Abstract
Objective: Debridement to remove necrotic and/or infected tissue and promote active healing remains a cornerstone of contemporary chronic wound management. While there has been a recent shift toward less invasive polymer-based debriding devices, their efficacy requires rigorous evaluation.
Approach: This study was designed to directly compare monofilament debriding devices to traditional gauze using a wounded porcine skin biofilm model with standardized application parameters. Biofilm removal was determined using a surface viability assay, bacterial counts, histological assessment, and scanning electron microscopy (SEM).
Results: Quantitative analysis revealed that monofilament debriding devices outperformed the standard gauze, resulting in up to 100-fold greater reduction in bacterial counts. Interestingly, histological and morphological analyses suggested that debridement not only removed bacteria, but also differentially disrupted the bacterially-derived extracellular polymeric substance. Finally, SEM of post-debridement monofilaments showed structural changes in attached bacteria, implying a negative impact on viability.
Innovation: This is the first study to combine controlled and defined debridement application with a biologically relevant ex vivo biofilm model to directly compare monofilament debriding devices.
Conclusion: These data support the use of monofilament debriding devices for the removal of established wound biofilms and suggest variable efficacy towards biofilms composed of different species of bacteria.
Keywords: : wound biofilm, debridement, Pseudomonas aeruginosa, Staphylococcus aureus

Matthew J. Hardman, PhD

Andrew J. McBain, PhD
Introduction
Nonhealing chronic wounds remain a major area of unmet clinical need, leading to increased patient morbidity and mortality, while imposing a significant financial burden on healthcare providers worldwide.1,2 Wound chronicity can arise through malfunction at any stage of repair and can be influenced by local (e.g., ischemia) and systemic factors (e.g., age, malnutrition, and disease), along with imbalances in cytokine levels and growth factors.1 One of the most discussed extrinsic causes of chronicity is the presence of infection, where opportunistic “critical” colonization of a wound by microorganisms can lead to the formation of a biofilm.3–5 Biofilms are becoming increasingly implicated in pathological healing due to their resistance to the host immune defence and transient responses to most therapeutic treatments.5,6 Established biofilms have a number of distinguishing characteristics that contribute to their resistant phenotype, most notably physiological and physiochemical effects resulting from encapsulation in a tough protective layer of extracellular polymeric substances (EPS).7
While current wound management involves a multidisciplinary team approach, debridement to remove necrotic, devitalized, and infected tissue remains a primary consideration in wound management.8 Historically, debridement has been suggested to promote healing by removing nonviable ischemic tissue and exposing underlying vascularized tissue, triggering endogenous reparative processes.9 More recently, it has been suggested that debridement may also promote healing through the physical disruption and removal of established wound biofilms.7
Clinical Problem Addressed
Traditional debridement techniques, including sharp excision and curettage can be invasive and painful, requiring professional administration.10 A number of alternative, less costly debridement modalities have been developed over recent years, such as autolytic and mechanical debridement. Classical mechanical debridement involves the removal of necrotic tissue by scrubbing and stripping with cotton gauze.11 However, scrubbing and stripping can be invasive, painful, and may scrub the wound of valuable repair cells, depending on wound etiology and nature.12 Notably, the efficacy of simple cotton gauze at removing wound biofilms remains largely untested.13 Recently, new, mechanical, polymer fiber debriding devices have emerged, designed to remove both established wound biofilms and devitalized tissue with minimal discomfort (e.g., Debrisoft®; Activa Healthcare, Debrimitt™; Crawford Healthcare Ltd.14).
In this study, we report a detailed side-by-side comparison to test the efficacy of monofilament debriding devices versus traditional gauze material in mechanical debridement of established ex vivo porcine wound biofilms.
Materials and Methods
Growth of biofilms
Single-colony inoculates of Pseudomonas aeruginosa (NCTC10781) and Staphylococcus aureus (NCTC13277) were grown overnight in Mueller Hinton (MH; Oxoid) broth, at 37°C with 220 rpm shaking (Innova 44; New Brunswick Scientific). The (O/N) cultures were diluted to obtain densities at c. 107 colony forming units per milliliter (CFU/mL), verified by spread plating and colony counts. Sterile filter membranes (0.45 μm thick; Merck Millipore Ltd.) were inoculated with 10 μL of diluted cultures and incubated on MH agar plates for 48 h at 37°C (as in Merritt et al.15). The resultant 48 h established biofilms were added to wounded porcine skin.
Porcine skin preparation
Porcine skin was collected of schedule 1 killed female pigs (weighing 43–45 kg) from the abattoir, epilated using dry razors and stored at −80°C until use. Defrosted skin was cut into 1.3 × 1.3 cm2 and wounded by completely removing the epidermis (No. 22 blades; Swann Norton). The skin squares were sequentially washed in sterile phosphate buffered saline containing 5 ug/L TWEEN 20 (PBST), 0.6% sodium hypochlorite (in PBST), and 70% ethanol (in PBST). Finally, tissue was washed thrice in sterile phosphate-buffered saline (PBS) to remove solvent residues (modified from Yang et al.16). To maintain a humid environment for bacterial growth, skin squares were placed in six-well plates on sterile PBS-soaked absorbent pads. A 48-h established biofilm was added to the wounded surface of each porcine biopsy, ensuring even coverage. Nonbiofilm controls were prepared in parallel. Biopsies were incubated at 37°C for an additional 24 h to allow further biofilm maturation and attachment to the wounded porcine skin.
