NMD) pathway and downregulated [12]; however, cryptic exons and NMD play also an important role in controlling the expression of naturally occurring transcripts [13] and differentiation stage-specific splicing switches, as exemplified by terminal stages of hematopoiesis [14,15]. In addition, cryptic splice sites may permit unproductive or partial spliceosome assemblies that may compete with natural splice sites, which may facilitate their accurate selection at a single-nucleotide resolution [16,17]. Cryptic splice sites can activate pseudoexons that limit gene expression (also known as poison or NMD switch exons), thus regulating the pool of mRNA isoforms and providing interesting targets for nucleic acid therapeutics [18]. However, potential exploitation of such strategies is in its infancy.
Splice-switching oligonucleotides (SSOs) are antisense reagents that modulate intron splicing by binding splice site recognition or regulatory sequences and competing with cis-elements or trans-acting factors for their targets [19–21]. They have been shown to restore aberrant RNA processing, modify the relative abundance of existing mRNA isoforms, or produce novel splice variants that are not normally expressed by the cell [20]. Most SSOs employed in preclinical and clinical development have targeted exonic sequences [19–21]. Whereas most exonic SSOs designed to induce exon skipping usually have a desired effect, functional intronic SSOs are more difficult to identify, unless they block access to intronic cryptic splice sites activated by a disease-causing mutation. First, a large fraction of intronic sequences may not affect RNA processing at all, despite the wealth of intronic auxiliary splicing motifs in the human genome [22]. In addition, a search for functional intronic SSOs that produce desirable RNA processing outcomes is usually inefficient and costly and may fail completely. For example, most SSOs systematically covering exon 7 of the SMN2 (survival of motor neuron 2) gene stimulated exon skipping, a prerequisite for antisense therapy of spinal muscular atrophy; however, ∼20% SSOs increased exon inclusion [23]. By contrast, stimulation of intron splicing was found only for ∼10% of SSOs targeting INS intron 1, while the majority failed to show this effect [24]. Third, introns are enriched for many repetitive elements that preclude the SSO use with endogenous targets. Identification of effective intronic SSOs may be facilitated by global pre-mRNA folding and ultraviolet cross-linking and immunoprecipitation studies that identify binding sites for components of the spliceosome [18,25] or the exon junction complex [26]. However, these binding sites may not reflect optimal antisense targets and their resolution may be insufficient. Thus, identification of functional intronic SSOs remains challenging.
Our RNA-Seq studies have recently revealed activation of an NMD switch exon (termed NSE) deep in ATM (ataxia-telangiectasia, mutated) intron 28 in cells depleted of each subunit of the auxiliary factor of U2 small nuclear RNP (U2AF) [18]. U2AF binds to polypyrimidine tracts coupled with highly conserved 3′ss AG dinucleotides at intron ends and this binding promotes U2 recruitment to the branch site and formation of lariat introns [27–29]. However, recent identification of a large number of exons that were activated in cells depleted of each U2AF subunit (U2AF35 and U2AF65) and exhibited a distinct 3′ss organization [30,31] suggested that a subset of both canonical and NSEs is repressed by U2AF, similar to exon-repressing and -activating activities found for a growing number of RNA-binding proteins [32,33]. The NSE levels were responsive to knockdown of additional splicing factors involved in 3′ss recognition and were influenced by two natural DNA variants, rs609261 and rs4988000, located in the NSE 3′ss and further downstream, respectively [18]. We have also identified SSOs that modulate NSE inclusion levels in the ATM mRNA by targeting NSE and its competing pseudoexon in the same intron [18]. The ATM NSE provides an interesting and promising target for anticancer therapy for several reasons: (1) the ATM kinase is activated in response to double-strand breaks, mobilizing an extensive signaling network with a broad range of targets and influencing cellular sensitivity to DNA-damaging agents [34]; (2) the U2AF-regulated exon usage in the ATM signaling pathway is centered on the MRN/ATM-CHEK2-CDC25 axis and preferentially involves transcripts implicated in cancer-associated gene fusions and chromosomal translocations [18]; and (3) ATM NSE activation limits expression of ATM protein in cells lacking each U2AF subunit [18]. However, optimal NSE SSOs have not been defined and their delivery to lymphoid cells has not been tested.
In the present study, we have systematically screened SSOs covering unique sequences in the entire intron 28 and identify additional SSOs that activate or repress NSE in vitro. In the same intron, we also identify transposed elements that influence NSE inclusion in mature transcripts. Finally, we show efficient NSE repression upon SSO delivery to embryonic and lymphoblastoid cell lines using chitosan-based nanoparticles.
Materials and Methods
Plasmid constructs
Splicing reporter constructs containing full ATM intron 28 and flanking exons were obtained by ligating a ∼3.5 kb polymerase chain reaction (PCR) amplicon into the HindIII/XbaI site of pCR3.1 (Invitrogen). PCR was carried out with amplification primers, ATM26 and ATM27 (Table 1), and DNA from human embryonic kidney HEK293 cells as a template. PCR employed the Phusion® High-Fidelity DNA Polymerase (Thermo Scientific) at the annealing temperature of 56°C and 1.5 mM MgCl2 for 30 cycles. Plasmids were propagated in Escherichia coli (strain DH5α). Plasmid DNA was extracted with the Gene JET Plasmid Miniprep kit (Thermo Scientific) and separated on 1.5% agarose gels to confirm the correct insert size following restriction enzyme digests prior to sequence validation. Deletion constructs (Fig. 1) were obtained by overlap extension PCR with mutagenic primers (Table 1) using the validated reporter plasmid with the full intron as a template. Each deletion insert was also fully sequenced to confirm the identity of intended changes and exclude undesired mutations. Hybrid ATM minigenes prepared by cloning ∼0.9-kb amplicons containing NSE and exon 29 into XhoI/XbaI sites of the U2AF1 construct were described previously [18].
