Abstract
Objective: Impaired wound healing in diabetic (DB) patients is a significant health problem; however, the roles that cytokines and innate immune cells contribute to this impaired healing are not completely understood.
Approach: A mouse model was used to compare the innate immune response during DB and normal wound healing. Two 5-mm full-thickness wounds were created on the dorsal skin of BKS.Cg-m+/+Leprdb/J (DB) and C57BL/6 (wild-type) mice. Innate immune cell markers and cytokine mRNA levels were measured in wound biopsies during the first week of healing.
Results: Innate immune cell influx (typified by the Gr-1 neutrophil marker and the Ym1 macrophage marker) was delayed in the DB wounds. Expression of the M2 macrophage-related genes, Ym1 and arginase 1, was significantly reduced in the DB wounds. PCR array analysis demonstrated altered cytokine expression in DB wounds. Most prominently, both interleukin (IL)-17 and IL-20 mRNA levels were significantly increased in the DB wounds.
Innovation: This is the first study to identify increased levels of IL-17 and IL-20 in DB wounds. These cytokines are also elevated in the inflammatory skin disorder, psoriasis; thus, they may be potential therapeutic targets to aid in DB wound healing.
Conclusion: The entire cytokine profile of DB wounds over the course of healing is not completely understood. This study suggests that the IL-17 and IL-20 families of cytokines should be further analyzed in the context of DB wound healing.
Keywords: : diabetes, cytokine, macrophage, wound healing

Laurie P. Shornick, PhD
Introduction
In normal wound healing, the first cells to be recruited to the wound are the polymorphonuclear cells (PMN). These cells decontaminate the wound by phagocytosing bacteria and they release chemokines and cytokines to recruit and activate macrophages in the wound. The macrophages also phagocytose bacteria, remove damaged tissue, and produce growth factors that stimulate angiogenesis, collagen deposition, and wound closure. Notably, depletion studies have suggested important roles for both PMN and macrophages in wound healing. Depletion of PMN was shown to accelerate both normal and diabetic (DB) wound healing.1 The consequence of macrophage depletion is dependent on the phase of healing. Selective macrophage depletion (utilizing transgenic mice expressing the diphtheria toxin receptor under the CD11b promoter) showed that during the early healing response macrophages are needed for granulation tissue formation and myofibroblast differentiation. Depletion of macrophages during mid-phase healing resulted in hemorrhage in the wound tissue.2
In addition to the phase of wound healing, the role of the macrophage may depend on the subtype of macrophage that is present in each wound. Several different phenotypes of activated macrophages have been identified, including classically activated macrophages (M1) and alternatively activated macrophages (M2) (reviewed in Refs.3–5). The M1 macrophages kill pathogens and clean the wound by removing dying cells and debris. They can also produce proteases, which may degrade the tissue. In contrast, interleukin (IL)-4 and IL-13 activate M2 macrophages, which express the mannose receptor, L-arginase 1, dectin-1, FIZZ1, and Ym1, and decreased expression of IL-1, IL-6, and tumor necrosis factor (TNF)-α.4,6–9 M2 macrophages secrete anti-inflammatory mediators such as TGF-β, IL-10, and IL-4, which may contribute to angiogenesis and the resolution of inflammation.8 Recently, it has been demonstrated that this macrophage categorization is oversimplified and that macrophage populations exist on a spectrum of phenotypes.10 In the present study, we used a mouse model of wound healing to analyze the time course of innate cell influx, macrophage phenotype, and cytokine profile of the DB wound.
Clinical Problem Addressed
Nonhealing chronic foot ulcers in DB patients lead to over 70,000 nontraumatic lower limb amputations per year in the United States.11 These amputations are expensive and greatly reduce the quality of life for the patients. Effective treatments for these chronic wounds will require a comprehensive understanding of the impaired innate immune mechanisms during DB wound healing.
Materials and Methods
Animals
The Saint Louis University Animal Care and Use Committee approved all animal procedures before the start of the study. Healthy, 8-week-old female adult BKS.Cg-m+/+Leprdb/J (DB) and wild-type C57BLKS/J (WT) mice were obtained from Jackson Laboratories. The animals were housed in the Department of Comparative Medicine animal facility at Saint Louis University School of Medicine on a 12-h light/12-h dark cycle. The animals were allowed to acclimate for 7 days upon arrival and had access to food and water ad libitum.
Murine wound model
We utilized an excisional mouse model of wound healing as previously described.12,13 A punch biopsy tool was used to create two 5-mm full-thickness wounds on the dorsal skin, and a ring-shaped silicone splint was applied to the skin 2–3 mm beyond the perimeter of the wound. The silicone splint was affixed with VetBond™ tissue adhesive and six interrupted sutures. The wounds were then dressed with moist gauze plus Tegaderm™. During recovery, the mice were placed in individual cages under a warming lamp.
