Abstract
Recent advances in understanding the activity and selectivity of kinase inhibitors and their relationships to protein structure are presented. Conformational selection in kinases is studied from empirical, data-driven and simulation approaches. Ligand binding and its affinity are, in many cases, determined by the predetermined active and inactive conformation of kinases. Binding affinity and selectivity predictions highlight the current state of the art and advances in computational chemistry as it applies to kinase inhibitor discovery. Kinome wide inhibitor profiling and cell panel profiling lead to a better understanding of selectivity and allow for target validation and patient tailoring hypotheses.
Keywords: kinase inhibitor, kinase inhibition profile, DFG in-out transition, conformational selection, cancer cell panel profiling
1. Introduction
Understanding the relationship of kinase targets in normal and disease states and their modulation by inhibitors stands at a crossroads in the discovery and delivery of new medicines. Overcoming key challenges such as target selection, pathway modulation, compound prioritization, understanding toxicity, biomarker selection and patient tailoring are key to the design of better treatments. Computational sciences, including bio, chemo, and structural informatics are increasingly indispensable in kinase discovery. Chemical and structural informatics streamline complexity, finding patterns in large data sets and generating testable hypotheses for experimentation. Several talks at the conference reported analyses of large datasets of structural and activity data of many compounds tested with many kinases. Alexander Baumann, Richard Engh and Thibault Varin described platforms for experimental activity profiling of compounds across large kinase panels, and computational methods to understand patterns of cross-reactivity across compound, kinase, and cell line dimensions. Eric Martin used arrays of kinase predictive models to estimate inhibition profiles when experimental data are incomplete or lacking. Henrik Moebitz and Stefan Knapp evaluated large databases of X-ray structures and compound activities to relate protein conformational states to binding affinity. Benoit Roux used physics-based molecular dynamics simulations rather than informatics to understanding relative energies of DFG-in and DFG-out kinase conformations. Valerio Berdini employed a chemistry-based approach to the conformation problem, building up ligands to DFG-in and DFG-out conformations from fragment-based starting points. Another aspect of kinase computation correlates chemical similarity with kinase potency to predict activity in lieu of or in advance or experiments. Thibault Varin, Eric Martin and Jens Meiler described ligand-based kinase inhibition and selectivity models, and their impact on drug discovery projects. In addition to their predictive, explanatory aspects, computational sciences play an increasing role in experiment design spanning a range from target hypotheses to compound design. This short review will briefly highlight some of these diverse approaches to computational kinase discovery presented at the conference.
2. Discussion
2.1 Conformation selection
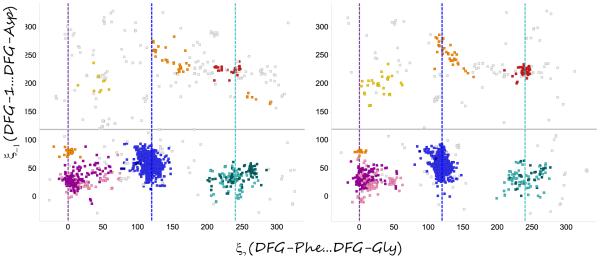
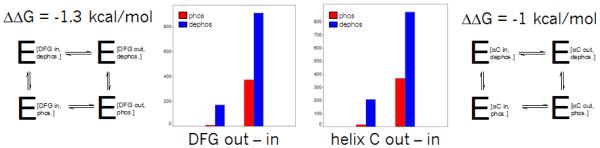
A number of speakers described the interplay between active-inactive kinase conformations, and ways to computationally analyze and address them. Most kinase can be activated by phosphorylation of the activation loop, causing a conformational shift of the DFG motif from the “out” to the “in” positions, bringing the catalytic ASP into position to interact with phosphates on the ATP and Magnesium ions to perform phosphate transfer. Henrik Moebitz presented a 3D alignment and structural clustering of all mammalian kinase X-ray conformations. The structures formed distinct clusters when plotted in 2 specialized graphs, a “DFG-plot” and a “Helix-C plot”, according to a few simple geometric criteria. The secret to getting distinct interpretable clusters was the identification of pseudo DFG torsions formed by sets of 4 consecutive alpha carbons, a measure of the torsion between two consecutive sidechains. These angles were divided into regions: FG-down/DFG-active/G-down, and DFG-in/out. The structures could then be plotted on the 2 graphs by adding a distance to helix-C, classified as in/dilated/out. Comparing two subsets of the PDB, prior and post June 2010, gave similar distributions of clusters (Fig. 1). Analyzing the populations provided estimates of the energy differences between kinase conformational states. One interesting conclusion was that phosphorylation shifts the relative balance between active and inactive conformation by 1 kcal/mole on average (Fig. 2). This observation can explain why type II inhibitors, which bind in the inactive (DFG-out) conformation, exhibit lower potency (by 10 fold on average), but measurable in biochemical (phosphorylated) kinase assays. He also observed that first-shell polar residues hinder the DFG transition. The research extends the long standing interest in classification of binding modes of kinase inhibitors[1].
