Abstract
Sendai virus (SeV), a murine paramyxovirus, has been used to study the induction of type I interferon (IFN) subtypes in robust quantities. Few studies have measured whether the IFN that SeV induces actually fulfills its intended purpose of interfering with virus-mediated effects in the cells in which it is produced. We determined the effects of IFN on SeV-mediated cytopathic effects (CPE) and the ability of IFN to protect against virus infection. SeV-induced biologically active IFN resulted in Jak/STAT activation and the production of a number of interferon-stimulated genes (ISGs). However, these responses did not inhibit SeV replication or CPE. This observation was not due to SeV effects on canonical IFN signaling. Furthermore, pretreating cells with type I IFN and establishing an antiviral state before infection did not mediate SeV effects. Therefore, the induction of canonical IFN signaling pathways and ISGs does not always confer protection against the IFN-inducing virus. Because type I IFNs are approved to treat various infections, our findings suggest that typical markers of IFN activity may not be indicative of a protective antiviral response and should not be used alone to determine whether an antiviral state against a particular virus is achieved.
Keywords: : interferon, Sendai virus, antiviral state, signaling
Introduction
Type I interferons (IFNs) are produced early upon virus infection as a first line of defense. In humans, at the protein level, type I IFNs consist of 12 different IFN-α subtypes, IFN-β, IFN-ɛ, IFN-κ, and IFN-ω. These IFNs induce hundreds of interferon-stimulated genes (ISGs), which inhibit various stages of virus replication as well as enhance the host−response. It has been reported that distinct type I IFN subtypes have differential capabilities to exert their antiviral effects in a cell type and virus-specific manner (Lavoie and others 2011; Sperber and others 1992).
Type I IFNs are induced upon stimulation of pattern recognition receptors (PRRs), including Toll-like receptors and cytosolic receptors, with their cognate pathogen-associated molecular patterns. Depending on which PRR is stimulated, a series of signaling pathways are activated that eventually converge upon the activation of various transcription factors, including IFN regulatory factors (IRFs) 3 and/or 7 and NF-kB. These activated transcription factors translocate into the nucleus where they induce the transcription of early type I IFN subtypes (Génin and others 2009). These subtypes are then secreted and bind to the type I IFN receptor (IFNAR) on cells in an autocrine and paracrine manner.
Receptor engagement with type I IFN leads to the phosphorylation and activation of the receptor-associated Janus kinases Jak1 and Tyk2. These kinases phosphorylate the receptor creating SH2 docking sites for STATs 1 and 2, where they in turn become phosphorylated. The phosphorylated STATs complex with IRF-9 to form the ISGF3 complex, which translocates into the nucleus and induces the transcription of hundreds of ISGs that are the hallmark of the antiviral response (Yoneyama and others 1996). The transcription factor IRF-7 also becomes induced during this phase of signaling and aids in the amplification of the IFN response by inducing the transcription of additional type I IFN subtypes (Marie and others 1998). This signaling occurs in a positive feedback loop until negative regulators of IFN signaling, for example, suppressors of cytokine signaling, are activated to down regulate the response (Sato and others 1998).
The murine paramyxovirus Sendai virus (SeV) has long been considered an extremely potent inducer of type I IFNs and was used in some of the pioneering studies that first isolated type I IFNs from human leukocytes and lymphoblastoid cells (Zoon and others 1978; Cantell and others 1981; Nyman and others 1998). Because of the large amounts of IFN produced upon SeV infection, it could be assumed that IFN plays a role in creating an antiviral environment that restricts SeV replication. However, there have been some conflicting reports concerning SeV and the role it plays in the IFN response.
Some groups have reported that the C protein encoded by SeV functions by inhibiting STAT phosphorylation and subsequent Jak/STAT signaling (Komatsu and others 2000; Gotoh and others 2003; Kato and others 2004). In this case, type I IFNs are induced but cannot signal, which may suggest that the large amounts of type I IFNs produced may be compensating for its restricted ability to signal. However, others have reported that a subset of genes that SeV induces are stimulated by type I IFN signaling downstream of the IFNAR, as deletion of Jak1, one of the Janus kinases associated with the IFNAR and necessary for downstream signaling, inhibited the induction of these genes (Elco and others 2005). This suggests that IFN signaling leading to the induction of ISGs is intact during SeV infection. Therefore, the effects of SeV-induced type I IFNs are unclear.
In this report, we sought to determine whether the type I IFN that is induced upon SeV infection played a role in creating an antiviral environment that restricted virus replication and enhanced cell survival. We looked at 3 different human cell lines, U937, a monocytoid line, A549, an epithelial cell line, and Namalwa, a B cell lymphoblastoid line, as well primary human monocytes. We found that SeV infection leads to the induction of biologically active IFN that was capable of signaling through the Jak/STAT pathway and inducing a number of ISGs and their respective proteins, indicating that viral proteins were not preventing their production. However, these ISGs were unable to protect the cells against SeV-induced cytopathic effects (CPE).
Pretreating the cells with type I IFNs was also unable to confer protection against SeV infection, even though protection against vesicular stomatitis virus (VSV) and/or encephalomyocarditis virus (EMCV) was apparent. Because Jak/STAT signaling and several IFN-stimulated genes and proteins were induced before virus infection, these results further indicate that the inability of IFN to protect against SeV infection was not due to a viral protein halting canonical IFN signaling. Taken together, these results indicate that markers of IFN signaling, such as Jak/STAT activation and the induction of ISGs and proteins, are not necessarily indicative of a functional antiviral response against certain viral infections. This could have major implications in determining the efficacy of type I IFN therapy for viral infections in the clinic.
Materials and Methods
Cell culture and reagents
U937, Namalwa, and A549 cell lines were obtained and confirmed from American Type Culture Collection (ATCC, Manasass, VA). These cell lines were maintained in RPMI 1640 + 10% fetal bovine serum (FBS) at 37°C, 5% CO2 and passaged every 3–4 days when the cell concentration reached about 1 × 106 cells/mL. Due to differences seen in virus-mediated effects in cells passaged more than 6 times, only cells within passage numbers 2 and 5 were used for experiments. Elutriated primary monocytes from anonymous healthy volunteers were obtained from the Department of Transfusion Medicine at the Clinical Center, National Institutes of Health. Only cells that had a purity of >90% were used for experiments. Monocytes were maintained in RPMI 1640 media +10% FBS at a concentration of 5 × 105 cells/mL for the duration of the experiments.
Purified IFN-α subtypes and recombinant IFN-ω were obtained from PBL Laboratories (Piscataway, NJ). IFN-β was obtained from Millipore (Temecula, CA). The neutralizing antibody to one subunit of the IFN-lambda receptor (IFNLR), human IL-10Rβ, was purchased from R&D Systems (Minneapolis, MN) and used to neutralize type III IFN subtypes. The IFNAR2 neutralizing antibody was produced by Precision Antibody (Columbia, MD) and used to neutralize type I IFN subtypes. Neutralizing antibody treatment of cells was performed as previously described at a concentration of 1:100 (antibody:total cell culture volume) for all experiments (Zaritsky and others 2015).
Viruses and infections
All infections were performed when the cells were between passages 2 and 5. Three independent biological replicates were performed for each infection.
Sendai virus
SeV Cantell strain was obtained from Charles River Laboratories (Frederick, MD) at a stock concentration of 4,000 hemagglutination units/mL (HA U/mL). Suspension cells (U937, Namalwa, and primary monocytes) were plated at 5 × 105 cells/mL just before infection. Adherent cells (A549) were plated at 5 × 105 cells/mL 4 h before infection to allow time for the cells to adhere. One hundred fifty HA unit/milliliter were used to infect cells in normal culture media, which equated to a multiplicity of infection (MOI) of 5, unless otherwise indicated. After 1 h, the virus was washed off with 1× phosphate-buffered saline (PBS) pH 7.4 and the media were changed to RPMI 1640 + 10% FBS.
The supernatants were sampled at the indicated time points and virus levels were quantitated using the HA assay. Briefly, 2-fold dilutions of the supernatants in 50 μL were made across a 96-well plate, and 50 μL of a solution of 0.5% chicken red blood cells (Fitzgerald, Acton, MD) were added. The plate was incubated at room temperature for about 60 min and the wells were assessed as either positive or negative for hemagglutinin activity. The virus titer was the reciprocal of the dilution in which HA occurred.
VSV and EMCV
VSV Indiana strain and EMCV (ATCC) were grown in Vero cells and titered via the plaque assay. The Vero cell supernatants were used to infect cells in RPMI 1640 + 10% FBS. The supernatants from the virus-infected cells were sampled at the indicated time points, and virus levels were quantitated via TCID50 assay. Briefly, Vero cells were plated in 96-well plates at a concentration of 3 × 105 cells/well (100 μL). After 4 h, 10 μL of the virus-infected cell supernatants were added to the first row of wells and then 10-fold serial dilutions were made down the plate. After 72 h, the cells were stained with crystal violet (Sigma, St. Louis, MO) to assess viability.
Antiviral assays
The supernatants from the various viral infections were incubated with 0.05% β-propiolactone (Sigma) for 16 h at 4°C to inactivate any virus that was present (Barrett and others 1984). The following day, the treated supernatants were incubated at 37°C for 2 h to inactivate the chemical. Controls were run to ensure that the β-propiolactone treatment did not affect cell viability or interfere with the antiviral activity of IFNs (Zaritsky and others 2015).
