Abstract
As the dynamic nature of progenitor genomes accompanies the speciation by interspecific hybridization, the extraction of the constituent subgenome(s) from a natural allopolyploid species of long history and then restitution of the progenitor(s) provides the unique opportunity to study the genome evolution and interplay. Herein, the A subgenome from the allotetraploid oilseed rape (Brassica napus L., AACC) was extracted through inducing the preferential elimination of C-subgenome chromosomes in intertribal crosses and the progenitor B. rapa was restituted (RBR). Then by crossing and backcrossing RBR with B. napus donor, the C subgenome was in situ dissected by adding each of its nine chromosomes to the extracted A subgenome and establishing the whole set of monosonic alien addition lines (MAALs). RBR from spring-type B. napus genotype “Oro” expressed a phenotype resembling some type of B. rapa never observed before, but showed a winter-type flowering habit. This RBR had weaker growth vigor and suffered more seriously from biotic and abiotic stresses compared with Oro. The phenotypes specific for these MAALs showed the location of the related genes on the particular C-subgenome chromosomes. These MAALs exhibited obviously different frequencies in homeologous pairing and transmission of additional C-subgenome chromosomes, which were associated with the distinct degrees of their relatedness, and even with the possible genetic regulation for meiotic pairing evolved in B. napus. Finally, large scaffolds undetermined for sequence assembly of B. napus were anchored to specific C-subgenome chromosomes using MAALs.
Keywords: allopolyploid, Brassica napus, B. rapa, alien additional lines, aneuploid
MANY angiosperms including important crops (bread wheat, cotton, oilseed rape, tobacco, etc.) are allopolyploid species that originated from interspecific hybridizations between two or more diploid ancestors followed by chromosome doubling (Ramsey and Schemske 1998; Levin 2003; Otto 2007; Doyle et al. 2008; Soltis and Soltis 2012). These allopolyploid crops outcompete their progenitors by their adaptability to a wide range of climatic conditions and higher yield and better quality of targeted products, under human domestication and improvement. To unravel the origin and structure of the component genomes in these crops has been the goal of the extensive genetic research over a long period. The hybridization events leading to the evolutionary origin of these allopolyploids were traditionally elucidated by the artificial synthesis of their counterparts from the crosses between the extant relatives of the presumed progenitors available or by observing the meiotic chromosome pairing in the hybrids between the natural allopolyploid and the possible progenitors (Kihara 1924; Nagaharu 1935). Alternatively, the genome of one progenitor (AA) was readily dissected by separating and adding its own chromosome to the genome of another progenitor (BB) (namely the alien addition lines), from successively backcrossing the natural or synthesized allopolyploid (AABB) to the other progenitor (BB). In another aspect, great efforts were made to develop the whole set of aneuploids for one allopolyploid, which gain or lose one particular chromosome or chromosome arm from the normal complement (nullisomics, monosomics, trisomics, telosomics, etc.) (Sears 1954). These alien additions and aneuploids contributed substantially and excellently to our early knowledge of genome structure and relationship in allopolyploids in tobacco (Clausen and Cameron 1944) and bread wheat (Sears 1954). Recently, the sequencing and assembling of the large and repetitive 17-Gb genome of bread wheat was realized by the chromosome-based strategy using the chromosome arms isolated from the complete set of double ditelosomic stocks (Mayer et al. 2014).