Table 1.
Cloning, RT-PCR, and Mutagenic Primers
| Primer | 5′-3′ Sequence |
|---|---|
| Cloning primers | |
| ATM26 | ataaagcttcttgttataaggttttgattcc |
| ATM27 | atatctagatgtacataccctgaaaagtcac |
| RT-PCR primers | |
| PL4 | agtcgaggctgatcagcgg |
| ATM-F | gagggtaccagagacagtgggatggc |
| ATM-R | ggctcatgtaacgtcatcaat |
| Mutagenic primers | |
| del-1F | atacaatttaccataatttacttttgaattatgtt |
| del-1R | aagtaaattatggtaaattgtatcatacattag |
| del-2F | ccttgccagaccagtttcctagttatctatattgaac |
| del-2R | taactaggaaactggtctggcaaggtggctta |
| del-3F | cttcaagggaccttggccgggtgcggtggct |
| del-3R | gcacccggccaaggtcccttgaagtttatctaa |
| del-4F | acacaaacaaagcttaggtttctttcttgtcaccttcta |
| del-4R | agaaagaaacctaagctttgtttgtgtgttttatacaa |
| del-5F | tgcctcatttacgtcatacaacttaatgatagacct |
| del-5R | ttaagttgtatgacgtaaatgaggcagggcaa |
| del-6F | tgatacaatttacctcatacaacttaatgatagacct |
| del-6R | attaagttgtatgaggtaaattgtatcatacattag |
RT-PCR, reverse transcription polymerase chain reaction; restriction sites are underlined.
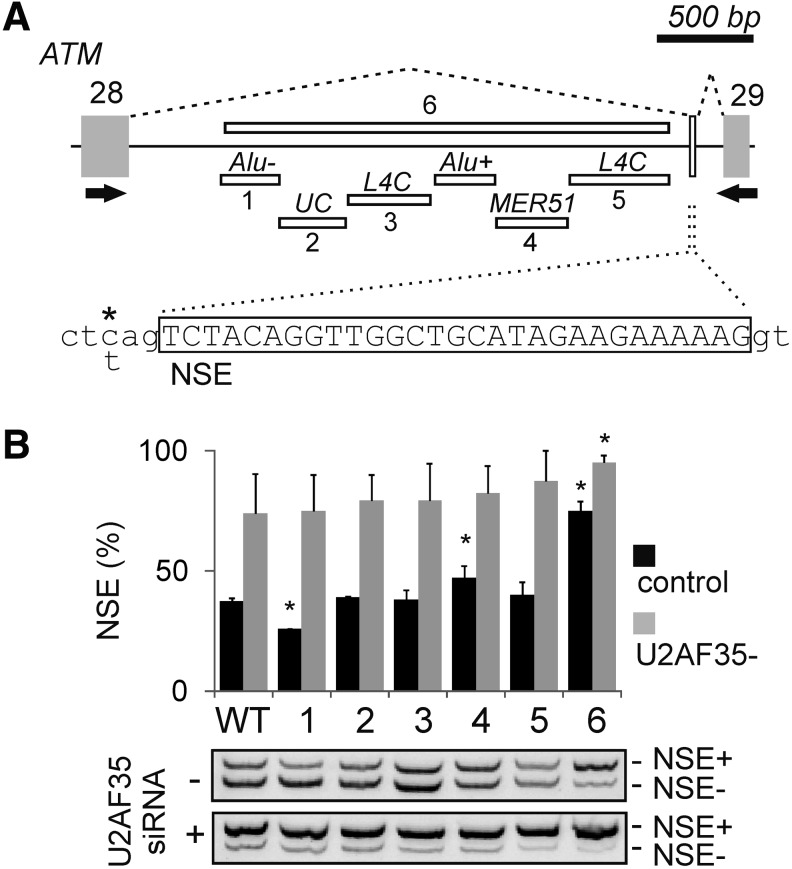
FIG. 1.
Identification of transposed elements in ATM intron 28 that influence NSE activation. (A) Location of transposed elements in intron 28 and schematics of NSE activation. Canonical exons [54] are shown as gray boxes, the NSE as a white box, introns flanking the NSE as horizontal lines, and their splicing by dotted lines. Deletions (numbered 1–6) of transposed elements are shown as horizontal white rectangles; UC, a unique sequence lacking recognizable transposons. Deletion numbers correspond to lanes in (B). RT-PCR primers are denoted by black arrows. A scale is at the top. The NSE sequence is boxed in the lower panel. Asterisk denotes the C/T variant rs609261 located at the NSE 3′ss; rs4988000 (not shown) is 64 bp downstream of the NSE 5′ss. (B) Deletion of antisense Alu and MER51 alters NSE inclusion levels. WT and mutated constructs [designated 1–6 in (A)] were transiently transfected into HEK293 cells (mock) depleted of U2AF35. NSE+/−, RNA products with/without NSE. Columns represent mean NSE inclusion (%), error bars are SDs of two independent transfection experiments. Asterisks denote P values <0.01 for comparisons with the WT. 5′ss, 5′ splice site; ATM, ataxia-telangiectasia, mutated; NSE, NMD switch exon; RT-PCR, reverse transcription polymerase chain reaction; SDs, standard deviations; WT, wild-type.
