Abstract
The main limitations of hematopoietic cord blood (CB) transplantation, viz, low cell dosage and delayed reconstitution, can be overcome by ex vivo expansion. CB expansion under conventional culture causes rapid cell differentiation and depletion of hematopoietic stem and progenitor cells (HSPCs) responsible for engraftment. In this study, we use combinatorial cell culture technology (CombiCult®) to identify medium formulations that promote CD133+ CB HSPC proliferation while maintaining their phenotypic characteristics. We employed second-generation CombiCult screens that use electrospraying technology to encapsulate CB cells in alginate beads. Our results suggest that not only the combination but also the order of addition of individual components has a profound influence on expansion of specific HSPC populations. Top protocols identified by the CombiCult screen were used to culture human CD133+ CB HSPCs on nanofiber scaffolds and validate the expansion of the phenotypically defined CD34+CD38lo/−CD45RA−CD90+CD49f+ population of hematopoietic stem cells and their differentiation into defined progeny.
Keywords: : CombiCult, high-throughput-screening, hematopoietic stem/progenitor cells, ex-vivo expansion, alginate encapsulation
Introduction
Hematopoietic stem cell transplantation (HSCT), the most successful and widely used stem cell therapy to date, is used to treat patients with hematological malignancies, bone marrow (BM) failure, severe anemia, metabolic storage diseases, immunodeficiencies, and some cancers following radio-chemo-therapeutic treatments. Cord blood (CB) has several advantages as the HSCT cell source over BM or mobilized peripheral blood (mPB) as it is tested and banked ahead of use and readily available, contains more immature HSCs, has extensive proliferative capacity that exceeds that of BM cells, shows less associated GvHD (graft-versus-host disease), and is not subject to donor attrition. Its main limitations are its delayed hematological engraftment (early neutrophil and platelet and longer term immune reconstitution), which is associated with defective adhesion receptor fucosylation or reduced homing/engraftment receptor expression on CB hematopoietic stem and progenitor cells (HSPCs) [1–4], the immaturity of cells, and the limited cell numbers in each CB unit [5,6]. A single CB unit is generally sufficient for pediatric, but not adult, transplant recipients [7]. CB HSPCs can also be used for generating mature blood cells ex vivo for transfusion into difficult-to-transfuse patients or modulating resistance to specific acquired infections and correcting monogenic gene disorders, in dividing HSCs, using new genome editing technologies [8–10].
To improve early hematological engraftment, extend the use of single CB unit therapy to adults, and develop uses for banked CB units in other therapies, different approaches to modulate the HSPC compartment in CB ex vivo have been explored [9,11,12]. Expansion of CB cells in the presence of factors known to enhance HSPC proliferation, such as the Notch ligand Delta 1 (DLL1), StemRegenin-1, and nicotinamide or coculture of CB cells with mesenchymal stromal cells, has been reported to increase the number of short-term repopulating cells and reduce the median time to neutrophil engraftment [11,13–17]. However, in these clinical trials, a second, unmanipulated CB unit with adequate cell numbers and/or the unmanipulated CD34− or CD133− fraction of the expanded CB unit was cotransplanted to ensure the presence of durable, long-term engrafting HSCs.
Progress in designing expansion protocols has coincided with progress in the identification of specific markers, which in combination allow significant enrichment of short and long-term hematopoietic repopulating cells (STRCs and LTRCs, respectively) as assessed in immunodeficient animal models. Although all HSCs reside in the CD133+ and most in the CD34+ fraction in hematopoietic tissues [18–22], efforts to define a set of markers, which identify the rare LTRCs, are still in progress, with novel discriminatory marker sets being identified [23].
In this study, we use Plasticell's proprietary combinatorial technology CombiCult to discover new protocols for the ex vivo expansion of human CB CD133-positive HSPCs. CombiCult (www.plasticell.co.uk) is a high-throughput bead-based technology, which allows the user to assay multiple media combinations simultaneously in small volumes [24,25]. This technology has previously been used to discover optimal protocols for the differentiation of pluripotent stem cells into terminally differentiated cells [26]. We show here that CombiCult can also be used to discover stem cell maintenance and expansion protocols. We have adapted the CombiCult platform to incorporate the use of nonadherent hematopoietic cells by a process of encapsulation in alginate hydrogel beads [27].
Specifically, we describe the identification of medium compositions that promote proliferation of phenotypically defined CB HSPCs, as assessed by the maintenance of expression of the HSC marker, CD133. When these medium compositions were used to expand CD133+ human CB cells on nanofiber scaffolds, they specifically promoted the expansion of the CD34+CD38low/−CD45RA−CD90+CD49f+ subpopulation, which is proposed as the most likely phenotype of the rare LTR cells [23].
Materials and Methods
Human CB cell collection, isolation, cryopreservation, and thawing of cryopreserved cells, as well as matrix design, are described in Supplementary Data 1 (Supplementary Data are available online at www.liebertpub.com/scd). These studies using human cells were conducted with NHSBT and Plasticell institutional R&D regulatory approval.
CombiCult screen
Encapsulation of cells in alginate beads
Before encapsulation, isolated CD133+ CB cells were harvested, counted, and analyzed by flow cytometry to quantify expression of CD133 and CD34. A total of 6 × 105 cells were resuspended in sodium alginate [2% solution in phosphate-buffered saline (PBS); Protanal FMC BioPolymer]. The alginate–cell suspension was encapsulated by electrospraying using an encapsulation system (Nisco Engineering AG) [28] equipped with a 0.5-mm needle at a rate of 5 mL/h and a voltage of 7.7 kV. The alginate beads were cross-linked using a 200 mM CaCl2 solution, then washed twice in Dulbecco's modified Eagle's medium (DMEM). The alginate beads formed and ranged in size from 300–400 μM. Under the conditions used, 0.6 mL of alginate produced ∼10,000 beads, averaging 60 cells/bead.
Split-pool
Beads were split into 10 tubes in equal amounts, resuspended in one of the 10 medium conditions for day 1 (Table 1), and plated into 10 wells of 25-well microbiological plates (Bibby Sterilin). Beads were incubated for 3 days (day 4) in the first set of conditions, after which the beads from each condition were tagged with a unique fluorescent tag using a modified alginate multilayering technique [27]. The beads were then pooled and split into 10 new conditions. This split-pool cycle was repeated once more on day 7. On day 10, beads were fixed in 4% PFA prepared in Dulbecco's phosphate-buffered saline with Ca2+ and Mg2+ (DPBS+).
Table 1.
Matrix Formulations for CombiCult Screen
| Condition | Cytokine | |||
|---|---|---|---|---|
| 1 | ANGPTL5 (500 ng/mL) | DLL1 (50 ng/mL) | Flt3L (100 ng/mL) | IGFBP2 (100 ng/mL) |
| 2 | ANGPTL5 (500 ng/mL) | DLL1 (50 ng/mL) | Flt3L (100 ng/mL) | OSM (50 ng/mL) |
| 3 | ANGPTL5 (500 ng/mL) | DLL1 (50 ng/mL) | IGFBP2 (100 ng/mL) | OSM (50 ng/mL) |
| 4 | ANGPTL5 (500 ng/mL) | FGF-1 (10 ng/mL) | IGFBP2 (100 ng/mL) | OSM (50 ng/mL) |
| 5 | ANGPTL5 (500 ng/mL) | FGF-1 (10 ng/mL) | IGFBP2 (100 ng/mL) | Wnt5a (500 ng/mL) |
| 6 | ANGPTL5 (500 ng/mL) | FGF-1 (10 ng/mL) | OSM (50 ng/mL) | Wnt5a (500 ng/mL) |
| 7 | ANGPTL5 (500 ng/mL) | Flt3L (100 ng/mL) | IGFBP2 (100 ng/mL) | OSM (50 ng/mL) |
| 8 | ANGPTL5 (500 ng/mL) | IGFBP2 (100 ng/mL) | OSM (50 ng/mL) | Wnt5a (500 ng/mL) |
| 9 | DLL1 (50 ng/mL) | Flt3L (100 ng/mL) | IGFBP2 (100 ng/mL) | OSM (50 ng/mL) |
| 10 | FGF-1 (10 ng/mL) | IGFBP2 (100 ng/mL) | OSM (50 ng/mL) | Wnt5a (500 ng/mL) |
Combinations of cytokines added to basal media comprising SCF, TPO, and heparin in StemSpan SFEM serum-free media.
ANGPTL5, angiopoietin-like 5; FGF-1, fibroblast growth factor-1; IGFBP2, insulin growth factor-binding protein 2; OSM, oncostatin M; SCF, stem cell factor; TPO, thrombopoietin.