U937 and A549 cells were plated in 96-well plates at a concentration of 3 × 104 cells/well (100 μL). After 4 h, cells were pretreated with 10 μL of the virus-inactivated supernatants or the indicated amount of purified IFN (PBL Assay Science, Piscataway, NJ) for 16–24 h at 37°C. The cells were then infected with either VSV or EMCV for 72 h, at which time cell viability was measured via crystal violet staining for adherent cells or the MTT assay for suspension cells. Briefly, MTT was dissolved in sterile 1× PBS pH 7.4 at a concentration of 5 μg/mL. Ten microliters of the MTT solution was added to each well, and the plates were incubated at 37°C for 4 h. The precipitate that formed was dissolved in 150 μL of acidified isopropanol, which was a stock consisting of 100 mL isopropanol + 660 μL concentrated HCl. The absorbance was then measured for both suspension and adherent cells at 570 nm using a Hidex Sense plate reader.
RNA isolation
At the indicated time points, the suspension cells (U937, Namalwa and primary monocytes) were pelleted by spinning at 1,000 rpm using a Beckman Coulter centrifuge at 4°C, washed 1× with PBS pH 7.4 and then resuspended in 350 μL of RLT lysis buffer from the Qiagen RNeasy plus mini kit (Qiagen, Germantown, MD). For adherent cells (A549), 350 μL of RLT buffer was added directly to the washed well and then transferred to a tube. RNA was isolated as per the manufacturer's instructions. The isolated RNA was quantified using the Nanodrop 1000 and then stored at −80°C until further use.
cDNA synthesis
A stock of cDNA was made using 1.5 μg of starting RNA. The RNA was reverse transcribed using the superscript III reverse transcriptase kit as per the manufacturer's instructions (Thermo Fisher, Waltham, MA). After reverse transcription, the cDNA was diluted to a concentration of 50 ng/10 μL in water. The cDNA was stored at −20°C until further use.
Quantitative polymerase chain reaction
A total of 10 μL of the cDNA stock (50 ng of starting RNA) was used for each quantitative polymerase chain reaction (qPCR) reaction. The cDNA was mixed with 10 μL of SensiFAST probe No-Rox TaqMan reaction mix (Bioline, Taunton, MA), 0.075 μL of forward and reverse primers (100 μM), and 0.025 μL of TaqMan probe (100 μM) in 96-well plates and then run according to the following protocol: 95°C for 5 min for polymerase activation, and then 40 cycles of denaturation at 95°C for 10 s followed by annealing/extension at 60°C for 50 s. For each reaction, the gene Hypoxanthine-guanine phosphoribosyltransferase (HPRT) was multiplexed with the genes of interest as an internal housekeeping control. Reactions were run on a CFX96 thermocycler (Bio-Rad, Hercules, CA) and analyzed using the CFX Manager software version 2.1 (Bio-Rad). All qPCR data are expressed as the % relative expression of HPRT.
Primer and probe sequences (5′-3′):
IFIT3: F- CACTGTCTTCCTTGAATAAGTTCC, R- AAGGAACAAATCAGCCTGGTCA, probe-56-FAM/CTGCCCTCT/ZEN/GTGTCTCTGGCTGTT; ISG15: F- CGAACTCATCTTTGCCAGTACA, R- GCCTTCAGCTCTGACACC, probe- 5HEX/CACCTGGAA/ZEN/TTCGTTGCCCGC; Mx1: F- CCACCCATATTTCAGGGATCTG, R- TCTGGTGAGTCTCCTTGATTTG, probe- TGTGTGATGAGCTCGCTGGTAAGTTT; IRF-7: F- TCCCCACGCTATACCATCTAC, R- GAAGACACACCCTCACGC, probe- TTCCAGCTTCACCAGGACCAGG; HPRT: F- GTATTCATTATAGTCAAGGGCATATCC, R- AGATGGTCAAGGTCGCAAG, probe- TGGTGAAAAGGACCCCACGAAGT.
Whole cell protein lysates
At the indicated time points, the cells were pelleted by spinning at 1,000 rpm using a Beckman Coulter Allegra X-15R centrifuge at 4°C for 5 min. The pellet was resuspended in 200 μL of radioimmunoprecipitation assay (RIPA) buffer [0.25% sodium deoxycholate, 0.1% sodium dodecyl chloride, 25 mM Tris (pH 7.4), 150 mM sodium chloride, 1 mM ethylenediaminetetraaceticacid (EDTA), and 1% NP40] with a 1:100 dilution of Halt protease and phosphatase inhibitors (Sigma). The samples were then sonicated for 5 s at an amplitude of 21% using a Vibra cell sonicator (Sonics, Newtown, CT) and then incubated on ice for 30 min. The lysates were spun at 14,000 rpm at 4°C for 20 min and quantitated using the BCA protein assay (Pierce, Waltham, MA) according to the manufacturer's instructions. Ten micrograms aliquots were stored at −80°C until further use.
Nuclear and cytoplasmic protein lysate preparation
The cells were harvested at the indicated time points and pelleted by spinning at 1,000 rpm at 4°C using a Beckman Coulter Allegra X-15R centrifuge. The supernatants were removed, and the pellets were washed 1× with PBS pH 7.4 at 4°C. The cells were then resuspended in 400 μL of the cytoplasmic buffer [10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT), and 0.05% NP40] with Halt protease and phosphatase inhibitors (Pierce) added just before use (1:100 for each). The lysates were incubated on ice for 15 min, and then spun at 12,000 rpm in a Beckman Coulter Microfuge 22R microcentrifuge at 4°C for 5 min.
The supernatants, which contained the cytoplasmic fractions, were transferred to a different tube, and the nuclear pellets were washed once with PBS pH 7.4. The nuclear pellets were lysed in 100 μL of the nuclear lysis buffer [5 mM HEPES pH 7.9, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 26% glycerol (v/v), and 300 mM NaCl] with Halt protease and phosphatase inhibitors (Pierce) added just before use. The nuclear lysates were sonicated for 5 s at an amplitude of 21%, incubated on ice for 30 min, and were spun at 12,000 rpm at 4°C. The supernatants, which contained the cleared nuclear lysates, were transferred to a different tube. Both the nuclear and cytoplasmic lysates were quantitated using the BCA protein assay (Pierce) according to the manufacturer's protocol and 10 ug aliquots were stored at −80°C until further use.
Western blot analysis
Ten micrograms aliquots of the protein lysates were used for western blot analysis. To each aliquot, 1 μL of reducing agent (Invitrogen, San Diego, CA) and the appropriate amount of 4× loading dye (Invitrogen) were added. The samples were then boiled for 5 min and loaded onto 10% bis-tris mini gels (Invitrogen). The gels were run at 200 V for 55 min and then transferred onto nitrocellulose membranes via the iBlot apparatus (Invitrogen) according to the manufacturer's protocol.
The membrane was then blocked in 5% milk in tris-buffered saline (pH 7.4) with 0.1% tween 20 (TBST) for 1 h at room temperature. The blots were then incubated overnight at 4°C in primary antibody that was diluted in 5% milk in TBST. The next morning, the blots were washed 3× for 10 min in TBST and then incubated in a horseradish peroxidase-conjugated secondary antibody of the appropriate isotype (Santa Cruz, Dallas, TX) diluted 1:5,000 in 5% milk in TBST for 1 h. The membranes were washed 3× for 10 min in TBST and then developed using the SuperSignal West Femto Maximum Sensitivity Substrate (Pierce). Chemiluminescence was visualized using the Fujifilm LAS-3000 camera.
Antibodies and dilutions
The following antibodies and dilutions were used for the western blot analyses:
Monoclonal rabbit anti-pIRF-3(S386; Abcam, Cambridge, MA): 1:5,000; monoclonal anti-IRF-3 (Cell Signaling Technologies, Danvers, MA): 1:1,000; polyclonal rabbit anti-IRF-7 (Santa Cruz Biotechnologies, Dallas, TX): 1:1,000; polyclonal rabbit anti-STAT1 (Santa Cruz Biotechnologies): 1:1,000; monoclonal mouse anti-pSTAT1(Y701; BD Transduction Laboratories, San Jose, CA): 1:5,000; polyclonal rabbit anti-STAT2 (Santa Cruz Biotechnologies): 1:1,000; polyclonal rabbit anti-pSTAT2(Y689; End Millipore, Billerica, MA): 1:2,000; monoclonal mouse anti-IRF-9(ISGF3γ; BD Transduction Laboratories): 1:1,000; monoclonal mouse anti-Actin (Cell Signaling Technologies): 1:5,000; polyclonal rabbit anti-β tubulin (Cell Signaling Technologies): 1:2,000; mouse monoclonal RNA polymerase II (Santa Cruz Biotechnologies): 1:1,000; rabbit polyclonal IFIT3 (Covance, Inc., Denver, PA) 1:1,000; rabbit polyclonal ISG15: 1:1,000 (Santa Cruz Biotechnologies); mouse polyclonal Mx1: 1:1,000 (Abnova, Taipei, Taiwan).