Fig. 1.
DFG-plots of two sets of the PDB, post (left, 2171 chains) and pre June 2010 (right, 1909 chains), showing similar distributions of clusters. The main clusters are active (blue), FG-down (margenta) and G-down (cyan). DFG-out clusters are found above the border of ξ−1 >100.
Fig. 2.
Estimation of conformational bias from phosphorylation. Based on the population shift between DFG in and out conformations, the stabilizing effect of phosphorylation can be estimated from these thermodynamic cycles.
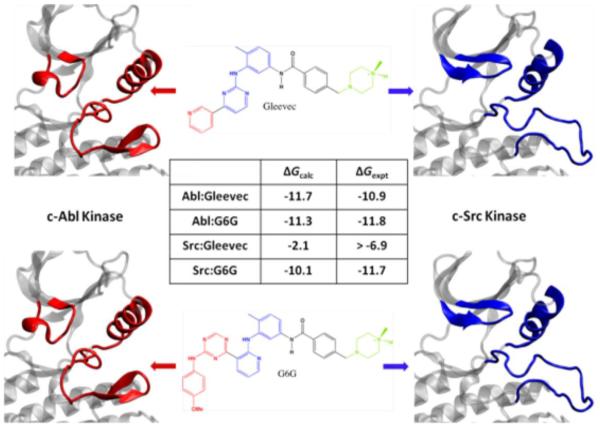
Aiming to understand energetic and conformational preferences leading to observed selectivity, Benoit Roux and his co-workers Yen-Lin Lin and Yilin Meng described free energy molecular dynamics simulations to directly calculate the binding affinity of Imatinib, which inhibits Abl and c-Kit, but not c-Src, even though all three have >30% sequence homology. He drove 2 pseudo dihedrals by umbrella sampling to get full 2D conformational free energy maps of the unphosphorylated proteins around the DFG motif region of the activation loop. The maps showed only 2 stable conformations: DFG-in and DFG-out. According to the umbrella sampling calculations, Abl kinase appears to be more stable in the DFG-in conformation by a modest 1.4 kcal/mol, while Src is more stable in the DGF-in by 5.4 kcal/mol, suggesting that the free energy cost of the DFG flip between these two kinases could be one determinant of type II selectivity [2]. The team also calculated the affinity of Imatinib to the binding pocket by using the “alchemical double decoupling” technique to “annihilate” the inhibitor from solution and grow it into the protein. They decomposed the total free energy difference, which agrees well with experiment (Fig. 3), into components by vanishing and growing the ligand one energy term at a time: first the repulsive part, then the van der Waals dispersion part, and finally the electrostatic part. This analysis, in close agreement with experimental data, correctly concluded that Imatinib binds Abl better, and identified the van der Waals dispersion term as the dominant energy component. In addition, the binding of an analog of Gleevec (G6G), which is equally potent for Abl and Src, was investigated and agreement with experiment in binding affinity was observed [2]. This additional set of free energy calculations further supported that both conformational selection and protein-ligand interaction are responsible for the specificity of Gleevec.
Fig. 3.
Free energy differences from simulations agree well with experiments[2].
Stefan Knapp presented a wide range of experimental studies investigating compounds which stabilize inactive conformations of kinases. In collaboration with Nathanael Grays laboratory, he reported that more than 200 kinases, covering all branches of the kinome, were found to be inhibited by a small set of type2 inhibitors, suggesting that a large fraction of kinases can be targeted by type II inhibitors [3]. Type II inhibitor structures are underrepresented in the protein data based (PDB) and several type-II inhibitors were co-crystallized with kinases for which no experimental type-II structure has been reported. These structures included CDK2. .