Mechanical debridement and sample collection
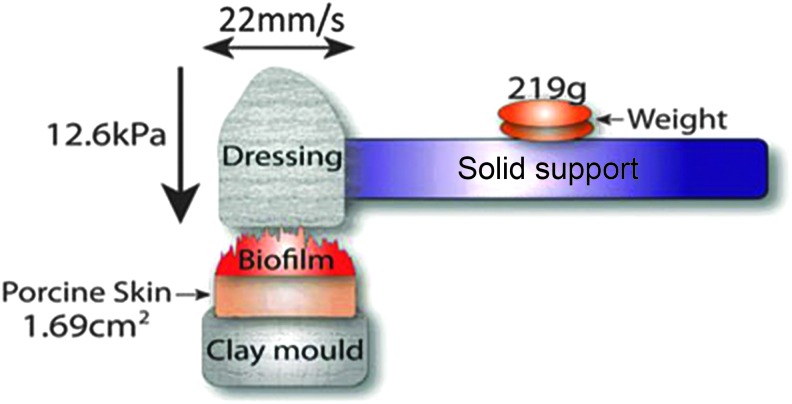
Debriding devices (gauze, Debrisoft®, and Debrimitt™) were cut into strips, firmly stapled to toothbrush heads, and loaded into an SD Mechatronik ZM-3 tooth brushing simulator (Fig. 1A). The porcine biofilm biopsies were glued onto custom fabricated clay moulds and mounted in the toothbrush simulator. Each sample was brushed in a linear manner for 50 cycles at 22 mm per second, with a travel length of 10 mm and a constant force of 12.6 kPa (Fig. 1B). Separate samples were collected for PrestoBlue™ cell viability analysis (n = 3), bacterial viability counts (n = 3), and scanning electron microscope (SEM) preparation (n = 2) and embedded in optimum cutting temperature (OCT) solution for histological analysis (n = 2). In addition, S. aureus and P. aeruginosa 48-h membrane biofilms (before addition to porcine skin) were also collected to confirm the presence of established biofilms (data not shown).
Figure 1.
Debriding apparatus. Samples were debrided using an SD Mechatronik ZM-3 toothbrush simulator (Germany). Diagram of the debriding experiment setup. Debriding devices (gauze, Debrisoft®, or Debrimitt™) were attached to solid supports and inserted into the simulator. Porcine skin biofilms were attached to custom fabricated clay moulds positioned directly beneath the debriding devices. A weight of 219 g was placed at the center of each brush, exerting a force of 12.6 kPa per sample. Debridement consisted of 50 linear strokes at a speed of 22 mm/s.
PrestoBlue™ in situ bacterial viability assay
Following debridement, the PrestoBlue™ Cell Viability Reagent (Invitrogen, Life Technologies) was applied evenly to the full surface of each tissue square and incubated for 3 min. Images were captured (Nikon D3200 Digital SLR camera with a Nikon AF-S DX Micro NIKKOR 40 mm f/2.8G Lens) and percentage cell viability calculated using ImagePro-Plus software (Media Cybernetics, Inc., Rockville, MD, USA).
Bacterial viability counts
Each sample was cut into small (<1 mm) pieces using an aseptic technique and sterile scalpel blades (No. 22 blades; Swann Norton), then vortexed in 1 mL MH broth with 5 mL sterile glass beads (Borosilicate, 3 mm diameter; Sigma-Aldrich) for 30 s. The resultant resuspended bacteria were serial diluted and plated in triplicate to determine viable CFUs per biofilm.
Scanning electron microscopy
Tissue samples (∼2 mm diameter), cut into trapezium shapes to identify orientation, were fixed for 4 h in 2% glutaraldehyde/paraformaldehyde in HEPES buffer, followed by transfer through an ETOH gradient (10%, 25%, 50%, 70%, 90%, and 3 × 100% for 15 min each). Critical point drying was performed using an E3000 (Quorum Technologies) in 100% ETOH. Samples underwent three exchanges of ETOH for liquid CO2 before heating and critical point drying. Samples were mounted onto stubs, placed in an argon vacuum, and sputter coated (SC7690; Quorum Technologies) with gold (Quanta Feg 250; FEI Company). Images were taken at three points per sample at a range of magnifications using a high-vacuum SEM. Gauze, Debrisoft®, and Debrimitt™ were also collected post-treatment, processed and dried for SEM as above, and imaged using low-vacuum SEM.
Histological analysis of frozen tissue sections
OCT embedded samples were cryosectioned at 12 μm (CM3050 S; Leica Biosystems) and stored at −80°C until use. Before staining, sectioned slides at −80°C were brought to −20°C and fixed with cold acetone for 2 min, which was then left to evaporate at room temperature (<20 min). Slides were immersed in PBS to remove any residual OCT. Modified Gram–Twort staining was achieved following Bancroft and Gamble,17 DPX mounted and imaged using an Eclipse E600 microscope (Nikon) and SPOT camera (Image Solutions, Inc.). Biofilm thickness was quantified from Gram–Twort images using ImageJ software (NIH). Concanavalin A (ConA) staining for EPS (Alexa Fluor® 488 conjugate, Molecular Probes™; Thermo Fisher Scientific) was performed O/N at 4°C. Sections were mounted in VECTASHIELD® with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories Ltd.) to counterstain bacterial DNA. Similarly, sections were stained with Acridine Orange (Sigma-Aldrich) to visualize bacteria on host tissue. Fluorescent in situ hybridization (FISH), using PNA FISH® kits (AdvanDx, Inc.) with P. aeruginosa and S. aureus probes, was used to show species specificity. Sections were imaged on an Olympus Snapshot fluorescence microscope using fluorescein isothiocyanate, DAPI, and TEXAS RED filters. S. aureus and P. aeruginosa membrane biofilms were embedded in OCT, sectioned, and then stained as above.
Statistical analyses
Statistical tests were performed on all S. aureus and P. aeruginosa quantitative data (PrestoBlue™ cell viability, viability counts, and biofilm thickness), where treatment (uninoculated skin, established “control” biofilms, gauze dressing, Debrisoft®, and Debrimitt™) was plotted against viability/abundance/thickness. For viability counts and PrestoBlue™ viability data, one-way analysis of variances (ANOVAs) were performed in R v3.2.2 (R Development Core Team) with accompanying Tukey post hoc analyses. Nonpaired T tests were performed for biofilm thickness data.