Mice were observed daily for general health, and wounds were harvested on days 2, 5, and 7 after injury. Blood samples were taken through tail vein punctures, and a Contour® blood glucose monitor and test strips (Bayer HealthCare, LLC, Mishawaka, IN) were used to measure glucose levels. Mice with blood glucose levels over 300 mg/dL were considered DB. To track wound healing, digital photographs were taken throughout the healing processes. Wound area was measured by digital planimetry using SigmaScan Pro 5.0 (SPSS Science, Chicago, IL), with calibrated digital photographs. To increase measurement accuracy, 10 different tracings were made with SigmaScan Pro 5.0 for each wound and then averaged.
Immunohistochemistry
Wound biopsies were fixed in 10% neutral buffered formalin for 72 h. Five-micrometer paraffin sections were incubated with primary antibody for 1 h at 25°C. Primary antibody binding was detected with biotinylated secondary antibody, either goat anti-rabbit or goat anti-rat (Vector Laboratories, Burlingame, CA), for 30 min at 25°C and the VECTASTAIN ABC-AP Kit (Vector Laboratories). Sections were stained with an alkaline phosphatase red substrate and counterstained with hematoxylin. Primary antibodies were rabbit anti-ECF-L (Ym1) (R&D Systems, Minneapolis, MN) and rabbit anti-Gr-1 (R&D Systems).
Gene expression analysis
RNA was isolated from wound biopsies at day 5 of wound healing using the PerfectPure RNA isolation kit for fibrous tissue (5 Prime, Inc., Gaithersburg, MD). RNA quality was assessed with the RT2 PCR Quality Control Kit (SABiosciences, Frederick, MD). An inflammatory pathway-focused real-time PCR array (Array #PAMM-011; SABiosciences) was utilized according to the manufacturer's directions, which determined mRNA levels for 84 different inflammatory cytokines and receptors. RNA samples were converted into cDNA using the RT2 First Strand Kit (SABiosciences). RT2 SYBR Green PCR Master Mix (SABiosciences) was added to the cDNA samples. The samples were added to RT2 Profiler PCR Arrays (SABiosciences) for mouse inflammatory cytokines and receptors. The arrays were run through an MJ Chromo 4 thermal cycler using the following protocol: denaturing at 95°C for 15 s, annealing at 55°C for 40 s, and amplification at 72°C for 30 s, for a total of 40 cycles. The genes represented on the PCR array include the following: Chemokine Genes: Ccl1, Ccl11, Ccl12, Ccl17, Ccl19, Ccl2, Ccl20, Ccl22, Ccl24, Ccl25, Ccl3, Ccl4, Ccl5, Ccl6, Ccl7, Ccl8, Ccl9, Cx3cl1, Cxcl1, Cxcl10, Cxcl11, Cxcl12 (Sdf1), Cxcl13, Cxcl15, Cxcl4, Cxcl5, Cxcl9, and Il13. Chemokine Receptors: Ccr1, Ccr2, Ccr3, Ccr4, Ccr5, Ccr6, Ccr7, Ccr8, Ccr9, Cxcr3, Il8rb, and Xcr1. Cytokine Genes: Ifng (IFNγ), Il10, Il11, Il13, Il15, Il16, Il17, Il18, Il1a, Il1b, Il1f6, Il1f8, Il20, Il3, Il4, Itgam, Itgb2, Lta, Ltb, Mif, Scye1, Spp1, Tgfb1, Tnf, and Cd40lg. Cytokine Receptors: Ifng (IFNγ), Il10ra, Il10rb, Il13, Il13ra1, Il1r1, Il1r2, Il2rb, Il2rg, Il5ra, Il6ra, Il6st, Tnfrsf1a (TNFR1), and Tnfrsf1b (TNFR2). Other genes involved in inflammatory response include Abcf1, Bcl6, Blr1, C3, Casp1, Crp, Il1r1, Il8rb, and Tollip. In addition to the 84 inflammatory cytokines and receptors, the arrays also have 5 housekeeping genes, 1 genomic DNA control, 3 reverse transcription controls, and 3 PCR controls.
The following primers were used to verify the expression of the target genes: Arginase: forward, 5′-CTC CAA GCC AAA GTC CTT AGA G-3′, reverse, 5′-AGG AGC TGT CAT TAG GGA CAT C-3′; CCL19: forward, 5′-GGG GTG CTA ATG ATG CGG AA-3′, reverse, 5′-CCT TAG TGT GGT GAA CAC AAC A-3′; CCL24: forward, 5′-ATT CTG TGA CCA TCC CCT CAT-3′, reverse, 5′-TGT ATG TGC CTC TGA ACC CAC-3′; CXCL9: forward, 5′-GGA GTT CGA GGA ACC CTA GTG-3′, reverse, 5′-GGG ATT TGT AGT GGA TCG TGC-3′; dectin-1: forward, 5′-GAC TTC AGC ACT CAA GAC ATC C-3′, reverse, 5′-TTG TGT CGC CAA AAT GCT AGG-3′; FIZZ1: forward, 5′- ATG AAC AGA TGG GCC TCC TG-3′, reverse, 5′-AGC CAC AAG CAC ACC CAG TAG-3′; IL-17A: forward, 5′-TTT AAC TCC CTT GGC GCA AAA-3′, reverse, 5′-CTT TCC CTC CGC ATT GAC AC-3′; IL-20: forward, 5′-TCT TGC CTT TGG ACT GTT CTC C-3′, reverse, 5′-GTT TGC AGT AAT CAC ACA GCT TC-3′; Mrc-1: forward, 5′-CTC TGT TCA GCT ATT GGA CGC-3′, reverse, 5′-CGG AAT TTC TGG GAT TCA GCT TC-3′; Ym1: forward, 5′-CAG GTC TGG CAA TTC TTC TGA A-3′, reverse, 5′-GTC TTG CTC ATG TGT GTA AGT GA-3′. The specificity of amplification was assessed for each sample by dissociation curve analysis and the size of the amplicon was confirmed by agarose gel electrophoresis. Relative expression of each gene was assessed through the Delta–Delta CT method using GAPDH as the housekeeping gene.