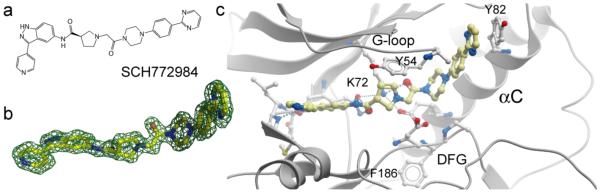
A unique binding mode was reported for the ERK inhibitor SCH772984, which bound in a so far unreported conformation to ERK1 and ERK2 (Fig. 4). In this novel binding mode, which would be impossible to predict with current computational approaches, the inhibitor induced a binding pocket between the P-loop and αC, forming a number of hydrogen bonds and aromatic stacking interactions with residues present in these structural elements. Binding of SCH772984 was associated with slow off rates in vitro as well as in cellular assays, whereas off-targets such as haspin and JNK interacted with the inhibitor in diverse but canonical type I binding modes and showed fast on and off-rates. Mutagenesis studies suggested that aromatic stacking interactions of residues located in αC, as well as the glycine rich loop, were important for the slow binding kinetics of this inhibitor. The novel binding pocket may offer an alternative design strategy for type II inhibitors. [4].
Fig. 4.
Novel binding mode of SCH772984 in ERK2. a: chemical structure of SCH772984. b: 2FoFc OMIT electron density map contoured at 2σ. c: Details of the interaction in ERK2. C.
Valerio Berdini used MELK kinase as an example of how medicinal chemistry can use fragment starting points to create insights into stabilizing unique kinase conformations [5, 6]. From 231 fragments that showed an effect in a protein melting-point screen, 144 confirmed in NMR. Subsequent X-ray crystallography showed 20 novel hinge binders. Isoquinoline fragments were optimized into both highly efficient (LE=0.54) type I ligands, and highly potent type II inhibitors. MELK has a large leucine gate keeper, and traditional type II linkers did not induce the DFG out conformation. On average, type I fragments and inhibitors had much higher ligand efficiencies, suggesting that the type II conformation in MELK is higher energy.
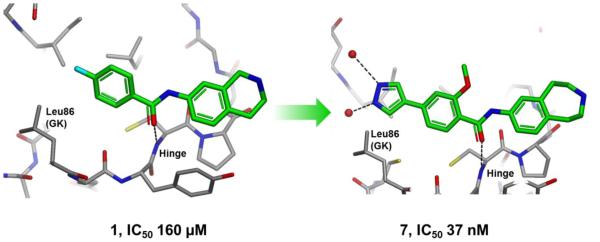
One of the Type I starting points was optimized into a selective Melk inhibitor that offered conformational selection for the MELK hinge region[5]. A path from an initial, relatively inefficient 160uM fragment with unique binding, to the optimized 37nM molecule with good selectivity, involved using a variety of structure based design tools and computational analog modeling to identify strong interactions with MELK (Fig. 5). Another approach utilizing the ASTEX structural informatics platform allowed for the rational design of a 19nM type II inhibitor, although with a less optimal selectivity profile[6]. In this approach, existing structural fragments of hinge binders, linkers and positively ionizable groups were combined to stabilize the type II MELK conformation. Structure-based design was employed together with computational tools in the course of project evolution.
Fig. 5.
A low affinity fragment hit was optimized by SBDD to a selective 37 nM tool compound.
2.2 Predictive modeling
Predictive models are widely used for virtual screening against kinase targets. Both ligand-based, structure-based and mixed models are used in an industrial setting to initiate and focus kinase inhibitor discovery efforts. Kinome-wide profiling data allow the creation and evaluation of computational models not only for activity but also selectivity predictions. Thibault Varin presented an application of ligand-based models in screening[7] campaigns at Lilly, and the discovery and initial optimization of selective RIO2 kinase inhibitors. Using chemical similarity, he selected from a set of virtual, robot-capable reactions a set of 8 compounds. These were robotically synthesized[8] and tested for activity[9]. Three showed activity improvement ranging from 2 to 10-fold from the initial hit.