Results
Comparative quantification of biofilm removal postdebridement
Biofilms of P. aeruginosa and S. aureus established on wounded porcine skin were subjected to controlled mechanical debridement with defined speed, duration, and pressure (Fig. 1). The biofilm remaining postdebridement was assessed by the following methods:
Surface area coverage of viable biofilm
PrestoBlue™ was used to directly visualize the surface area of porcine skin with the remaining viable biofilm versus untreated established biofilms (Fig. 2A; ANOVA: F3,11 = 8, p = 0.004). In this study, Debrimitt™ and Debrisoft® significantly reduced viable P. aeruginosa biofilm surface coverage compared to control biofilms that underwent no debridement (Debrimitt™, 95% confidence interval [CI 23.15–108.13], p = 0.003; Debrisoft®, 95% CI [12.45–97.43, p = 0.01). Of note, no significant difference was found between the gauze dressing and control P. aeruginosa biofilms (95% CI [−2.49–82.49], p = 0.067). A similar trend of surface area debridement efficacy was observed with S. aureus biofilms, although the data failed to reach significance (ANOVA: F3,8 = 3.16, p = 0.086).
Figure 2.
Assessment of debridement efficacy. (A) Quantification of biofilm coverage using a resazurin-based cell viability stain. In each case, debridement reduced biofilm coverage which achieved statistical significance for Debrisoft and Debrimitt application to P. aeruginosa biofilms. (B) Viable counting of bacteria remaining indicates statistically significant reductions in colony forming units following each treatment. Monofilament debridement outperformed traditional gauze. Graphs show means; error bars represent SEM. *p < 0.05; **p < 0.01; ***p < 0.001.
Quantification of biofilm bacteria
Resuspending biofilm bacteria to gain quantitative viability data (Fig. 2B) revealed significantly reduced viable P. aeruginosa following debridement (ANOVA: F3,20 = 67.43, p < 0.001). Debrimitt™ treatment appeared most effective, resulting in a 6 log10 reduction (p < 0.001) versus control biofilms. Debrisoft® and gauze debridement both led to 5 log10 reductions in viable P. aeruginosa (p < 0.001 and p < 0.001, respectively). S. aureus viability was also significantly reduced following debridement (Fig. 2B; ANOVA: F3,11 = 81.91, p < 0.001). In this study, both Debrimitt™ and Debrisoft® treatment led to a 7 log10 reduction in S. aureus compared to control biofilms (p < 0.001). By contrast, treatment with a gauze dressing gave only a 5 log10 reduction in S. aureus (p < 0.001). Overall, the data suggest that all three debridement modalities remove viable biofilms of S. aureus and P. aeruginosa from porcine wound tissue, with Debrimitt™ proving most effective.
Visualizing established biofilms in situ
Gram–Twort stain
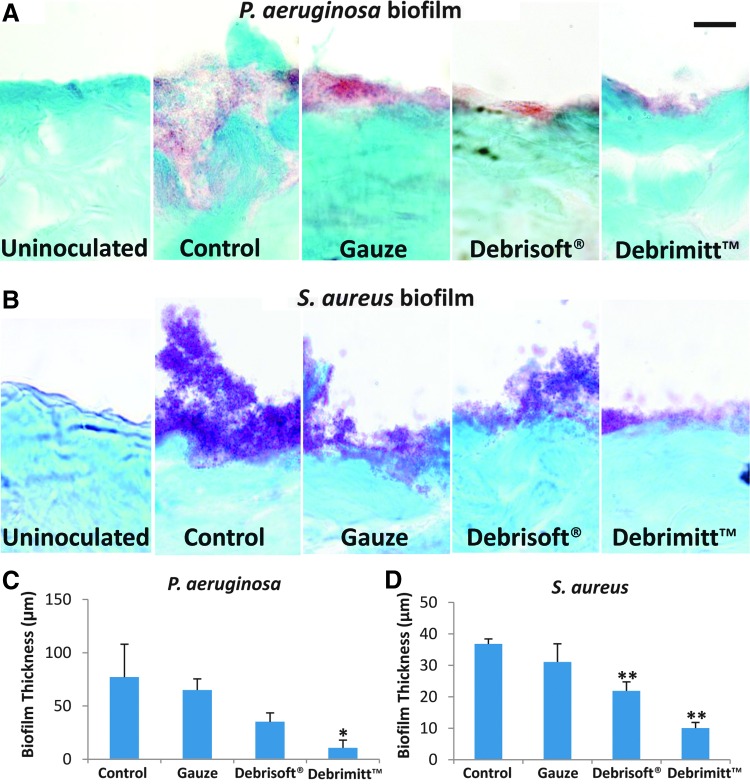
Frozen histological sections subjected to Gram–Twort staining confirmed both the presence and identity (i.e., gram positive vs. gram negative) of P. aeruginosa (pink stain) and S. aureus (purple stain) biofilms on the porcine wound surface (Fig. 3). Control biofilms of gram-negative P. aeruginosa appeared as a pink continuous mass in the wounded porcine skin, mirroring the staining found in the P. aeruginosa membrane biofilms (data not shown). Following debridement, the amount (thickness) of visible bacteria decreased, with greatest reduction in visible biofilm in the Debrimitt™-treated group (T9 = 2.28, p < 0.05; Fig. 3C). Note, staining was absent from nonbiofilm, untreated porcine skin. S. aureus, a gram-positive bacterium stained purple with the Gram–Twort stain, was readily apparent on the surface of each control porcine wound biofilm and mimicked the staining of S. aureus membrane biofilms (data not shown). Thick aggregated clumps of S. aureus were readily visualized in nondebrided established biofilm tissue. These clumps of bacteria were greatly diminished following debridement, with the greatest reduction in biofilm thickness following monofilament debridement (Debrisoft® and Debrimitt™, p < 0.01; Fig. 3D).
