ABSTRACT
To effectively infect cells, Lassa virus needs to switch in an endosomal compartment from its primary receptor, α-dystroglycan, to a protein termed LAMP1. A unique histidine triad on the surface of the receptor-binding domain from the glycoprotein spike complex of Lassa virus is important for LAMP1 binding. Here we investigate mutated spikes that have an impaired ability to interact with LAMP1 and show that although LAMP1 is important for efficient infectivity, it is not required for spike-mediated membrane fusion per se. Our studies reveal important regulatory roles for histidines from the triad in sensing acidic pH and preventing premature spike triggering. We further show that LAMP1 requires a positively charged His230 residue to engage with the spike complex and that LAMP1 binding promotes membrane fusion. These results elucidate the molecular role of LAMP1 binding during Lassa virus cell entry and provide new insights into how pH is sensed by the spike.
IMPORTANCE Lassa virus is a devastating disease-causing agent in West Africa, with a significant yearly death toll and severe long-term complications associated with its infection in survivors. In recent years, we learned that Lassa virus needs to switch receptors in a pH-dependent manner to efficiently infect cells, but neither the molecular mechanisms that allow switching nor the actual effects of switching were known. Here we investigate the activity of the viral spike complex after abrogation of its ability to switch receptors. These studies inform us about the role of switching receptors and provide new insights into how the spike senses acidic pH.
INTRODUCTION
Viral hemorrhagic fevers (VHFs) are a devastating global health problem with a significant yearly death toll. One of the causative agents of VHFs in Africa is Lassa virus (LASV), which belongs to the Arenaviridae family. LASV is an enveloped, negative-strand RNA virus that naturally resides in rodents from the Mastomys genus (1) and can spread to humans (2). The primary cellular receptor of LASV is α-dystroglycan (α-DG) (3, 4), which is recognized by a trimeric class 1 viral glycoprotein complex (spike complex) on the viral surface (5, 6). Following successful attachment to α-DG on cells, LASV is internalized via macropinocytosis (7), and the spike complex facilitates membrane fusion at the acidic environment of a late endosomal compartment (5, 8).
The LASV spike complex is expressed as a glycoprotein precursor (GPC) that is cleaved into three segments by a signal peptidase and subtilisin-kexin-isozyme 1/site 1 protease (9). The functional spike complex consists of a receptor-binding subunit (GP1), a membrane-anchored fusion protein (GP2), and a unique and stable structured signal peptide (SSP) (10). Three fully processed glycoprotein moieties comprise the functional trimeric spike complex. It was recently shown that successful infection by LASV requires it to switch in a pH-dependent manner from the primary α-DG receptor to lysosome-associated membrane protein 1 (LAMP1) (11). LAMP1 (also called CD107a) is a major constituent of the lysosome membrane (12) and is thought to play an important role in maintaining lysosome integrity (13). Only a basal level of LAMP1 could be detected on the plasma membrane for some cell lines, and it is typically restricted to lysosomes and, to some degree, late endosomes (14, 15).
We recently solved the crystal structure of GP1 from LASV (16). This structure revealed a unique cluster of three histidine residues that are highly conserved among the Old World arenaviruses. This histidine triad is located on the surface of the protein, and we showed that mutating each of the histidine residues to tyrosine either reduces (H92Y) or completely prevents (H93Y and H230Y) binding to LAMP1 (16). Thus, the histidine triad constitutes part of the LAMP1-binding site of Lassa virus GP1, providing a possible explanation for the pH dependence of LAMP1 binding. However, we do not know the molecular role of this histidine triad for the function of the spike, and we also do not know why binding to LAMP1 is needed during infection. Here we investigate the modulation of the spike's membrane fusion activity as well as the entire process of infection upon mutating the histidine triad and abrogating the interaction with LAMP1. Our studies reveal important mechanistic roles of the histidine triad in pH sensing and triggering of the spike complex and provide a mechanistic role for binding of LAMP1.
MATERIALS AND METHODS
Materials.
HEK293T cells (human embryonic kidney cells; ATCC CRL-11268) were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with minimal essential medium (MEM) nonessential amino acids (Biological Industries), 1% (vol/vol) 200 mM l-glutamine solution (Biological Industries), 1% (vol/vol) Pen-Strep solution (10,000 U/ml penicillin, 10 mg/ml streptomycin; Biological Industries), and 10% (vol/vol) fetal bovine serum (FBS; Gibco), here referred to as 293 full medium (293 FM). The GP2-293 retroviral packaging cell line (TaKaRa, Clontech) was maintained in DMEM with the same supplements as those described above, except for the addition of 1% (vol/vol) 100 mM sodium pyruvate (Biological Industries) instead of nonessential amino acids. Vero cells were maintained in RPMI medium (Gibco) supplemented with 10% FBS and 1% Pen-Strep. Anti-LAMP1 antibody (CD107a) was obtained from Millipore, anti-green fluorescent protein (anti-GFP) antibody was from MBL, anti-α-tubulin clone DM1A was from Millipore, anti-transferrin receptor (CD71) was from Thermo Fisher Scientific, anti-α-dystroglycan (IIH6 C4) was obtained from the DSHB hybridoma bank, and anti-Flag polyclonal antibody (pAb) was from Thermo Fisher. Anti-rabbit and anti-mouse IgGs conjugated to horseradish peroxidase (HRP) were obtained from Jackson, and Strep-Tactin conjugated to HRP was purchased from Iba. A monoclonal antibody against Lassa virus GP1 (Ab482) (16) was prepared in our laboratory from a hybridoma.
Construction of expression vectors.
The LASV GPC coding DNA sequence was chemically synthesized (Genescript) and subcloned into the pcDNA3.1 expression vector by use of BamHI-NotI restriction sites. Mutated variants of LASV GPC were generated by PCR, using Kapa HiFi DNA polymerase (Kapa Biosystems) according to instructions of the QuikChange site-directed mutagenesis manual. GP1LASV-Fc fusion proteins were generated as previously described (16). Point mutations of histidine residues 92, 93, and 230 of Lassa virus GP1 were introduced by PCR mutagenesis. Flag-tagged and Strep-tagged Lassa virus GPCs were prepared by amplifying the GPC sequence with primers containing a C-terminal Flag/Strep tag and cloning it into pcDNA3.1 by use of BamHI-NotI restriction sites. LAMP1-mGFP was a gift from Esteban Dell'Angelica (Addgene plasmid 34831) (17). LAMP1-pENTR221 was obtained from the Weizmann Institute Forscheimer plasmid bank and was subcloned into pcDNA3.1 by use of BamHI-NotI restriction sites. LAMP1-Δ384 mGFP and LAMP1-Δ384-pcDNA mutants were generated using PCR-based site-directed mutagenesis. The pEGFP-N1 vector (Clontech) was a kind gift from Y. Yarden (Weizmann Institute of Science).
Cell fusion assay.