Data analysis
The data were screened before analysis for accuracy and normality. Group differences were investigated with appropriate parametric statistical tests. If parametric assumptions were violated, equivalent nonparametric statistical tests were used. An alpha of p < 0.05 was used throughout the analysis.
Results
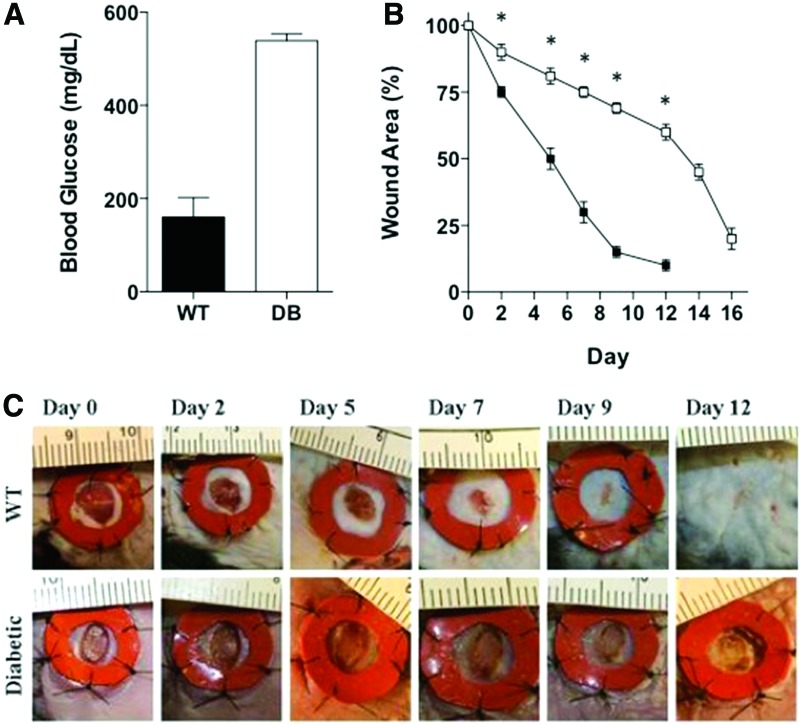
Punch biopsy wounds (5 mm) were created on the backs of DB and control mice (WT). The DB mice (mean = 538 mg/dL, SD = 19.29) had a statistically significant higher blood glucose level than WT mice (mean = 160.3, SD = 51.35) (z = −2.93, p < 0.05) (Fig. 1A). As described previously, the DB mice (n = 8) had delays in wound healing compared with WT mice (n = 8) [13]. On day 9, the WT wounds were 85% closed compared with only 31% closure in the DB wounds. By day 12, WT wounds were closed, while the DB wounds remained open. (Fig. 1B). Figure 1C shows a significant delay in healing in the DB mice. Larger wounds were observed in DB mice compared with WT mice on day 2 (t[18] = 13.90, p < 0.001), day 5 (t[18] = 22.92, p < 0.001), day 7 (t[18] = 34.82, p < 0.001), day 9 (t[18] = 53.74, p < 0.001), and day 12 (t[18] = 60.2, p < 0.001) after initial wounding.
Figure 1.