Figure 3.
High-power modified Gram–Twort stained images confirm the extent of biofilm removal. (A) P. aeruginosa biofilm (red) was evident at the wounded surface in all but the untreated skin. All three debriding treatments reduced biofilm, with the most efficient removal following Debrimitt™ treatment. (B) S. aureus biofilm (purple) was also clearly attached to the surface of the wounded porcine skin. Again, debridement treatment substantially reduced the surface biofilm. Note, bacteria were observed not just at the surface but also integrated into the host tissue. (C) P. aeruginosa biofilm thickness was significantly reduced with Debrimitt debridement. (D) S. aureus biofilm thickness was significantly reduced with monofilament debridement. Date shows mean ± standard error of the mean. *p < 0.05; **p < 0.01. Bar (A) = 14.4 μm.
ConA, Acridine Orange, and FISH staining
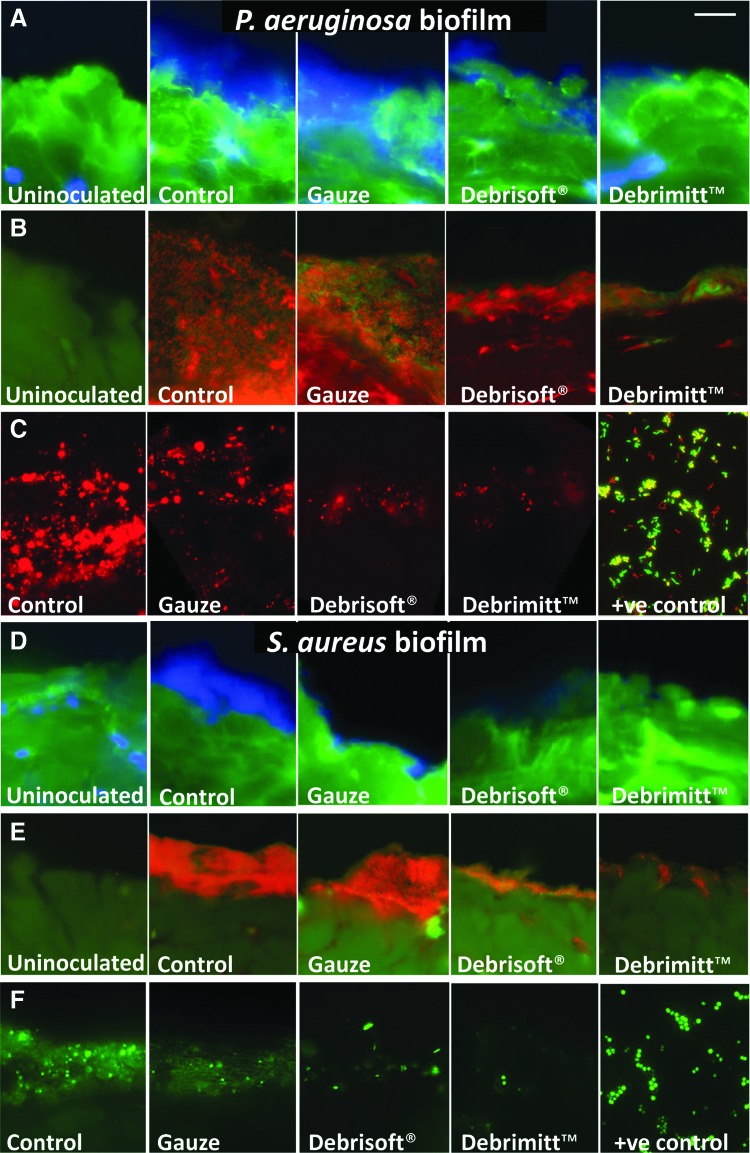
Next biofilms were stained with a combination of ConA and DAPI counterstain (Fig. 4). Labeled lectins, such as fluor-conjugated ConA, which interacts with carbohydrates, are often used to indicate biofilm formation.18 However, EPS staining can vary due to biofilm-specific variation in polymeric substances.19 The bacteria within the biofilm mass could be visualized by DAPI (blue) (Fig. 4A, D), Acridine Orange (Fig. 4B, E), and FISH staining (Fig. 4C, F). Control P. aeruginosa and S. aureus biofilms were both prominent across the surface of the porcine wound. In keeping with the Gram–Twort staining, less biofilm was observed following each debridement treatment, with greatest reduction in the Debrimitt™ treatment group. Note, no biofilm mass (blue DAPI staining, orange/red Acridine Orange staining, or FISH staining) was observed in any of the uninoculated porcine skin samples (Fig. 4).
Figure 4.
ConA, Acridine Orange, and FISH stained images confirm the extent of biofilm removal. (A) P. aeruginosa biofilms stain for bacterial EPS (green; ConA) with prominent DAPI (blue) bacteria visible at the porcine wound surface. (B) Acridine Orange staining for bacteria (red/orange) at the surface layers of the porcine wounds, with green host tissue. (C) FISH of P. aeruginosa (red) from the treatments, including a positive (+ve) slide containing P. aeruginosa (red), Escherichia coli (green), and Klebsiella pneumoniae (yellow). Debridement reduces apparent biofilm mass with Debrisoft® and Debrimitt™ showing the most effective removal. (D) S. aureus biofilms exhibit DAPI stained bacterial mass (blue). (E) Acridine Orange staining for bacteria (red/orange) at the wound surface. (F) FISH of S. aureus (green) from the treatments, including a positive (+ve) slide containing S. aureus (green). S. aureus bacteria are prominent in nondebrided control samples, which substantially reduced following debridement, particularly following monofilament debridement treatments (Debrisoft® and Debrimitt™). Bar (A) = 12.7 μm. ConA, concanavalin A; DAPI, 4′,6-diamidino-2-phenylindole; EPS, extracellular polymeric substances; FISH, fluorescent in situ hybridization.