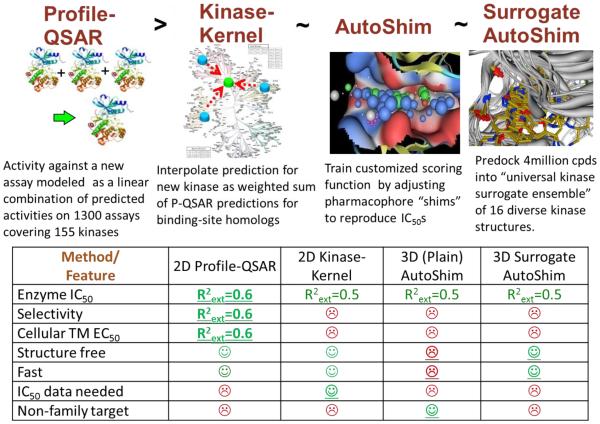
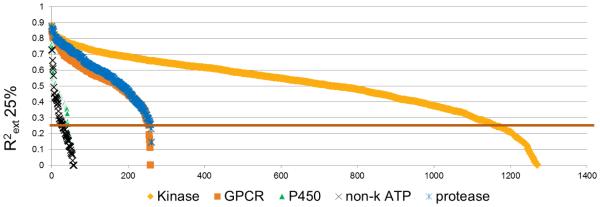
Eric Martin described a collection of empirical protein-family virtual screening (PFVS) models (Fig. 6) which combine extensive IC50 and structural data from all historical kinase projects to produce predictive activity and selectivity models for both biochemical and cellular assays of new kinases, with accuracy comparable to experimental high-throughput screens.[10] He described numerous case studies where accurate prediction of biochemical and cellular selectivity identified starting points for medicinal chemistry and tool compounds that validated, or in several instances invalidated, newly proposed drug targets. Hit rates were consistently 25% to 80%, even for novel scaffolds completely unrelated to the known inhibitors.
Fig. 6.
Four modeling methods comprising Protein-Family Virtual Screening
2.3 Biochemical and cellular assay panels and computational target identification
Panels of biochemical and cellular assays [11] have been used in multiple ways to understand potential uses of kinase inhibitors to treat various cancers. The signatures together with Genetic backgrounds, mRNA expression levels and shRNA data have in turn been used to understand compound signatures, with the goal of creating patient tailoring hypotheses.
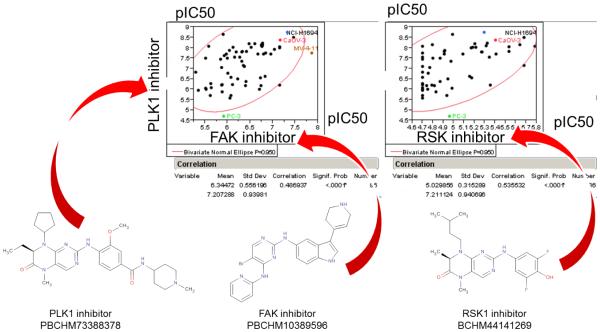
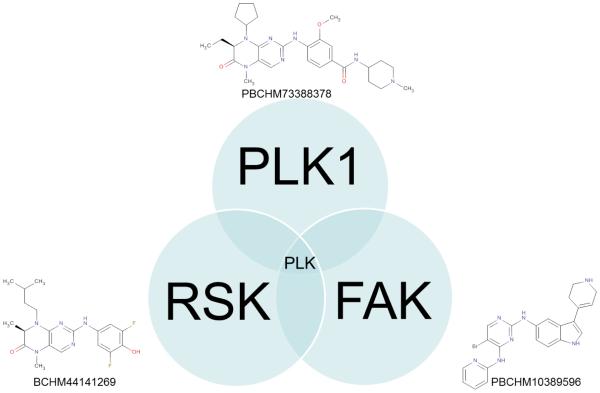
Thibault Varin showed how one could utilize kinase inhibitor profiles to elucidate reasons for cell panel signature similarity. In several cases, a target hypothesis could be derived, indicating and confirming the role of PLK1[12] in cell proliferation. He presented comparisons of compounds based on the activity on large cancer cell sensitivity and kinase affinity panels. By integrating these two compound profiles, he showed that the target toward which a compound has been historically optimized doesn’t necessarily drive the cellular activity of a given cell line or even of the overall cancer cell line panel. A RSK (BI-D1870) and a FAK (PBCHM10389596) inhibitor were reported to have a similar cancer cell sensitivity panel as a PLK1 (BI-2536) inhibitor. These compounds both showed affinity for PLK1 in the kinase panels (Fig. 7 & 8) (Kinomescan, DiscoverX).