HEK293T cells were seeded into a 24-well plate precoated with poly-l-lysine (Sigma). Seeded cells were transfected 24 h later with 0.2 μg of Lassa virus GPC plasmid DNA by use of linear polyethylenimine (PEI) (25 kDa; Polysciences). Twenty-four hours after transfection, cells were rinsed once with 293 FM supplemented with 20 mM morpholinoethanesulfonic acid (MES; Acros Organics) that was titrated to pH 5.5 or lower. Cells were then incubated with the same medium for 10 min or as indicated, followed by washing and incubation with 293 FM for 2 h at 37°C. Following incubation, cells were fixed in prewarmed 3.7% formaldehyde solution (J. T. Baker) in phosphate-buffered saline (PBS) (Biological Industries) for 10 min. Phase images of syncytia were taken at a magnification of ×10 by using a phase microscope. Syncytia were manually spotted and selected on images. The boundaries and coverage area were automatically traced and calculated using the Versatile Wand tool of ImageJ (18).
Pseudoviral particle production and RT-qPCR quantification.
Lassa virus GPC-pseudotyped viruses were produced by transfecting the retroviral transfer vector pQCXIP-Vx3K0-mEGFP (Addgene plasmid 35527; a gift from Robert Cohen) (19) along with the Lassa virus GPC-pcDNA envelope plasmid or its mutant derivatives into the GP2-293 retroviral packaging cell line (Clontech). Briefly, GP2-293 cells were seeded at 4 × 106 to 5 × 106 in 100-mm culture dishes. The next day, cells were transfected with the retroviral vector and the envelope plasmid at a 1:1 ratio by using the calcium phosphate transfection method. After 5 h, the transfection medium was replaced with GP2-293 FM, and supernatants containing viral particles were harvested 48 h later. For viral quantification, reverse transcription-quantitative PCR (RT-qPCR) was used to determine the RNA levels of the murine enhanced green fluorescent protein (mEGFP) reporter gene. Media were centrifuged at 19,000 × g at 4°C for 10 min, and 50-μl aliquots from the supernatants were treated with 2 μg/ml RNase A (Bio Basic Inc.) for 10 min at room temperature. To inhibit RNase A activity, 40 U of RNasin RNase inhibitor (Promega) was added, and the reaction mixture was incubated for 10 min at 37°C. RNA was then extracted from viral particles by use of an RNeasy minikit (Qiagen). To eliminate remnants of the mEGFP-containing retroviral vectors, RNA was treated with RNase-free DNase I (NEB) for 10 min at 37°C, followed by heat inactivation at 75°C for 10 min. Thereafter, cDNA was prepared from RNA by use of a high-capacity cDNA reverse transcription kit (Applied Biosystems). cDNA was diluted 1:40 and subjected to qPCR using specific primers for EGFP and Fast SYBR green master mix (Applied Biosystems).
Infectivity assays.
HEK293T or Vero cells were seeded onto poly-l-lysine-precoated coverslips in 24-well plates. Cells were infected 24 h later with 2 ml pseudovirus-containing supernatant, and the medium was replaced after 24 h. At 48 h postinfection, cells were fixed with prewarmed 3.7% formaldehyde solution in PBS and mounted on slides using fluorescence mounting medium with DAPI (4′,6-diamidino-2-phenylindole) (GBI Labs). Raw images were taken in a fluorescence microscope at a magnification of ×10 and further processed using ImageJ. For quantification of infection events, HEK293T cells were seeded and infected in a 6-well plate. At 48 h postinfection, cells were detached using trypsin-EDTA solution (BI), resuspended in DMEM, and analyzed using an LSR II flow cytometer (BD Biosciences) to detect GFP-positive cells and determine the infectivity levels.
LAMP1 pulldown assay.
Wild-type (WT) GP1 and the various mutants fused to Fc were expressed by transfection of plasmid DNA into HEK293T cells in monolayers by use of linear PEI 25-kDa transfection reagent (Polysciences), and fusion protein-containing media were collected after 5 days and assessed for expression of secreted protein by dot blotting using anti-human Fc. GP1-Fc proteins were captured from the medium by incubation with protein A Sepharose beads (Santa Cruz Biotechnology) at 4°C for 1 h, followed by 3 washing steps in NETI buffer (50 mM Tris-HCl, pH 8.0, 150 mM sodium chloride, 1 mM EDTA, 0.5% [vol/vol] octylphenoxypolyethoxyethanol [IGEPAL; Sigma]). Beads were then incubated with whole-cell lysates from HEK293T cells prepared in NETI buffer and adjusted to pH 5.0 at 4°C for 1 h. Proteins were eluted in 20 mM Tris-HCl, pH 8.0, 150 mM sodium chloride. Eluted protein samples were precipitated using cold acetone. Protein pellets were recovered in sample buffer and subjected to SDS-PAGE and immunoblot analysis.
Biotinylation and pulldown of cell surface proteins.
To detect surface levels of Lassa virus GPC complexes, we cotransfected HEK293T cells with GFP (pEGFP-N1) and Lassa virus GPC-pCDNA3.1 or its mutant variants by using PEI. At 48 h postinfection, cells were washed twice with ice-cold PBS and were gently agitated for 30 min at 4°C with 0.25 mg/ml EZ-Link sulfo-NHS-SS-biotin (Thermo Scientific) dissolved in ice-cold PBS. To quench the reaction, cells were washed with cold PBS followed by a cold Tris-buffered saline (TBS) wash. Cells were then lysed using Triton lysis buffer, and the lysate was centrifuged at 10,000 × g for 2 min. Biotin-labeled proteins were then isolated from clarified supernatant by batch purification by agitation with 50 μl of NeutrAvidin beads (Thermo Scientific). Biotinylated proteins were eluted by boiling of beads with dithiothreitol (DTT)-containing sample buffer. To assess the levels of GPC, eluted samples were subjected to SDS-PAGE and immunoblotting with anti-Lassa virus GP1, anti-transferrin receptor, or anti-GFP, as a control for nonsurface proteins.
α-DG pulldown assay.
HEK293 cells were transfected with Flag-tagged Lassa virus GPC and its mutant variants. At 48 h posttransfection, whole-cell lysates were prepared using Triton lysis buffer. Lysates were clarified by centrifugation, and supernatants were incubated with EZview Red anti-Flag beads (Sigma-Aldrich). Beads were then washed and incubated with whole-cell lysates of nontransfected cells prepared in NETI buffer. Beads were washed, and proteins were eluted using a 3× Flag peptide (Sigma-Aldrich). Eluents were boiled in sample buffer and subjected to SDS-PAGE and immunoblotting using anti-α-DG and anti-Flag.
Generation of GFP Δ384-LAMP1 stable cell line.
HEK293T cells were transfected with the ΔΑ384-LAMP1-mGFP or pEGFP-N1 vector. At 48 h posttransfection, the medium was replaced with selective antibiotic-containing medium (800 μg/ml G418). Cells were grown in the presence of the antibiotic for 2 weeks. Resistant colonies of stable cells were collected as a polyclonal cell line.
Pseudovirus cell attachment assay.
Lassa pseudoviruses were treated with 2 μg/ml RNase A and DNase I for 10 min at 37°C, followed by treatment with RNasin RNase inhibitor (Promega) for 10 min at 37°C. Treated pseudoviruses were then diluted 1:10 in DMEM and applied to cells on ice. Following 30 min of incubation, cells were washed twice with PBS and lysed for RNA preparation and RT-qPCR with primers against GFP and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). For each experiment, the 2ΔΔCT value was calculated for each sample, and expression was normalized to that of the WT.
Immunofluorescence assay.