Results
SeV induces biologically active type I IFN in human cell lines and primary monocytes
We first determined whether SeV infection induced the production of biologically active IFN in different human cell types, including primary monocytes and the following 3 cell lines: U937, a monocytoid cell line, A549, an epithelial cell line, and Namalwa, a B lymphoblastoid line. These cell types were used to determine whether the ability of SeV to induce type I IFNs was cell type dependent.
The cells were infected with SeV (150 HA U/mL corresponding to a MOI of 5) for either 16 or 24 h and then the supernatants were harvested. The virus that was present in the supernatants was inactivated using β-propiolactone (Barrett and others 1984), and then the supernatants were used to pretreat fresh cells for 24 h. These cells were then infected with EMCV or VSV for 72 h, and the ability of the supernatants to protect against EMCV/VSV-mediated CPE was assessed via an MTT assay for cell viability.
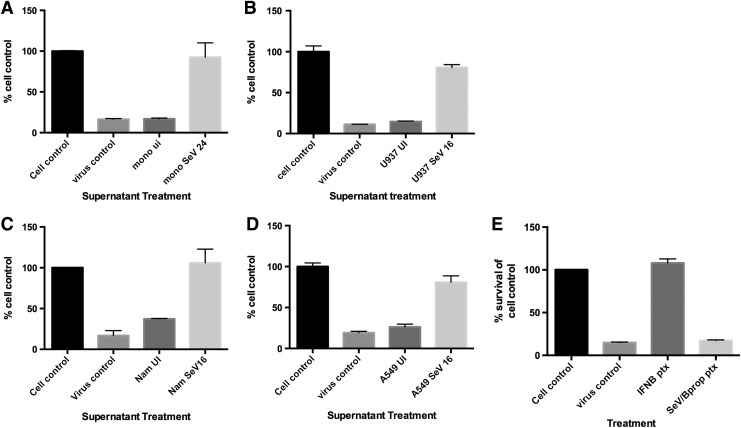
For all cell types, we found that supernatants from the SeV-infected but not uninfected cells were able to protect against EMCV/VSV-mediated CPE, indicating the presence of type I IFNs (Fig. 1A–D). Similar results were obtained when all cells were infected with 15 HA U/mL of SeV (MOI 0.5) (data not shown), indicating that even low levels of SeV can induce biologically active IFN. Supernatants from cells treated with inactivated SeV by β-propiolactone were not protective against VSV/EMCV-mediated CPE, indicating that the inactivated SeV did not induce IFN in the cells during the antiviral assay (Fig. 1E).
FIG. 1.
Multiple cell types produce biologically active type I IFNs upon SeV infection. (A) Supernatants from primary monocytes that were either UI or infected with SeV (150 HA U/mL) for 24 h were treated with β-propiolactone to inactivate the virus and then used to pretreat fresh U937 cells for 24 h. The U937 cells were then infected with VSV for 72 h and cell viability was measured via MTT assay. Cell viability is shown as the % of cell control. Cell control: UI, untreated. Virus control: VSV infected, untreated. Supernatants from UI or SeV infected (B) U937, (C) Namalwa, or (D) A549 cells for 16 h were treated with β-propiolactone to inactive the virus and then used to pretreat fresh U937 cells for 24 h. The U937 cells were then infected with EMCV for 72 h and cell viability was measured via MTT assay. Cell viability is shown as the % of cell control. Cell control: UI, untreated. Virus control: EMCV infected, untreated. Data shown are from 3 independent biological replicates. (E) U937 cells were pretreated with IFN-β (100 U/mL) or supernatants from U937 cells treated with β-propiolactone-inactivated SeV (150 HA U/mL) for 24 h. The U937 cells were then infected with EMCV for 72 h and cell viability was measured via MTT assay. Cell control: UI, untreated. Virus control: EMCV infected, untreated, IFNB ptx: pretreated with IFN-β; SeV/Bprop ptxt: treated with inactivated SeV by β-propiolactone. SeV, Sendai virus; IFN, interferon; UI, uninfected; HA, hemagglutination; VSV, vesicular stomatitis virus; EMCV, encephalomyocarditis virus.
SeV-induced type I IFN activates Jak/STAT signaling and ISG induction
Because many viruses have evolved to interfere with the innate immune response and several studies have reported that SeV viral proteins are able to interfere with IFN signaling (Komatsu and others 2000; Gotoh and others 2003; Kato and others 2004), we assessed the regulation of IRF proteins that are involved in type I IFN induction and if the SeV-induced type I IFNs were able to signal through the IFNAR to activate an antiviral response during virus infection. Cells were infected with SeV for 4, 8, 16, and 24 h, and protein and RNA were harvested to measure levels of IRFs, Jak/STAT signaling modulators, and IFN induced genes and proteins.
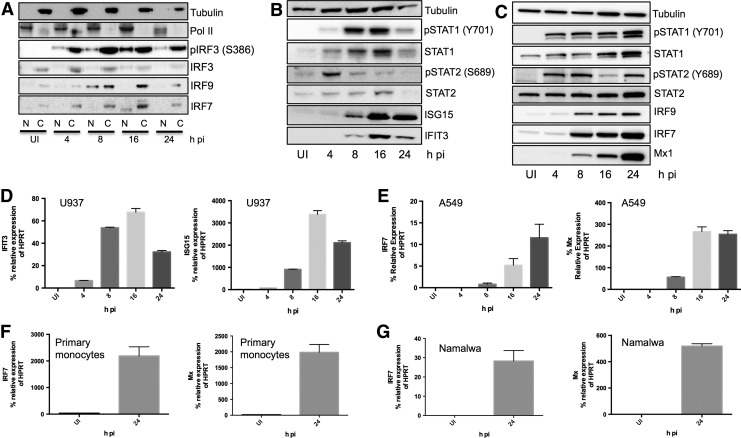
We measured the expression of phosphorylated and total IRF-3, which has been shown to induce early type I IFN subtypes in response to virus infection (Génin and others 2009), and several signaling proteins that are activated upon Jak/STAT signaling downstream of the IFNAR, including pSTAT1(Y701), pSTAT2(Y689), IRF-9 and IRF-7. In U937 cells, we found that SeV infection led to the phosphorylation and nuclear translocation of IRF-3 at 4 h postinfection (pi) and the induction and nuclear accumulation of IRF-9 at 8 h and IRF-7 at 16 h pi (Fig. 2A). Nuclear IRF-7 appears to be at a slightly lower molecular weight than the form found in the cytoplasm, most likely due to the protein modifications that occur to enable nuclear trafficking.
FIG. 2.
SeV-induced IFN production activates Jak/STAT signaling and ISG induction. (A) U937 cells were infected with SeV (150 HA U/mL) for 4, 8, 16, and 24 h. Nuclear and cytoplasmic protein extracts were prepared and western blots probing for pIRF-3(S386), IRF-3, IRF-7, and IRF-9 were performed. RNA pol II and β-tubulin were the nuclear and cytoplasmic loading controls, respectively. N, nuclear, C, cytoplasmic. (B) U937 cells were infected with SeV (150 HA U/mL) for 4, 8, 16, and 24 h and whole cell protein lysates were prepared. Western blots probing for pSTAT1(Y701), STAT1(total), pSTAT2(Y689), STAT2(total), ISG15, and IFIT3, and tubulin as the loading control were performed. (C) A549 cells were infected with SeV (150 HA U/mL) for 4, 8, 16, and 24 h and whole protein lysates were prepared. Western blots probing for pSTAT1(Y701), STAT1(total), pSTAT2(Y689), STAT2(total), IRF-9, IRF-7, and Mx1, and tubulin as the loading control were performed. (D) ISG15 and IFIT3 mRNA expression in U937 cells infected with SeV (150 HA U/mL) for 4, 8, 16, and 24 h. (E) IRF-7 and Mx1 mRNA expression in A549 cells infected with SeV (150 HA U/mL) for 4, 8, 16, and 24 h. IRF-7 and Mx1 mRNA induction in (F) primary monocytes and (G) Namalwa cells infected with SeV (150 HA U/mL) for 24 h. All gene induction data are expressed as the % of relative HPRT expression. All data are representative of 3 independent biological replicates. UI, uninfected, untreated. IRF, IFN regulatory factor; ISG, interferon-stimulated gene; HPRT, hypoxanthine-guanine phosphoribosyltransferase.
We also found that SeV infection led to the phosphorylation of both STATs 1 and 2, with increased STAT2 phosphorylation at 4 h followed by increased STAT1 phosphorylation at 8 and 16 h pi (Fig. 2B). Phosphorylation and expression levels of STAT2 decreased at 24 h pi. However, this decrease in STAT2 expression at 24 h was also observed when U937 cells were infected with EMCV and VSV (data not shown). Namalwa cells, on the other hand, expressed pSTAT2 and STAT2 at 24 h pi with SeV, EMCV, and VSV (data not shown), indicating STAT phosphorylation and expression levels in response to virus infection may be cell type dependent. In A549 cells, SeV infection led to phosphorylation of STATs 1 and 2 along with induction of STATs 1 and 2, IRF-9, and IRF-7 (Fig. 2C). Together, these data indicate that Jak/STAT signaling downstream of IFNAR was activated by SeV-induced type I IFNs.