Wound healing is delayed in DB mice compared with WT mice. (A) Blood glucose levels in WT and DB mice. (B) Wound area in WT (closed squares, n = 8) and DB (open squares, n = 8) wounds across time. Each time point is the mean wound area percentage of the original wound. *p < 0.05. Error bars represent standard deviations. (C) Representative wounds were photographed throughout healing. DB, diabetic; WT, wild-type. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
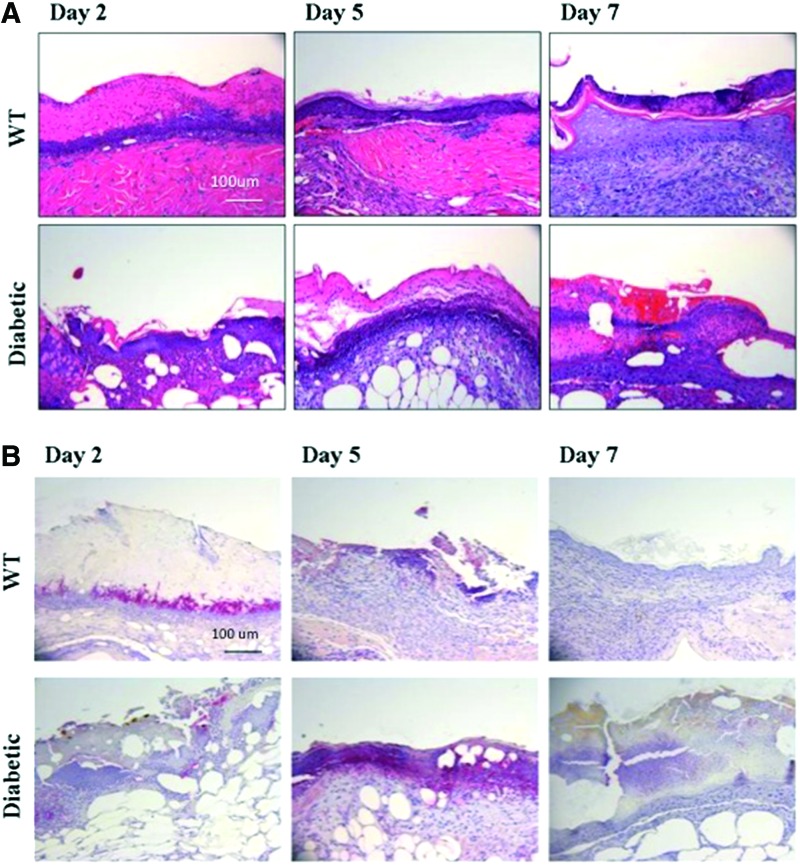
Inflammatory cells were observed in the WT mouse wounds on the second day of healing based on hematoxylin and eosin staining. However, these cells did not appear in the DB wounds until the fifth day of wound healing (Fig. 2A). To determine if the neutrophil population was delayed in migrating into the wound, we performed immunostaining for the neutrophil marker, Gr-1. WT wounds showed the presence of Gr-1+ cells on day 2, but the Gr-1+ cells were no longer detectable on days 5 and 7. In contrast, the Gr-1+ cell influx was not observed in the DB wounds until day 5 and were still present on day 7 (Fig. 2B)
Figure 2.
(A) Hematoxylin and eosin staining. (B) Anti-Gr-1 immunostaining of wound tissue between WT and DB mice across days 2, 5, and 7. Primary anti-Gr-1 antibody was detected using an alkaline phosphatase red substrate and counterstained with hematoxylin. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
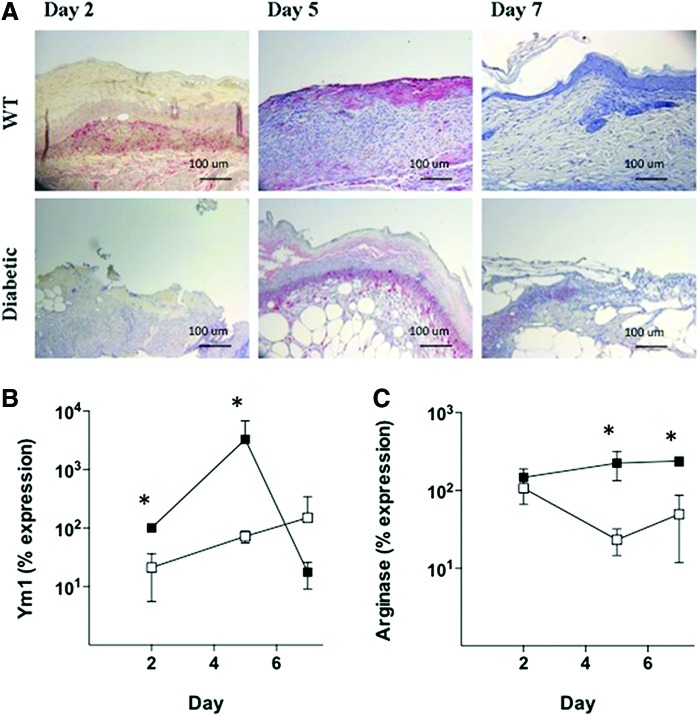
To examine M1 (classical) and M2 (alternative) macrophage recruitment, we performed immunohistochemistry and quantitative PCR (qPCR) for macrophage markers. Results showed that there was no significant difference in the expression of the M1 markers, Mac-3 and F4/80, between WT and DB wounds during the first week of healing (data not shown). To examine M2 (alternative) macrophage recruitment, we used the markers, Ym1, arginase 1, FIZZ, dectin-1, and Mrc-1. As shown in Fig. 3A, Ym1+ cells were present in WT wounds on days 2 and 5; subsequently, these cells were no longer detectable on day 7. In the DB wounds, Ym1+ cells were not detectable on day 2; however, they were present on day 5 and still detectable on day 7. This was consistent with real-time PCR for Ym1, which showed that Ym1 mRNA expression peaked on day 5 in WT wounds. In contrast, Ym1 mRNA expression in DB wounds was significantly lower on both days 2 (z = 1.97, p < 0.05) and 5 (z = 1.97, p < 0.05) postwounding (Fig. 3B). In addition, there was significantly lower arginase mRNA expression in the DB wounds at these times (Fig. 3C). The other markers of M2 macrophages (FIZZ, dectin-1, Mrc-1) were not significantly different between WT and DB wounds (data not shown).