Fig. 7.
Similarity of PLK1 (BI-2536), RSK (BI-D1870) and FAK (PBCHM10389596) inhibitors cancer cell sensitivity profiles. Whereas these three compounds were developed and optimized for different targets, they show a similar activity on a large cancer cell sensitivity panel.
Fig. 8.
The PLK1, RSK and FAK inhibitors are not fully selective and all inhibit also PLK1. As PLK1 is an essential gene, this could explain the similar activity of these compounds on the cancer cell sensitivity panel profile.
Eric Martin applied Protein Family Virtual Screening (PFVS) (see above) to predict the IC50s for 3 million compounds against 2000 biochemical and cellular assays (Fig. 9)[7][13]. These were applied to predicting polypharmacology, modes-of-action for phenotypic screens, toxicity profiling, and selecting commercial compounds with diverse selectivity profiles for chemical archive enhancement.
Fig. 9.
IC50 Prediction vs. Experimental correlations for 25% held-out test sets for 2000 Profile-QSAR models covering 5 protein families. Predictions for 3,000,000 compounds have been pre-calculated and stored.
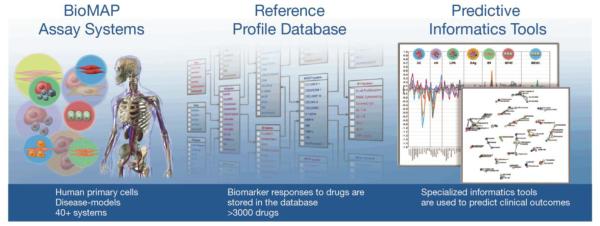
Alexander Baumann described the extension of the well-established kinomescan methodology to the bromodomain (BRD) family. Screening kinase inhibitor libraries against a bromodomain panel identified several established kinase inhibitors that were cross-reactive and might be repurposed as kinase-BRD dual inhibitors. He also introduced the“BioMAP” technology platform that provides a measure of overall phenotypic response of compounds under disease-like conditions, and identifies clinically relevant activities across a broad protein biomarker panel. The BioMAP systems are stimulated primary human cell types and co-cultures designed to recapitulate the complex signaling networks and microenvironment in diseased human tissue (Fig. 10)[14]. The resulting biomarker fingerprints are useful for identifying modes of action and toxicity profiling. The biomarker fingerprints from the kinase/BRD dual inhibitors showed a hybrid of both mechanisms [15].
Fig. 10.
The BioMAP system showing Human Primary cells Disease models on the left, biomarker responses to >3000 drugs stored in the database in the middle and a variety of specialized informatics tools with ability to predict clinical outcomes from data.
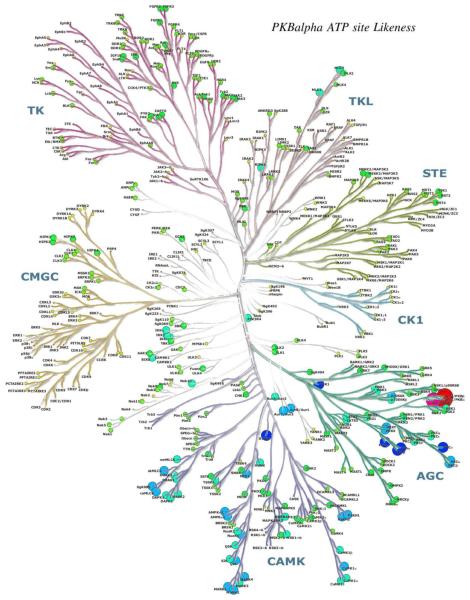
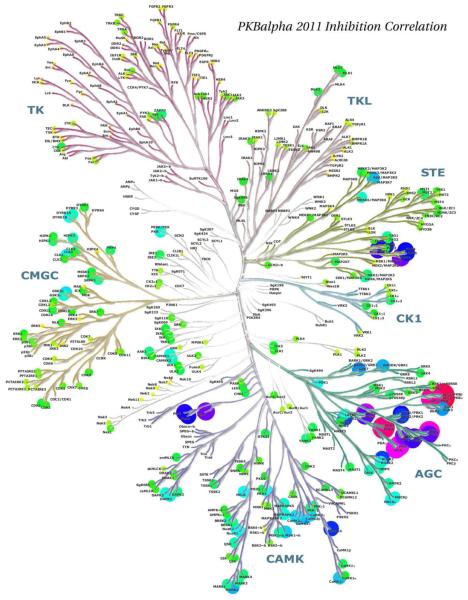
Richard Engh examined methods to evaluate protein kinase target similarities with the aim to compare information types hierarchically. At the simplest level, "pseudosequence" similarities were calculated based on sequences chosen to represent binding site residues (Fig. 11). Statistically, these corresponded quite well with experimental inhibition profiles from Ambit 2011 data [16], especially for tyrosine kinase targets (Fig. 12). Such analyses support the use of surrogate kinases in structure-based drug discovery [17], and may aid in choosing focused screening libraries for repurposing or retargeting known compounds.