RNA and protein ISG induction was also detected upon SeV infection. In U937 cells, IFIT3 and ISG15 protein was upregulated at 8 h pi (Fig. 2B) and RNA was upregulated at all time points tested (Fig. 2D). Increased expression of Mx1 and IRF-7 was detected in U937 cells (data not known) (Zaritsky and others 2015) and in A549 cells at 8 h pi with SeV (Fig. 2C, E). We also detected the induction of Mx1 and IRF-7 in primary monocytes along with Namalwa cells that were infected with SeV for 24 h, indicating functional IFN signaling in these cell types (Fig. 2F, G). These results demonstrate that SeV infection is able to activate Jak/STAT signaling and induce several antiviral ISGs and proteins.
SeV-induced IFNs and type I IFN subtypes are unable to protect against SeV infection
Because SeV was able to activate the canonical IFN signaling pathway, induce ISGs, and produce biologically active IFN in the cell types tested, we sought to determine whether this IFN was functional in protecting cells against the infecting virus, SeV. We first pretreated cells with β-propiolactone-treated supernatants from the 4 SeV-infected cell types (U937, A549, Namalwa, and primary monocytes infected with 150 HA U/mL of SeV) to prime the fresh cells with the specific IFNs that SeV induces. After 24 h of pretreatment, the cells were infected with SeV for 72 h (150 HA U/mL), and then cell viability was measured to determine whether inducing the IFN response before SeV infection could protect against SeV-mediated CPE.
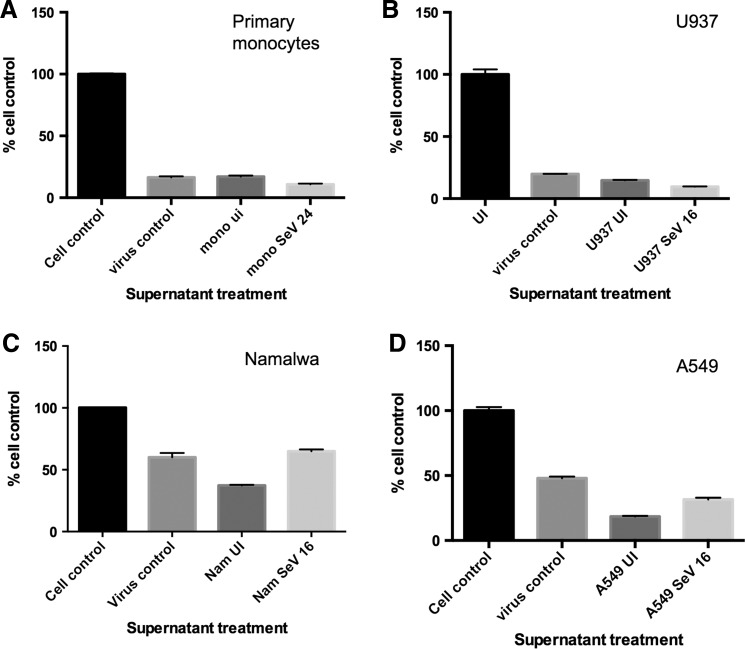
We found that none of the supernatants from any of the cells types were able to protect against SeV-mediated CPE (Fig. 3A–D), which is in contrast to our results in Fig. 1, where we found that these same supernatants were able to protect against VSV and/or EMCV-mediated CPE. These results indicate that SeV-induced biologically active IFN that was able to activate the canonical IFN pathway and ISGs was unable to protect against the very virus that is responsible for its induction, even when that antiviral response is initiated before infection. These results also suggest that the inability of IFN signaling to protect against SeV is not due to viral proteins blocking Jak/STAT signaling, since inducing the IFN-mediated antiviral response before virus infection did not result in protection.
FIG. 3.
Type I IFNs induced by SeV do not protect against SeV-induced CPE. (A) Supernatants from primary monocytes that were either UI or infected with SeV (150 HA U/mL) for 24 h were treated with β-propiolactone to inactivate the virus and then used to pretreat fresh U937 cells for 24 h. The U937 cells were then infected with SeV (150 HA U/mL) for 72 h and cell viability was measured via MTT assay. Results are shown as the % of cell control. Supernatants from (B) U937, (C) Namalwa, or (D) A549 cells that were UI or infected with SeV for 16 h were treated with β-propiolactone to inactive the virus and then used to pretreat fresh U937 cells for 24 h. The cells were then infected with SeV (150 HA U/mL) for 72 h and cell viability was measured via MTT assay. Results are shown as the % of cell control and are representative of 3 independent biological replicates. CPE, cytopathic effects.
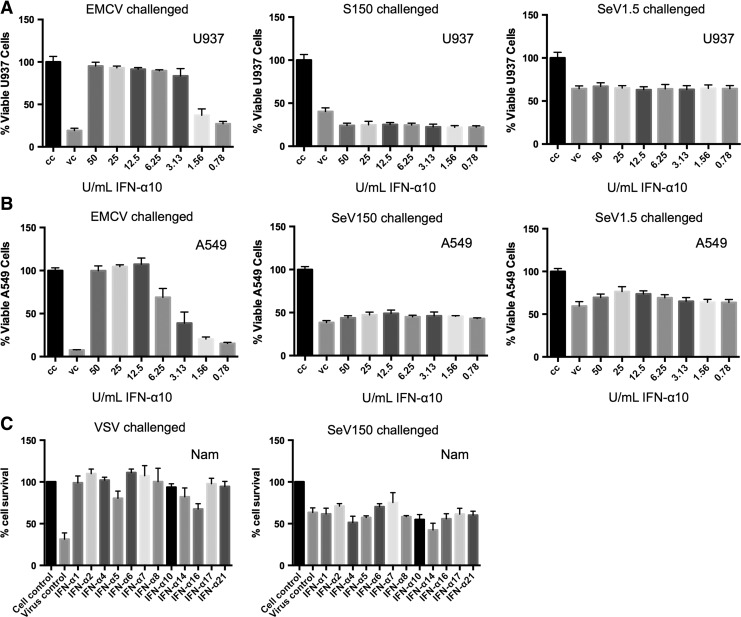
These observations were recapitulated when U937, A549, and Namalwa cells were pretreated with purified type I IFN subtypes before infection. U937 and A549 cells were pretreated with serial dilutions of purified type I IFN subtypes for 24 h and were then challenged for 72 h with EMCV (MOI 0.5) or SeV (150 or 1.5 HA U/mL corresponding to MOIs of 5 and 0.05). We found that type I IFN subtype pretreatment was able to protect cells against EMCV-mediated CPE in a dose-dependent manner (Fig. 4A, B). Cells that were pretreated with IFN and challenged with SeV, however, were not protected against SeV-mediated CPE at either MOI (Fig. 4A, B). Shown are results from pretreatment with purified IFN-α10, but similar results were seen with pretreatment with all IFN-α subtypes, IFN-β, and IFN-ω (data not shown).
FIG. 4.
Type I IFN pretreatment does not inhibit SeV-induced CPE. (A) U937 and (B) A549 cells were pretreated with serial dilutions of IFN-α10 (ranging from 0.78 to 50 U/mL) for 24 h and then UI or infected with either EMCV (MOI 0.5) or SeV (150 or 1.5 HA U/mL) for 72 h. Cell viability was measured via MTT assay (U397) or crystal violet staining (A549). CC: untreated, UI; VC: untreated virus infected. (C) Namalwa cells were pretreated with 100 U/mL of all 12 IFN-α subtypes for 24 h and then infected with either VSV (MOI 0.5) or SeV (150 HA U/mL) for 72 h. Cell viability was measured via MTT assay. Results show cell viability as % of cell control. Cell Control: UI, untreated; virus control: virus infected, untreated. MOI, multiplicity of infection.
Protection against VSV-mediated CPE, but not SeV, was also seen in Namalwa cells pretreated with 100 U/mL type I IFN subtypes (Fig. 4C). It should be noted that 10,000 U/mL of the IFN subtypes were used to pretreat the cell types, which were still unable to protect against SeV-mediated CPE (data not shown). Interestingly, IFN pretreatment and SeV infection (150 HA U/mL) of U937 cells led to decreased cell viability compared to the untreated SeV-infected virus control cells (Fig. 4A). Together, the data show that IFN pretreated cells that were challenged with SeV result in the same as or even more cell death than the untreated SeV-infected cells, indicating type I IFNs do not protect cells from SeV infection and may contribute to SeV-mediated CPE in certain cell types.
Blocking IFNAR signaling during SeV infection in untreated or IFN pretreated cells has no effect on SeV-induced CPE
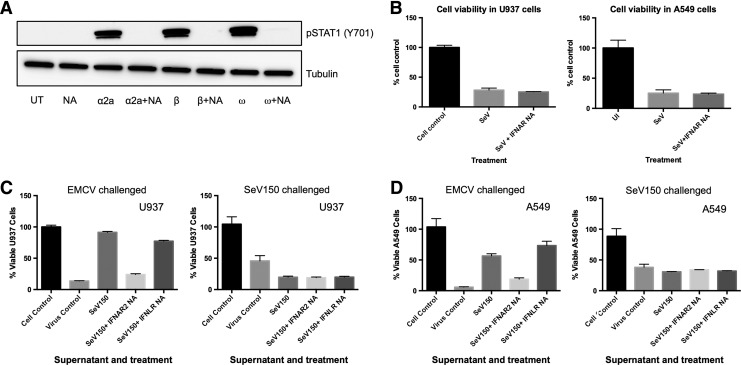
We next determined whether IFNAR signaling, which led to the IFN-induced response that was activated upon SeV infection, served a functional purpose in protecting cells against virus-induced CPE. To test this, U937 and A549 cells were infected with SeV (150 HA U/mL) in the presence or absence of a blocking antibody to IFNAR2. This antibody successfully inhibits phosphorylation of STAT1(Y701) following treatment with 1000 U/mL of IFN-α2a, IFN-β, and IFN-ω (Fig. 5A).