HEK293T cells were seeded onto poly-l-lysine-precoated coverslips in 24-well plates. Cells were fixed 24 h later with prewarmed 3.7% formaldehyde solution in PBS and blocked with 3% bovine serum albumin (BSA) in PBS. Cells were stained with anti-LAMP1 and fluorescein isothiocyanate (FITC)-conjugated wheat germ agglutinin (WGA). Cells were imaged at a magnification of ×100 by using an Olympus IX83 microscope coupled to a Yokogawa CSU-W1 spinning disc confocal scanner. Images were processed using ImageJ.
Detection of incorporated spikes into pseudoviruses.
To analyze the incorporation of Lassa virus GP into pseudoviruses, we followed a previously described protocol (20). C-terminally Flag-tagged Lassa virus GPC-pseudotyped viruses were prepared as we previously mentioned, without the addition of serum to the medium. Supernatants containing the viruses were collected at 48 h posttransfection, clarified by centrifugation at 200 × g for 5 min, filtered through a 0.45-μm filter, and clarified again by centrifugation at 19,000 × g for 10 min. Clarified viral stocks were then layered onto a cushion of 20% sucrose in PBS and centrifuged at 367,000 × g for 30 min. Pelleted pseudoviruses were lysed in Triton buffer, and Flag-GP2 was immunoprecipitated using anti-Flag beads. Proteins were eluted from beads by boiling of beads in sample buffer and were subjected to SDS-PAGE and immunoblotting with anti-Flag.
RESULTS
Spike-mediated cell fusion.
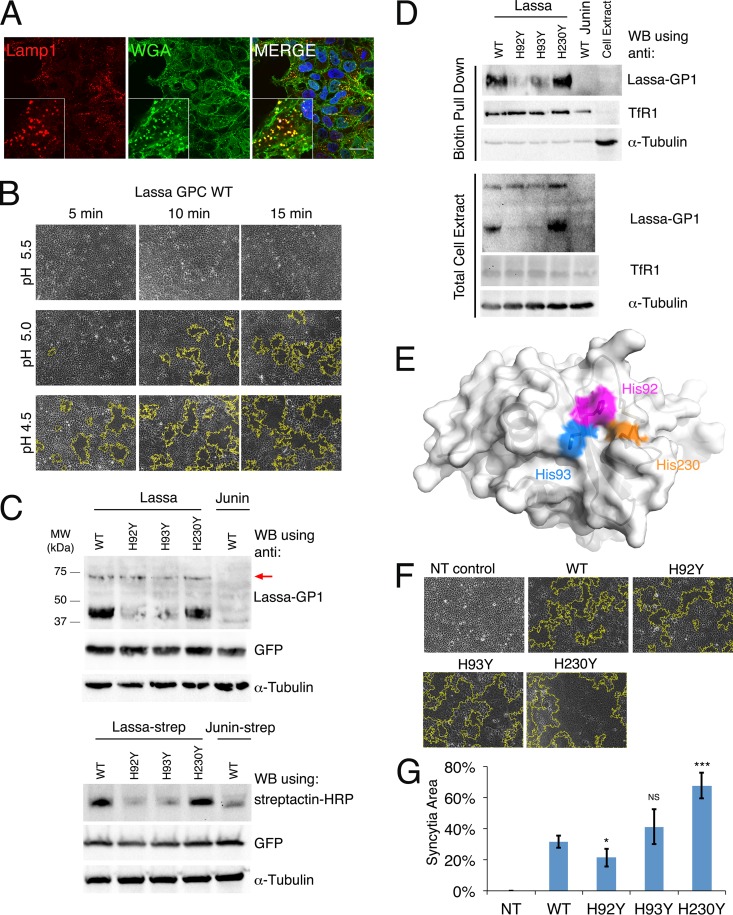
Cell fusion events result in the formation of multinucleated cells (syncytia) that are easily identified by simple bright-field light microscopy. We monitored syncytium formation in HEK293T cells following transfection with the spike GPC gene, as an indicator of the fusogenic activity of the LASV spike complex. HEK293T cells naturally present the LASV receptor α-DG on the plasma membrane (21), but not other molecules that may serve as entry receptors for LASV (22). To verify that these cells indeed display a negligible level of LAMP1 on the plasma membrane, as expected (14, 15), we used confocal microscopy with immunofluorescence labeling to visualize the cellular localization of LAMP1 (Fig. 1A). LAMP1 in HEK293T cells appeared inside the cells, with a punctate staining pattern (Fig. 1A). LAMP1 staining colocalized with wheat germ agglutinin staining only at intracellular compartments, not at the plasma membrane (Fig. 1A).
FIG 1.
Fusogenic activity of histidine triad mutants. (A) Confocal microscopy imaging of HEK293T cells. DAPI staining is shown in blue, anti-LAMP1 staining is shown in red, and wheat germ agglutinin staining is shown in green. Bar, 20 μm. Insets show higher-magnification views of a selected region. (B) Cell fusion assay using the WT spike complex of LASV. Transiently transfected, confluent HEK293T cells were exposed to the indicated pH values for 5, 10, and 15 min. Bright-field images of the cells are shown. Syncytial boundaries were traced using ImageJ and are marked with yellow lines. (C) Western blot (WB) analysis of HEK293T cell lysates for cells transfected with the various WT and mutated LASV spikes (upper panel) and the same spikes fused to a Strep tag at the C terminus (lower panel). Transfection of the spike complex from Junin virus is shown as a negative control (upper panel) and as a reference with a Strep tag (lower panel). A plasmid encoding GFP was cotransfected to monitor transfection efficiency. For the anti-Lassa virus GP1 antibody, higher-molecular-weight bands are visible and correspond to unprocessed GPCs (marked with a red arrow). As a loading control, we stripped each membrane twice and used it for detection of α-tubulin and GFP. (D) Pulldown assay of cell surface biotinylated proteins. Cells were transfected with the indicated GPC and exposed to sulfo-NHS-SS-biotin to specifically tag proteins at the cell surface. Cells were lysed, and the levels of GP1 in the total cell extract (lower panel) and the biotinylated pulldown fraction (upper panel) were analyzed. Levels of transferrin receptor 1 (TfR1), which resides on the cell surface, were monitored to control for the efficiency of the biotinylation reactions and for the load levels. The levels of α-tubulin were monitored to detect the exposure of cytoplasmic content to the biotinylating reagent. (E) Histidine triad on the surface of Lassa virus GP1 (PDB ID 4ZJF), shown in magenta, blue, and orange for His92, His93, and His230, respectively. (F) Cell fusion assay with pH 5.0 buffer for 10-min exposures of transiently transfected HEK293T cells to the WT, H92Y, H93Y, and H230Y spike complexes of LASV. Fusion of nontransfected cells (NT) is shown as a negative control. Representative images from multiple experimental repeats are shown. (G) Areas of syncytia as fractions of the total area of the field of view were measured using ImageJ and averaged over 6 independent experiments. Error bars show standard deviations. Statistical significance, determined using Student's t test, is indicated as follows: NS, not significant; *, P < 0.05; and ***, P < 0.005.