Fig. 11.
Disk sizes and colors depict pseudosequence similarities to PKB alpha
Fig. 12.
Disk sizes and colors depict correlation of AMBIT 2011 inhibition profiles of protein kinase targets with those of PKB alpha. Note similarities to Fig 11, pseudosequence similarities.
At a higher level of information content, similarity analyses of target proteins would involve comparisons of X-ray structures. Currently, comprehensive integration of structural information is not practically possible: there is too much unpredictable structural variability, the distributions of structural states are strongly influenced by crystallization conditions[18], and experimental binding data are highly dependent on assay conditions, which in turn are often not accessible to data mining tools. On the other hand, some specific areas are well supported, including reliable clustering of key structural states (notably DFG and C-helix states, see many other talks from IPK2014 and other references, including[19]). For well characterized diseases, the data increasingly enable targeted pharmacology by identifying key determinants of target similarities, possibly combined with "orthogonal" dissimilarities.
2.4 Future directions – introducing crowd sourcing into drug design
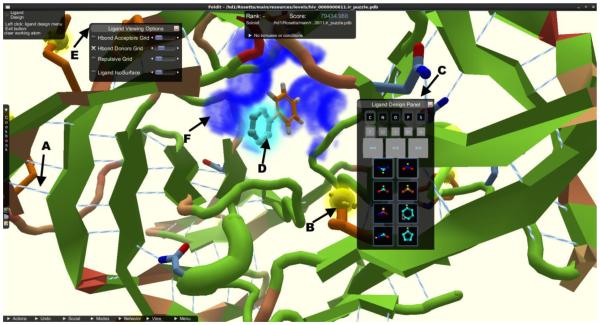
Jens Meiler showed principles and application examples of the RosettaLigand [20] and BCL::Cheminformatics [21] computational software packages, developed across academic institutions, highlighting both structure-based and ligand-based drug design (Fig. 13). A fascinating application of Rosetta is the computer game Foldit [22], with over 200,000 users. The goal of the game is to predict the structure of a protein. In addition to educational value, it is an example of crowd sourcing to solve challenging scientific problems. The crowd sourcing approach is similarly being translated into drug discovery with a drug design component of Foldit. One application of the game was to one of malaria’s essential kinases PKG [23], the X-ray structure of which was revealed at the meeting. For ligand-based drug design the QSAR application BCL::Cheminformatics[21] was introduced which converted a ligand structure into a property vector of charge, shape, or H-bonds donors and acceptors. Chirality was included by a signed volume using a right-hand rule[24]. A QSAR model was trained using an artificial neural network. In several example applications the hit-rate in virtual screening increased by factors of 15-50 over conventional diversity approaches.
Fig. 13.
Screenshot of Foldit, from the new Rosetta Ligand application, a crowd-sourced multiplayer game adopted for ligand design. A) Hydrogen bond contacts are shown in light-blue/white lines and B) surface exposed hydrophobic residues are shown as yellow blobs. C) The ligand design panel is the control center for players and allows them to choose from a variety of fragments, bond manipulations, and element modifications to design the ligand in the protein binding pocket. D) When players hover over a fragment, a ghost view of the new fragment is drawn at the attachment point (light blue glowing fragment). E) The ligand viewing options menu allows players to turn off QSAR grids calculated for hydrogen bond acceptors and donors F) (shown as dark blue fog) or repulsion for the ligand, with slider bars on the side to adjust the alpha of the drawn fogs. Players can turn on the protein isosurface or a ligand centric view that draws the isosurface around the ligand, allowing for advance spatial alignment of the ligand in the binding pocket.