FIG. 5.
Inhibiting canonical type I IFN signaling during SeV infection does not affect SeV-mediated CPE. (A) U937 cells were treated with 1000 U/mL of IFN-α2a, IFN-β, or IFN-ω for 1 h in the presence or absence of the IFNAR2 neutralizing antibody. Western blot analysis was performed to detect phosphorylated STAT1(Y701) and tubulin was used as the loading control. UT: untreated; NA: cells treated with IFNAR2 neutralizing antibody alone; α2a: IFN-α2a treated; α2a + NA: cells treated with both antibody and IFN-α2a; β: IFN-β treated; β + NA: cells treated with both antibody and IFN-β; ω: IFN-ω treated; and ω + NA: cells treated with both antibody and IFN-ω. (B) U937 (panel 1) and A549 cells (panel 2) were infected with SeV (150 HA U/mL) in the presence or absence of IFNAR2 neutralizing antibody. Cell viability was measured 72 h pi. Data are representative of 3 independent biological replicates. Supernatants from cells infected with SeV (150 HA U/mL) for 16 h were inactivated (β-propiolactone) and used to pretreat fresh (C) U937 and (D) A549 cells for 24 h in the presence or absence of the IFNAR or IFNLR neutralizing antibodies. The U937 cells were then infected with either EMCV or SeV (150 HA U/mL) for 72 h and cell viability was measured via MTT assay (U937) or crystal violet assay (A549). Results are shown as the % of cell control. SeV150: virus-inactivated supernatant from SeV-infected cells (150 HA U/mL); SeV150 + IFNAR2 NA: treated with virus-inactivated supernatant from SeV-infected cells (150 HA U/mL) in the presence of IFNAR2-neutralizing antibody; SeV150 + IFNLR NA: treated with virus-inactivated supernatant from SeV-infected cells (150 HA U/mL) in the presence of IFNLR-neutralizing antibody. IFNAR, IFN receptor; IFNLR, IFN-lambda receptor.
We found that in both cell types, blocking IFNAR2 signaling during SeV infection did not affect cell viability (Fig. 5B). Similar results were obtained in cells infected with 15 and 1.5 HA U/mL (data not shown). These results indicate that although SeV induces an antiviral response through the Jak/STAT pathway, blocking this pathway during infection has no protective effect on cell viability.
As previously described in Figs. 3 and 4, the IFN-induced antiviral response that was initiated before SeV infection was also unable to provide protection against SeV-induced CPE; therefore, we blocked the type I and type III IFN receptors before the antiviral assays to determine whether canonical IFN signaling had any effect on cell viability. IFNAR and IFNLR neutralizing antibodies were used to treat fresh cells 1 h before 24 h pretreatment with virus-inactivated supernatants from cells infected with SeV (150 HA U/mL). Cells were then challenged with EMCV or SeV for 72 h and cell viability was assessed via the MTT assay (U937) or crystal violet staining (A549).
Blocking the IFNLR did not affect viability in either cell type that was challenged with EMCV, but protection was lost when the IFNAR2 was blocked (Fig. 5C, D). Lack of protection following IFNAR2 neutralization demonstrates that cells infected with SeV primarily induce biologically active type I IFN rather than type III IFN during SeV infection and that type I IFN is responsible for protection against EMCV-mediated CPE. In addition, blocking type I and III canonical signaling pathways in A549 cells that are pretreated and challenged with SeV (MOI 5) does not affect viability, although U937 cells still exhibit increased CPE (Fig. 5C, D). It is clear that type I IFN pretreatment before SeV infection causes decreased U937 cell viability, but these IFNs do not signal canonically through IFNAR2 to affect CPE.
Type I IFNs affect SeV replication in a cell type-dependent manner
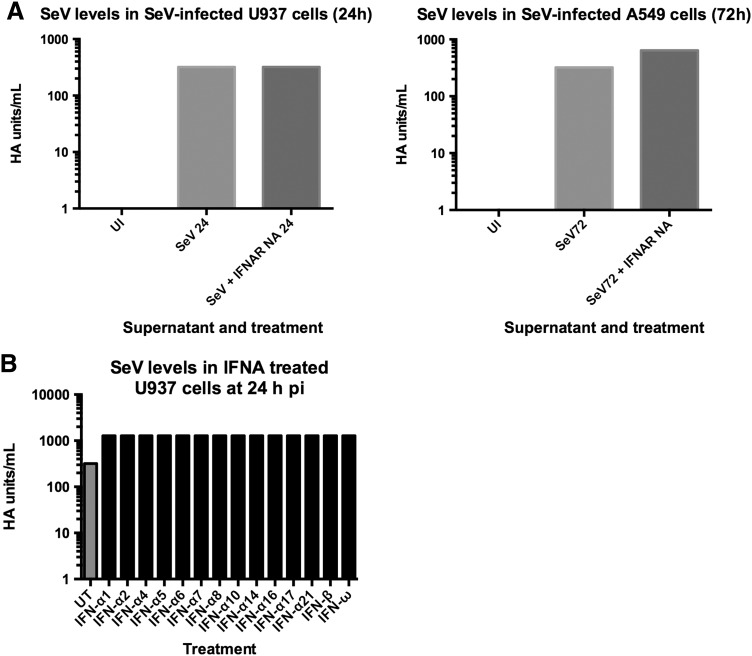
Because the IFN-induced antiviral response that is activated by SeV infection is not functional in protecting against CPE, we determined whether the IFN-induced antiviral state was functional in restricting virus replication. SeV virus production was measured in U937 and A549 cells treated with and without the IFNAR2 neutralizing antibody via HA assays with the supernatants of SeV-infected cells (150 HA U/mL). The U937 and A549 cell supernatants were sampled after SeV-induced CPE was observed (24 and 72 h pi, respectively). We found that blocking IFNAR2 signaling slightly increased SeV production in A549 cells (Fig. 6A). Even though SeV induces an antiviral response through the Jak/STAT pathway, blocking this pathway in U937 cells had no effect on virus production during infection (Fig. 6A).
FIG. 6.
The effect of IFNAR signaling on SeV production is cell type dependent. (A) SeV virus levels were measured via hemagglutination assay in U937 cells (panel 1) and A549 cells (panel 2) at 24 and 72 h pi with SeV (150 HA U/mL), respectively. The data are expressed as HA U/mL. Two biological replicates were performed and resulted in the same titer. (B) U937 cells were pretreated with all 12 IFN-α subtypes, IFN-β or IFN-ω (100 U/mL) for 24 h. Cells were then infected with SeV (150 HA U/mL) for 24 h. Hemagglutination assay was performed to measure SeV virus production in the cell supernatants. Data are expressed as HA U/mL. UT: untreated, virus infected cell supernatant. pi, postinfection.
It was possible that the inability of the IFN-induced state to protect against SeV replication was due to the virus replicating more rapidly than the antiviral response was occurring; in other words, the IFN response was too far behind virus replication to have an observable effect. To determine whether this was the case, we pretreated U937 cells with type I IFNs before infection and assessed SeV production by a HA assay.
We found that pretreatment did not reduce SeV production by 24 h pi and, interestingly, there was a half log increase in virus production in U937 cells that were pretreated with type I IFN subtypes 24 h before infection (Fig. 6B). Shown are results from cells infected with 150 HA U/mL; similar results were obtained in cells infected with 15 HA U/mL (data not shown). These results indicate that while SeV is a potent inducer of type I IFNs, these IFNs are not only unable to protect against SeV-mediated CPE in all cell types tested, but IFN pretreatment leads to increased CPE and SeV production in U937 cells.