Immunostaining of alginate beads
Fixed beads were washed in DPBS+, permeabilized with 0.1% Triton X-100 in DPBS+, blocked in 3% FCS in DPBS+ (blocking buffer), and incubated with primary antibodies against CD133 (1:200 dil; rabbit polyclonal; Biorbyt Ltd.) and Ki67 (1:50 dil; mouse monoclonal; BD Bioscience) in blocking buffer for 2 days at 4°C, followed by washing in DPBS+. Beads were then incubated with secondary antibodies (goat anti-rabbit Alexa Fluor 594 and goat anti-mouse Alexa Fluor 488) in blocking buffer, both at 1:500 dilution overnight at 4°C. The beads were then washed and analyzed.
Hit detection and isolation
Hit beads were isolated manually using a Nikon Eclipse 2000-S inverted epifluorescence microscope equipped with filter sets for visualization of TRITC, DAPI, GFP-B (all from Nikon Ltd.), and Cy5.5 (Chroma Technology).
Tag analysis
Individually isolated beads were dissolved to release tags by overnight incubation in Trypsin/EDTA at 37°C and then resuspended in 0.1% Tween in PBS.
Tags used in the experiment comprised 30 unique populations of inert fluorescent microspheres (Plasticell Ltd.). Discrete populations in the size range of 1–10 μm were assigned according to diameter (three size groups: S, M, L) and fluorescence intensity (10 gradations for each size group). Tags were analyzed using FACSCanto II flow cytometer equipped with 488, 635, and 405 nm lasers (Red PMT 710/50 nm; Becton Dickinson). Before tag analysis, a reference tag set was used to establish side and forward scattering gates and to calibrate fluorescence intensity of each tag set. Tag identification was performed using Ariadne™ bioinformatics software (Plasticell Ltd.). Fluorescence histograms of each scattering gate and FSC/Red fluorescence plots were used to identify clusters of events with correlated scattering and fluorescence parameters, which are mapped to cell culture media. Clusters were identified when they contained ≥5 events with near identical scattering and fluorescence values. If two or more clusters of events were identified, which mapped to media from the same split and/or the signal-to-noise ratio was too low, the hit was not recorded.
Ariadne and component analysis
Probability values for the occurrence of given events by chance were obtained from computer simulation experiments. A Mersenne Twister [29] random number generator was used to output uniformly distributed 32-bit integers, which were scaled to cover 10,000, 1,000, or 100 possible pathways, when simulating common cell culture media on three of three splits or two of three splits. Simulation begins by setting tally counters associated with each pathway to zero. Event probabilities were computed by repeating the process 100 million times and dividing the number of positive results by the total number of simulations, resulting in probability values accurate to eight decimal places.
Validation of CombiCult-derived protocols using alginate encapsulation
Validations were carried out in alginate beads using the same encapsulation technique described above. CD133+ CB cells isolated and thawed as described above were incubated for 3 days in thawing media. A total of 1 × 106 cells were encapsulated in 1 mL of alginate as above. Beads obtained were distributed evenly into tubes, and each tube was resuspended in media corresponding to the condition at stage 1 to be tested. Beads from each tube were plated onto two wells of a 25-well suspension plate and incubated for 3 days. On D4, media were decanted, beads washed in DMEM, and media for stage 2 were added. This was repeated again on D7. On day 10, beads were washed, fixed in 4% PFA, and immunostained with antibodies against CD133 and Ki67 (as in section “immunostaining of alginate beads”). Fluorescent beads were analyzed using a large particle sorter (COPAS; Union Biometrica) and photographed using epifluorescence microscopy with a Nikon Eclipse 2000 inverted microscope and NIS-elements software.
Scaffold-based cell culture to assess CombiCult results on human HSPC subsets
The expansion protocols were further evaluated in a previously established culture system comprising human CB CD133+ HSPCs cultured on a 3D nanofiber scaffold in serum-free StemSpan ACF media [30,31]. Magnetic activated cell sorting (MACS) separated and thawed CD133+ HSPCs from human CB were first cultured in StemSpan ACF media (Stem Cell Technologies) in the basal cytokine stem cell factor (SCF; 100 ng/mL) and thrombopoietin (TPO; 20 ng/mL; both from R&D Systems) for 18–22 h in 96-well round bottom plates (Bibby Sterilin) at 20,000 cells/100 μL (termed day 0). On day 1, cells were harvested and quantified by flow cytometry using CountBright beads (Thermo Fisher) and DAPI staining (Supplementary Fig. S1). Cells were plated at a density of 2,500 cells/well in 24-well 3D scaffold (Nanex) plates (Compass Biomedical) in 1 mL medium containing the different cytokine cocktails identified in the CombiCult screen. Media were replaced with fresh media containing different cytokine combinations (all from R&D Systems) on days 2 and 5. Details of cytokine changes for each protocol are listed in Supplementary Tables S1, S2 and S3. On day 8, cells were analyzed by flow cytometry for viable nucleated cell count using CountBright beads and DAPI staining and phenotypically characterized for HSPC subsets using nine-color flow cytometry on the BD LSRII as described below and in Chang et al. [32].
Antibodies and flow cytometry
For assessing cell purity or HSPC content, following MACS purification, CD133+ HSPCs were stained with CD133-APC or -PE and DAPI as described previously [30–32]. Multicolor flow cytometry characterization of CD133+ human CB-derived HSPCs was based on the method described by Notta et al. [23]. The following antibodies were used: CD34 AF700 (581), CD45RA APC-H7 (HI100), CD38 PE-TxR (HIT2), CD90 PE (5E10), CD123 PerCP-Cy5.5 (6H6), CD49f PE-Cy7 (GoH3), and a panel of lineage markers conjugated to PE-Cy5: CD2 (RPA-2.10), CD3 (HIT3a), CD4 (RPA-T4), CD7 (CD7-6B7), CD8a (RPA-T8), CD10 (HI10a), CD11b (ICRF44), CD14 (61D3), CD19 (HIB19), CD20 (2H7), CD56 (B159), and CD235ab (HIR2). Briefly, cells were resuspended in human FcR blocking reagent diluted in MACS buffer (both from Miltenyi Biotec) and incubated for 10 min at 4°C. The cells were then incubated with a mixture of fluorescently labeled antibodies diluted in MACS buffer (Miltenyi Biotec) for 20 min on ice. Cells were washed once, resuspended in MACS buffer, and acquired immediately on an LSRII flow cytometer (BD Biosciences). DAPI was added at 100 ng/mL directly before acquisition to distinguish live and dead cells. Data were analyzed on FlowJo (TreeStar, Inc.). HSPC stem and progenitor subsets were determined by gating on single viable Lin− cells and defined as HSC: Lin−CD38−CD34+CD45RA−CD90+CD49f+; MPP: Lin−CD38−CD34+ CD45RA−CD90−; CLP: Lin−CD38−CD34+ CD45RA+CD90−; CMP: Lin−CD38+CD34+CD45RA−CD123+; GMP: Lin−CD38+CD34+ CD45RA+CD123+; and MEP: Lin−CD38+CD34+CD45RA−CD123−. Details of flow cytometry configuration are listed in the MIFlowCyt file (Supplementary Data 2 and Supplementary Tables S4–S8). Statistical analyses used one-way analysis of variance (ANOVA) with Tukey's post hoc testing and were performed using GraphPad Prism version 6.03 (Graphpad Software, Inc.). P values <0.05 were considered statistically significant.
Results
CombiCult screen
CombiCult system
CombiCult is a bead-based screening technology that allows miniaturization and multiplexing of large numbers of stepwise cell culture experiments, increasing throughput by orders of magnitude. Briefly, beads seeded with stem cells are shuffled randomly through multiple predetermined combinations of cell culture medium formulations using a split-pool process analogous to that used in combinatorial chemistry. Each cell culture medium is spiked with a distinctive fluorescent tag that attaches to the bead substrate, allowing us to track the history of each bead. Following the split-pool process, beads are assayed to identify those on which stem cells have a specific phenotype of choice (hits). Hits are isolated using a large particle flow sorter or manually under a fluorescence microscope and the beads are then digested to release the fluorescent tags accumulated during the course of the experiment. Tags are analyzed using a flow cytometer to deconvolute the cell culture history of beads and thereby deduce the combination of media, which results in the desired phenotype. A customized bioinformatics program (Ariadne) is used to collate data and perform statistical analysis to predict the most robust and effective protocols. Finally, a subset of candidate protocols is validated to quantitate cell yield and study lineage markers and functional attributes of the resulting cells.