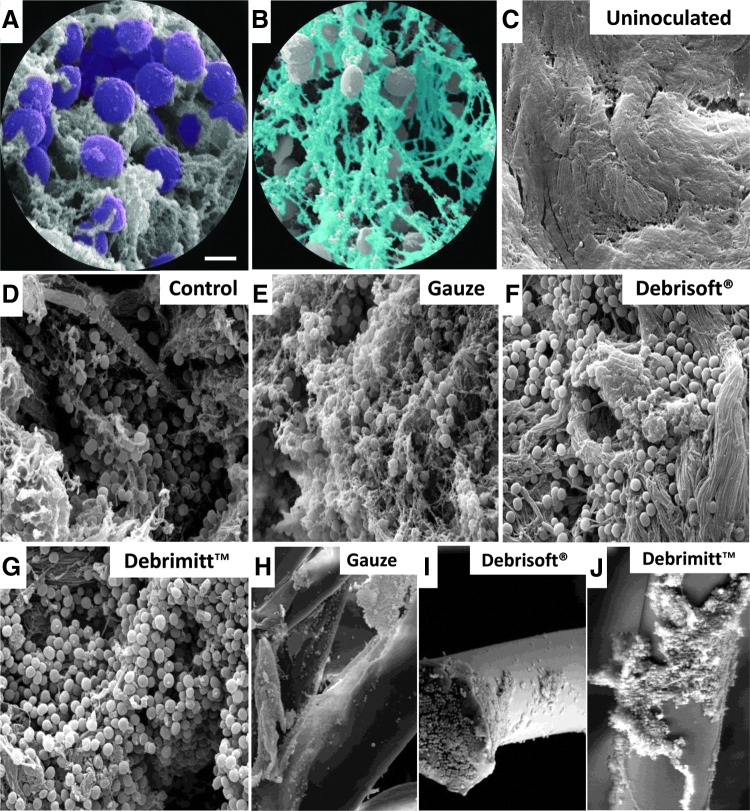
Scanning electron microscopy
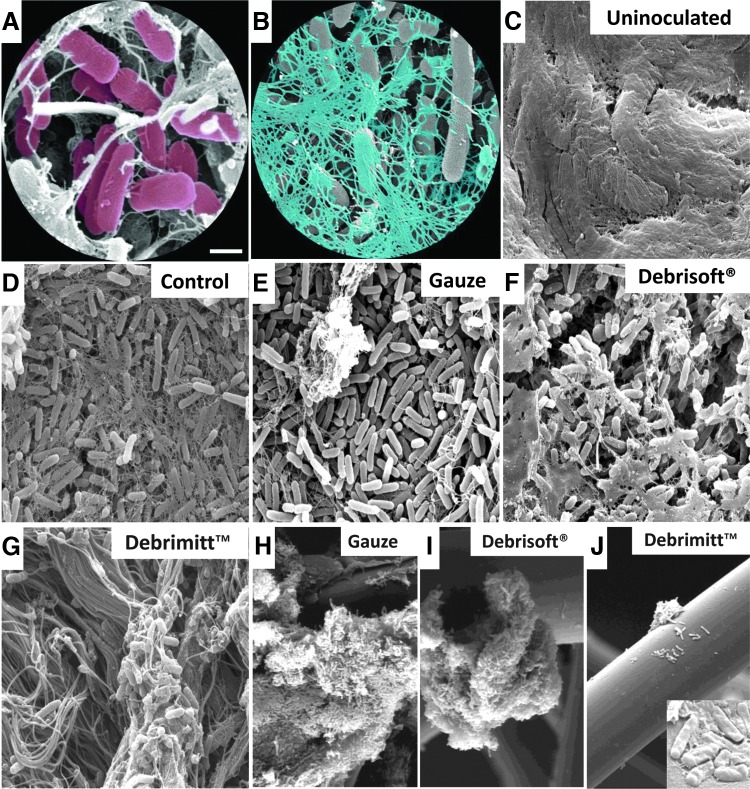
Finally, samples were subjected to SEM to directly visualize bacteria/tissue interaction. Control P. aeruginosa biofilm-treated skin (Fig. 5D) was covered with abundant rod-shaped P. aeruginosa (pseudocolored pink; Fig. 5A), encased in a stringy dehydrated EPS (pseudocolored blue; Fig. 5B), indicative of biofilm formation. As expected, uninoculated porcine wound samples lacked a visible bacterial biofilm (Fig. 5C). Interestingly, the surface of gauze debrided samples retained virtually full coverage of bacteria, but exhibited less EPS. A substantial proportion of the bacteria and EPS were removed by both Debrisoft® and Debrimitt™ treatments, with remaining bacteria mainly residing within the porcine wound tissue (Fig. 5F, G). SEM imaging of the gauze, Debrisoft®, and Debrimitt™ following application revealed bacterial removal in large clumps, which remain on the fibers of the devices (Fig. 5H, J). Moreover, at high resolution, the P. aeruginosa bacteria retained on the Debrisoft® and Debrimitt™ fibers appeared deformed and compressed compared to the P. aeruginosa removed by the gauze (Fig. 5J, inset). This suggests that debridement using Debrisoft® or Debrimitt™ may adversely affect bacterial viability.
Figure 5.
SEM images confirm P. aeruginosa biofilm removal following debridement. (A) Pseudocolored rod-shaped P. aeruginosa (pink). (B) Pseudocolored stringy EPS (blue). (C) Uninoculated porcine wound. (D) Control. (E) Gauze debrided. (F) Debrisoft® debrided. (G) Debrimitt™ debrided. (H–J) Direct imaging of postdebridement devices reveal attached clumps of bacteria, with altered morphology [inset, (J)]. Bar (A) = 1 μm (A, B), 5 μm (C–G, J; inset), and 20 μm (H–J). SEM, scanning electron microscopy.