3 Conclusions and summary
These computational presentations illustrated many recent advances in understanding the activity and selectivity of kinase inhibitors and their relationships to protein structure. The approaches ranged from highly empirical to purely physics-based. They ranged from mining large activity and structure databases, to simulating the physics of ligand binding into active and inactive conformations, to probing conformational energetics by synthesizing related ligands designed to bind alternate conformations. The presentations helped to move our understanding of the chemistry, physics, biochemistry and biology of kinase structure and activity, and illustrated how that understanding impacts drug discovery programs. It is also our view that computational methods will facilitate drug discovery/development against kinases of eukaryotic pathogens for which there is limited experimental information available, an additional theme of the conference. Hopefully these highlights have whetted your appetite to dig further into some of these topics in the accompanying articles in this issue.
Highlights.
Conformational stabilization affects binding modes and selectivity of inhibitors
Computational predictive models aid in hypothesis generation and screening
Whole Kinome and cell panel profiling allows for targeted kinase drug discovery
4 Acknowledgements
The authors would like to thank the IPK2014 organizing committee for their continued support of this conference series and Dr. Steven Combs for his contributions and research on drug design in Foldit.
Abbreviations
- Abl
Abelson murine leukemia
- MD
Molecular Dynamics
- Melk
Maternal embryonic leucine zipper kinase
- Kit
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
- JNK
c-Jun N-terminal kinase
- PLK
Polo Like Kinase
- CDK
Cyclin Dependent Kinase
- ERK
Extracellular-Signal-Regulated Kinase
- Pdb
Protein Data Bank
- NMR
Nuclear Magnetic Resonance
5 References
- [1].Cowan-Jacob SW, Mobitz H, Fabbro D. Structural biology contributions to tyrosine kinase drug discovery. Curr Opin Cell Biol. 2009;21:280–287. doi: 10.1016/j.ceb.2009.01.012. [DOI] [PubMed] [Google Scholar]
- [2].Lin Y-L, Meng Y, Huang L, Roux B. Computational Study of Gleevec and G6G Reveals Molecular Determinants of Kinase Inhibitor Selectivity. J Am Chem Soc. 2014;136:14753–14762. doi: 10.1021/ja504146x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhao Z, Wu H, Wang L, Liu Y, Knapp S, Liu Q, Gray NS. Exploration of Type II Binding Mode: A Privileged Approach for Kinase Inhibitor Focused Drug Discovery? ACS Chemical Biology. 2014;9:1230–1241. doi: 10.1021/cb500129t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chaikuad A, Tacconi EMC, Zimmer J, Liang Y, Gray NS, Tarsounas M, Knapp S. A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat Chem Biol. 2014;10:853–860. doi: 10.1038/nchembio.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Johnson CN, Berdini V, Beke L, Bonnet P, Brehmer D, Coyle JE, Day PJ, Frederickson M, Freyne EJ, Gilissen RA, Hamlett CC, Howard S, Meerpoel L, McMenamin R, Patel S, Rees DC, Sharff A, Sommen F, Wu T, Linders JT. Fragment-based discovery of type I inhibitors of maternal embryonic leucine zipper kinase. ACS Med Chem Lett. 2015;6:25–30. doi: 10.1021/ml5001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Johnson CN, Adelinet C, Berdini V, Beke L, Bonnet P, Brehmer D, Calo F, Coyle JE, Day PJ, Frederickson M, Freyne EJ, Gilissen RA, Hamlett CC, Howard S, Meerpoel L, Mevellec L, McMenamin R, Pasquier E, Patel S, Rees DC, Linders JT. Structure-Based Design of Type II Inhibitors Applied to Maternal Embryonic Leucine Zipper Kinase. ACS Med Chem Lett. 2015;6:31–36. doi: 10.1021/ml5001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gao C, Cahya S, Nicolaou CA, Wang J, Watson IA, Cummins DJ, Iversen PW, Vieth M. Selectivity Data: Assessment, Predictions, Concordance, and Implications. J. Med. Chem. 2013;56:6991–7002. doi: 10.1021/jm400798j. [DOI] [PubMed] [Google Scholar]