Second-generation CombiCult for use with non-adherent cell types
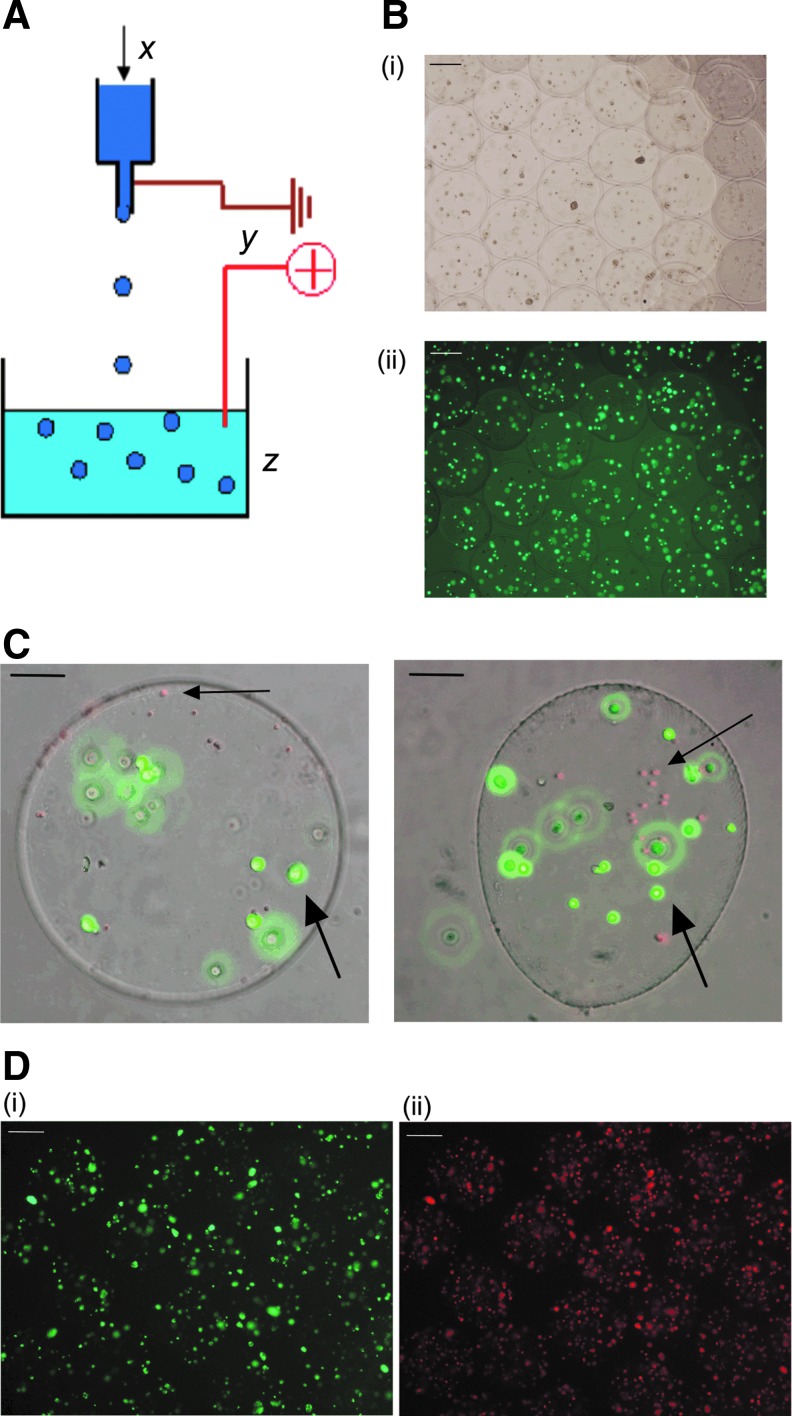
CombiCult was initially developed for use with anchorage-dependent cells attached to solid microcarriers [24–26]. As suspension cells do not adhere strongly to microcarriers, it was necessary to entrap cells inside beads, which could be shuffled between the different medium conditions and also labeled with fluorescent tags. To do this, we developed a protocol for cell encapsulation in alginate beads produced using electrospraying (Fig. 1a). Parameters for encapsulation, such as alginate concentration, viscosity, and bead size, were optimized for this CombiCult screen (see the Materials and Methods section for details).
FIG. 1.
Alginate encapsulation using electrospraying. (A) Diagram of electrospraying technique used to make alginate beads (B). Representative micrograph of CB cells encapsulated in alginate (i) bright-field, (ii) calcein-AM staining, showing the cells are viable. (C) Composite image of alginate beads containing cells visualized with calcein (green) and tagged with red fluorescent tags (shown by large and small arrows respectively). (D) Micrograph showing CB cells encapsulated in alginate and stained with the antibodies against (i) Ki67 [Alexa Fluor 488 (green)] and (ii) CD133 [Alexa Fluor 594 (red)]. CB, cord blood. Scale bars = 100 micrometres.
Cells encapsulated in alginate were analyzed for viability and proliferative capacity (Fig. 1b). Layer-by-layer tagging, previously developed by Plasticell Ltd., [27] was optimized for use with CD133+ CB cells encapsulated in alginate and is illustrated in Fig. 1c.
An immunocytochemistry assay was developed to detect screening hits based on the proliferation marker, Ki67, and the HSC maker, CD133. The antigen recognized by the antibody, Mab-Ki67, was chosen as it is a nuclear protein strictly associated with cell cycle progression and is only expressed in cells undergoing mitosis, while absent from quiescent cells [33]. As a marker of stemness, we used the cell surface antigen CD133, which in hematopoietic tissues and blood, is selectively expressed on HSCs that are highly enriched for LTRCs and their immediate progeny (Fig. 1d).
CombiCult matrix development
Cytokines and growth factors used to design the CombiCult screening matrix were chosen for their reported effects on the maintenance and expansion of CB HSCs. The basal media used contained essential cytokines for the survival of hematopoietic cells. In particular, we included our observation that the addition of SCF and TPO served as an important starting point in our formulation of an optimal cocktail for CB HSPC expansion [30]. It has been reported that the fms-related tyrosine kinase 3 ligand (Flt3L) is essential for expansion of early stem cells [CD34+ Thy-1 (CD90)+ CD45+] [34,35]. However, other studies suggest that it can be substituted for by fibroblast growth factor-1 (FGF-1), which has a greater mitogenic effect and at the same time preserves the LTR ability of HSCs [36]. Expansion of CB CD133+ cells in the presence of angiopoietin-like 5 (ANGPTL5) and insulin growth factor-binding protein 2 (IGFBP2) has a beneficial effect on the capacity of cells for long-term engraftment in animal models [37]. In turn, oncostatin M (OSM) counteracts the loss of repopulating activity seen in HSCs during SCF-driven proliferation [38]. It is also reported that interplay between niche-mediated activities of Notch and Wnt signaling controls the balance between CB stemness and proliferation [39,40]. Therefore, the Notch ligand, DLL1, and the noncanonical Wnt pathway activator, Wnt5a, were included in the screening matrix, in addition to Flt3L, FGF-1, ANGPTL5, IGFBP2, and OSM, and the basal cocktail of SCF, TPO, and heparin.
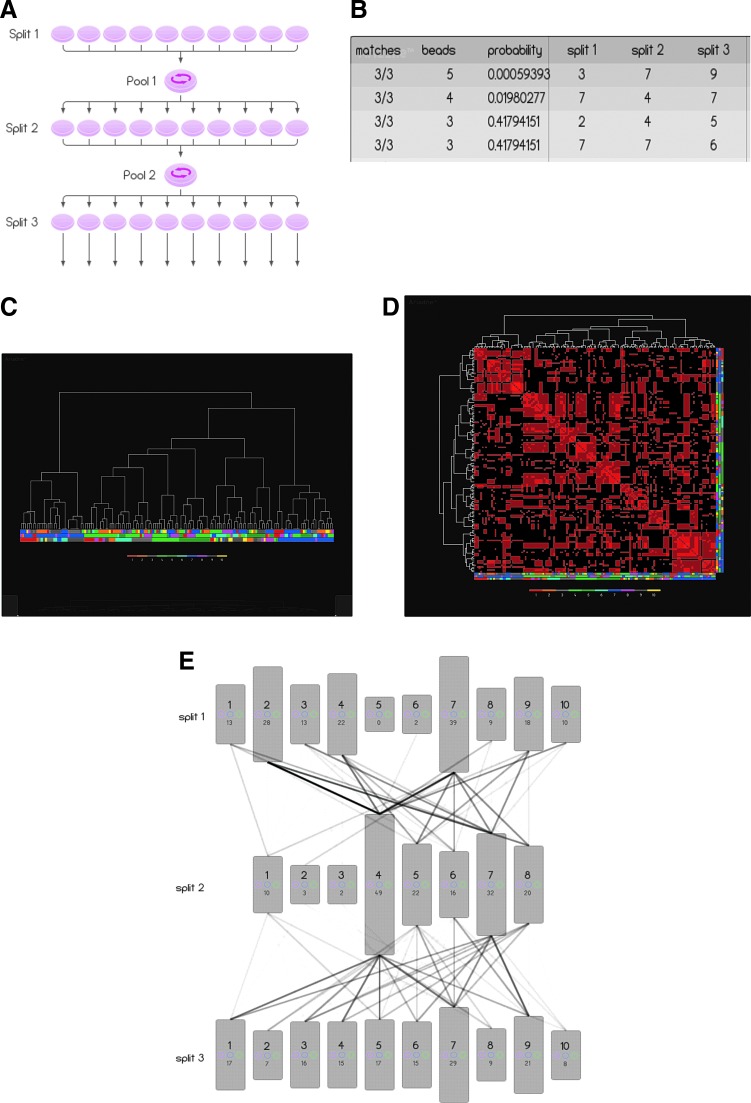
For optimization of the CombiCult screening matrix, we applied a mathematical algorithm (see Supplementary Experimental Procedures section), which rationalizes the distribution of medium components among the different formulations to minimize component bias. To create 10 different medium compositions, specific rules were introduced to ensure that certain components were never used together or that at least one component from a given class was always included. These 10 medium compositions were then repeated in each of three cycles of serial cell culture (Table 1; Fig. 2a).