Splice-switching oligonucleotides
To test the effect of SSOs on both the endogenous and the exogenous ATM pre-mRNA, SSOs were designed to avoid transposed elements in intron 28. These elements were identified in the human reference sequence (hg19) with the crossmatch and slow options of the RepeatMasker web server (v.4.0.2; www.repeatmasker.org) [35] and confirmed in our constructs using the same method. The SSOs comprehensively covered three unique regions in ATM intron 28 (termed A, B, and AN, Fig. 2), avoiding only homopolymeric tracts. SSOs (Eurofins) were 2′-O-methyl modified at each ribose and by phosphorothioates at each end linkage to ensure adequate stability for the ex vivo screening. The GC content of SSOs was at least 24% (mean 31%) and their average length was ∼20 nt. SSOs were diluted in double-distilled water and quantified using Nanodrop (Thermo Scientific). Their normalized aliquots were stored at −80°C.
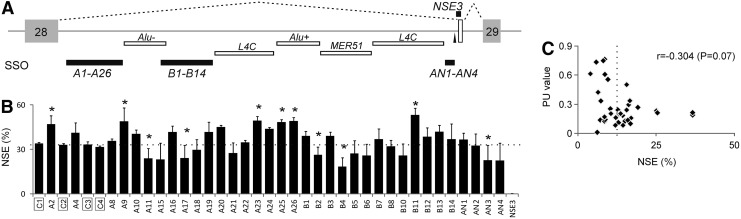
FIG. 2.
Identification of intronic SSOs that activate or repress NSE. (A) Location of tested SSOs in intron 28 relative to transposed elements (for legend, see Fig. 1A). The branch point sequence (GGCTGAT; branch point adenosine is underlined) of NSE is denoted by a vertical arrowhead. (B) Intronic SSOs that alter NSE inclusion in exogenous transcripts. SSOs are at the bottom. Multiple controls are boxed. SSO sequences are in Table 2. The average NSE inclusion in controls is denoted by a dotted line, error bars are SDs of two independent transfection experiments. Columns represent mean NSE inclusion levels, asterisks show significant P values. (C) SSOs targeting single-stranded regions tended to repress endogenous NSE. r, Pearson correlation coefficient. The P value is in brackets. SSOs, splice-switching oligonucleotides.
Cell cultures and transfections of minigene constructs
HEK293 cells were maintained in standard culture conditions in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% (v/v) of bovine calf serum. Cells were seeded at 50% confluency 24 h before transfections. Transfections of wild-type and deletion constructs were carried out in 12- or 24-well plates using jetPRIME (Polyplus) according to the manufacturer's recommendations, as described [30]. Briefly, 250 ng of plasmid DNA was mixed with 1.3 μL of jetPRIME in 75 μL of the jetPRIME buffer and incubated at room temperature for 15 min before adding into cell cultures. Each SSO was transfected with and without the full-length ATM construct using the same transfection reagent (2.5 μL) to a final concentration of 50 nM in each well. The cells were harvested 24 h later for total RNA extraction with TRI-Reagent (Ambion). Replicate experiments were carried out 1 week later.
Determination of PU values
The PU (probability of unpaired) values estimate RNA single-strandedness using the equilibrium partition function by considering all possible RNA structures of short sequences, permitting their comparison at each nucleotide position [36]. Higher PU values indicate a higher single-strandedness of an RNA motif [36]. The PU values were computed as described [36] using the three unique intronic regions and their 30-bp flanks as an input. PU values for each position of an SSO target were averaged and the means were correlated with SSO-induced NSE inclusion levels.
Preparation of stearylated trimethyl chitosan
Trimethyl chitosan, originally derived from ultrapure chitosan obtained from Agaricus bisporus, was provided by KitoZyme. Purified products had the number average molecular weight (Mn) of 43.3 ± 5.5 kDa and the polydispersity index (Mw/Mn) of 2.4 ± 0.3, as determined by gel permeation chromatography in a 0.33 M NaCH3COOH/0.28 M CH3COOH eluent at a flow rate of 1 mL/min. The degrees of acetylation and quaternization, as determined by the Fourier-transform infrared spectroscopy and 1H-nuclear magnetic resonance spectroscopy (1H NMR) [37], respectively, were 11.1% ± 0.9% and 30.1% ± 4.6%. Trimethyl chitosan was functionalized with N-succinimidyl stearate (Santa Cruz Biotechnologies), as previously described [37], achieving a final degree of substitution of 2.1% ± 0.6% (mol %), as determined by 1H NMR.
Formation and delivery of nanocomplexes
The nanocomplexes were prepared by mixing equal volumes (30 μL) of SSO and polymer solutions, as described previously [37]. Briefly, SSOs were diluted in buffer A [20 mM HEPES, pH 7.3, 5% (w/v) glucose] and supplemented with 1 M Na2SO4 to a final concentration of 50 mM. Both the polymer and SSO solutions were heated at 60°C for 5 min before mixing with vortex at 1,000 rpm for 15 s. The tested complexes were prepared with molar ratios of quaternized amines (N) to phosphate groups (P) of 20, 40, and 80 (N/P ratios), as previously optimized for the first SSOs [37]. Their hydrodynamic diameter was between 110 and 130 nm for the N/P ratios between 20 and 80. The complexes were allowed to stabilize at room temperature for 30 min before adding to 240 μL DMEM without serum and antibiotics. A final concentration of SSOs in chitosan-containing cultures was 300 nM. Twenty-four hours after addition of complexes, 300 μL of the culture medium with serum/antibiotics was added to HEK293 or VAVY cells. The cells were harvested 24 h later for total RNA extraction. Replicate experiments were carried out 1 week later.