Similarly, S. aureus (pseudocolored purple, Fig. 6A) formed an extensive covering on the surface of control biofilm-treated porcine wounds (Fig. 6D), this time encased in a more globular EPS (pseudocolored blue; Fig. 6B). As with P. aeruginosa, S. aureus porcine biofilm skin subjected to gauze debridement retained many S. aureus bacteria with prominent EPS remaining (Fig. 6E). By contrast, Debrisoft® and Debrimitt™ debridement appeared to remove the majority of surface EPS (Fig. 6F, G). Again, S. aureus bacteria were clearly visible on the monofilament device fibers following treatment (Fig. 6H, J). Interestingly, S. aureus appeared to be removed in large EPS encapsulated clumps, particularly evident following debridement with Debrimitt™.
Figure 6.
SEM images confirm S. aureus biofilm removal following debridement. (A) Pseudocolored coccoid-shaped S. aureus (purple). (B) Pseudocolored globular EPS (blue). (C) Uninoculated porcine wound. (D) Control. (E) Gauze debrided. (F) Debrisoft® debrided. (G) Debrimitt™ debrided. (H–J) Direct imaging of postdebridement devices reveals attached clumps of bacteria. Bar (A) = 1 μm (A, B), 5 μm (C–G), and 20 μm (H–J).
Discussion
Chronic wounds, often refractory to current treatments, remain a huge challenge in the clinic.3 Bacterial infection has long been implicated in impaired wound repair, where some studies suggest chronicity is prolonged by established bacterial biofilms comprising pathogens such as P. aeruginosa and S. aureus.3,20,21 Simple yet efficient removal of wound biofilms should therefore be particularly beneficial to promote wound repair. In this study, we have established an ex vivo single-species porcine biofilm model using P. aeruginosa and S. aureus. Using this model, and carefully controlled application parameters, we show that new polymer debriding devices are more effective at removing established wound biofilms than standard gauze. Indeed, combination of quantitative and morphological assessments revealed that the polymer debriding device Debrimitt™ was most effective at removing established biofilms from porcine wound tissue.
While virulence is associated with biofilm prevalence, bacterial loads of greater than 105 bacteria per gram have been linked to nonhealing pathophysiology.22 Four log10 is also a key measure for bacterial reduction, being the FDA threshold criteria for dressing/device antibacterial efficacy claims. Indeed, it has been suggested that a 5 log10 reduction in wound bacteria would require 17 generations to recover, equating to around 12 h in a wound environment.23 Therefore, utilizing monofilament debriding products may open a time-dependent therapeutic window for cotreatment and biofilm eradication.24 In this study, biofilm viability plate spread counts indicate that monofilament debridement reduced wound biofilm bacterial loads by up to 7 log10 (against S. aureus biofilms), equating to over 16 h recovery time (based on in vitro planktonic culture growth rates).
A major strength of this study is the use of ex vivo porcine skin for biofilm establishment. Tissue studies have been indicated as more adequate than in vitro models for testing the significance of clinical treatments4 as they provide the opportunity for bacteria to interact with host tissues (reviewed in Roberts et al.25). The ex vivo porcine wound model used here provides a close physiological comparison to human wound tissue26 using clinically relevant bacteria.21 Indeed, SEM analysis reveals that in our biofilm model, bacteria are closely integrated into the porcine tissue, depositing EPS that form an interface with the host dermal fibers. In vivo models will provide a further level of relevance, allowing the contribution of host factors such as ischemia and necrosis on biofilm to be assessed.27 However, as animal studies are expensive, time consuming, and technically demanding, we suggest that ex vivo tissue models allow easier control of experimental variables and assessment of outcomes.16
By virtue of their protective EPS matrix, bacteria within complex biofilms benefit from increased resistance to treatments and host immunity versus planktonic species.28 The mechanism of EPS function has begun to be investigated. For example, inhibiting synthesis of the EPS component, exopolysaccharide alginate, in P. aeruginosa biofilms potentiated phagocytosis by activated human leukocytes.29 Further studies inhibiting EPS demonstrate reduced antibiotic resistance28 or complete inability to form biofilms (reviewed in Flemming and Wingender30). Interestingly, virulent biofilm-forming P. aeruginosa is known to synthesize exogenous polysaccharides for encapsulation and annealment to host tissues through carbohydrate-modulating genes such as algC31. Modulations in genes coding for motility and aggregation have been demonstrated in biofilm-forming S. aureus (reviewed in Jefferson32). Gene expression profiling of planktonic bacteria compared to biofilms has identified multiple genes which contribute to the sessile, antibiotic-resistant nature of biofilms.33,34
Despite these studies, identification of bacterial biofilms in wound tissue remains an area of contention.35 In this study, we used 48 h ex vivo established biofilms of P. aeruginosa and S. aureus based on previous published data.4,16 In vitro studies indicate P. aeruginosa biofilm growth within 10 h22 and certainly by 24 h.36 Similarly, S. aureus, known to establish more slowly than P. aeruginosa, formed biofilms in 24 h when grown on MH substrate.37 Although the patterns of EPS expression and the distinct polymers produced differ between species,38 SEM imaging of mature biofilms has previously depicted EPS as long strings (P. aeruginosa30) or cloud-like clumps (S. aureus,4) surrounding the bacteria within a biofilm. In this study, we demonstrate the same morphological features in our 48 h established ex vivo porcine skin biofilms.