- [8].Godfrey AG, Masquelin T, Hemmerle H. A remote-controlled adaptive medchem lab: an innovative approach to enable drug discovery in the 21st Century. Drug Discovery Today. 2013;18:795–802. doi: 10.1016/j.drudis.2013.03.001. [DOI] [PubMed] [Google Scholar]
- [9].DiscoverX. KINOMEscan-Word's Largest Kinase Assay Panel. 2014;2014 [Google Scholar]
- [10].Martin E, Mukherjee P. Kinase-Kernel Models: Accurate In silico Screening of 4 Million Compounds Across the Entire Human Kinome. J. Chem. Inf. Model. 2012;52:156–170. doi: 10.1021/ci200314j. [DOI] [PubMed] [Google Scholar]
- [11].Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- [12].Barr FA, Sillje HHW, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–441. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- [13].Martin E, Mukherjee P, Sullivan D, Jansen J. Profile-QSAR: A Novel meta-QSAR Method that Combines Activities across the Kinase Family To Accurately Predict Affinity, Selectivity, and Cellular Activity. J Chem Inf Model. 2011;51:1942–1956. doi: 10.1021/ci1005004. [DOI] [PubMed] [Google Scholar]
- [14].Berg EL, Yang J, Melrose J, Nguyen D, Privat S, Rosler E, Kunkel EJ, Ekins S. Chemical target and pathway toxicity mechanisms defined in primary human cell systems. J Pharmacol Toxicol Methods. 2010;61:3–15. doi: 10.1016/j.vascn.2009.10.001. [DOI] [PubMed] [Google Scholar]
- [15].Ciceri P, Muller S, O'Mahony A, Fedorov O, Filippakopoulos P, Hunt JP, Lasater EA, Pallares G, Picaud S, Wells C, Martin S, Wodicka LM, Shah NP, Treiber DK, Knapp S. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat Chem Biol. 2014;10:305–312. doi: 10.1038/nchembio.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- [17].Aberg E, Lund B, Pflug A, Gani OA, Rothweiler U, de Oliveira TM, Engh RA. Structural origins of AGC protein kinase inhibitor selectivities: PKA as a drug discovery tool. Biol Chem. 2012;393:1121–1129. doi: 10.1515/hsz-2012-0248. [DOI] [PubMed] [Google Scholar]
- [18].Oliveira TM, Ahmad R, Engh RA. VX680 binding in Aurora A: pi-pi interactions involving the conserved aromatic amino acid of the flexible glycine-rich loop. J Phys Chem A. 2011;115:3895–3904. doi: 10.1021/jp108286r. [DOI] [PubMed] [Google Scholar]
- [19].Gani OABSM, Narayanan D, Engh RA. Evaluating the Predictivity of Virtual Screening for Abl Kinase Inhibitors to Hinder Drug Resistance. Chem. Biol. Drug Des. 2013;82:506–519. doi: 10.1111/cbdd.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lemmon G, Meiler J. Rosetta Ligand Docking with Flexible XML Protocols. In: Baron R, editor. Computational Drug Discovery and Design. Vol. 819. Springer; New York: 2012. pp. 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Butkiewicz M, Lowe EW, Jr., Mueller R, Mendenhall JL, Teixeira PL, Weaver CD, Meiler J. Benchmarking ligand-based virtual High-Throughput Screening with the PubChem database. Molecules. 2013;18:735–756. doi: 10.3390/molecules18010735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Khatib F, DiMaio F, Cooper S, Kazmierczyk M, Gilski M, Krzywda S, Zabranska H, Pichova I, Thompson J, Popović Z, Jaskolski M, Baker D. Crystal structure of a monomeric retroviral protease solved by protein folding game players. Nat Struct Mol Biol. 2011;18:1175–1177. doi: 10.1038/nsmb.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hopp CS, Flueck C, Solyakov L, Tobin A, Baker DA. Spatiotemporal and functional characterisation of the Plasmodium falciparum cGMP-dependent protein kinase. PLoS One. 2012;7:e48206. doi: 10.1371/journal.pone.0048206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sliwoski G, Lowe EW, Butkiewicz M, Meiler J. BCL::EMAS--enantioselective molecular asymmetry descriptor for 3D-QSAR. Molecules. 2012;17:9971–9989. doi: 10.3390/molecules17089971. [DOI] [PMC free article] [PubMed] [Google Scholar]