Analysis of spliced products and NSE measurements
Total RNA samples from chitosan experiments were extracted with the RNeasy kit (Qiagen). RNA was quantified using Nanodrop (Thermo Scientific) and 1 μg of total RNA was reverse transcribed with the Moloney murine leukemia virus reverse transcriptase (Promega) and oligo-d(T) primers. Exogenous complementary DNA (cDNA) samples were amplified using primers, PL4 and ATM-F, and endogenous products were amplified with primers, ATM-F and ATM-R (Table 1), and GoTaq polymerase (Promega) for 28 cycles (the annealing temperature was 56°C at 1.5 mM MgCl2). Spliced products were separated on 1.5% agarose and then on 6% polyacrylamide gels. The signal intensities were measured in polyacrylamide gels using FluorQuant and Phoretix software packages (Nonlinear Dynamics, Inc.) to obtain the percentage of NSE inclusion in polyadenylated RNAs.
Branch site prediction
The NSE branch point was predicted using the SVM-BP finder (available at http://regulatorygenomics.upf.edu/Software/SVM_BP/). The SVM score of the indicated NSE branch site was 1.3.
Statistical analysis
Descriptive statistics were computed with Stat200 (BioSoft). Mean NSE inclusion levels were compared using t-tests; the indicated P values are two-tailed.
Results
SSOs targeting either 3′ or 5′ss of the ATM NSE efficiently repress this exon in a haplotype-dependent manner [18]. To facilitate identification of optimal intronic SSOs that increase NSE inclusion in mature transcripts, we first prepared splicing reporter constructs with the entire ATM intron 28 (Fig. 1A). The construct was obtained by PCR using the HEK293 DNA as a template. The reference sequence (hg19) of human intron 28 is ∼3.1 kb long, which is similar to the average human intron [38]. About 64% of this intron is occupied by transposed elements, filling completely its middle part, except for a ∼350 bp region in the 5′ half of the intron and exonic flanks (Fig. 1A). Plasmid DNA sequencing revealed the same organization of transposed elements without any additional transposon copies. It also showed the C and G alleles at rs4988000 and rs609261, respectively, indicating that the construct contained the haplotype most permissive for NSE inclusion in the ATM mRNA [18]. Following transfections into HEK293 cells, total RNA was extracted and reverse transcribed before amplification with a combination of a vector primer PL4 (Table 1) and an exon primer [18] (Fig. 1A). Examination of spliced products showed that most transcripts entirely lacked intronic sequences (NSE−), whereas ∼36% of the mRNA contained NSE (NSE+, Fig. 1B, lane 1). This fraction was slightly higher than for a hybrid ATM reporter examined previously [18], consistent with the presence of auxiliary splicing sequences in newly cloned intronic segments.
To determine the importance of transposed elements for NSE inclusion, we individually deleted each transposon from intron 28 (deletions 1–5, Fig. 1A). We also deleted a large middle part of the intron along with all transposons, leaving the NSE and its upstream sequences intact, including the predicted branch site (∼75% of the intron, deletion 6). Transfection of validated mutated constructs, which all had identical genotypes to the wild-type construct at rs4988000 and rs609261, revealed that the large deletion promoted NSE-containing transcripts (deletion 6, Fig. 1B). Deletion of the MER51 element increased NSE inclusion to a lesser extent. In contrast, deletion of the antisense Alu inhibited NSE, while deletion of long interspersed repeats (deletions 3 and 5) or a unique intronic segment (deletion 2) had no effect on NSE inclusion. The variability of NSE inclusion levels was higher following transfections of the same plasmids into cells depleted of U2AF35, with a significant increase of NSE levels maintained only for deletion 6 (Fig. 1B), consistent with a major stress component of NSE responses [18].
We next designed a series of SSOs targeting three intronic regions that have unique sequences in the genome (termed A, B, and AN) while avoiding a predicted branch site upstream of NSE (Fig. 2A and Table 2). Each SSO was modified with 2′-O-methyl at each ribose and phosphorothioate at each end linkage to ensure their RNase H resistance and sufficient stability in transient transfections. As a positive control, we used SSO-NSE3, which was highly efficient in blocking the NSE 3′ss [18]. As negative controls, we employed a series of scrambled SSOs and SSOs targeting other genes, including INS [24] and BTK [39] (Table 2), which were not expressed in HEK293 cells, as confirmed by our RNA-Seq data [30]. Each SSO was individually transfected with or without the wild-type ATM construct. Measurements of spliced products revealed that SSO-NSE3 yielded the most efficient NSE repression, as expected (Fig. 2B). About a half of tested SSOs significantly altered NSE inclusion levels compared with controls, with similar numbers of repressor and activator SSOs. Pearson correlation coefficient between replicate transfections was highly significant, reaching 0.88 (P < 10−8); however, the overall correlation between exogenous and endogenous NSE levels was only 0.35 (P < 0.01).
Table 2.