The inability of IFNs to protect against SeV infection is independent of virus concentration
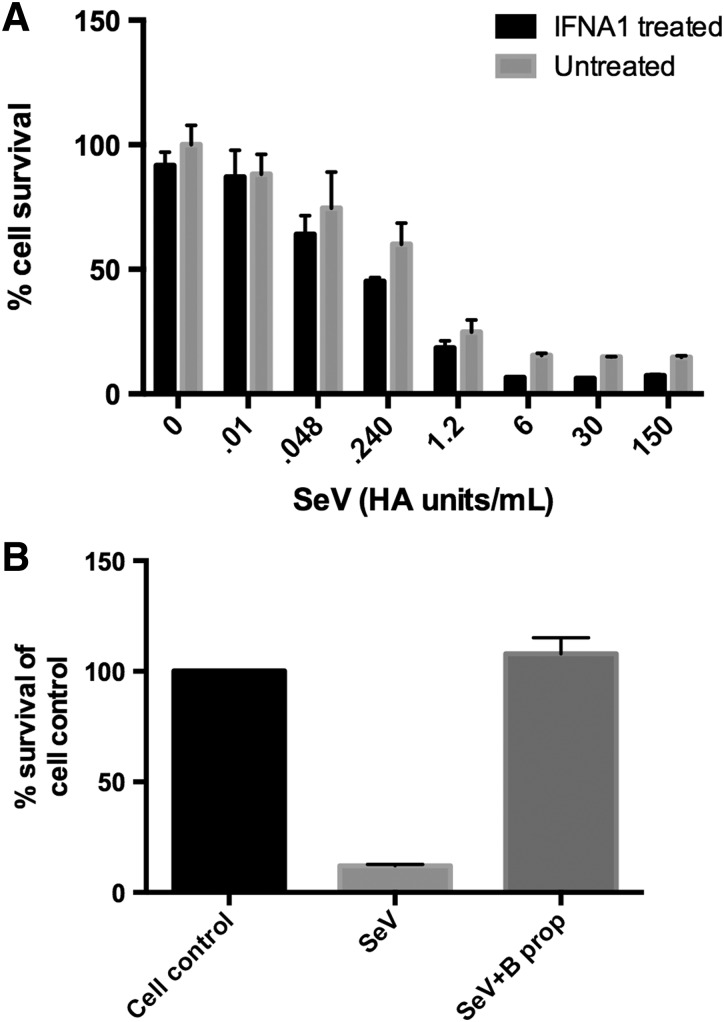
To examine whether the MOI of SeV infection was important in determining the effects of IFN, we repeated the IFN pretreatment experiments with serial dilutions of SeV in IFN treated and untreated cells. We found that even reducing the amount of SeV by 4-logs did not change the inability of IFN to protect against SeV-mediated CPE. In addition, the amount of infecting SeV did not alter the decrease in U937 cell viability observed following pretreatment with 100 U/mL type I IFN pretreatment and SeV infection (Fig. 7A). Results are shown for pretreatment with IFN-α1, but treatment with all subtypes had comparable results (data not shown). Since IFN pretreatment and infection with 1.5 HA U/mL SeV did not result in increased CPE in U937 cells in Fig. 4A, it is likely that higher concentrations of IFN are needed before SeV infection to cause the increase in virus-induced CPE.
FIG. 7.
Decreasing the amount of infecting virus does not result in IFN-mediated protection against SeV-induced CPE. (A) U937 cells were pretreated with 100 U/mL of type I IFNs for 24 h and then infected with serial 2-fold dilutions of SeV, ranging from 0 to 150 HA U/mL of SeV for 72 h. Cell viability was measured via MTT assay. Results are shown for IFN-α1 pretreatment, but all IFN-α and IFN-β subtypes were tested with similar results. (B) Virus preparations were inactivated using β-propiolactone. U937 cells were then infected with SeV or treated with inactivated SeV (β-propiolactone) for 72 h. Cell viability was measured via MTT assay and results are represented as % cell control.
To ensure that nothing present in the virus preparation was responsible for the cell death that resulted upon infection, we inactivated SeV in our preparation via treatment with β-propiolactone and then treated the cells for 72 h with the inactivated virus. We found that upon infection with SeV that was not inactivated, about 20% of the cells were viable; however, treating cells with the inactivated virus resulted in no change in viability compared to the untreated, uninfected cell control (Fig. 7B). These results indicate that active virus replication is necessary to cause CPE rather than a component in the virus preparation.
Discussion
The results from this study indicate that the ability of a cell to induce type I IFNs and antiviral genes and their respective proteins upon virus infection does not always result in protection against virus replication and CPE. In the case of SeV infection, it is unclear exactly what role the induced IFN is playing, as neither blocking IFNAR2 signaling in infected cells nor pretreating cells with IFN to prime an antiviral response resulted in any protective effect against SeV.
Interestingly, we discovered that IFN pretreatment of the monocytoid cell line, U937, followed by SeV infection leads to increased CPE compared to cells that were untreated and infected with SeV. Blocking canonical type I and type III IFN signaling before IFN pretreatment and SeV infection still resulted in decreased cell viability, indicating an alternative IFN binding site or mechanism by which IFN may be enhancing CPE in cells that are infected with SeV.
A number of studies provide evidence and suggest that IFN-α may interact with receptors other than the IFNAR. For example, Uddin and others (1995, 1997) have shown that both insulin and IFN-α can induce tyrosine phosphorylation of the insulin receptor substrate-1 (IRS-1), which can affect cell survival, growth, and metabolism (Franceschini and others 2011; Oka and others 2011). It is important to note that the IRS-pathway is independent of the STAT1-pathway (Uddin and others 1997), which could explain why IFNAR2 neutralization might not affect IRS signaling. Microarray results from U937 cells that were infected with SeV show an upregulation of signaling pathways related to insulin and diabetes at 24 h pi (GEO accession number GSE67198), which provides more evidence of noncanonical IFN signaling that could involve the insulin receptor and/or the IRS pathway.
Other IFN signaling mechanisms may be occurring to induce the increased CPE we observed following IFN pretreatment and SeV infection. Hu and others (1993) have suggested multiple IFN binding sites while determining the sensitivity of U937 cells to the antiproliferative effects of IFN-α. A direct correlation was found between the antiproliferative effects and binding affinities of certain IFN-α components. IFN-α component o, however, was shown to have a low binding affinity to the type I IFN receptor, but induced a high antiproliferative effect, suggesting the involvement of a binding site other than the IFNAR or a multicomponent receptor to induce inhibition of U937 cell growth (Hu and others 1993).
In our studies, U937 cells that were pretreated with IFN and infected with SeV may have been more sensitive to the antiproliferative effects of IFN than untreated, infected cells. Moreover, the existence of an alternative IFN binding site could explain why IFNAR2 neutralization before IFN pretreatment and SeV infection had no effect on CPE. Although U937 cells were the only cell type tested to exhibit decreased cell viability upon IFN pretreatment and SeV infection, none of the cell types were able to elicit effective antiviral responses that could protect cells from the infecting virus, SeV.
The lack of protection following IFN treatment and SeV infection has also been reported in mice. Pretreatment of mouse thymic epithelial cells with IFN-β for 24 h before SeV infection did not result in inhibition of SeV replication (Morales and others 2015). These in vitro results were also recapitulated in vivo, where SeV-infected IFNAR−/− mice displayed the same or extremely similar viral loads as wild-type mice infected with SeV (López and others 2006). This is in contrast to mice infected with influenza B virus, where deletion of the IFNAR resulted in a dramatic increase in virus replication (Lai and others 2009). These reports and our studies suggest that IFN production and the establishment of an antiviral response may not necessarily be indicative of efficient viral clearance or have a functional antiviral effect.
In addition, generic markers such as Jak/STAT activation and induction of a few ISGs may not be sufficient to conclude whether a successful antiviral state is achieved. The type of antiviral response that is necessary to protect against SeV infection may be much more specific than the canonical markers that are usually measured. Several reports have demonstrated that different cell types can elicit distinct antiviral responses when infected with a particular virus (Sperber and others 1992; Lavoie and others 2011; Zhao and others 2012). Other cell types that were not tested in this study might express a much more specific profile of antiviral genes and proteins that are needed to induce an effective antiviral response against SeV infection.
Typically, when IFN treatment or production is ineffective against restricting virus replication, it is likely the case that the virus has developed strategies to circumvent the innate immune response, such as inactivating necessary signaling modulators or shutting down global transcription or translation (Levy and García-Sastre 2001). In our studies we found that IRF and Jak/STAT signaling pathways were activated upon SeV infection, leading to the transcription and translation of several ISGs, including IFIT3, ISG15, Mx1, and IRF-7, which indicates that viral proteins were not inactivating these processes.
Other studies have also reported the ability of SeV-induced IFN to signal, as deletion of Jak1, a major tyrosine kinase necessary for IFN signaling through the IFNAR, resulted in the downregulation of a subset of ISGs during SeV infection (Elco and others 2005). This is in contrast to several studies found in the literature, where groups have reported that the SeV C protein interferes with Jak/STAT signaling by either binding to STAT proteins and preventing their activation or targeting STATs for degradation (Komatsu and others 2000; Young and others 2000; Strähle and others 2003; Kato and others 2004).
Although we detected phosphorylation and expression of STATs 1 and 2 in SeV-infected cells, a loss of STAT2 phosphorylation and expression was observed at 24 h pi with SeV in U937 cells. It is possible that this loss represents an inhibition in STAT signaling by SeV; however, we observed a loss of STAT2 expression in U937 cells at 24 h pi with VSV and EMCV and this loss was not detected in other cell types that were infected with SeV, VSV, or EMCV (data not shown). Thus, it is likely that cell type-specific signaling kinetics and negative feedback mechanisms affect STAT expression at 24 h pi.
One theory to reconcile the conflicting literature regarding SeV effects on STAT proteins may involve the extent through which the viral proteins inhibit Jak/STAT signaling. Even if SeV proteins were inhibiting STAT signaling in our studies, this inhibition is not complete, as we clearly still see enough signaling to induce type I IFNs, ISGs, and the activation of IRF and STAT proteins to levels far beyond those in uninfected cells.