A second important characteristic of established biofilms is the formation of microcolonies (shown by SEM37). Maintaining close proximity allows bacteria in biofilm microcolonies to exchange genetic information and make use of chemical signaling (quorum sensing19) to increase virulence.39 Our SEM data reveal that in the ex vivo porcine model, both P. aeruginosa and S. aureus establish EPS encapsulated microcolonies that are characteristic of established biofilms.4 Interestingly, our data agree with previous SEM studies suggesting that monofilament polymer devices structurally integrate wound debris.40 In the current study, we observed clumps of aggregated P. aeruginosa in all three test groups, while clumped aggregates of S. aureus were visible only on the monofilament devices (Fig. 6). Finally, our inclusion of uninoculated porcine skin for SEM analysis confirmed that wounded skin had been adequately cleaned to remove endogenous bacteria before establishment of species-specific biofilms. When combined with Gram–Twort, Con A, Acridine Orange, and FISH staining, to indicate the regional extent of biofilm presence and removal, our data indicate (i) established biofilm presence before debridement and (ii) improved efficacy of monofilament debridement versus standard gauze.
In summary, we describe a 48-h ex vivo porcine skin biofilm model that exhibits many features of the established biofilm phenotypes, including EPS production. Using this model, we demonstrate the efficacy of monofilament debridement as a clear alternative to more invasive debridement techniques currently used in the clinic. Quantitative assessment revealed the monofilament debriding device, Debrimitt™, to provide the greatest reduction in biofilm bacterial load. Intriguingly, our data suggest that monofilament devices effectively remove both bacteria and EPS and that the mechanical debridement process may damage bacteria in situ, further influencing viability. Our study highlights further opportunities to carefully and reproducibly test the efficacy of mechanical debridement in vivo and in clinical contexts.
Innovation
This is we believe, the first study to directly compare the efficacy of monofilament debriding devices for the removal of established biofilm from skin. We report standardized conditions for the application and testing of debriding devices and a portfolio of assessments to quantitatively and qualitatively monitor biofilm removal. Finally, our data reveal differential efficacy toward defined bacterial species.
Key Findings.
• Monofilament debridement efficiently removes established bacterial biofilms ex vivo.
• S. aureus and P. aeruginosa biofilms are differentially susceptible to debridement.
• Debridement removes both EPS and wound bacteria.
Abbreviations and Acronyms
- ANOVA
analysis of variance
- CFU
colony forming units
- ConA
concanavalin A
- DAPI
4′,6-diamidino-2-phenylindole
- EPS
extracellular polymeric substances
- FISH
fluorescent in situ hybridization
- MH
Mueller Hinton
- O/N
overnight
- OCT
optimum cutting temperature
- PBS
phosphate-buffered saline
- SEM
scanning electron microscopy
Acknowledgments and Funding Sources
This work was funded by research grants from the MRC, Innovate UK (Knowledge Transfer Partnership), and Crawford Healthcare Ltd. The authors wish to thank Tobias Starborg in the Faculty of Life Sciences, EM Facility for their assistance and the Wellcome Trust for equipment grant support to the EM Facility.
Author Disclosure and Ghostwriting
C.S. is an employee of Crawford Healthcare Ltd. The content of this article was expressly written by the authors listed. No ghostwriters were used to write this article.
About the Authors
Holly N. Wilkinson, MRes, is studying for a PhD in the Healing Foundation Centre at The University of Manchester. Andrew J. McBain, PhD, is a Professor of Microbiology at the University of Manchester. He leads a research group that focuses on the interaction of microorganisms with the human body in health and disease, and on clinical and environmental biofilms. Christian Stephenson, BSc, has a background in Chemistry with extensive experience in medical devices product development and wound management. Matthew J. Hardman, PhD, formerly the Edmond de Rothschild Senior Fellow in Ageing Research, is currently a senior lecturer in the Healing Foundation Centre at The University of Manchester. He received his PhD in Skin Developmental Biology under the supervision of Professor Carolyn Byrne at The University of Manchester. The primary focus of his research is elucidating molecular and cellular correlates to pathological wound healing.
References
- 1.Harding K, Morris H, Patel G. Science, medicine, and the future: healing chronic wounds. BMJ 2002;324:160–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watson JM, Kang'ombe AR, Soares MO, Chuang L-H, Worthy G, Bland JM, et al. . Use of weekly, low dose, high frequency ultrasound for hard to heal venous leg ulcers: the VenUS III randomised controlled trial. BMJ 2011;342:d1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis SC, Martinez L, Kirsner R. The diabetic foot: the importance of biofilms and wound bed preparation. Curr Diab Rep 2006;6:439–445 [DOI] [PubMed] [Google Scholar]
- 4.Davis SC, Ricotti C, Cazzaniga A, Welsh E, Eaglstein WH, Mertz PM. Microscopic and physiologic evidence for biofilm‐associated wound colonization in vivo. Wound Repair Regen 2008;16:23–29 [DOI] [PubMed] [Google Scholar]
- 5.James GA, Swogger E, Wolcott R, Secor P, Sestrich J, Costerton JW, et al. . Biofilms in chronic wounds. Wound Repair Regen 2008;16:37–44 [DOI] [PubMed] [Google Scholar]
- 6.Monroe D. Looking for chinks in the armor of bacterial biofilms. PLoS Biol 2007;5:e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nusbaum AG, Gil J, Rippy MK, Warne B, Valdes J, Claro A, et al. . Effective method to remove wound bacteria: comparison of various debridement modalities in an in vivo porcine model. J Surg Res 2012;176:701–707 [DOI] [PubMed] [Google Scholar]