FIG. 2.
CombiCult screen and Ariadne analysis. (A) CombiCult matrix scheme showing the split-pool schedule and number of conditions sampled. Ariadne analysis of the CombiCult screen showing (B) the fingerprint analysis: each row of the table represents a group of hits clustered according to protocol similarity. Column 1 identifies the number of culture conditions the group has in common; column 2, the number of hits included in the group; and column 3 the probability of the group occurring by chance. Columns 4–6 identify common media in the groups. (C) Hierarchical clustering analysis of 124 unique protocols derived from the screen, showing all protocol clusters. Each node at the bottom of the dendrogram (leaf node) corresponds to a hit bead. The associated protocol is denoted by the column of four colors directly below the node, specifying the media sampled in splits 1–3. The legend at the bottom of the figure specifies the color used to denote the cell culture media in each split. (D) The similarity matrix comprising a pairwise comparison of all protocols. Each column and each row corresponds to a protocol. The brightness of each cell in the matrix is proportional to the number of identical cell culture media shared by the two protocols. The brightest cell corresponds to identical protocols, while a black cell corresponds to two protocols with no common media. The diagonal row of cells (from the top left to bottom right) corresponds to protocols being compared with themselves. Protocol families with high internal homology appear as bright red squares. (E) Schematic diagram illustrating an overlay of all protocols deconvoluted from 154 hits. The height of boxes representing each cell culture medium is proportional to the number of hits generated by that medium (written at the bottom of each box). The opacity of the linkage lines is proportional to the number of hits generated by specific medium combinations—the darkest line corresponds to 12 hits.
CombiCult screen results
Screening of 10,000 beads that had been cultured in 1,000 possible combinations of cell culture media resulted in isolation of 220 hits positive for both Ki67 and CD133 (2.2% of beads). Complete tagging data were obtained for 154 beads (70%), and from these, 124 unique protocols were identified. Comparison of the protocols deduced from these hits was performed using Ariadne bioinformatics software. Each of the medium combinations tested by CombiCult was sampled by multiple beads, allowing statistical analysis of results. In the present experiment, where 10,000 beads were used to test 1,000 protocols, each medium sequence (protocol) was sampled by an average of 10 independent beads, therefore maximally efficient protocols should return multiple hits. We identified four protocols that were sampled by multiple beads (Fig. 2b): protocols, 379 (ie, media 3 stage 1, media 7 stage 2, media 9 stage 3) (5 repeats), 747 (4 repeats), 245, and 776 (3 repeats). We found no multiple hits that had been cultured in the same medium in all three stages of expansion. However, some hit protocols, such as 747, had near-identical media for all stages, but for the substitution of Flt3L for FGF-1 in stage 2. Moreover, the majority of the top protocols were in fact asymmetrical, with different components used at each stage of the expansion protocol.
Since similarity among the putative protocols discovered by combinatorial cell culture likely indicates protocol efficacy, we used hierarchical clustering analysis to group protocols (pathway diagram Fig. 2c) and then scored a pairwise comparison of each (similarity matrix Fig. 2d). In this way, we identified several protocol clusters with substantial internal homology all centered on multiple hit beads. Three main clusters can be seen in the similarity matrix (Fig. 2d). One revolves around media 7 in stage 2 (top left corner); a larger less compact cluster (center) shares media 4 in stage 2 and a third cluster (bottom right) features media 7 in stages 1 and 3. This type of analysis allowed us to identify media 7 as the most favorable combination of medium components for HSPC expansion. Media 4 is similar in composition to media 7, but for the substitution of Flt3L by FGF-1, two factors which may be interchangeable. In addition to cluster analysis, Ariadne also displays the results as a pathway diagram (Fig. 2e), which illustrates not only the most important links between medium composition at different stages but also the frequency in which given media are used. The pathway diagram in Fig. 2e highlights the importance of stage 2, demonstrating that at this stage, the presence of factors with mitogenic activity, such as Flt3L and FGF-1, and the absence of factors such as DLL1 (conditions 4–8) are essential for expansion. The unequal distribution of hit beads among the different conditions in stages 1 and 2 also suggests that these two stages are more important in determining cell expansion than the late stage.
Component analysis
Component analysis measures the usage of each medium component in screening hits, considering the number of times it is used in the matrix. Flt3L and IGFBP2 were the most frequently used factors. However, we also observed that some cytokines and growth factors occurred preferentially at specific stages of expansion. For example, Flt3L was most frequently present in the first and also final stages of expansion, while DLL1 seemed to be more important at the beginning of the expansion process, but in fact seldom found in the second stage, while FGF-1 was preferentially used in stage 2.
A discrete K-medoids clustering algorithm [41] was also applied to the dataset to find commonly used components. The components comprising the top cluster center are illustrated in the first row of Table 2, which coincide with the components used in the media in protocol 747, which was one of the highest ranking protocols indicated by Ariadne analysis. The resulting conditions at each stage were then stacked to highlight component commonality (Table 2).
Table 2.
Component Analysis Based on the K-Medoids Algorithm
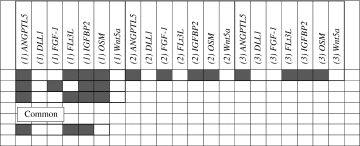 |
Stacked splits for central bead.
Validations
Based on the different types of result analyses discussed above, 13 protocols were chosen for further study. Protocol validations were carried out in alginate beads to reproduce the conditions used in the CombiCult screen. Protocols 1–6 (Supplementary Table S1; associated with Fig. 3) were chosen based on Ariadne pathways/probability analysis, protocols 7–9 based on Ariadne component analysis, and protocols 12 and 13 based on the K-medoids algorithm described earlier.