Splice-Switching Oligoribonucleotides
| SSO | 5′-3′ Sequencea |
|---|---|
| A2 | aacuuaaagguuauaucuc |
| A4 | uauaaauacgaauaaaucga |
| A8 | cauggguuggcuaugcuag |
| A9 | caacacgacauaaccaaa |
| A10 | aagccaaucagagggagaca |
| A11 | aacauuucuauuuaguuaaaagc |
| A15 | ucguguauuacaacaguuaa |
| A16 | caaccaguuugcauucgu |
| A17 | uuaguauuccuugacuuua |
| A18 | uucuguacacuguuuaguauucc |
| A19 | gaagagggagugaagguu |
| A20 | aaagcuuggugagauuga |
| A21 | uuucuugaaaaguggaaagcuug |
| A22 | uggaaugagggacgguuguuuuuc |
| A23 | gguaugagaacuauagga |
| A24 | aaacaaacagcaggguau |
| A25 | gguaauaagugucacaaa |
| A26 | guaucauacauuagaagg |
| B1 | ucaaaaguaaauuauggucu |
| B2 | gacugguaaauaauaaacauaauuc |
| B3 | aaauguauacuggagaagacu |
| B4 | auauauuagagauacaucagcc |
| B5 | gacaaacauuuaaugaauacucaa |
| B6 | uugacuccuucuuuugacaaacau |
| B7 | uuuaaauccuuccuuacuu |
| B8 | gauuauaaaacaaacgaagc |
| B10 | uguuuuaauauaaguugcuucaa |
| B11 | uguggggugaccacagcuu |
| B12 | ucccuuacuuauauccaa |
| B13 | ccaaguuugguuacuuauc |
| B14 | gaaguuuaucuaauauugacc |
| AN1 | ggucuaucauuaaguuguauga |
| AN2 | uuaaauaagacuucaggucua |
| AN3 | uuagagaaucauuuuaaauaagac |
| AN4 | cuuaauccaauucuucaauuuuag |
| C1 | aggugcucgcgggugg |
| C2 | guugcaaauuucuucaaauc |
| C3 | agcuggggccuggggu |
| C4 | ggaaacuugcccguguucca |
Each SSO was modified with 2′-O-methyl at each ribose and phosphorothioate at each end linkage.
SSO, splice-switching oligonucleotide.
Experiments in Fig. 1 showed that NSE inclusion is controlled by distant splicing regulatory sequences within and outside transposons. Splicing enhancer and silencer motifs in their natural pre-mRNA context occur preferentially in single-stranded regions [36], suggesting that they are more accessible to RNA-binding proteins or other ligands that control exon selection. Preferential targeting of SSOs to unpaired regions could thus improve our search strategy. To test this assumption, we correlated endogenous NSE inclusion levels after the SSO treatment with their average PU values (Fig. 2C). These values estimate single-strandedness of their RNA targets using an equilibrium partition function, with higher values signaling a higher probability of single-stranded conformation [36]. Interestingly, SSO targets with higher average PU values tended to induce exon skipping, suggesting that efficient blocking of unpaired interactions as far as 2 kb from the exon can impair its inclusion levels in the mRNA.
The experiments described above identified a small set of intronic SSOs that increased NSE inclusion in exogenous and endogenous mRNAs and that might serve as gene-specific repressors since NSE can limit ATM expression through NMD [18]. ATM repression by NSE-activating SSOs might be advantageous for cancer treatment by inhibiting the double-strand break signaling pathway and radiosensitization [40]. To test if ATM SSOs can be delivered to cells that have much lower transfection efficiency than HEK293 cells, we employed a stearylated trimethylated chitosan (TMC-SA). Chitosan is a natural copolymer of d-glucosamine and N-acetyl-d-glucosamine known for biocompatibility, biodegradability, and low toxicity and immunogenicity [41,42]. When trimethylated, chitosan acquires a permanent positive charge that improves its solubility at neutral pH [41]. Stearylation was found to be necessary for formation of stable nanocomplexes with SSOs and their transfection activity [37] in the HeLa/pLuc705 system, which makes use of a luciferase gene interrupted by a mutated HBB intron [43].
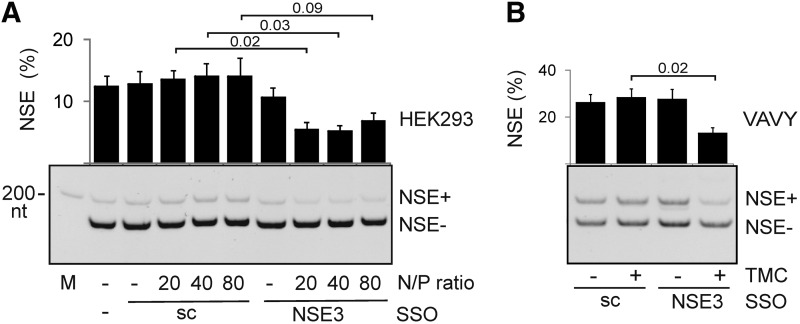
We first tested if TMC-SA can facilitate delivery of SSO-NSE3 into HEK293 cells. Figure 3A shows reduction of NSE levels following exposure to the TMC-SA/SSO-NSE3 nanocomplexes compared with a complexed scrambled SSO. This decline was significant for the TMC-SA/SSO-NSE3 (N/P) ratios 20 and 40. NSE reduction was also apparent when comparing NSE inclusion in cells exposed to uncomplexed SSO-NSE3, consistent with their significant uptake by this highly transfectable cell line. However, the decline of NSE levels was smaller for TMC-SA/SSO-NSE3 than for the same oligo transfected with jetPrime to the same cell line at a lower final concentration. Finally, significant NSE repression upon exposure to the TMC-SA/SSO-NSE3 nanocomplexes was observed also for a lymphoblastoid cell line where uncomplexed SSO-NSE3 failed to reduce NSE altogether (Fig. 3B).