Figure 3.
Analysis of M2 macrophage markers during wound healing. (A) Ym1 immunostaining of wound tissue between WT and DB mice across days 2, 5, and 7. Primary anti-Ym1 antibody was detected using an alkaline phosphatase red substrate and counterstained with hematoxylin. (B) Real-time PCR of Ym1 mRNA expression in WT (closed squares) and DB (open squares) wounds. (C) Real-time PCR of arginase mRNA expression in WT (closed squares) and DB (open squares) wounds. *p < 0.05. Error bars represent standard deviations. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
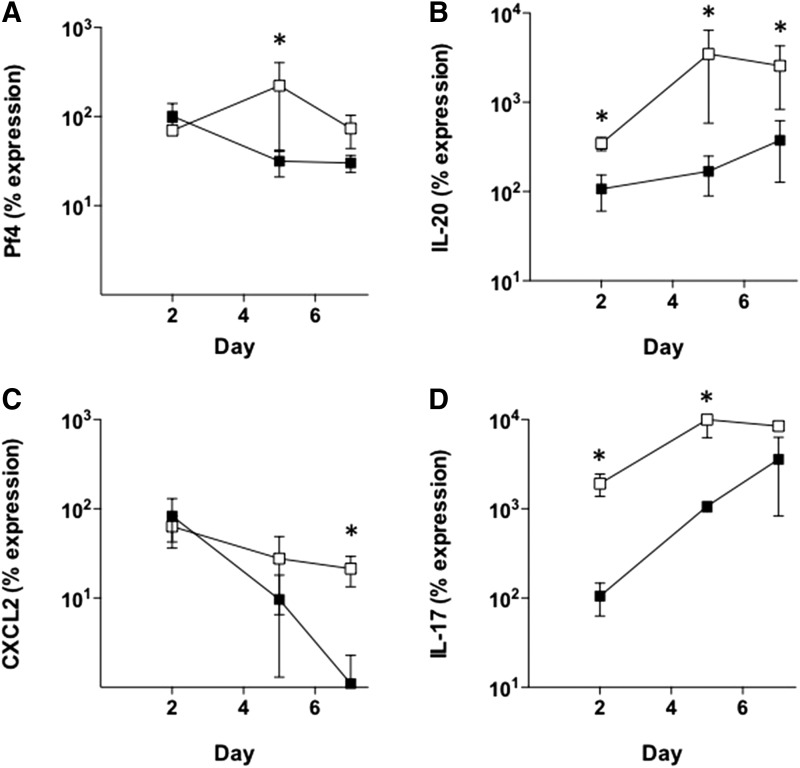
Because expression of both the neutrophil marker, Gr-1, and the M2 macrophage marker, Ym1, appeared to be delayed in DB wounds, we hypothesized that cytokine signals may be altered as well. To analyze the expression of many cytokine genes at one time, we utilized a real-time PCR array to quantitatively measure mRNA levels for 84 different inflammatory cytokines and receptors in wound biopsies at day 5 of healing. The results showed that several genes were statistically different between DB and WT wounds. The following genes were expressed at a significantly higher level in WT (n = 4) compared with DB (n = 4) wounds using an independent t-test: CXCL9 (−2.95-fold, p = 0.0195), CCL24 (−3.35-fold, p = 0.0327), IL-13 (−3.8-fold, p = 0.0231), and CCL19 (−4.09-fold, p = 0.0046). In contrast, two genes, Pf4 and IL-20, were expressed at a significantly higher level in DB wounds compared with WT. Pf4 was 2.17-fold higher in DB wounds (p = 0.0323) and IL-20 was 9.75-fold higher in DB wounds (p = 0.0348).
To validate the PCR array data, mRNA expression for each gene was analyzed over the course of wound healing using real-time PCR. The results showed that there was no significant difference in the expression of CXCL9, CCL24, IL-13, and CCL19 between WT and DB wounds (data not shown). In contrast, the differences in Pf4 and IL-20 were confirmed by real-time PCR. The time course of Pf4 expression is shown in Fig. 4A. Pf4 mRNA expression was similar on days 2 and 7; however, there was a significantly higher level of Pf4 mRNA on day 5 of healing in the DB wounds (Fig. 4A). The time course showed that IL-20 expression is significantly higher in DB wounds (n = 4) at days 2, 5, and 7 compared with WT (n = 4) (Fig. 4B).