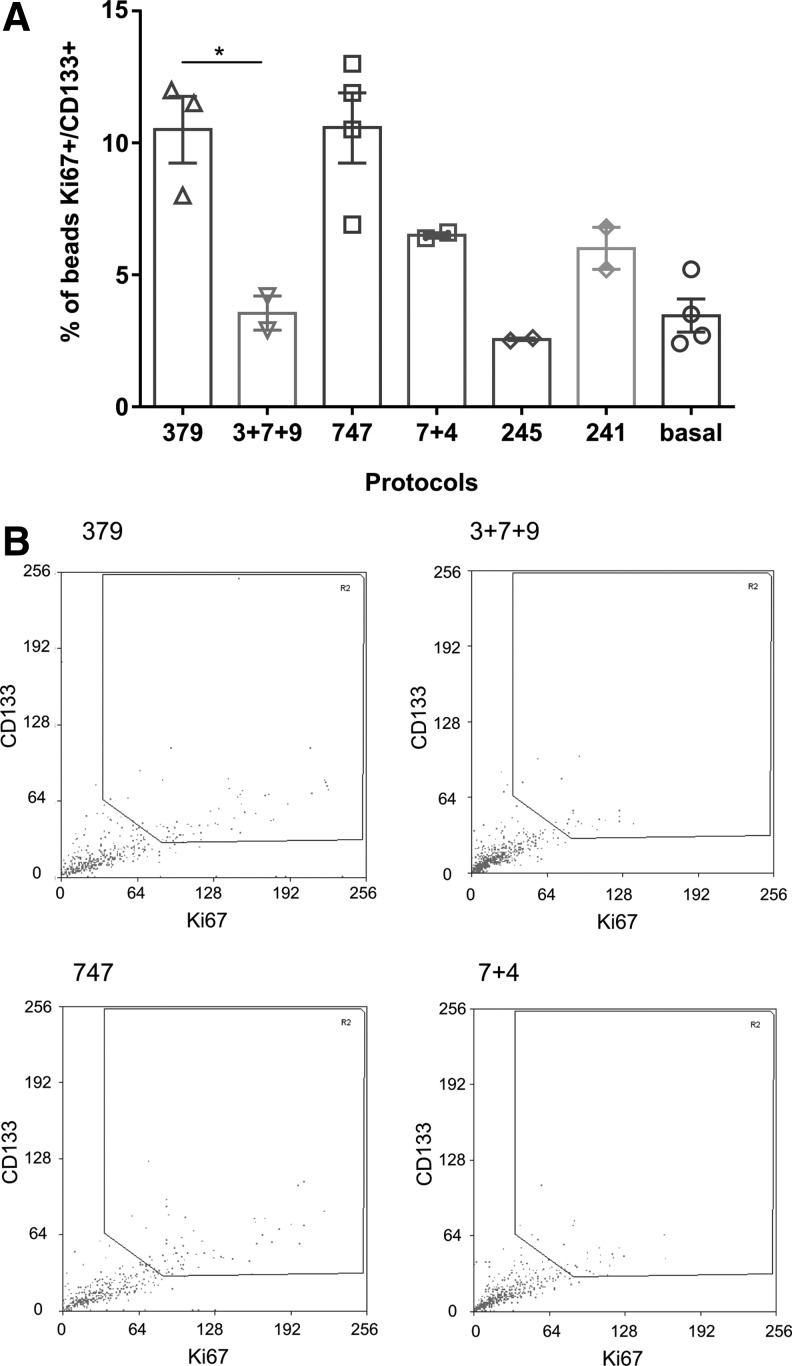
FIG. 3.
Validation of top ranked protocols in alginate beads. Expansion of CD133+ cells in alginate beads using the top ranking protocols was evaluated using large particle flow cytometric analysis (COPAS). (A) Percentage of beads containing cells positive for Ki67 and CD133 with total fluorescence values above the threshold is shown. Values are mean ± SEM for two to four technical replicates with a pool of three independent CB donors. Statistical analysis was performed in GraphPad Prism using one-way ANOVA and Tukey's ad hoc test, and P < 0.05 was considered as statistically significant (*P < 0.05). (B) Flow cytometry plots from large particle sorter (COPAS) showing the gating strategy and the distribution of beads in two top ranking protocols (379 and 747 as a serial or mix protocol). ANOVA, analysis of variance.
For the validations, MACS-enriched CB CD133+ cells were encapsulated in alginate and expanded according to the 13 protocols listed in Supplementary Table S1, as described in the Materials and Methods section. Quantification of positive beads was carried out using a large particle sorter (COPAS; Union Biometrica). To set up a threshold gate, a negative control consisting of alginate-encapsulated cells grown in StemSpan SFEM together with the basal cocktail of SCF, TPO, and heparin only and stained with the markers, Ki67 and CD133, was run. From the results of the first round of validations, the top four protocols were repeated using CB CD133+ cells from different CB donors encapsulated in alginate. We also investigated if the order of addition of components had an impact on the expansion of CD133+ cells. To this end, we compared the stepwise protocols (ie, culture in a series of media or serial protocols) with culture in a single formulation comprising a mix of the components present in all media used in the three stages of expansion (mixed protocols—Supplementary Table S2). The highest ranking protocols by Ariadne analysis (747 and 379) gave the highest percentage of beads containing proliferating CD133+ cells (Ki67+/CD133+ of 10.5%, compared with only 3.5% in the basal medium). Furthermore, there was a statistically significant difference (P < 0.05) in the proportion of positive beads between the serial protocol and the mixed formulations, with the media added in series outperforming the mixed protocols (Fig. 3a). Interestingly, the asymmetrical protocol 379 was three times more efficient at expanding CD133+ cells when different media were applied in sequence compared with using a single medium containing a mixture of all components, emphasizing the importance of the order of addition of individual components and the timing of exposure to each medium composition.
Solid scaffolds for HSC expansion
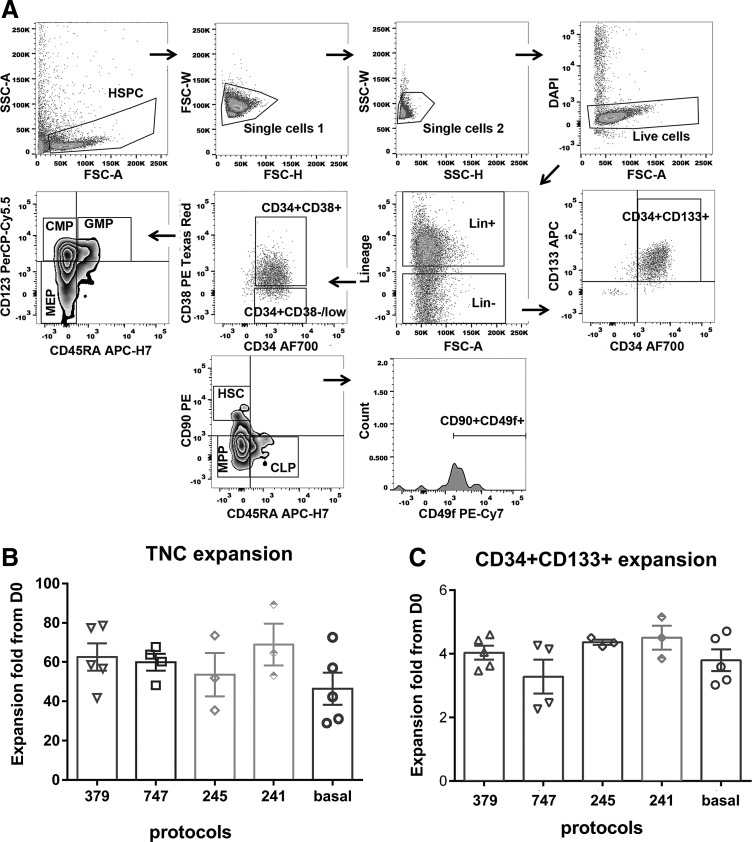
The four protocols identified as the best performing ones in the above alginate validations were next evaluated in a previously established 3D nanofiber scaffold system [30,31]. Cryopreserved CD133+ cells isolated from three to five separate CB donors were thawed, cultured overnight in SCF and TPO, and then on day 0 seeded onto 24-well Nanex 3D nanofiber-coated plates at a density of 2,500 cells/mL. Cells were then expanded using the serial protocols, 379, 747, 241, and 245 (described in Supplementary Table S3; associated with Fig. 4), with medium changes at days 2 and 5. On days 0 and 8, viable nucleated cells were quantified using CountBright beads to evaluate total cell expansion. The proportion of HSPCs in the expanded cells was determined by multicolor flow cytometric analysis [23,30–32,42]. Figure 4A shows the flow cytometric gating strategy used for these experiments.
FIG. 4.
Strategy for flow cytometric gating of human CB CD133+ subsets. (A) Representative flow cytometric gating strategy is shown for human CB CD133+ cells selected at day 0 of culture. Single, live (top density plots), and lineage-negative (Lin−) cells were discriminated into two subsets based on their expression of CD34 and CD38 or analyzed for CD34+CD133+ cell content. The CD34+CD38−/low population was further distinguished based on CD90 and CD45RA staining to distinguish HSC, MMP, and CLP. In addition, CD90+CD45RA− HSCs were characterized for CD49f expression. The CD34+CD38+ population was segregated into the progenitor subsets, GMP, CMP, and MEP, based on their expression of CD123 and CD45RA (Supplementary Data 2; MIFlowCyt configuration file). (B, C) Fold expansion of (B) total nucleated cells and (C) Lin−CD34+CD133+ cells after 8-day culture of 2,500 human CB CD133+ cells on 3D nanofiber scaffolds. Cytokines were added by medium change for the protocols shown (379, 747, 245, 241, and basal) at specific days of culture as described in Table 2. Individual biological replicates and average values are shown. Values are mean ± SEM for three to five donors (biological replicates). Statistical analysis was performed in GraphPad Prism using one-way ANOVA and Tukey's ad hoc test and P < 0.05 was considered as statistically significant.