We first optimized the cell fusion assay by using the wild-type spike complex from LASV (Fig. 1B). Three buffer conditions, adjusted to different pH values, and the incubation times in these respective buffers were tested. Syncytium formation was monitored using bright-field imaging. This experiment revealed a clear correlation between syncytial area and acidity levels as well as between syncytial area and incubation times (Fig. 1B). Whereas no syncytia formed at pH 5.5, pH 5.0 was sufficient to induce cell fusion after incubation for 10 min, and robust cell fusion at pH 4.5 was already observed after 5 min of incubation (Fig. 1B). Thus, the fraction of syncytial area as we measure it is indicative of the fusogenic activity of the spike, and we can modulate this activity by using different buffer conditions. The results from this experiment generally agree with previous studies (23, 24) reporting that fusion starts at pHs of <5.0 and that maximal activity is obtained at pH 4.0 or pH 3.0 when cells are exposed to the low-pH buffer for 15 min (23) or 2 min (24), respectively.
Cell fusion activity of histidine triad mutants.
To reveal the functional role of the histidine triad, we constructed three mutated GPC genes, each bearing a single histidine-to-tyrosine point mutation. We already demonstrated that in the context of isolated GP1, these mutations give rise to properly folded, soluble, and secreted proteins with circular dichroism (CD) spectra similar to that of GP1WT, which are equally recognized by an anti-GP1 antibody (Ab482) (16). In the current study, we tested the influence of these mutations on the expression and maturation of the complete spike complex. For this purpose, we transiently transfected HEK293T cells with vectors encoding the WT and mutated GPCs and monitored the levels of matured spikes by using our anti-Lassa virus GP1 monoclonal antibody (Fig. 1C). GPCH230Y was present at a level similar to that of GPCWT, as can be judged from the GP1 levels (Fig. 1C). Mutating His92 and His93, on the other hand, resulted in a decrease in GP1 levels (Fig. 1C). Since the levels of the GPC precursors were similar for all mutants (Fig. 1C, upper bands at ∼75 kDa), we wanted to see if the lower GP1 levels were due to enhanced GP1 dissociation or to overall decreased levels of mature spikes. We fused Strep tags to the C termini of the GPC variants and monitored the levels of the GP2 subunits (Fig. 1C). As observed with the levels of GP1, the His92 and His93 mutations reduced the amount of GP2, whereas the His230 mutation did not. Next, we tested whether these mutations interfered with the ability of the spikes to reach the cell surface. Cell surface proteins were biotinylated by exposing intact cells to sulfo-NHS-SS-biotin, which reacts with exposed amine groups. We then pulled down the biotinylated fraction and analyzed for the presence of GP1 (Fig. 1D). All mutated GPCs were present at the cell surface, in the same relative quantities as those observed in the total cell extracts (Fig. 1C). As expected, the band corresponding to the precursor GPC was visible only in the total cell extract. Thus, mutating His92 and His93 reduces the overall levels of mature spikes compared to those with GPCWT but does not prevent cell surface presentation or selectively increase GP1 dissociation from the spikes. The differences in expression levels indicate that changes at His230 are better tolerated than changes at His92 and His93 (Fig. 1E), probably due to a structural function of His92 and His93 in the context of the native GPC. Regardless of the differences in levels of matured spikes, all mutants were able to induce cell fusion after 10 min of incubation at pH 5.0 (Fig. 1F). By repeating these assays and calculating the fractions of syncytial area, we could quantify the activities of the mutated spikes (Fig. 1G). Despite their relatively low expression levels, GPCH92Y and GPCH93Y achieved fusion levels comparable to or slightly greater than that of GPCWT. In this assay, GPCH230Y displayed significantly enhanced activity compared to that of GPCWT (Fig. 1G). The ability of the various GPCs to facilitate cell fusion may imply that the mutated GPCs retained their capacity to interact with their cellular receptor.
Cell infection mediated by mutated spike complexes.
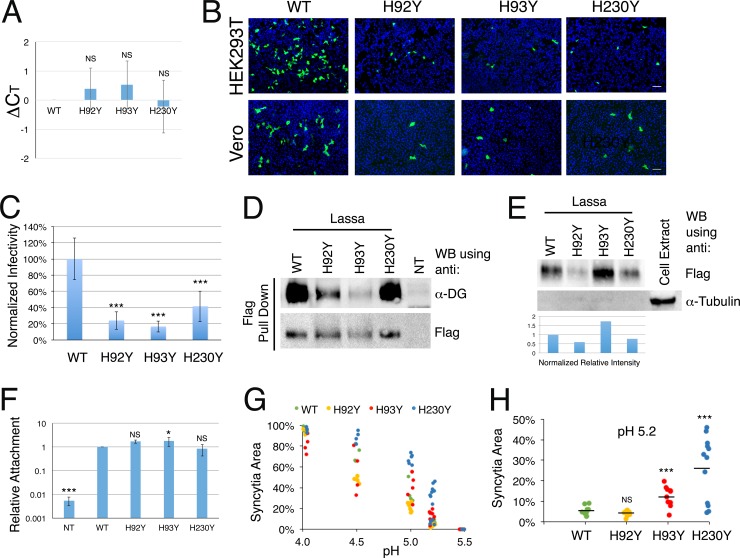
We next evaluated the ability of the histidine triad mutants to facilitate infection. Using a retroviral packaging cell line (GP2-293; Clontech), we generated pseudotyped viruses carrying the desired GPC and a GFP reporter gene. We used RT-qPCR to quantify the RNA levels of the GFP reporter gene as a means to measure the levels of the pseudoviruses. Infectivity in this regard cannot be used for quantification due to the potential differences in activity of the various GPCs. We first treated the pseudovirus stock solutions with RNase A and DNase I to eliminate nucleic acid contaminations that are not protected inside virus-like particles. Analyzing the number of cycles needed to cross the detection threshold (CT) revealed maximal averaged differences (ΔCT) of less than one cycle between the reporter gene levels in the mutated pseudoviral stocks and those for the WT (Fig. 2A), which translates to average <2-fold differences in the RNA levels. The average ΔCT values were obtained by determining individual ΔCT values for several independently produced batches of pseudoviruses. The efficiency of pseudovirus production as measured by CT values varied according to the passage number of the packaging cell line, but the differences (ΔCT) between viruses that were produced at the same time were minute (Fig. 2A). Equal volumes of all pseudoviral stocks were applied to HEK293T and Vero cells, and GFP-positive cells were visualized 48 h later using fluorescence microscopy (Fig. 2B). Compared to that of GPCWT, the histidine triad mutants exhibited a noticeably reduced capacity to infect cells (Fig. 2B). Quantification of infected HEK293T cells by fluorescence-activated cell sorter (FACS) analysis further demonstrated that difference (Fig. 2C).
FIG 2.