Figure 4.
Time course of cytokine mRNA expression in WT (closed squares) and DB (open squares) wounds. (A) Pf4, (B) IL-20, (C) CXCL2, and (D) IL-17. *p < 0.05. Error bars represent standard deviations. IL, interleukin.
Finally, the expression of the neutrophil chemokines, CXCL2 and IL-17, was also altered in the DB wounds. Figure 4C shows that CXCL2 decreases over time in the WT wounds. CXCL2 also decreases in the DB wound, but has significantly higher expression on day 7 compared with WT. In contrast, IL-17 levels increase in WT wounds over the time course of healing; however, the IL-17 levels are significantly higher on days 2 and 5 in the DB wounds (Fig. 4D).
Discussion
Our results demonstrated a significantly increased time to wound closure in DB mice that was consistent with previous reports of impaired wound healing in DB mice.14,15 This impaired wound closure was accompanied by an initial delay of Gr-1+ neutrophils into the wounds. Similarly, Nguyen et al. showed that in the TallyHo mouse model of diabetes, full-thickness excisional wounds containing Staphylococcus aureus biofilms had reduced neutrophil myeloperoxidase activity on postwounding day 3.14 Although we did not inoculate the wounds with bacteria, our previous studies have shown the presence of the biofilm producer Staphylococcus xylosus in our model.13 In contrast, other studies have shown that DB wounds in db/db mice have an increased and persistent presence of Gr-1+ neutrophils at 13 days postwounding.16 Together, this suggests that the processes of both early infiltration and later clearance of neutrophils are affected in the DB wound environment.
There was also a delay in the presence of Ym1+ M2 (alternative) macrophages in the DB wounds. The macrophages enter the wound after the neutrophils and they can differentiate into either an M1 (classical) or M2 (alternative) phenotype. In our model, the M1 markers, Mac-3 and F4/80, were similar in both levels of expression and time course in WT and DB wounds; however, the expression levels of Ym1 and arginase 1 mRNA were significantly reduced in the DB wounds. Ym1 is a secreted chitinase-like protein expressed by M2 cells. It is associated with a type 2 immune response and is upregulated during parasitic infection. The type 2 response is believed to play a role in healing and repair after a helminth infection.17,18 Arginase 1, the precursor of nitric oxide, is upregulated in M2 macrophages exposed to IL-4 and IL-13, and its expression associated with angiogenesis, reepithelialization, and granulation tissue formation.19 The observed reduced arginase mRNA expression in DB wounds may contribute to the impairment of these processes. Dectin-1 and Mrc-1 are also both used as M2 markers, but there was no difference in their expression between WT and DB wounds.20,21 This reduced expression of alternatively activated macrophage markers is comparable with observations of an increased M1/M2 ratio in DB wounds of humans and mice.22–25 Because neutrophils and macrophages play both proinflammatory and anti-inflammatory roles during the course of normal wound healing, it will be important to more fully characterize the functional and migrational time course of these cells in the DB wound.
Two chemoattractants, Pf4 and CXCL2 (MIP-2), were expressed at higher levels in DB wounds. Pf4 was higher on postwounding day 5, and CXCL2 was higher on day 7. Both chemokines had similar levels of expression in WT and DB wounds on postwounding day 2, suggesting that these particular signals were not the cause of the delayed recruitment of neutrophils or macrophages to the DB wounds. Thus, these delays may be due to other defects in adhesion molecule expression, chemokine ligand or receptor expression, or chemokine signaling pathways.
Finally, we observed that the proinflammatory cytokines, IL-17 and IL-20, were both significantly higher in DB wounds. The proinflammatory cytokine, IL-17, is associated with several chronic inflammatory diseases, including psoriasis, rheumatoid arthritis, and multiple sclerosis.26 Production of IL-17 by CD4+ Th17 cells, CD8+ T cells, γδ T cells, NKT cells, innate lymphoid cells, and neutrophils is important in host defense against pathogens. Interestingly, overexpression of Ym1 during helminth infection in mice caused increased IL-17 expression. In the helminth model, the increased IL-17 stimulated the accumulation of neutrophils and aided in control of infection.27 In contrast, our results showed that although IL-17 expression was increased in DB wounds, it was associated with decreased expression of Ym1, suggesting that Ym1 may not be a critical regulator of IL-17 expression during DB wound healing.
IL-20 expression was also significantly higher in DB wounds on days 2, 5, and 7 compared with WT wounds. Overexpression of IL-20 in transgenic mice causes symptoms similar to psoriasis, including thickened epidermis and hyperkeratosis. Indeed, both human psoriatic lesions and mouse models of psoriasis are associated with high expression levels of IL-20. In mice, intradermal injections of IL-20 induced psoriatic lesions; in humans, IL-20 expression decreases during the treatment and resolution of psoriasis.28,29
Both IL-17 and IL-20 play a significant role in the pathogenesis of psoriasis.29,30 Drugs blocking IL-17 are effective in treating psoriasis,31,32 and the pharmacokinetics of anti-IL-20 antibodies are also being tested in humans for treatment of psoriasis or rheumatoid arthritis.33 Together, these findings may suggest a relationship between certain skin pathologies and increased IL-17 and IL-20 expression. Consequently, these cytokines may also be useful targets for therapies aimed at improving DB wound healing.