Over the 8-day period, all protocols, including the basal media, supported the expansion of total nucleated cells. Expansion of total nucleated cells ranged from 48- to 68-fold from starting cell numbers at day 0, with all four protocols performing better than the basal control, although these differences were not statistically significant (Fig. 4B). When we investigated the expansion of the Lin−CD34+CD133+ HSPC subset, the fold expansion for these cells for 379, 747, 245, 241, 379, and basal protocols was 4.0 ± 0.2, 3.3 ± 0.5, 4.4 ± 0.1, 4.5 ± 0.4, and 3.8 ± 0.3, respectively (mean ± SEM; Fig. 4C), which again did not show statistically significant differences between treatment groups. However, when we subsequently distinguished the Lin−CD38−CD34+CD45RA−CD90+CD49f+ HSC subset based on the multicolor phenotypic analysis of Notta et al. [23], the CombiCult-derived protocols showed a clear advantage over the basal media alone. While in the basal media the fold expansion of this HSC population only reached 3.4 ± 0.7, expanding cells with protocols, 379, 747, 245, and 241, increased this phenotypically defined HSC population by 10.3 ± 1.3, 7.8 ± 1.3, 8.7 ± 0.6, and 9.1 ± 1.8-fold, respectively (Fig. 5A). One-way ANOVA analysis following multiple comparisons suggests that there are significant differences between protocols, 379, 245, and 241, against the basal protocol as the multiplicity-adjusted P values are 0.0014, 0.0320, and 0.0214, respectively. We extended these studies to examine the effect of these protocols on more lineage-committed cell subsets. As indicated in Fig. 4A, cells were segregated phenotypically into MPP (Multipotent Progenitor, Lin−CD38−CD34+CD45RA−CD90−), CLP (Common Lymphoid Progenitor, Lin-CD38−CD34+CD45RA+CD90−), CMP (Common Myeloid Progenitor, Lin−CD38+CD34+CD45RA−CD123+), GMP (Granulocyte–Monocyte Progenitor, Lin−CD38+CD34+CD45RA+CD123+), and MEP (Megakaryocyte–Erythroid Progenitor, Lin−CD38+CD34+CD45RA−CD123−) subsets. Figure 5B shows that the 379 protocol, where the highest HSC subset expansion was observed, demonstrated lower average levels of MPP fold expansion when compared with all other expansion conditions tested, although this did not reach statistical significance. In contrast, the 245 protocol, which contains Wnt5a as an additional growth factor, supported the highest average MPP, CLP, and MEP fold expansion over 8 days of culture (Fig. 5B, C, E, respectively). Basal conditions were on average less supportive of CMP and MEP expansion (Fig. 5C, E, respectively) and more permissive for GMP expansion (Fig. 5F) than the other protocols tested from the CombiCult screen (Fig. 5).
FIG. 5.
Expansion of human CB CD133+ cell subsets on 3D nanofiber scaffolds. Histograms show the fold change in phenotypically defined HSC (A), MPP (B), CLP (C), CMP (D), MEP (E), and GMP (F) after 8 days of culture commencing from 2,500 human CB CD133+ cells plated onto 3D nanofiber scaffolds at day 0. Cytokines were added by medium change for the protocols shown (379, 747, 245, 241, and basal) at specific days of culture as described in Table 2. Individual biological replicates and average values are shown. Values are mean ± SEM for three to five donors (biological replicates). Statistical analysis was performed in GraphPad Prism using one-way ANOVA and Tukey's ad hoc test and P < 0.05 was considered as statistically significant (**P < 0.01, *P < 0.05).
Discussion
CB offers an alternative supply of HSCs for transplantation and has several clinical and logistical advantages over more conventional BM and mPB. Nevertheless, single CB grafts contain limited cell numbers and CB HSCT is associated with delayed early and long-term hematological engraftment [5–7]. Indeed, it has been reported that CB units contain 25- to 50-fold fewer early engrafting neutrophil and platelet progenitor cells than mPB [43]. Delays in early neutrophil recovery post-CB HSCT can be overcome by using in vitro pretreatment strategies to either increase specific progenitor cell numbers [11,13–17] or to improve their adhesive, homing, and BM retention properties [2–4,44–46]. CB can be used as a source of stem cells to generate blood cells ex vivo for use in difficult-to-transfuse patients and also for gene editing to modify their phenotype (eg, to make the HSPCs resistant to HIV infection) or correct defective genes in monogenic diseases [8,9,10,12,47], therefore it is important to enhance HSPC numbers in CB while maintaining potentiality. Different approaches to increase HSPC numbers in single CB units include use of cytokine cocktails, stromal coculture, chemicals, and small molecules [13,14,16,17,34–38,40, 42,45,48–53]. Additionally, high-throughput approaches have been used in the past to identify molecules that promote the expansion of HSPCs such as UM271 [54], PGE2 [46], and SR1 [55], which are now being tested in clinical trials [15].
The screening methodology reported here not only allows the identification of individual compounds but also tests combinations of factors that may have complementary activities. In addition, CombiCult is capable of determining the importance of sequential exposure of cells to different signaling inputs. To test the benefits of this method, we designed a pilot study, in which we used a limited number of cytokines and ligands that had been shown individually or in combination to expand hematopoietic cells. In this CombiCult screen, 10 unique mixes of 4 cytokines and/or ligands were tested in 1,000 possible combinations leading to the discovery of 124 positive protocols. Importantly, among these, we discovered new combinations of factors that were powerful at promoting the proliferation of the CD133+ HSPC subpopulations. Furthermore, we demonstrated that serial protocols, where components are added in a particular order and with specific timing, are superior to mixes where all components are applied to cells simultaneously, suggesting that the order of addition of individual components has a profound influence on cell signaling and therefore affects cell fate. As an interesting example, our high-throughput screen revealed that activation of the Notch pathway (DLL1) at the start and end of the expansion process has a positive effect, as in protocol 379, while the presence of DLL1 in stage 2 was detrimental to proliferation and therefore does not feature among positive protocols. Moreover, cells continuously exposed to this Notch ligand, as in the 3 + 7 + 9 mix, fail to reach the expansion levels achieved in the serial protocol. On the other hand, mitogenic signals mediated by Flt3L or FGF-1 were found to be crucial for the second stage of expansion. Although both FGF-1 and Flt3L have been shown to preserve the long-term repopulating activity of HSCs in culture [34–36], our screen shows that they have differential effects at the three stages of expansion. FGF-1 has unfavorable effects when present at the beginning and end of the expansion, but is preferentially used instead of Flt3L in the second stage. When both Flt3L and FGF-1 are used together (as in 7 + 4 mix), proliferation of CD133+ cells diminishes compared with the serial 747 protocol. CombiCult technology therefore not only permits an assessment of the serial addition of different cytokine cocktails, but also provides the opportunity to rapidly assess the dose response to cytokines or small molecules, which may alter HSPC responses [56]. We therefore suggest that this differential exposure to growth factors may more closely reflect the mechanisms by which BM hematopoietic stem cell niches dynamically regulate hematopoiesis [57] and where over 1011 human blood cells are generated on average each day. Notably, such effective hematopoiesis has not yet been replicated with current ex vivo expansion protocols for single CB units.
High-throughput screening for expansion of HSCs has also been hampered by the lack of availability of singular markers, which define the true HSC population. Although CD34 has been traditionally used to mark the HSPC compartment, substantial evidence indicates that expression of CD133 correlates better with long-term repopulation ability than CD34 [18,21], an observation supported by the fact that CB has higher proportions of CD133+ cells than mPB and BM [19,49]. The discrimination of a more defined population using the markers Lin−CD38−CD34+CD45RA−CD90+CD49f+ [23], which closely correlates with long-term repopulation ability, can only be achieved by multifactorial flow cytometry. However, this type of analysis is hard to implement in a high-throughput manner unless highly specialized equipment is used [58]. Our results support the view that the CD133+ compartment contains all the LTR HSCs as choosing protocols, which preferentially amplified this population, singled out protocols that also amplify the more restricted Lin−CD38−CD34+CD45RA−CD90+CD49f+ population. As an alternative in their high-throughput screen, Fares et al. [54] analyzed a library of 5,280 compounds for their ability to expand human CD34+CD45RA−mPB HSPCs (that were less limited in cell number than those from single CB donors examined here) using flow cytometry before in vivo confirmation in NSG mice. In our study, analysis of more differentiated subpopulations of CB hematopoietic progenitor cells revealed that different protocols preferentially expand certain precursor populations. For example, protocol 245, which did not favor HSC expansion, promoted proliferation of the MPP population, and conversely protocol 379 favored expansion of phenotypically defined HSCs at the expense of MPP amplification. As expected, in basal media, a higher proportion of more committed progenitors such as GMPs were observed. These studies provide the basis for selecting protocols, which can be used to confirm the expansion of repopulating HSPCs in surrogate in vivo models or their differentiation into specific progenitor subsets.