Infectivity and pH-dependent fusion by histidine triad mutants. (A) RNA quantification by RT-qPCR analysis of the indicated pseudoviral stocks. We produced 4 different batches of pseudoviral stocks. For each batch of viruses, we extracted RNA and performed reverse transcription and qPCR analysis. The CT value for GPCWT was subtracted from the values measured for the mutated GPCs from the same batch to give the net differences (ΔCT) in RNA levels. ΔCT values were averaged, and error bars show standard deviations. (B) HEK293T (upper row) and Vero (lower row) cells were exposed to equal volumes of the pseudoviral stocks, and positively infected cells that expressed the GFP reporter gene were visualized by fluorescence microscopy. Nuclei were stained with DAPI. Bars, 50 μm. Infectivity experiments were repeated multiple times, and representative images are shown. (C) Infected HEK293T cells were analyzed using FACS to quantify the levels of infection. We repeated the quantification 6 times, using 4 different batches of pseudoviral stocks. The averaged infectivity of GPCWT was defined as 100% to normalize the results. Averaged relative infection levels of the indicated mutants are shown. Error bars show standard deviations, and statistical significance compared to the WT is indicated with asterisks (***, P < 0.005; Student's t test). (D) Pulldown of α-DG by GPC-Flag variants, detected using anti-Flag M2 resin. Loading volumes were adjusted to have similar levels of Flag. Nontransfected cells (NT) were used as a negative control. (E) Incorporation of spike complexes into pseudoviruses. Flag-tagged spikes that were incorporated into pseudoviruses were detected using anti-Flag antibody following isolation of the pseudoviruses by ultracentrifugation and capture by anti-Flag beads. Equal volumes of the lysed pseudovirus solutions were analyzed by Western blotting for the presence of tubulin as a control for cell-derived contaminations. The anti-Flag signals from two Western blot analyses were quantified using densitometry and normalized to the RNA levels of the reporter gene in the concentrated pseudovirus samples as determined using RT-qPCR. The normalized intensities are displayed relative to the WT level. (F) Cell attachment assay. Equal dilutions of the indicated pseudoviruses that were validated to have the same CT by RT-qPCR were applied to cells, and the levels of attached viruses were quantified by monitoring GFP levels by RT-qPCR after lysis of the cells. The levels of GAPDH were also quantified using RT-qPCR to serve as a reference. The relative and normalized levels of pseudoviruses bearing either mutated or WT GPC were obtained by calculating ΔΔCT (see Materials and Methods). Nontreated (NT) cells were used as a negative control and indicate detection of background levels. The experiment was independently repeated three times, and average values are shown. Error bars show standard deviations, and Student's t test results indicate statistical significance compared to the WT, as follows: NS, not significant; *, P < 0.05; and ***, P < 0.005. (G) Quantification of cell fusion assay results for GPCWT, GPCH92Y, GPCH93Y, and GPCH230Y at various pH values. For each mutant, individual experimental values for the fraction of syncytial area coverage are shown. (H) Cell fusion by GPCWT, GPCH92Y, GPCH93Y, and GPCH230Y at pH 5.2. Individual experimental values are shown for each variant, and the horizontal line shows the average value (***, P < 0.005; Student's t test).
The observed decrease in cell infectivity prompted us to test various aspects of the production of the pseudoviruses and of the GPCs that may account for it. We first tested if the mutations in the histidine triad abrogated the ability of the spikes to attach to α-DG. We constructed GPCs with a Flag tag attached at the C terminus, performed a pulldown assay using M2 anti-Flag beads, and used Western blot analysis to detect α-DG (Fig. 2D). For this analysis, we adjusted the loading volumes to have comparable amounts of GPCs as estimated using anti-Flag antibody (Fig. 2D). All the GPC mutants retained some capacity to interact with α-DG (Fig. 2D), which agrees with their ability to promote cell-cell fusion (Fig. 1F and G). Although it is not a quantitative method, this Western blot analysis further indicates that whereas GPCH230Y effectively pulled down α-DG, like GPCWT, the interactions of GPCH92Y and GPCH93Y with α-DG were impaired. This observation supports the above-mentioned notion that the H92Y and H93Y mutations may undesirably affect the structure of the spike. We next addressed the possible caveat that mutations in the histidine triad affect the incorporation of the spikes into pseudoviruses. We first pelleted pseudoviruses by ultracentrifugation but failed to detect the spike complexes by using the anti-GP1 antibody. We thus generated pseudoviruses using the Flag-tagged GPCs, pelleted them by ultracentrifugation, and used anti-Flag to detect the incorporated GP2 proteins (Fig. 2E). The mutations introduced into the histidine triad did not prevent the incorporation of the spike complexes into the pseudoviruses (Fig. 2E). We further quantified and averaged the intensities of the anti-Flag signals from two independent analyses and normalized them to the RNA levels of the pelleted pseudoviruses as a proxy for the incorporation levels of the GPCs (Fig. 2E). The incorporation levels of the various mutated GPCs relative to that of GPCWT, as estimated from this analysis, were in the range of ±50% and were not correlated with the infectivity levels of the pseudoviruses (Fig. 2C). Since cell attachment may involve factors other than α-DG (22), we further wanted to compare the abilities of the WT and mutated GPCs to mediate attachment to cell surfaces. For that purpose, we generated various pseudovirus stocks, verified the equal RNA levels (data not shown), diluted the stocks, and applied them to cells in monolayers for 30 min of incubation on ice. After incubation and washing, we lysed the cells and quantified the RNA levels (Fig. 2F). The mutations in the histidine triad did not affect the attachment of the pseudoviruses to cells compared to that with GPCWT (Fig. 2F). Based on the above-mentioned results and since GPCH230Y and GPCWT were expressed at similar levels (Fig. 1C), there is a bona fide impairment in infectivity that is mediated by the GPCH230Y mutated spike, and perhaps by the other two mutants as well.
Histidine triad mutants are more sensitive to acidic pH.
The apparent increase in membrane fusion activity of GPCH93Y and GPCH230Y (Fig. 1F and G) prompted us to further investigate the molecular roles of these histidines during spike-mediated and pH-dependent membrane fusion. First, we examined the pH-dependent fusion activity of the histidine triad mutants (Fig. 2G). In comparison to the WT spike complex, the onset of triggering by acidification for GPCH93Y and GPCH230Y occurred at a higher pH; at pH 5.2, GPCH230Y and, to some degree, GPCH93Y showed significant fusion activity with a stochastic appearance, whereas GPCWT and GPCH92Y consistently showed very low levels of fusion (Fig. 2H). At pH 5.0, the areas of syncytia formed by GPCH93Y and GPCH230Y were larger (Fig. 2G), as already reported above (Fig. 1F), but at a sufficiently low pH (i.e., pH 4.0) the fusogenic activity by all the spikes was saturated and approached complete coverage of the field of view by syncytia (Fig. 2G). Thus, GPCH93Y and GPCH230Y are more sensitive to acidic pH than the WT spike complex, with a higher probability of being activated at a less acidic pH. Importantly, the intrinsic abilities to sense pH and to induce membrane fusion were maintained in all mutants.
His230 functions as a negative regulator of fusion.