Innovation
This is the first study to examine cytokine expression by PCR microarray in DB mouse wounds as well as the first to identify significant increases in both IL-17 and IL-20 expression in the DB wounds. Recent advances in psoriatic treatments have focused on targeted therapeutics to reduce IL-17 and IL-20 production and may thereby play some role in DB wound treatment.
Key Findings.
• Gr-1+ neutrophils and Ym1+ macrophages were delayed in migrating into DB wounds.
• Ym1 and arginase 1 expression was reduced in DB wounds.
• IL-17 and IL-20 expression levels significantly increased in DB wounds.
Abbreviations and Acronyms
- DB
diabetic
- DNA
deoxyribonucleic acid
- ECF-L
eosinophil chemotactic factor-L
- FIZZ
found in inflammatory zone
- IL
interleukin
- Mrc
macrophage mannose receptor
- PCR
polymerase chain reaction
- Pf4
platelet factor 4
- PMN
polymorphonuclear cells
- RNA
ribonucleic acid
- SD
standard deviation
- TNF
tumor necrosis factor
- WT
wild-type
Acknowledgments and Funding Sources
The authors would like to thank Dr. John Long, Nancy Roth, and Frank Strebeck in the Department of Comparative Medicine at Saint Louis University for their guidance and technical assistance. P.J.F. received funding from the American Association for Wound Care Research Scholarship Award.
Author Disclosure and Ghostwriting
No competing financial interests exist. The content of this article was expressly written by the authors listed. No ghostwriters were used to write this article.
About the Authors
Phillip J. Finley, PhD, earned a PhD in Integrated and Applied Sciences from Saint Louis University. He is currently a Senior Research Scientist at Mercy Hospital in Springfield, Missouri, where he oversees the Division of Trauma and Burn Research. Cory E. DeClue, MS, received his MS in the Department of Biology at Saint Louis University. Scott A. Sell, PhD, is currently an Assistant Professor in Biomedical Engineering at Parks College of Engineering, Aviation, and Technology. He received his PhD in Biomedical Engineering from Virginia Commonwealth University. Joseph M. DeBartolo, MD, received a BS in Biology from Saint Louis University and his medical degree from the Chicago Medical School at Rosalind Franklin University. Laurie P. Shornick, PhD, is an Associate Professor in the Department of Biology at Saint Louis University and she holds a secondary appointment in the Department of Molecular Microbiology and Immunology at Saint Louis University. She received her PhD in Molecular Cell Biology from Washington University in Saint Louis and was a Parker B. Francis Pulmonary Research Fellow. The primary focus of her research is on the role of epithelial tissues in the innate immune response utilizing models of wound healing and viral respiratory infection.
References
- 1.Dovi JV, He LK, DiPietro LA. Accelerated wound closure in neutrophil-depleted mice. J Leukoc Biol 2003;73:448–455 [DOI] [PubMed] [Google Scholar]
- 2.Lucas T, Waisman A, Ranjan R, et al. . Differential roles of macrophages in diverse phases of skin repair. J Immunol 2010;184:3964–3977 [DOI] [PubMed] [Google Scholar]
- 3.Mosser DM. The many faces of macrophage activation. J Leukoc Biol 2003;73:209–212 [DOI] [PubMed] [Google Scholar]
- 4.Goerdt S, Politz O, Schledzewski K, et al. . Alternative versus classical activation of macrophages. Pathobiology 1999;67:222–226 [DOI] [PubMed] [Google Scholar]
- 5.Mege JL, Mehraj V, Capo C. Macrophage polarization and bacterial infections. Curr Opin Infect Dis 2011;24:230–234 [DOI] [PubMed] [Google Scholar]
- 6.Gordon S. Alternative activation of macrophages. Nat Rev Immunol 2003;3:23–35 [DOI] [PubMed] [Google Scholar]
- 7.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 2004;25:677–686 [DOI] [PubMed] [Google Scholar]
- 8.Eming SA, Werner S, Bugnon P, et al. . Accelerated wound closure in mice deficient for interleukin-10. Am J Pathol 2007;170:188–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiemessen MM, Jagger AL, Evans HG, van Herwijnen MJ, John S, Taams LS. CD4+CD25+Foxp3+ regulatory T cells induce alternative activation of human monocytes/macrophages. Proc Nat Acad Sci U S A 2007;104:19446–19451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 2012;122:787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Control CFD. National diabetes fact sheet. 2007. www.cdc.gov/diabetes/pubs/pdf/ndfs_2007.pdf (last accessed November15, 2008)