In summary, the development of new generation of CombiCult screening technology by merging it with encapsulation of nonadherent cells in hydrogel beads not only led to the discovery of new medium formulations for maintenance and expansion of naïve CB HSC cells but also demonstrated that expansion of hematopoietic cells is a multistage process in which exposure to different cytokine combinations over time influences HSC self-renewal and differentiation ex vivo. This technology will now be used to screen a larger set of cytokines and small molecules to identify optimal combinations that support the expansion of HSCs from CB and BM.
Supplementary Material
Acknowledgments
The authors would like to dedicate this article to the memory of and acknowledge Dr. Abdel Elamin for his expert assistance in isolating CD133+ cells. The authors would also like to thank Sandy Britt, Jan Walton, and Yvonne Caffrey and the NHS Cord Blood Bank for provision of CB units and Drs. David Rodham and Bill Thompson for their project management. These studies have received grant support from Technology Strategy Board and Innovate UK, the National Institutes of Health Research under its Program Grants Scheme (RP-PG-0310-1003), the University of Oxford RDM award, and NHS Blood and Transplant.
Author Disclosure Statement
The views expressed in this publication are those of the authors and not necessarily those of NHS, NHS Blood and Transplant, NIHR, or the Department of Health. The authors (B.K., P.P., P.H., F.G., M.v.d.G., Y.Z., and S.M.W.) have no conflict of interests or any disclosures to declare. The authors (M.T., D.H., T.W., L.H., and Y.C.) are or have been employees of Plasticell Ltd.
References
- 1.Hidalgo A. and Frenette PS. (2005). Enforced fucosylation of neonatal CD34+ cells generates selectin ligands that enhance the initial interactions with microvessels but not homing to bone marrow. Blood 105:567–575 [DOI] [PubMed] [Google Scholar]
- 2.Kahn J, Byk T, Jansson-Sjostrand L, Petit I, Shivtiel S, Nagler A, Hardan I, Deutsch V, Gazit Z, et al. (2004). Overexpression of CXCR4 on human CD34+ progenitors increases their proliferation, migration, and NOD/SCID repopulation. Blood 103:2942–2949 [DOI] [PubMed] [Google Scholar]
- 3.Ohno N, Kajiume T, Sera Y, Sato T. and Kobayashi M. (2009). Short-term culture of umbilical cord blood-derived CD34 cells enhances engraftment into NOD/SCID mice through increased CXCR4 expression. Stem Cells Dev 18:1221–1226 [DOI] [PubMed] [Google Scholar]
- 4.Xia L, McDaniel JM, Yago T, Doeden A. and McEver RP. (2004). Surface fucosylation of human cord blood cells augments binding to P-selectin and E-selectin and enhances engraftment in bone marrow. Blood 104:3091–3096 [DOI] [PubMed] [Google Scholar]
- 5.Ballen KK, Gluckman E. and Broxmeyer HE. (2013). Umbilical cord blood transplantation: the first 25 years and beyond. Blood 122:491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watt SM. (2014). Umbilical cord blood banking. In: Reference Module in Biomedical Sciences. Elsevier Press, Kidlington, UK: Article 04238.2014 [Google Scholar]
- 7.Michel G, Galambrun C, Sirvent A, Pochon C, Bruno B, Jubert C, Loundou A, Yakoub-Agha I, Milpied N, et al. (2016). Single versus double-unit cord blood transplantation for children and young adults with acute leukemia or myelodysplastic syndrome. Blood 127:3450–3457 [DOI] [PubMed] [Google Scholar]
- 8.Engert A, Balduini C, Brand A, Coiffier B, Cordonnier C, Dohner H, de Wit TD, Eichinger S, Fibbe W, et al. (2016). The European hematology association roadmap for European hematology research: a consensus document. Haematologica 101:115–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, Moi D, Mazzieri R, Bonini C, et al. (2014). Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pernet O, Yadav SS. and An DS. (2016). Stem cell-based therapies for HIV/AIDS. Adv Drug Deliv Rev 1:187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baron F, Ruggeri A. and Nagler A. (2016). Methods of ex vivo expansion of human cord blood cells: challenges, successes and clinical implications. Expert Rev Hematol 9:297–314 [DOI] [PubMed] [Google Scholar]
- 12.Darghouth D, Giarratana M-C, Oliveira L, Jolly S, Marie T, Boudah S, Mario N, Junot C, Douay L. and Romeo P-H. (2016). Bio-engineered and native red blood cells from cord blood exhibit the same metabolomic profile. Haematologica 101:e220–e222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lima M, McNiece I, Robinson SN, Munsell M, Eapen M, Horowitz M, Alousi A, Saliba R, McMannis JD, et al. (2012). Cord-blood engraftment with ex vivo mesenchymal-cell coculture. N Engl J Med 367:2305–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delaney C, Heimfeld S, Brashem-Stein C, Voorhies H, Manger RL. and Bernstein ID. (2010). Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med 16:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horwitz ME, Chao NJ, Rizzieri DA, Long GD, Sullivan KM, Gasparetto C, Chute JP, Morris A, Mcdonald C, et al. (2014). Umbilical cord blood expansion with nicotinamide provides long-term multilineage engraftment. J Clin Investig 124:3121–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horwitz ME. (2016). Ex vivo expansion or manipulation of stem cells to improve outcome of umbilical cord blood transplantation. Curr Hematol Malig Rep 11:12–18 [DOI] [PubMed] [Google Scholar]
- 17.Wagner JE, Brunstein CG, Boitano AE, DeFor TE, McKenna D, Sumstad D, Blazar BR, Tolar J, Le C, et al. (2015). Phase I/II trial of StemRegenin-1 expanded umbilical cord blood hematopoietic stem cells supports testing as a stand-alone graft. Cell Stem Cell 18:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Wynter EA, Buck D, Hart C, Heywood R, Coutinho LH, Clayton A, Rafferty JA, Burt D, Guenechea G, et al. (1998). CD34+AC133+ cells isolated from cord blood are highly enriched in long-term culture-initiating cells, NOD/SCID-repopulating cells and dendritic cell progenitors. Stem Cells 16:387–396 [DOI] [PubMed] [Google Scholar]
- 19.McGuckin CP, Pearce D, Forraz N, Tooze JA, Watt SM. and Pettengell R. (2003). Multiparametric analysis of immature cell populations in umbilical cord blood and bone marrow. Eur J Haematol 71:341–350 [DOI] [PubMed] [Google Scholar]
- 20.Radtke S, Görgens A, Kordelas L, Schmidt M, Kimmig KR, Köninger A, Horn PA. and Giebel B. (2015). CD133 allows elaborated discrimination and quantification of haematopoietic progenitor subsets in human haematopoietic stem cell transplants. Br J Haematol 169:868–878 [DOI] [PubMed] [Google Scholar]
- 21.Takahashi M, Matsuoka Y, Sumide K, Nakatsuka R, Fujioka T, Kohno H, Sasaki Y, Matsui K, Asano H, Kaneko K. and Sonoda Y. (2014). CD133 is a positive marker for a distinct class of primitive human cord blood-derived CD34-negative hematopoietic stem cells. Leukemia 28:1308–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J. and Buck DW. (1997). AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood 90:5002–5012 [PubMed] [Google Scholar]
- 23.Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I. and Dick JE. (2011). Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science 333:218–221 [DOI] [PubMed] [Google Scholar]
- 24.Choo Y. (2002). Cell culture. Patent US8,114,669 EP1551954
- 25.Choo Y. (2008). Use of combinatorial screening to discover protocols that effectively direct the differentiation of stem cells. In: Stem Cell Research and Therapeutics. Shi DOCY, ed. Springer Science + Business Media, Dordrecht, Netherlands: pp 227–250 [Google Scholar]
- 26.Tarunina M, Hernandez D, Johnson CJ, Rybtsov S, Ramathas V, Jeyakumar M, Watson T, Hook L, Medvinsky A, Mason C. and Choo Y. (2014). Directed differentiation of embryonic stem cells using a bead-based combinatorial screening method. PLoS One 9:e104301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choo Y, Johnson CJ, Odenwalder PK. and Jayasinghe. SN. (2011). Nested cell encapsulation. Patent WO 2011/047870