Type I IFNs, aside from having antiviral and antiproliferative capabilities, have also been reported to exhibit antitumor and immunomodulatory properties. Studies have demonstrated that type I IFNs can signal through the IFNAR to induce ISGs that lead to caspase-dependent apoptosis (Balachandran and others 1998, 2000; Miyake and others 2012). However, it is unlikely that the SeV-induced IFNs are primarily causing cell death through this mechanism in our studies because blocking IFNAR2 did not affect the viability of cells that were infected with SeV. The cell types we studied most likely induced apoptosis through IFN-independent mechanisms in response to SeV infection.
SeV-induced apoptosis was shown to require IRF-3 expression and activation through linear ubiquitination at specific lysine residues, a process referred to as RIG-I-like receptor-induced IRF-3-mediated pathway of apoptosis (RIPA) (Chattopadhyay and others 2016). IFN signaling was shown to have no effect on this process, as Jak knockout cells that were infected with SeV still induced RIPA (Peters and others 2008). In RIPA, IRF-3 interacts with the proapoptotic protein BAX, at the BH3 domain of IRF-3, and translocates into the mitochondria to induce apoptosis (Chattopadhyay and others 2010, 2011).
We detected the phosphorylation and nuclear accumulation of IRF-3 in our studies, but mitochondrial translocation and BAX interaction were not assessed. Thus, IRF-3 activation by SeV may have led to RIPA and the induction of IFN and ISGs (Zaritsky and others 2015). IRF-3, therefore, is a complex regulator of SeV-induced antiviral and apoptotic responses and more studies are needed to assess its roles in IFN-treated and SeV-infected cells. IFNs are also able to mediate cell death by stimulating immune cells to kill other cells.
Previous studies have shown that priming monocytes with specific stimuli, one of which is type I IFN, results in killing of tumor cell lines through mechanisms that have yet to be elucidated (Baron and others 2011; Nakashima and others 2012; Johnson and others 2015). It is possible that factors in the cell supernatants in combination with IFN-α treatment and SeV infection were able to stimulate the U937 monocytoid cells to kill neighboring tumor cells and cause a decrease in total U937 cell viability in a similar manner.
Type I IFNs have also been shown to affect the cytokine and chemokine milieus in virus infections. For example, SeV infected human macrophages, neutralization of type I IFNs resulted in the downregulation of IP-10, MCP-1, and MCP-3 expression (Matikainen and others 2000), which can have major effects on adaptive immunity. Furthermore, type I IFN treatment of monocyte-derived dendritic cells enhances the expression of chemokines and chemokine receptors CCR5 and CCR7 and leukocyte adhesion molecule LFA-1, which in turn affects the ability of dendritic cells to transmigrate to the lymph nodes (Paquette and others 1998; Santini and others 2000; Parlato and others 2001; Rouzaut and others 2010).
Therefore, the large amounts of type I IFNs produced during SeV infection may indirectly serve a functional purpose by modulating chemokine signaling in a system and affecting adaptive immune responses or pathological outcomes. In an in vitro system where only one cell type is present, as was the case in our experiments, this effect may not be apparent, but in an in vivo system where virus infection results in cellular trafficking and pathological changes in tissues, the effects of type I IFNs may be better exemplified.
In SeV-infected mice, for example, the induction of ISG15 had no effect on virus replication, but it did contribute to improving pathological changes in the diseased airways of the lung (Elco and others 2005). Interestingly, when a double knockout was performed for the IFN-induced antiviral protein IFIT2 and the IFNAR, mice were less susceptible to SeV pathogenesis than the IFIT2−/− mice, which suggests IFN signaling through the IFNAR plays a pathological role in mice that are infected with SeV (Wetzel and others 2014).
In IFNAR−/− mice that also lack transcriptional activity of IRF-3, as a result of selective mutations at transcriptionally active sites, RIPA was shown to have protective effects against SeV by reducing viral burden (Chattopadhyay and others 2016). These studies indicate that type I IFN signaling and ISG expression can have effects other than the typical antiviral outcomes and that IFN-independent processes have the ability to elicit antiviral effects. Therefore, the activation of type I IFN-induced signaling pathways and IFN-mediated ISG induction may not always be suggestive of protective responses against virus infection.
Even though SeV infection is not preventing canonical Jak/STAT signaling in our studies, it is possible that the apparent lack of type I IFN protection against SeV infection is due to viral inhibition of noncanonical, Jak/STAT-independent signaling pathways that are activated by type I IFNs. Type I IFNs have been reported to differentially activate ISGs through the MAPK and PI3K pathways in addition to the canonical Jak/STAT pathway (Uddin and others 1999; Kato and others 2004; Kaur and others 2008a, 2008b). We have also previously found that the different signaling pathways that IFNs can induce lead to the upregulation of different sets of ISGs (Zaritsky and others 2015). Non-Jak/STAT proteins that have been implicated in IFN-induced ISG expression include Mnk kinases, mTOR, and SKAR (Joshi and others 2009; Kaur and others 2012; Beauchamp and others 2013; Kroczynska and others 2014).
There are most likely many other IFN-induced signaling pathways that differentially induce ISGs in response to different viral infections that are as yet undiscovered. It is possible that these noncanonical pathways are responsible for inducing the particular sets of ISGs that are necessary for protecting against SeV infection, but virus infection is either directly or indirectly leading to their inhibition. Because most studies only look at canonical signaling to determine antiviral activity, a more in-depth analysis studying both the canonical and noncanonical pathways leading to ISG induction in SeV infection may be necessary to determine the role of IFNs in the antiviral response. Our results clearly indicate that typical markers of IFN signaling in virally infected cells do not necessarily correlate with antiviral activity or increased cell survival.
In conclusion, we found in our studies that although SeV is an extremely potent inducer of biologically active type I IFNs as determined by the ability of the supernatants to protect against VSV or EMCV infection, these IFNs are unable to protect against the very virus that is responsible for their induction. This is not due to a viral protein blocking IFN from Jak/STAT signaling or inducing several of the well-known antiviral genes and proteins.
Our findings reveal that typical markers of IFN activity may not be useful in determining whether a functional antiviral state, which inhibits virus replication and enhances cell survival, is actually induced. In other words, even with the typical markers of an antiviral response, such as IFN production, activation of Jak/STAT signaling modulators, and ISG transcription and/or translation, it does not necessarily mean that virus infection and pathological effects will be restricted. In some cases, treatment with IFN may even contribute to the CPE of virus infection, as we observed in U937 cells. This could have major implications in determining the safety and effectiveness of IFN treatment in patients with certain viral infections. It is therefore necessary to ensure a treatment regimen that is tailored and specific for the particular virus at hand.
Acknowledgments
We would like to thank the Cytokine Biology Section at NIAID for their discussion and help with this project.
This work was supported by the Intramural Research Training Award (IRTA) Program and the Division of Intramural Research (DIR) at the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH).
Author Disclosure Statement
No competing financial interests exist.