- 12.Galiano RD, Michaels J, Dobryansky M, Levine JP, Gurtner GC. Quantitative and reproducible murine model of excisional wound healing. Wound Repair Regen 2004;12:485–492 [DOI] [PubMed] [Google Scholar]
- 13.Finley PJ, Huckfeldt RE, Walker KD, Shornick LP. Silver dressings improve diabetic wound healing without reducing bioburden. Wounds 2013;25:293–301 [PubMed] [Google Scholar]
- 14.Nguyen KT, Seth AK, Hong SJ, et al. . Deficient cytokine expression and neutrophil oxidative burst contribute to impaired cutaneous wound healing in diabetic, biofilm-containing chronic wounds. Wound Repair Regen 2013;21:833–841 [DOI] [PubMed] [Google Scholar]
- 15.Scherer SS, Pietramaggiori G, Mathews JC, Chan R, Fiorina P, Orgill DP. Wound healing kinetics of the genetically diabetic mouse. Wounds 2008;20:18–28 [PubMed] [Google Scholar]
- 16.Wetzler C, Kampfer H, Stallmeyer B, Pfeilschifter J, Frank S. Large and sustained induction of chemokines during impaired wound healing in the genetically diabetic mouse: prolonged persistence of neutrophils and macrophages during the late phase of repair. J Invest Dermatol 2000;115:245–253 [DOI] [PubMed] [Google Scholar]
- 17.Nair MG, Cochrane DW, Allen JE. Macrophages in chronic type 2 inflammation have a novel phenotype characterized by the abundant expression of Ym1 and Fizz1 that can be partly replicated in vitro. Immunol Lett 2003;85:173–180 [DOI] [PubMed] [Google Scholar]
- 18.Raes G, De Baetselier P, Noel W, Beschin A, Brombacher F, Hassanzadeh Gh G. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J Leukoc Biol 2002;71:597–602 [PubMed] [Google Scholar]
- 19.Witte MB, Barbul A, Schick MA, Vogt N, Becker HD. Upregulation of arginase expression in wound-derived fibroblasts. J Surg Res 2002;105:35–42 [DOI] [PubMed] [Google Scholar]
- 20.Brown GD, Taylor PR, Reid DM, et al. . Dectin-1 is a major beta-glucan receptor on macrophages. J Exp Med 2002;196:407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SJ, Ruiz N, Bezouska K, Drickamer K. Organization of the gene encoding the human macrophage mannose receptor (MRC1). Genomics 1992;14:721–727 [DOI] [PubMed] [Google Scholar]
- 22.Sindrilaru A, Peters T, Wieschalka S, et al. . An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J Clin Invest 2011;121:985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goren I, Muller E, Schiefelbein D, et al. . Systemic anti-TNFalpha treatment restores diabetes-impaired skin repair in ob/ob mice by inactivation of macrophages. J Invest Dermatol 2007;127:2259–2267 [DOI] [PubMed] [Google Scholar]
- 24.Khanna S, Biswas S, Shang Y, et al. . Macrophage dysfunction impairs resolution of inflammation in the wounds of diabetic mice. PLoS One 2010;5:e9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirza R, Koh TJ. Dysregulation of monocyte/macrophage phenotype in wounds of diabetic mice. Cytokine 2011;56:256–264 [DOI] [PubMed] [Google Scholar]
- 26.Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol 2014;14:585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutherland TE, Logan N, Ruckerl D, et al. . Chitinase-like proteins promote IL-17-mediated neutrophilia in a tradeoff between nematode killing and host damage. Nat Immunol 2014;15:1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stenderup K, Rosada C, Worsaae A, et al. . Interleukin-20 plays a critical role in maintenance and development of psoriasis in the human xenograft transplantation model. Br J Dermatol 2009;160:284–296 [DOI] [PubMed] [Google Scholar]
- 29.Wang F, Lee E, Lowes MA, et al. . Prominent production of IL-20 by CD68+/CD11c+ myeloid-derived cells in psoriasis: gene regulation and cellular effects. J Invest Dermatol 2006;126:1590–1599 [DOI] [PubMed] [Google Scholar]
- 30.Durham LE, Kirkham BW, Taams LS. Contribution of the IL-17 pathway to psoriasis and psoriatic arthritis. Curr Rheumatol Rep 2015;17:55. [DOI] [PubMed] [Google Scholar]
- 31.Langley RG, Elewski BE, Lebwohl M, et al. . Secukinumab in plaque psoriasis—results of two phase 3 trials. N Engl J Med 2014;371:326–338 [DOI] [PubMed] [Google Scholar]
- 32.Leonardi C, Matheson R, Zachariae C, et al. . Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N Engl J Med 2012;366:1190–1199 [DOI] [PubMed] [Google Scholar]
- 33.Lundblad MS, Overgaard RV, Gothberg M, Fjording MS, Watson E. Clinical pharmacokinetics of the anti-interleukin-20 monoclonal antibody NNC0109-0012 in healthy volunteers and patients with psoriasis or rheumatoid arthritis. Adv Ther 2015;32:228–238 [DOI] [PubMed] [Google Scholar]