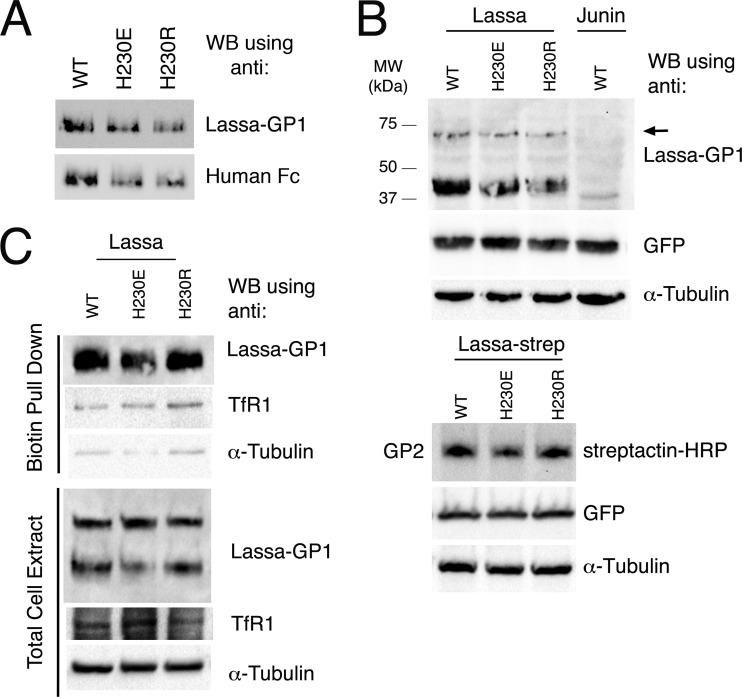
Histidine residues have an imidazole side chain with a theoretical pKa of ∼6.0. Below this pH, they favor the protonated, positively charged state. From our above-mentioned results, eliminating the potential to bear a positive charge in His93 and His230 by mutating them to tyrosine seems to increase the propensity of the spikes for triggering. To further investigate this notion, we mutated His230 to both a positively charged arginine and a negatively charged glutamate. The decision to mutate His230 was based on our observation that this residue better tolerates alterations than His93 (Fig. 1C). First, by expressing GP1H230E-Fc and GP1H230R-Fc fusion proteins, we were able to verify that GP1 recognition by the anti-GP1 antibody (Ab482) was not affected by these mutations (Fig. 3A). We then transiently transfected HEK293T cells with vectors encoding the mutated GPCs, lysed them, and probed for the levels of expression of the new His230 mutants by monitoring GP1 levels as well as GP2 levels, using Strep-tagged versions of the GPCs (Fig. 3B). GPCH230E and GPCH230R displayed levels of expression comparable to that of GPCWT (Fig. 3B). We further verified that these mutants reached the cell surface by using biotinylation of the surface proteins and a subsequent pulldown assay (Fig. 3C). Next, we tested the fusogenic activity of the mutants. The positively charged GPCH230R mutant showed a significantly delayed onset of fusion compared to GPCWT (Fig. 4A and B). At pH 5.0, only minor fusion activity was observed with GPCH230R, whereas GPCWT displayed substantial fusion activity. At pH 4.5, GPCH230R catalyzed massive cell fusion, indicating that this mutation affected the pH triggering point but not the fusogenic activity itself. Surprisingly, the negatively charged GPCH230E mutant became hypersensitive to acidic pH, as significant cell fusion was already induced at pH 5.5 (Fig. 4A and B).
FIG 3.
Expression and surface display of GP1H230E and GP1H230R. (A) GP1WT, GP1H230E, and GP1H230R fused to the Fc portion of a human IgG were transiently expressed in HEK293T cells, and equal volumes of medium were subjected in duplicate to SDS-PAGE followed by Western blotting, using the anti-LASV GP1 antibody (Ab482) for one half of the membrane and anti-human Fc as a loading control for the second half of the membrane. (B) Western blot analysis using anti-GP1 antibody, showing the expression levels of GPCH230E and GPCH230R relative to that of GPCWT following transient transfection of HEK293T cells (upper panel) and by following the levels of GP2 using Strep-tagged versions of the spikes (lower panel). A plasmid encoding GFP was cotransfected to monitor transfection efficiency, and GFP and α-tubulin (as a loading control) were detected on the same membrane after stripping. An arrow indicates unprocessed GPCs. (C) Pulldown assay of cell surface biotinylated proteins, similar to that in Fig. 1D. Cell surface proteins were biotinylated as described above, and levels of GP1 in the total cell extract (lower panel) and in the biotinylated pulldown fraction (upper panel) were analyzed on one single membrane, resulting in identical exposures of the two panels to the various antibodies. After stripping of the membrane, levels of TfR1 and α-tubulin were also monitored as controls.
FIG 4.
Molecular function of the histidine triad. (A) Cell fusion assay with GPCWT, GPCH230E, and GPCH230R at various pH values. (B) Quantification of the fractions of syncytial area coverage by GPCWT, GPCH230E, and GPCH230R. Values are the means from three independent experiments, and error bars show standard deviations. ***, statistically significant (P < 0.005; Student's t test) differences compared to GPCWT; NS, not significant. (C) Pulldown assay from HEK293T cell lysate at pH 5.0 by the indicated GP1-Fc fusion proteins, using protein A-conjugated beads. Western blotting was used to detect LAMP1 and Fc, as a loading control. (D) RNA quantification by RT-qPCR, performed similarly to the procedure for Fig. 2A. We produced 4 different batches of pseudoviral stocks, extracted RNA, and performed reverse transcription and qPCR analysis. The CT value for GPCWT was subtracted from the values measured for the mutated GPCs from the same batch to calculate the ΔCT value. ΔCT values were averaged, and error bars show standard deviations. (E) Quantification of infection in HEK293T cells by FACS analysis, performed similarly to the procedure for Fig. 2C. Data were collected from 6 independent experiments conducted with the 4 batches of pseudoviruses. The average infectivity of GPCWT was defined as 100% infectivity to normalize the results, and relative average infection levels for the indicated mutants are shown. Error bars show standard deviations, and statistical significance is indicated as follows: ***, P < 0.005 (Student's t test); NS, not significant. (F) Pulldown of α-DG by GPC-Flag variants, with detection by use of anti-Flag M2 resin, as in Fig. 2D. Nontransfected cells (NT) were used as a negative control. (G) Detection of incorporation of Flag-GPCs into pseudoviruses following isolation via ultracentrifugation, performed similarly to the procedure for Fig. 2E. Normalized intensities of anti-Flag signals were calculated as described for Fig. 2E. (H) Quantification of attachment of the indicated pseudoviruses to cells by RT-qPCR as described for Fig. 2F. (I) Quantification of syncytial area following cell fusion, using a stable cell line expressing LAMP1-Δ384 or a control cell line and conducted at pH 5.3. Averaged values of syncytial areas for 3 independent experiments (each consisting of 2 or 3 technical replicates) are shown. Statistical significance is indicated by asterisks (***, P < 0.005; Student's t test) and was calculated for each GPC based on the values obtained for the control and the LAMP1-Δ384 cell lines.
A positive charge at position 230 is required for LAMP1 binding and for efficient cell infectivity.
Mutating His230 to a tyrosine prevented LAMP1 binding to GP1 (16). We were intrigued to see whether it was the tyrosine substitution per se that interfered with binding or rather the elimination of the positive charge at this position. To address this question, we performed a pulldown assay using protein A beads at pH 5.0 in the presence of the GP1WT-Fc, GP1H230E-Fc, or GP1H230R-Fc fusion protein (Fig. 4C). GP1H230R-Fc was able to effectively pull down LAMP1 similarly to GP1WT-Fc, but GP1H230E-Fc was not (Fig. 4C). Next, we tested the ability of pseudoviruses bearing GPCH230E and GPCH230R to infect cells. We used the same packaging cell line as that mentioned above to produce pseudoviruses bearing GPCH230E and GPCH230R and used RT-qPCR to measure their RNA content compared to that of GPCWT-containing viruses (Fig. 4D). The variations in RNA levels between the pseudoviruses were <2-fold (average ΔCT < 1) across different batches of viruses (Fig. 4D). We used these pseudoviruses to infect HEK293T cells and monitored positively infected cells by FACS analysis (Fig. 4E). A negative charge at position 230 (H230E) was deleterious for infectivity, and only low levels of positive cells were identified (Fig. 4E). On the other hand, a positive charge at position 230 (GPCH230R) allowed efficient infection that was close to the level seen for pseudoviruses with GPCWT (Fig. 4E). Importantly, the mutations of His230 to glutamic acid and arginine did not change the ability of the spikes to interact with α-DG (Fig. 4F) or to be incorporated into pseudoviruses (Fig. 4G). Furthermore, we did not observe any change in the ability of these pseudoviruses to attach to cell surfaces compared to that of GPCWT (Fig. 4H).