FIG. 3.
TMC-SA-assisted delivery of SSO-NSE3 to human cell lines leads to NSE repression. (A) NSE inclusion in HEK293 cells is inhibited upon exposure of SSO-NSE3/TMC-SA nanocomplexes. Sc, a scrambled control with the same modification. M, size marker. Error bars denote SDs of two transfection experiments. P values are shown at the top for the indicated comparisons. (B) NSE repression in VAVY cells exposed to the SSO-NSE3/TMC-SA nanocomplexes. TMC-SA, stearylated trimethylated chitosan.
Discussion
In this work, we have shown for the first time that a chitosan-based delivery system for intronic SSOs can repress an NSE (Fig. 3). Our results also demonstrate the first transposed elements that promote or repress inclusion of an NSE in mature transcripts (Fig. 1).
Alu sequences have a propensity to exonize through 3′ss or 5′ss activation [4,44] or auxiliary splicing motifs [17,45] upon a single mutation. These events contribute significantly to human morbidity [46]. In addition, they can be exonized by outlying deletions and cause genetic disease [47], indicating that they can promote inclusion of remote intronic sequences in mature transcripts. Although the exact mechanism of such distant effects is not understood, the secondary structure of these GC-rich transcripts is likely to play a major role [46,48]. However, mutation-induced exonization has been shown for all other classes of transposed elements, including more ancient, short interspersed elements termed mammalian interspersed repeats [46]. In the present study, an intronic transposed element with the highest similarity to MER51A (MEdium Reiterated frequency repeat, family 51, [49]) repressed NSE, acting as a buffer to counteract the Alu-mediated NSE activation (Fig. 1A, B). The ATM MER51 is also relatively GC-rich (∼44%), which could facilitate intramolecular interactions with GC-rich Alus during cotranscriptional folding. The element contains several inverted repeats that might form stable hairpins, exposing purine-rich loops (Fig. 4) that could act as splicing regulatory motifs. These stem-loops should be examined in future studies to identify distant interactions underlying the MER-mediated exonization. About 250,000 copies of recognizable MER sequences were estimated to exist in the human genome [38,50] and many were found in mature transcripts of protein-coding genes, contributing to the diversity of protein interactions [51]. A mutation-induced MER exonization was also shown to cause Gitelman syndrome [52]. The 3′ part of MER51 is similar to long terminal repeats of retroviruses (Fig. 4) [49], which account for ∼15% of transposon-mediated exonization events leading to human genetic disease [46]. The origin of most MERs was placed after the decline of mammalian interspersed repeats before the spread of Alus, coinciding with the expansion of mammals and suggesting that MERs may offer insights into early mammalian radiation [49]. Taken together, our results suggest that the interplay of transposed elements in long introns could influence inclusion levels of many NSEs, fine-tuning gene expression.
FIG. 4.
Inverted repeats in the MER51 element of ATM intron 28. The alignment was carried out by RepeatMasker [35]. v, transversions; i, transitions. Putative purine-rich loops (highlighted in gray) are flanked by inverted repeats (underlined) that may form stable base-pairing interactions during transcription. The long terminal repeat homology region originally described for the MER51 family [49] is in italics. The aligned segment corresponds to deletion 4 shown in Fig. 1A. The MER51A consensus sequence is in the antisense orientation.
We have also identified candidate sequence-specific ATM inhibitors that act by promoting a regulated NSE important for ATM expression (Fig. 2). ATM inhibitors sensitize cancer cells to cytotoxic therapy that induces double-strand breaks, including local radiotherapy, an integral part of treatment regimens of many cancer types [53]. Although chemical ATM inhibitors showed great promise, their undesired pharmacokinetic profiles, high toxicity, or poor efficacy has hampered their progression into the clinic [53]. In contrast, SSOs target unique sequences in the human genome, can be delivered using natural biodegradable compounds (Figs. 1–3), and their mechanism of action is better defined. In addition, the availability of NSE-activating and -repressing SSOs may offer an opportunity to titrate gene expression more accurately than less specific chemical inhibitors, assuming their efficient delivery to newly identified intron 28 targets. Although NSE is included in natural transcripts at low levels, it can be dramatically upregulated in response to various stimuli [18,54], requiring further studies into NSE regulation. Recently, a gene-specific antisense inhibition of NMD employed SSOs targeting exon junction complex deposition sites, thus permitting NMD repression without relying on skipping of a PTC-containing exon [26]. The two approaches, the former relying on intronic sequence and the latter on exonic targets, might complement each other in the future to expand the repertoire of antisense strategies that inhibit NMD.
The average length of SSOs employed in our screening was close to the minimum for unique targets (Table 2). Shorter SSOs may induce more off-target effects than longer SSOs, which could contribute to the observed low correlation between NSE inclusion levels in endogenous and exogenous transcripts. Apart from the possible suboptimal target specificity, intron 28 splicing and NSE inclusion can be influenced by adjacent introns that were absent in exogenous transcripts. In addition, intron 28 splicing may not be strictly cotranscriptional [18]. Furthermore, distinct promoters of exo- and endogenous transcripts could be associated with dissimilar RNA folding or its kinetics, further contributing to the low correlation. Nevertheless, our study clearly demonstrates a wealth of candidate intronic target sites for functional SSOs, in agreement with a high information content of human intronic auxiliary splicing sequences. This content is higher than in lower organisms, which have smaller introns with a lower regulatory potential for alternative splicing [22].