- 28.Arumuganathar S, Suter N, Walzel P. and Jayasinghe SN. (2009). Aerodynamically assisted jetting and threading for processing concentrated suspensions containing advanced structural, functional and biological materials. Biotechnol J 4:64–72 [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto M. and Nishimura T. (1998). Mersenne twister: a 623-dimensionally equidistributed uniform pseudo-random number generator. ACM Trans Model Comput Simul 8:3–30 [Google Scholar]
- 30.Gullo F, Van Der Garde M, Russo G, Pennisi M, Motta S, Pappalardo F. and Watt S. (2015). Computational modeling of the expansion of human cord blood CD133+ hematopoietic stem/progenitor cells with different cytokine combinations. Bioinformatics 31:2514–2522 [DOI] [PubMed] [Google Scholar]
- 31.Pepperell EE. and Watt SM. (2013). A novel application for a 3-dimensional timelapse assay that distinguishes chemotactic from chemokinetic responses of hematopoietic CD133(+) stem/progenitor cells. Stem Cell Res 11:707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C-H, Hale SJ, Cox CV, Blair A, Kronsteiner B, Grabowska R, Zhang Y, Cook D, Khoo CP, et al. (2016). Junctional adhesion molecule-A is highly expressed on human hematopoietic repopulating cells and associates with the key hematopoietic chemokine receptor cxcr4. Stem Cells 34:1664–1678 [DOI] [PubMed] [Google Scholar]
- 33.Schluter C, Duchrow M, Wohlenberg C, Becker MHG, Key G, Flad HD. and Gerdes J. (1993). The cell proliferation-associated antigen of antibody Ki-67: a very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J Cell Biol 123:513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Felice L, Di Pucchio T, Breccia M, Agostini F, Mascolo MG, Guglielmi C, Ricciardi MR, Screnci M, Tafuri A, Carmini D. and Arcese W. (1998). Flt3L enhances the early stem cell compartment after ex vivo amplification of umbilical cord blood CD34+ cells. Bone Marrow Transplant 22 (Suppl. 1):S66–S67 [PubMed] [Google Scholar]
- 35.Haylock DN, Horsfall MJ, Dowse TL, Ramshaw HS, Niutta S, Protopsaltis S, Peng L, Burrell C, Rappold I, Buhring HJ. and Simmons PJ. (1997). Increased recruitment of hematopoietic progenitor cells underlies the ex vivo expansion potential of FLT3 ligand. Blood 90:2260–2272 [PubMed] [Google Scholar]
- 36.Yeoh JSG, van Os R, Weersing E, Ausema A, Dontje B, Vellenga E. and de Haan G. (2006). Fibroblast growth factor-1 and -2 preserve long-term repopulating ability of hematopoietic stem cells in serum-free cultures. Stem Cells 24:1564–1572 [DOI] [PubMed] [Google Scholar]
- 37.Zhang CC, Kaba M, Iizuka S, Huynh H. and Lodish HF. (2008). Angiopoietin-like 5 and IGFBP2 stimulate ex vivo expansion of human cord blood hematopoietic stem cells as assayed by NOD/SCID transplantation. Blood 111:3415–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oostendorp RAJ, Gilfillan S, Parmar A, Schiemann M, Marz S, Niemeyer M, Schill S, Hammerschmid E, Jacobs VR, Peschel C. and Götze KS. (2008). Oncostatin M-mediated regulation of KIT-Ligand-induced extracellular signal-regulated kinase signaling maintains hematopoietic repopulating activity of Lin−CD34+CD133+ cord blood cells. Stem Cells 26:2164–2172 [DOI] [PubMed] [Google Scholar]
- 39.Dahlberg A, Woo S, Delaney C, Boyle P, Gnirke A, Bock C, Bernstein BE, Meissner A, Gottardo R. and Bernstein ID. (2015). Notch-mediated expansion of cord blood progenitors: maintenance of transcriptional and epigenetic fidelity. Leukemia 29:1948–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang J, Zhao M, Zhang A, Yu M, Lin X, Wu M, Wang X, Lu H, Zhu S, et al. (2010). Characterization of a GSK-3 inhibitor in culture of human cord blood primitive hematopoietic cells. Biomed Pharmacother 64:482–486 [DOI] [PubMed] [Google Scholar]
- 41.Park H-S. and Jun C-H. (2009). A simple and fast algorithm for K-medoids clustering. Expert Syst Appl 36:3336–3341 [Google Scholar]
- 42.van der Garde M, van Hensbergen Y, Brand A, Slot MC, de Graaf-Dijkstra A, Mulder A, Watt SM. and Zwaginga JJ. (2015). Thrombopoietin treatment of one graft in a double cord blood transplant provides early platelet recovery while contributing to long-term engraftment in NSG mice. Stem Cells Dev 24:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung AMS, Nguyen LV, Carles A, Beer P, Miller PH, Knapp DJHF, Dhillon K, Hirst M. and Eaves CJ. (2013). Analysis of the clonal growth and differentiation dynamics of primitive barcoded human cord blood cells in NSG mice. Blood 122:3129–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cutler C, Multani P, Robbins D, Kim HT, Le T, Hoggatt J, Pelus LM, Desponts C, Chen Y-B, et al. (2013). Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood 122:3074–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, Kim HT, Nellore A, Patsoukis N, Petkova V, McDonough S, Politikos I, Nikiforow S, Soiffer R, et al. (2014). Prostaglandin E2 promotes survival of naive UCB T cells via the Wnt/beta-catenin pathway and alters immune reconstitution after UCBT. Blood Cancer J 4:e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.North TE, Goessling W, Walkley CR, Lengerke C, Kamden R, Lord AM, Weber GJ, Bowman TV, Jang I, et al. (2007). Prostaglandin E2 regulates vertebrate haematopoetic stem cell homeostasis. Nature 447:1007–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lombardo A. and Naldini L. (2014). Genome editing: a tool for research and therapy: targeted genome editing hits the clinic. Nat Med 20:1101–1103 [DOI] [PubMed] [Google Scholar]
- 48.Dahlberg A, Brashem-Stein C, Delaney C. and Bernstein ID. (2014). Enhanced generation of cord blood hematopoietic stem and progenitor cells by culture with StemRegenin1 and Delta1(Ext-IgG.). Leukemia 28:2097–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drake AC, Khoury M, Leskov I, Iliopoulou BP, Fragoso M, Lodish H. and Chen J. (2011). Human CD34+ CD133+ hematopoietic stem cells cultured with growth factors including Angptl5 efficiently engraft adult NOD-SCID Il2rγ-/- (NSG) mice. PLoS One 6:e18382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lechman ER, Gentner B, Ng SWK, Schoof EM, van Galen P, Kennedy JA, Nucera S, Ciceri F, Kaufmann KB, et al. (2016). miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell 29:214–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peled T, Mandel J, Goudsmid RN, Landor C, Hasson N, Harati D, Austin M, Hasson A, Fibach E, Shpall EJ. and Nagler a. (2004). Pre-clinical development of cord blood-derived progenitor cell graft expanded ex vivo with cytokines and the polyamine copper chelator tetraethylenepentamine. Cytotherapy 6:344–355 [DOI] [PubMed] [Google Scholar]
- 52.Piacibello W, Gammaitoni L, Bruno S, Gunetti M, Fagioli F, Cavalloni G. and Aglietta M. (2000). Negative influence of IL3 on the expansion of human cord blood in vivo long-term repopulating stem cells. J Hematother Stem Cell Res 9:945–956 [DOI] [PubMed] [Google Scholar]
- 53.Rappold I, Watt SM, Kusadasi N, Rose-John S, Hatzfeld J. and Ploemacher RE. (1999). Gp130-signaling synergizes with FL and TPO for the long-term expansion of cord blood progenitors. Leukemia 13:2036–2048 [DOI] [PubMed] [Google Scholar]
- 54.Fares I, Chagraoui J, Gareau Y, Gingras S, Ruel R, Mayotte N, Csaszar E, Knapp DJHF, Miller P, et al. (2014). Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 345:1509–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, et al. (2010). Aryl hydrocarbon receptor antagonist promote the expansion of human hematopoetic stem cells. Science 329:1345–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Famili F, Brugman MH, Taskesen E, Naber BEA, Fodde R. and Staal FJT. (2016). High levels of canonical Wnt signaling lead to loss of stemness and increased differentiation in hematopoietic stem cells. Stem Cell Reports 6:652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu VWC. and Scadden DT. (2016). Heterogeneity of the bone marrow niche. Curr Opin Hematol 23:331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathews LA, Keller JM, McKnight C, Michael S, Shinn P, Liu D, Staudt LM, Thomas CJ. and Ferrer M. (2013). Multiplexing high-content flow (HCF) and quantitative high-throughput screening (qHTS) to identify compounds capable of decreasing cell viability, activating caspase 3/7, expressing annexin V, and changing mitochondrial membrane integrity. Curr Protoc Chem Biol 5:195–212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.