- 8.Velnar T, Bailey T, Smrkolj V. The wound healing process: an overview of the cellular and molecular mechanisms. J Int Med Res 2009;37:1528–1542 [DOI] [PubMed] [Google Scholar]
- 9.Stephen-Haynes J, Thompson G. The different methods of wound debridement. Br J Community Nurs 2007;12(Suppl 3):S6–S16 [PubMed] [Google Scholar]
- 10.Armstrong DG, Lipsky BA. Diabetic foot infections: stepwise medical and surgical management. Int Wound J 2004;1:123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis R, Whiting P, ter Riet G, O'Meara S, Glanville J. A rapid and systematic review of the clinical effectiveness and cost-effectiveness of debriding agents in treating surgical wounds healing by secondary intention. Health Technol Assess 2001;5:1–131 [DOI] [PubMed] [Google Scholar]
- 12.Fonder MA, Lazarus GS, Cowan DA, Aronson-Cook B, Kohli AR, Mamelak AJ. Treating the chronic wound: a practical approach to the care of nonhealing wounds and wound care dressings. J Am Acad Dermatol 2008;58:185–206 [DOI] [PubMed] [Google Scholar]
- 13.Stang D. Is the scalpel the only way to debride? Diabetic Foot J 2013;16:74–78 [Google Scholar]
- 14.Bahr S, Mustafi N, Hättig P, Piatkowski A, Mosti G, Reimann K, et al. . Clinical efficacy of a new monofilament fibre-containing wound debridement product. J Wound Care 2011;20:242–248 [DOI] [PubMed] [Google Scholar]
- 15.Merritt JH, Kadouri DE, O'Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol 2005;1:1B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Phillips PL, Sampson EM, Progulske‐Fox A, Jin S, Antonelli P, et al. . Development of a novel ex vivo porcine skin explant model for the assessment of mature bacterial biofilms. Wound Repair Regen 2013;21:704–714 [DOI] [PubMed] [Google Scholar]
- 17.Bancroft JD, Gamble M. Theory and Practice of Histological Techniques. London, UK: Churchill Livingstone, 2008 [Google Scholar]
- 18.Chen M-Y, Lee D-J, Tay J-H, Show K-Y. Staining of extracellular polymeric substances and cells in bioaggregates. Appl Microbiol Biotechnol 2007;75:467–474 [DOI] [PubMed] [Google Scholar]
- 19.Lawrence JR, Swerhone GD, Topp E, Korber DR, Neu TR, Wassenaar LI. Structural and functional responses of river biofilm communities to the nonsteroidal anti‐inflammatory diclofenac. Environ Toxicol Chem 2007;26:573–582 [DOI] [PubMed] [Google Scholar]
- 20.Bjarnsholt T, Kirketerp‐Møller K, Jensen PØ, Madsen KG, Phipps R, Krogfelt K, et al. . Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen 2008;16:2–10 [DOI] [PubMed] [Google Scholar]
- 21.Gjødsbøl K, Christensen JJ, Karlsmark T, Jørgensen B, Klein BM, Krogfelt KA. Multiple bacterial species reside in chronic wounds: a longitudinal study. In Wound J 2006;3:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison‐Balestra C, Cazzaniga AL, Davis SC, Mertz PM. A wound‐isolated Pseudomonas aeruginosa grows a biofilm in vitro within 10 hours and is visualized by light microscopy. Dermatol Surg 2003;29:631–635 [DOI] [PubMed] [Google Scholar]
- 23.Phillips PL, Yang Q, Davis S, Sampson EM, Azeke JI, Hamad A, et al. . Antimicrobial dressing efficacy against mature Pseudomonas aeruginosa biofilm on porcine skin explants. Int Wound J 2013;12:469–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz G, Phillips P, Yang Q, Stewart P. Biofilm maturity studies indicate sharp debridement opens a time-dependent therapeutic window. J Wound Care 2010;19:320. [DOI] [PubMed] [Google Scholar]
- 25.Roberts AE, Kragh KN, Bjarnsholt T, Diggle SP. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol 2015;427:3646–3661 [DOI] [PubMed] [Google Scholar]
- 26.Meyer W. The skin of domestic mam-mals as a model for the human skin, with special reference to the domestic pig. Curr Probl Dermatol 1978;7:39–52 [DOI] [PubMed] [Google Scholar]
- 27.Phillips P, Yang Q, Gibson D, Schultz G. Assessing the bioburden in poorly healing wounds. Wounds International. 2011;2:17–19 [Google Scholar]
- 28.Davenport EK, Call DR, Beyenal H. Differential protection from tobramycin by extracellular polymeric substances from Acinetobacter baumannii and Staphylococcus aureus biofilms. Antimicrob Agents Chemother 2014;58:4755–4761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-γ-mediated macrophage killing. J Immunol 2005;175:7512–7518 [DOI] [PubMed] [Google Scholar]
- 30.Flemming H-C, Wingender J. The biofilm matrix. Nature Rev Microbiol 2010;8:623–633 [DOI] [PubMed] [Google Scholar]
- 31.Davies D, Geesey G. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol 1995;61:860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jefferson KK. What drives bacteria to produce a biofilm? FEMS Microbiol Lett 2004;236:163–173 [DOI] [PubMed] [Google Scholar]
- 33.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, et al. . Gene expression in Pseudomonas aeruginosa biofilms. Nature 2001;413:860–864 [DOI] [PubMed] [Google Scholar]
- 34.Mah T-F, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003;426:306–310 [DOI] [PubMed] [Google Scholar]
- 35.Spichler A, Hurwitz BL, Armstrong DG, Lipsky BA. Microbiology of diabetic foot infections: from Louis Pasteur to ‘crime scene investigation’. BMC Med 2015;13:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes‐Jørgensen A, Molin S, et al. . Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 2003;48:1511–1524 [DOI] [PubMed] [Google Scholar]
- 37.Nishimura S, Tsurumoto T, Yonekura A, Adachi K, Shindo H. Antimicrobial susceptibility of Staphylococcus aureus and Staphylococcus epidermidis biofilms isolated from infected total hip arthroplasty cases. J Orthop Sci 2006;11:46–50 [DOI] [PubMed] [Google Scholar]
- 38.López D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol 2010;2:a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schierle CF, De la Garza M, Mustoe TA, Galiano RD. Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair Regen 2009;17:354–359 [DOI] [PubMed] [Google Scholar]
- 40.Haemmerle G, Duelli H, Abel M, Strohal R. The wound debrider: a new monofilament fibre technology. Br J Nurs 2011;20:35–36 [DOI] [PubMed] [Google Scholar]