Although SSO-NSE3 and other SSOs can repress endogenous NSE-containing mRNAs (Fig. 2B, C) [18] and NMD transcripts with the relative abundance as low as ∼1% can contribute to the mRNA consumption [55], it remains to be tested if their reduction can lead to a sustained increase of ATM protein levels in normal cells. Unlike cells depleted of U2AF subunits where NSE inclusion is high (Fig. 1) and ATM signal on immunoblots was increased upon a single SSO transfection [18], the low endogenous NSE levels in normal cells would necessitate a prolonged and repeated SSO exposure and more sensitive assays, ideally coupled with functional analysis of ATM signaling targets. Newly identified SSOs might also alleviate phenotypic consequences of leaky ataxia-telangiectasia (A-T) alleles in a mutation-independent manner, especially in homozygous A-T patients carrying the C allele at rs609261, which facilitates 3′ss recognition of the NSE [18]. Since chitosan-based nanoparticles have been shown to penetrate the blood–brain barrier and accumulate in cerebellum [56], SSO-NSE3 and other NSE repressors might have a future therapeutic potential in slowing down the progression of cerebellar symptoms of a subset of A-T patients.
Acknowledgments
The authors wish to thank Professor Steven Marsh (UCL and the Anthony Nolan Trust) for a generous gift of the VAVY cell line. This work was funded by Bloodwise (grant 12060 to I.V. and N.C.P.C), Santa Casa da Misericórdia de Lisboa—Prémio Melo e Castro (grant MC-1068-2015 to A.P.P.), and Fundação para a Ciência e Tecnologia (grant SFRH/BPD/108738/2015 to P.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
A part of this work is subject to a UK patent application by the University of Southampton.
References
- 1.Wahl MC, Will CL. and Luhrmann R. (2009). The spliceosome: design principles of a dynamic RNP machine. Cell 136:701–718 [DOI] [PubMed] [Google Scholar]
- 2.Le Hir H, Nott A. and Moore MJ. (2003). How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 28:215–220 [DOI] [PubMed] [Google Scholar]
- 3.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP. and Burge CB. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lev-Maor G, Sorek R, Shomron N. and Ast G. (2003). The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300:1288–1291 [DOI] [PubMed] [Google Scholar]
- 5.Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J. and Lopez AJ. (2005). Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics 170:661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang D. and Wilusz JE. (2014). Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28:2233–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palazzo AF. and Lee ES. (2015). Non-coding RNA: what is functional and what is junk? Front Genet 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maniatis T. and Reed R. (2002). An extensive network of coupling among gene expression machines. Nature 416:499–506 [DOI] [PubMed] [Google Scholar]
- 9.Shcherbakova I, Hoskins AA, Friedman LJ, Serebrov V, Correa IR, Jr, Xu MQ, Gelles J. and Moore MJ. (2013). Alternative spliceosome assembly pathways revealed by single-molecule fluorescence microscopy. Cell Rep 5:151–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun H. and Chasin LA. (2000). Multiple splicing defects in an intronic false exon. Mol Cell Biol 20:6414–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busslinger M, Moschonas N. and Flavell RA. (1981). Beta+thalassemia: aberrant splicing results from a single point mutation in an intron. Cell 27:289–298 [DOI] [PubMed] [Google Scholar]
- 12.Kervestin S. and Jacobson A. (2012). NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol 13:700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peccarelli M. and Kebaara BW. (2014). Regulation of natural mRNAs by the nonsense-mediated mRNA decay pathway. Eukaryot Cell 13:1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, et al. (2013). Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154:583–595 [DOI] [PubMed] [Google Scholar]
- 15.Pimentel H, Parra M, Gee SL, Mohandas N, Pachter L. and Conboy JG. (2016). A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res 44:838–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemeroff ME, Utans U, Kramer A. and Krug RM. (1992). Identification of cis-acting intron and exon regions in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting presplicing complexes. Mol Cell Biol 12:962–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei H. and Vorechovsky I. (2005). Identification of splicing silencers and enhancers in sense Alus: a role for pseudo-acceptors in splice site repression. Mol Cell Biol 25:6912–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kralovicova J, Knut M, Cross NC. and Vorechovsky I. (2016). Exon-centric regulation of ATM expression is population-dependent and amenable to antisense modification by pseudoexon targeting. Sci Rep 6:18741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aartsma-Rus A. and van Ommen GJ. (2007). Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA 13:1609–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauman J, Jearawiriyapaisarn N. and Kole R. (2009). Therapeutic potential of splice-switching oligonucleotides. Oligonucleotides 19:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilton SD. and Fletcher S. (2010). Splice modification to restore functional dystrophin synthesis in Duchenne muscular dystrophy. Curr Pharm Des 16:988–1001 [DOI] [PubMed] [Google Scholar]
- 22.Lim LP. and Burge CB. (2001). A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci U S A 98:11193–11198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hua Y, Vickers TA, Baker BF, Bennett CF. and Krainer AR. (2007). Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol 5:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kralovicova J, Lages A, Patel A, Dhir A, Buratti E, Searle MS. and Vorechovsky I. (2014). Optimal antisense target reducing INS intron 1 retention is adjacent to a parallel G quadruplex. Nucleic Acids Res 42:8161–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, et al. (2016). Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13:508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomakuchi TT, Rigo F, Aznarez I. and Krainer AR. (2016). Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat Biotechnol 34:164–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruskin B, Zamore PD. and Green MR. (1988). A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52:207–219 [DOI] [PubMed] [Google Scholar]