References
- Balachandran S, Kim CN, Yeh WC, Mak TW, Bhalla KN, Barber GN. 1998. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J 17:6888–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Roberts PC, Kipperman T, Bhalla KN, Compans RW, Archer DR, Barber GN. 2000. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/caspase-8 death signaling pathway. J Virol 74(3):1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron S, Finbloom J, Horowitz J, Bekisz J, Morrow A, Zhao T, Fey S, Schmeisser H, Balinksy C, Miyake K, Clark C, Zoon K. 2011. Near eradication of clinically relative concentrations of human tumor cells by interferon-activated monocytes in vitro. J Interferon Cytokine Res 31(7):569–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AD, Hunt N, Dimmock NJ. 1984. A rapid method for the inactivation of virus infectivity prior to assay for interferons. J Virol Methods 8:349–351 [DOI] [PubMed] [Google Scholar]
- Beauchamp EM, Platanias LC. 2013. The evolution of the TOR pathway and its role in cancer. Oncogene 32:3923–3932 [DOI] [PubMed] [Google Scholar]
- Cantell K, Hirvonen S, Kauppinen HL, Myllylä G. 1981. Production of interferon in human leukocytes from normal donors with the use of Sendai virus. Methods Enzymol 78(Pt A):29–38 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Kuzmanovic T, Zhang Y, Wetzel JL, Sen GC. 2016. Ubiquitination of the transcription factor IRF-3 activates RIPA, the apoptotic pathway that protects mice from viral pathogenesis. Immunity 44:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Marques JT, Yamashita M, Peters KL, Smith K, Desai A, Williams BRG, Sen GC. 2010. Viral apoptosis is induced by IRF-3-mediated activation of Bax. EMBO J 29:1762–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Yamashita M, Zhang Y, Sen GC. 2011. The IRF-3/Bax mediated apoptotic pathway, activated by viral cytoplasmic RNA and DNA, inhibits virus replication. J Virol 85(8):3708–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elco CP, Guenther JM, Williams BRG, Sen GC. 2005. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not toll-like receptor 3. J Virol 79:3920–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini L, Realdon S, Marcolongo M, Mirandola S, Bortoletto G, Alberti A. 2011. Reciprocal interference between insulin and interferon-alpha signaling in hepatic cells: a vicious circle of clinical significance? Hepatology 54(2):484–494 [DOI] [PubMed] [Google Scholar]
- Génin P, Lin R, Hiscott J, Civas A. 2009. Differential regulation of human interferon A gene expression by interferon regulatory factors 3 and 7. Mol Cell Biol 29:3435–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh B, Takeuchi K, Komatsu T, Yokoo J. 2003. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J Virol 77:3360–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Gan Y, Liu J, Miller D, Zoon KC. 1993. Evidence for multiple binding sites for several components of human lympphoblastoid interferon-α. J Biol Chem 268(17):12591–12595 [PubMed] [Google Scholar]
- Johnson CL, Green DS, Zoon KC. 2015. Human monocytes in the presence of interferons alpha2a and gamma are potent killers of serous ovarian cancer cell lines in combination with paclitaxel and carboplatin. J Interferon Cytokine Res 35(1):55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S, Kaur S, Redig AJ, Goldsborough K, David K, Ueda T, Watanabe-Fukunaga R, Baker DP, Fish EN, Fukunaga R, Platanias LC. 2009. Type I interferon (IFN)-dependent activation of Mnk1 and its role in the generation of growth inhibitory responses. Proc Natl Acad Sci U S A 106(29):12097–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Cortese-Grogan C, Moyer SA, Sugahara F, Sakaguchi T, Kubota T, Otsuki N, Kohase M, Tashiro M, Nagai Y. 2004. Characterization of the amino acid residues of sendai virus C protein that are critically involved in its interferon antagonism and RNA synthesis down-regulation. J Virol 78(14):7443–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Dolniak B, Joshi S, Majchrzak-Kita B, Baker DP, Hay N, Fish EN, Platanias LC. 2008a. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci U S A 105(12):4808–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Joseph AM, Majchrzak-Kita B, Eklund EA, Verma A, Brachmann SM, Fish EN, Platanias LC. 2008b. Dual regulatory roles of phosphatidylinositol 3-kinase in IFN signaling. J Immunol 181(10):7316–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Majchrzak-Kita B, Baker DP, Su B, Fish EN, Platanias LC. 2012. Regulatory effects of mTORC2 complexes in type I IFN signaling and in the generation of IFN responses. Proc Natl Acad Sci U S A 109(20):7723–7728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T, Takeuchi K, Yokoo J, Tanaka Y, Gotoh B. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J Virol 74:2477–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroczynska B, Mehrotra S, Majchrzak-Kita B, Arslan AD, Altman JK, Stein BL, McMahon B, Kozlowski P, Kahle PJ, Eklund EA, Fish EN, Platanias LC. 2014. Regulatory effects of SKAR in interferon a signaling and its role in the generation of type I IFN responses. Proc Natl Acad Sci U S A 111(31):11377–11382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C, Struckhoff JJ, Schneider J, Martinez-Sobrido L, Wolff T, García-Sastre A, Zhang DE, Lenschow DJ. 2009. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J Virol 83:1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie TB, Kalie E, Crisafulli-Cabatu S, Abramovich R, Digioia G, Moolchan K, Pestka S, Schreiber G. 2011. Binding and activity of all human alpha interferon subtypes. Cytokine 56:282–289 [DOI] [PubMed] [Google Scholar]
- Levy DE, García-Sastre A. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev 12:143–156 [DOI] [PubMed] [Google Scholar]
- López CB, Yount JS, Hermesh T, Moran TM. 2006. Sendai virus infection induces efficient adaptive immunity independently of type I interferons. J Virol 80:4538–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie I, Durbin JE, Levy DE. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J 17:6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matikainen S, Pirhonen J, Miettinen M, Lehtonen A, Govenius-Vintola C, Sareneva T, Julkunen I. 2000. Influenza A and sendai viruses induce differential chemokine gene expression and transcription factor activation in human macrophages. Virol J 276:138–147 [DOI] [PubMed] [Google Scholar]
- Miyake K, Bekisz J, Zhao T, Clark CR, Zoon KC. 2012. Apoptosis-inducing factor (AIF) is targed in IFN-α2a-induced Bid-mediated bak activation in ovarian cancer cells. Biochim Biophys Acta 1823(8):1378–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales DJ, Monte K, Sun L, Struckhoff JJ, Agapov E, Holtzman MJ, Stappenbeck TS, Lenschow DJ. 2015. Novel mode of ISG15-mediated protection against influenza A virus and Sendai virus in mice. J Virol 89:337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima H, Miyake K, Clark CR, Bekisz J, Finbloom J, Husain SR, Baron S, Puri RK, Zoon KC. 2012. Potent antitumor effects of combination therapy with IFNs and monocytes in mouse models of established human ovarian and melanoma tumors. Cancer Immunol Immunother 61(7):1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman TA, Tölö H, Parkkinen J, Kalkkinen N. 1998. Identification of nine interferon-alpha subtypes produced by Sendai virus-induced human peripheral blood leucocytes. Biochem J 329(Pt 2):295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka R, Hiroi N, Shigemitsu R, Sue M, Oshima Y, Yoshida-Hiroi M. 2011. Type I diabetes mellitus associated with pegylated interferon-a plus ribavirin treatment for chronic hepatitis C: case report and literature review. Clin Med Insights Endocrinol Diabetes 4:39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette RL, Hsu NC, Kiertscher SM, Park AN, Tran L, Roth MD, Glaspy JA. 1998. Interferon-alpha and granulocyte-macrophage colony-stimulating factor differentiate peripheral blood monocytes into potent antigen-presenting cells. J Leukoc Biol 64:358–367 [DOI] [PubMed] [Google Scholar]
- Parlato S, Santini SM, Lapenta C, Di Pucchio T, Logozzi M, Spada M, Giammarioli AM, Malorni W, Fais S, Belardelli F. 2001. Expression of CCR-7, MIP-3beta, and Th-1 chemokines in type I IFN-induced monocyte-derived dendritic cells: importance for the rapid acquisition of potent migratory and functional activities. Blood 98:3022–3029 [DOI] [PubMed] [Google Scholar]
- Peters K, Chattopadhyay S, Sen GC. 2008. IRF-3 activation by Sendai virus infection is required for cellular apoptosis and avoidance of persistence. J Virol 82(7):3500–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzaut A, Garasa S, Teijeira A, González I, Martinez-Forero I, Suarez N, Larrea E, Alfaro C, Palazón A, Dubrot J, Hervás-Stubbs S, Melero I. 2010. Dendritic cells adhere to and transmigrate across lymphatic endothelium in response to IFN-α. Eur J Immunol 40:3054–3063 [DOI] [PubMed] [Google Scholar]
- Santini SM, Lapenta C, Logozzi M, Parlato S, Spada M, Di Pucchio T, Belardelli F. 2000. Type I interferon as a powerful adjuvant for monocyte-derived dendritic cell development and activity in vitro and in Hu-PBL-SCID mice. J Exp Med 191:1777–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka T. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor-7. FEBS Lett 441:106–110 [DOI] [PubMed] [Google Scholar]
- Sperber SJ, Gocke DJ, Haberzettl C, Kuk R, Schwartz B, Pestka S. 1992. Anti-HIV-1 activity of recombinant and hybrid species of interferon-alpha. J Interferon Res 12:363–368 [DOI] [PubMed] [Google Scholar]
- Strähle L, Garcin D, Le Mercier P, Schlaak JF, Kolakofsky D. 2003. Sendai virus targets inflammatory responses, as well as the interferon-induced antiviral state, in a multifaceted manner. J Virol 77:7903–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin S, Fish EN, Sher D, Gardziola C, Colamonici O, Kellum M, Pitha PM, White MF, Platanias LC. 1997. The IRS-pathway operates distinctively from the stat-pathway in hematopoietic cells and transduces common and distinct signals during engagement of the insulin or interferon a receptors. Blood 90(7):2574–2582 [PubMed] [Google Scholar]
- Uddin S, Majchrzak-Kita B, Woodson J, Arunkumar P, Alsayed Y, Pine R, Young PR, Fish EN, Platanias LC. 1999. Activation of the p38 mitogen-activated protein kinase by type I interferons. J Biol Chem 274:30127–30131 [DOI] [PubMed] [Google Scholar]
- Uddin S, Yenush L, Sun X, Sweet ME, White MF, Platanias LC. 1995. Interferon-a engages the insulin receptor substreate-1 to associate with the phosphatidylionisitol 3′-kinase. J Biol Chem 270(27):15938–15941 [DOI] [PubMed] [Google Scholar]
- Wetzel JL, Fensterl V, Sen GC. 2014. Sendai virus pathogenesis in mice is prevented by Ifit2 and exacerbated by interferon. J Virol 88(23):13593–13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fukhara Y, Sato M, Ozato K, Fujita T. 1996. Autocrine amplification of type I interferon gene expression mediated by interferon stimulated gene factor 3 (ISGF3). J Biochem 120:160–169 [DOI] [PubMed] [Google Scholar]
- Young DF, Didcock L, Goodbourn S, Randall RE. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virol J 269:383–390 [DOI] [PubMed] [Google Scholar]
- Zaritsky LA, Bedsaul JR, Zoon KC. Virus multiplicity of infection affects type I interferon subtype induction profiles and interferon-stimulated genes. J Virol 89(22):11534–11548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Zhang J, Phatnani H, Scheu S, Maniatis T. 2012. Stochastic expression of the interferon-B gene. PLoS Biol 10(1): e1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoon KC, Buckler CE, Bridgen PJ, Gurari-Rotman D. 1978. Production of human lymphoblastoid interferon by Namalva cells. J Clin Microbiol 7:44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]