LAMP1 binding triggers the GPC spike complex.
Our results indicate that LAMP1 requires a positive charge at position 230 that can equally be provided by either a histidine or arginine residue (Fig. 4C). This observation strongly implies that LAMP1 donates a negative charge for this interaction. Positioning a negative charge in the vicinity of His230 may emulate to some degree the H230E mutant, which has a higher propensity to be triggered at moderate pH values (Fig. 4B). To investigate this notion, we generated a stable cell line that expresses a LAMP1-Δ384 mutant that ectopically accumulates on the cell surface (25) and is conjugated to GFP at its cytoplasmic C terminus. In conjunction, we generated a similar cell line that expresses only GFP as a control. We performed cell fusion assays using these cell lines and found that in the presence of overexpressed LAMP1 on the cell surface, the probability of both GPCWT and GPCH230R being triggered at pH 5.3 was significantly higher than that for control fusion in the GFP control cell line (Fig. 4I).
DISCUSSION
The crystallographic structure of GP1 from LASV that we previously determined allowed us to identify a unique cluster of three histidines that contribute to the interaction with LAMP1 (16). Cell fusion assays clearly showed that tyrosine mutations in this histidine triad in the context of the complete spike complex do not hinder pH-induced membrane fusion (Fig. 1F). Since the histidine triad mutants have an impaired capability to bind LAMP1 (16), we can conclude that LAMP1 binding is not required for spike fusogenic activity per se. Unlike the fusogenic activity, disrupting the ability of the spikes to engage LAMP1 lowered but did not completely abrogate their capacity to mediate infection (Fig. 2B). Thus, although it is not required for membrane fusion, LAMP1 binding is needed for efficient infection.
Compared to GPCWT, two of the histidine triad mutant spikes displayed enhanced fusogenic activity (Fig. 1F and G), which could be explained by a higher sensitivity to acidic pH (Fig. 2G and 3B). This observation suggests a possible regulatory role for these histidines during the triggering of the spike. Surface-exposed histidine residues have the potential to assume a positive charge at an acidic pH, with a theoretical pKa of ∼6.0. Accordingly, as the pH in the endocytic pathways drops, the histidine triad assumes some partial positive charge. Given the higher propensity of GPCH93Y and GPCH230Y to be triggered by acidic pH, a positive charge on His93 or His230 might act as a negative regulator that inhibits premature triggering. Indeed, when we mutated His230 to a positively charged arginine, it significantly lowered the pH needed for triggering of the spike (Fig. 4A and B). Thus, His230 is a negative regulator that inhibits premature triggering when protonated and assumes a positive charge upon acidification.
The conformation of GP1 in the context of the native spike complex may differ from the currently known crystallographic structure of Lassa virus GP1 due to conformational changes (16). The reduced levels of matured spikes that resulted from mutation of His92 and His93 (Fig. 1C) indicate a possible structural role for these residues in the native spike. In a recently described structure of the GPC protein from lymphocytic choriomeningitis virus (LCMV) in a prefusion conformation (26), the histidines that are homologous to His92 and His93 are in a slightly different orientation compared to that seen for isolated Lassa virus GP1 and have close contacts with other residues. Although LCMV enters cells independently of LAMP1 (11, 26), the GP1 subunit of LASV may adopt a prefusion conformation similar to that observed in the LCMV structure. If this is true, it may explain the lower tolerance for mutation of His92 and His93. The putative structural role of His92 and His93 in the context of native GPC complicates their functional analysis by mutagenesis as was done in the case of His230.
How might His230 regulate the triggering of the spike? As reported before, the optimal pH for membrane fusion by the LASV spike complex measured on the cell surface is pH 4.0 (23) or pH 3.0 (24), which is low compared to that for other viruses (23) and is even below the pH of the lysosome (27). Although we do not know the actual pKa of His230, it is likely that the surface-exposed His230 residue starts to become protonated and charged at or even slightly above pH 6.0. When charged, His230 may lower the propensity (pKa) of nearby histidine residues from the triad (or others in the context of the native spike) to assume a positive charge by making it energetically less favorable (28). The involvement of other histidine residues in this process is further supported by the fact that a sufficiently low pH can effectively override the inhibition that a positive charge at position 230 imposes on the spike complex. In such a scenario, a negatively charged group that will interact with His230 is likely to mask the positive charge and raise the pKa of adjacent histidine residues, which would allow them to protonate in a sharp transition. This hypothesis will need to be investigated further. Other sites on the spike may take part in pH sensing and triggering. In this regard, Lys33 from the stable signal peptide was shown to be important for pH triggering in Junin virus (29), but GP1 from Junin virus lacks histidines equivalent to the histidine triad of LASV.
We demonstrated that effective LAMP1 binding requires a positive charge at position 230 in the context of GP1 (Fig. 4C) and that histidine as well as arginine may equally provide this charge. Thus, it is likely that LAMP1 donates a negatively charged group for this interaction. Hence, positively charged His230 inhibits spike triggering until it is neutralized by a countercharge from LAMP1 that promotes triggering even at the late endosome, where the acidity is well above the optimal pH for fusion. This notion is further supported by the fact that a negatively charged glutamate at position 230 strongly potentiates pH-induced triggering, to the point that robust cell fusion is observed even at pH 5.5 (Fig. 4A and B). Indeed, ectopically expressing LAMP1 on the cell surface enabled cell fusion by GPCWT and GPCH230R of LASV at a pH that was otherwise not acidic enough for effective triggering of fusion (Fig. 4I).
The first encounter of LASV with LAMP1 in an acidic environment will happen in the late endosomes, where 5% to 8% of the total LAMP1 resides (15). In the vicinity of the endosomal membrane, multiple binding events between spikes and LAMP1 molecules may occur. This, consequently, would cause multiple spikes to be triggered. Notably, multiple spikes need to act in a concerted fashion to efficiently catalyze membrane fusion (30). Thus, the binding of LAMP1 and the release of His230 inhibition could serve as the molecular mechanism that allows multiple spikes to be triggered in a coordinated fashion. The low infectivity of viruses with GPCH230Y and GPCH230E mutants (Fig. 2C and 4E) may accordingly result from premature and noncoordinated spike triggering. In this regard, a recent electron microscopy (EM) study demonstrated that the spike complex of LASV undergoes some conformational changes when transitioned from neutral pH to pH 5.0 (6). This observation, together with our data, suggests that the spikes are initially primed but are arrested during the gradual acidification in the endocytic pathway, in a process that may involve conformational changes of GP1 that would allow LAMP1 binding and may exclude α-DG (16). After priming, triggering may then be achieved either by binding to LAMP1 or by a sufficiently low and nonphysiologic pH. This series of events provides a mechanistic explanation for the ability of LASV to enter cells via late endosomes that are not acidic enough to reach the optimal triggering pH as determined on the cell surface.