- 28.Zamore PD. and Green MR. (1989). Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A 86:9243–9247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S, Romfo CM, Nilsen TW. and Green MR. (1999). Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832–835 [DOI] [PubMed] [Google Scholar]
- 30.Kralovicova J, Knut M, Cross NC. and Vorechovsky I. (2015). Identification of U2AF(35)-dependent exons by RNA-Seq reveals a link between 3′ splice-site organization and activity of U2AF-related proteins. Nucleic Acids Res 43:3747–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao C, Yang B, Wu T, Huang J, Tang P, Zhou Y, Zhou J, Qiu J, Jiang L, et al. (2014). Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol 21:997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G. and Smith CW. (2010). Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol 17:1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M, Jr, and Fu XD. (2013). Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell 50:223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiloh Y. and Ziv Y. (2013). The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 14:197–210 [PubMed] [Google Scholar]
- 35.Smith AF, Hubley R. and Green P. (1996). RepeatMasker Open-3.0
- 36.Hiller M, Zhang Z, Backofen R. and Stamm S. (2007). Pre-mRNA secondary structures influence exon recognition. PLoS Genet 3:e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno PM, Santos JC, Gomes CP, Varela-Moreira A, Costa A, Leiro V, Mansur H. and Pêgo AP. (2016). Delivery of splice switching oligonucleotides by amphiphilic chitosan-based nanoparticles. Mol Pharm 13:344–356 [DOI] [PubMed] [Google Scholar]
- 38.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, et al. (2001). Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- 39.Kralovicova J, Hwang G, Asplund AC, Churbanov A, Smith CI. and Vorechovsky I. (2011). Compensatory signals associated with the activation of human GC 5′ splice sites. Nucleic Acids Res 39:7077–7091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rainey MD, Charlton ME, Stanton RV. and Kastan MB. (2008). Transient inhibition of ATM kinase is sufficient to enhance cellular sensitivity to ionizing radiation. Cancer Res 68:7466–7474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buschmann MD, Merzouki A, Lavertu M, Thibault M, Jean M. and Darras V. (2013). Chitosans for delivery of nucleic acids. Adv Drug Deliv Rev 65:1234–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomes CP, Lopes CDF, Moreno PM, Varela-Moreira A, Alonso MJ. and Pêgo AP. (2014). Translating chitosan to clinical delivery to nucleic acid-based drugs. MRS Bull (Cambridge University Press) 39:60–70 [Google Scholar]
- 43.Kang SH, Cho MJ. and Kole R. (1998). Up-regulation of luciferase gene expression with antisense oligonucleotides: implications and applications in functional assay development. Biochemistry 37:6235–6239 [DOI] [PubMed] [Google Scholar]
- 44.Sorek R, Lev-Maor G, Reznik M, Dagan T, Belinky F, Graur D. and Ast G. (2004). Minimal conditions for exonization of intronic sequences: 5′ splice site formation in Alu exons. Mol Cell 14:221–231 [DOI] [PubMed] [Google Scholar]
- 45.Lei H, Day INM. and Vorechovsky I. (2005). Exonization of AluYa5 in the human ACE gene requires mutations in both 3′ and 5′ splice sites and is facilitated by a conserved splicing enhancer. Nucleic Acids Res 33:3897–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vorechovsky I. (2010). Transposable elements in disease-associated cryptic exons. Hum Genet 127:135–154 [DOI] [PubMed] [Google Scholar]
- 47.Nozu K, Iijima K, Ohtsuka Y, Fu XJ, Kaito H, Nakanishi K. and Vorechovsky I. (2014). Alport syndrome caused by a COL4A5 deletion and exonization of an adjacent AluY. Mol Genet Genomic Med 2:451–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY. and Ast G. (2008). Intronic Alus influence alternative splicing. PLoS Genet 4:e1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jurka J, Kapitonov VV, Klonowski P, Walichiewicz J. and Smit AF. (1996). Identification of new medium reiteration frequency repeats in the genomes of primates, rodentia and lagomorpha. Genetica 98:235–247 [DOI] [PubMed] [Google Scholar]
- 50.Kaplan DJ, Jurka J, Solus JF. and Duncan CH. (1991). Medium reiteration frequency repetitive sequences in the human genome. Nucleic Acids Res 19:4731–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levy A, Sela N. and Ast G. (2008). TranspoGene and microTranspoGene: transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res 36:D47–D52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vorechovsky I. (2010). MER91B-assisted cryptic exon activation in Gitelman syndrome. Pediatr Res 67:444–445 [DOI] [PubMed] [Google Scholar]
- 53.Min J, Guo K, Suryadevara PK, Zhu F, Holbrook G, Chen Y, Feau C, Young BM, Lemoff A, et al. (2016). Optimization of a novel series of ataxia-telangiectasia mutated kinase inhibitors as potential radiosensitizing agents. J Med Chem 59:559–577 [DOI] [PubMed] [Google Scholar]
- 54.Coutinho G, Xie J, Du L, Brusco A, Krainer AR. and Gatti RA. (2005). Functional significance of a deep intronic mutation in the ATM gene and evidence for an alternative exon 28a. Hum Mutat 25:118–124 [DOI] [PubMed] [Google Scholar]
- 55.Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M. and Smith CW. (2005). Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans 33:457–460 [DOI] [PubMed] [Google Scholar]
- 56.Yuan ZY, Hu YL. and Gao JQ. (2015). Brain localization and neurotoxicity evaluation of polysorbate 80-modified chitosan nanoparticles in rats. PLoS One 10:e0134722. [DOI] [PMC free article] [PubMed] [Google Scholar]