ACKNOWLEDGMENTS
We thank Joseph Georgeson for helping us to record confocal microscopy images.
Ron Diskin is the incumbent Tauro Career Development Chair in Biomedical Research.
The Diskin lab is supported by research grants from the Enoch Foundation, the Abramson Family Center for Young Scientists, Rudolfine Steindling, and the I-CORE Program of the Planning and Budgeting Committee and The Israel Science Foundation (grant 1775/12). This research was further supported by a grant from the Mizutani Foundation for Glycoscience.
The α-DG (IIH6) antibody developed by Kevin P. Campbell was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa.
REFERENCES
- 1.Oldstone MB. 2002. Arenaviruses. I. The epidemiology molecular and cell biology of arenaviruses. Introduction. Curr Top Microbiol Immunol 262:V–XII. [PubMed] [Google Scholar]
- 2.Oldstone MB. 2002. Arenaviruses. II. The molecular pathogenesis of arenavirus infections. Introduction. Curr Top Microbiol Immunol 263:V–XII. [PubMed] [Google Scholar]
- 3.Cao W, Henry MD, Borrow P, Yamada H, Elder JH, Ravkov EV, Nichol ST, Compans RW, Campbell KP, Oldstone MB. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 4.Kunz S, Rojek JM, Perez M, Spiropoulou CF, Oldstone MB. 2005. Characterization of the interaction of Lassa fever virus with its cellular receptor alpha-dystroglycan. J Virol 79:5979–5987. doi: 10.1128/JVI.79.10.5979-5987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eschli B, Quirin K, Wepf A, Weber J, Zinkernagel R, Hengartner H. 2006. Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J Virol 80:5897–5907. doi: 10.1128/JVI.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Sun Z, Pryce R, Parsy ML, Fehling SK, Schlie K, Siebert CA, Garten W, Bowden TA, Strecker T, Huiskonen JT. 2016. Acidic pH-induced conformations and LAMP1 binding of the Lassa virus glycoprotein spike. PLoS Pathog 12:e1005418. doi: 10.1371/journal.ppat.1005418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oppliger J, Torriani G, Herrador A, Kunz S. 2016. Lassa virus cell entry via dystroglycan involves an unusual pathway of macropinocytosis. J Virol 90:6412–6429. doi: 10.1128/JVI.00257-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasqual G, Rojek JM, Masin M, Chatton JY, Kunz S. 2011. Old World arenaviruses enter the host cell via the multivesicular body and depend on the endosomal sorting complex required for transport. PLoS Pathog 7:e1002232. doi: 10.1371/journal.ppat.1002232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasquato A, Burri DJ, Traba EG, Hanna-El-Daher L, Seidah NG, Kunz S. 2011. Arenavirus envelope glycoproteins mimic autoprocessing sites of the cellular proprotein convertase subtilisin kexin isozyme-1/site-1 protease. Virology 417:18–26. doi: 10.1016/j.virol.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 10.Burri DJ, da Palma JR, Kunz S, Pasquato A. 2012. Envelope glycoprotein of arenaviruses. Viruses 4:2162–2181. doi: 10.3390/v4102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jae LT, Raaben M, Herbert AS, Kuehne AI, Wirchnianski AS, Soh TK, Stubbs SH, Janssen H, Damme M, Saftig P, Whelan SP, Dye JM, Brummelkamp TR. 2014. Virus entry. Lassa virus entry requires a trigger-induced receptor switch. Science 344:1506–1510. doi: 10.1126/science.1252480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winchester BG. 2001. Lysosomal membrane proteins. Eur J Paediatr Neurol 5(Suppl A):11–19. doi: 10.1053/ejpn.2000.0428. [DOI] [PubMed] [Google Scholar]
- 13.Eskelinen EL. 2006. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med 27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Amos B, Lotan R. 1990. Modulation of lysosomal-associated membrane glycoproteins during retinoic acid-induced embryonal carcinoma cell differentiation. J Biol Chem 265:19192–19198. [PubMed] [Google Scholar]
- 15.Lippincott-Schwartz J, Fambrough DM. 1987. Cycling of the integral membrane glycoprotein, LEP100, between plasma membrane and lysosomes: kinetic and morphological analysis. Cell 49:669–677. doi: 10.1016/0092-8674(87)90543-5. [DOI] [PubMed] [Google Scholar]
- 16.Cohen-Dvashi H, Cohen N, Israeli H, Diskin R. 2015. Molecular mechanism for LAMP1 recognition by Lassa virus. J Virol 89:7584–7592. doi: 10.1128/JVI.00651-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falcon-Perez JM, Nazarian R, Sabatti C, Dell'Angelica EC. 2005. Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J Cell Sci 118:5243–5255. doi: 10.1242/jcs.02633. [DOI] [PubMed] [Google Scholar]
- 18.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sims JJ, Scavone F, Cooper EM, Kane LA, Youle RJ, Boeke JD, Cohen RE. 2012. Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat Methods 9:303–309. doi: 10.1038/nmeth.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wool-Lewis RJ, Bates P. 1998. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol 72:3155–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rojek JM, Campbell KP, Oldstone MB, Kunz S. 2007. Old World arenavirus infection interferes with the expression of functional alpha-dystroglycan in the host cell. Mol Biol Cell 18:4493–4507. doi: 10.1091/mbc.E07-04-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimojima M, Stroher U, Ebihara H, Feldmann H, Kawaoka Y. 2012. Identification of cell surface molecules involved in dystroglycan-independent Lassa virus cell entry. J Virol 86:2067–2078. doi: 10.1128/JVI.06451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klewitz C, Klenk HD, ter Meulen J. 2007. Amino acids from both N-terminal hydrophobic regions of the Lassa virus envelope glycoprotein GP-2 are critical for pH-dependent membrane fusion and infectivity. J Gen Virol 88:2320–2328. doi: 10.1099/vir.0.82950-0. [DOI] [PubMed] [Google Scholar]
- 24.Cosset FL, Marianneau P, Verney G, Gallais F, Tordo N, Pecheur EI, ter Meulen J, Deubel V, Bartosch B. 2009. Characterization of Lassa virus cell entry and neutralization with Lassa virus pseudoparticles. J Virol 83:3228–3237. doi: 10.1128/JVI.01711-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohrer J, Schweizer A, Russell D, Kornfeld S. 1996. The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J Cell Biol 132:565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hastie KM, Igonet S, Sullivan BM, Legrand P, Zandonatti MA, Robinson JE, Garry RF, Rey FA, Oldstone MB, Saphire EO. 2016. Crystal structure of the prefusion surface glycoprotein of the prototypic arenavirus LCMV. Nat Struct Mol Biol 23:513–521. doi: 10.1038/nsmb.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellman I, Fuchs R, Helenius A. 1986. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem 55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- 28.Pace CN, Grimsley GR, Scholtz JM. 2009. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem 284:13285–13289. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.York J, Nunberg JH. 2009. Intersubunit interactions modulate pH-induced activation of membrane fusion by the Junin virus envelope glycoprotein GPC. J Virol 83:4121–4126. doi: 10.1128/JVI.02410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrison SC. 2008. Viral membrane fusion. Nat Struct Mol Biol 15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]