As the most economically valuable species in the Brassicaceae family, the allotetraploid oilseed rape (Brassica napus L., 2n = 4x = 38, AACC) was formed only ∼7500 yr ago by the hybridization between the ancestors of two extant diploids B. rapa L. (2n = 2x = 20, AA) and B. oleracea L. (2n = 2x = 18, CC) (Nagaharu 1935; Chalhoub et al. 2014). The resynthesized and natural B. napus has been used as one model to investigate the genomic alterations and interaction at different levels during the allopolyploidization process (Albertin et al. 2006; Gaeta et al. 2007; Szadkowski et al. 2010; Xiong et al. 2011; Zhou et al. 2011; Cui et al. 2012; Chalhoub et al. 2014; Zhang et al. 2015a). The genome dissection of the extant B. oleracea was achieved by adding its individual chromosomes to B. rapa, following the synthesis of B. napus and backcrossing to B. rapa (Quiros et al. 1987; McGrath and Quiros 1990; Prakash 2009; Geleta et al. 2012; Heneen et al. 2012). But the genome dissection for B. rapa by such a crossing scheme was rarely reported; likely the crossability between B. napus and B. oleracea was quite low. For the small and similar size of the chromosomes (Prakash 2009; Heneen et al. 2012) and the later development of chromosome-specific cytological markers (Xiong and Pires 2011), the dissection of the A and C genomes in natural B. napus was not achieved by the development of the complete set of additions. One nullisomic of B. napus with the loss of the C2 chromosome established up to now showed much reduced plant stature and shorter growth period and also the genome-wide change of gene expression (Zhu et al. 2015).
As these allopolyploids experienced the evolutionary history of thousands or millions of years, their component genomes diverged from those of the extant descendants of their progenitors (Brenchley et al. 2012; Paterson et al. 2012; Chalhoub et al. 2014; Li et al. 2014; Zhang et al. 2015b). The extent and pattern of the genomic changes seemed to be affected by age, genome relatedness, and existence or absence of the genetic system regulating the chromosome pairing, such as the Ph1 loci in wheat. In B. napus, abundant homeologous exchanges including crossovers and noncrossovers between two subgenomes were revealed to occur, by the sequence comparisons with extant B. rapa and B. oleracea (Wang et al. 2011b; Liu et al. 2014; Chalhoub et al. 2014), likely resulting from the close relatedness between the two progenitors (Prakash 2009) and from the absence of the Ph1-type system to suppress the homeologous recombination. So the possibility should exist that the exact founder phenotypes of the progenitors for one polyploid might be different from those we studied today, due to evolution of the diploid lineages subsequent to the allopolyploidization event (Buggs et al. 2014). Then, the extraction of the constituent subgenome(s) of a given allopolyploid to restitute an independent organism through certain experimental procedures provides the unique opportunity to investigate the genome evolution and interplay. The study of the AABB component extracted from bread wheat (AABBDD) provided novel clues for the modifications in phenotype, karyotype, and gene expression due to a history at the allohexaploid level (Kerber 1964; Zhang et al. 2014).
The A subgenome was previously extracted from one B. napus cultivar “Oro” and the ancestral B. rapa was restituted (RBR Oro) by inducing the preferential loss of the C-genome chromosomes in intertribal crosses with Isatis indigotica (2n = 14) (Tu et al. 2010). The cultivar was likely representative of one origin event for B. napus, as it was bred from the landrace “Liho” in Germany and kept the commonest type of plastid haplotypes among a wide range of B. napus accessions (Allender and King 2010). In this study, Oro was successively backcrossed to the RBR Oro to dissect the C subgenome in the A-subgenome background of the same origin and to establish a complete set of monosomic alien addition lines (MAALs) with one of its nine chromosomes. With the C-subgenome chromosome-specific gene markers designed from the draft sequences of B. napus (Chalhoub et al. 2014), the whole set of additions was distinguished and established, which inversely helped to anchor some undetermined scaffolds to specific chromosomes. The successful in situ dissection of the C subgenome in natural B. napus not only provides the unique genetic stock for analyzing its structure and function but also advances our insights into the genome change and interplay during the evolution of this allotetraploid species. In addition, the feasibility of the restitution of ancestral B. rapa was further confirmed in the intertribal crosses between different B. napus genotypes and another crucifer Crambe abyssinica, and the cytological process of the successive elimination of C-subgenome chromosomes was further elucidated.
Materials and Methods
Plant materials
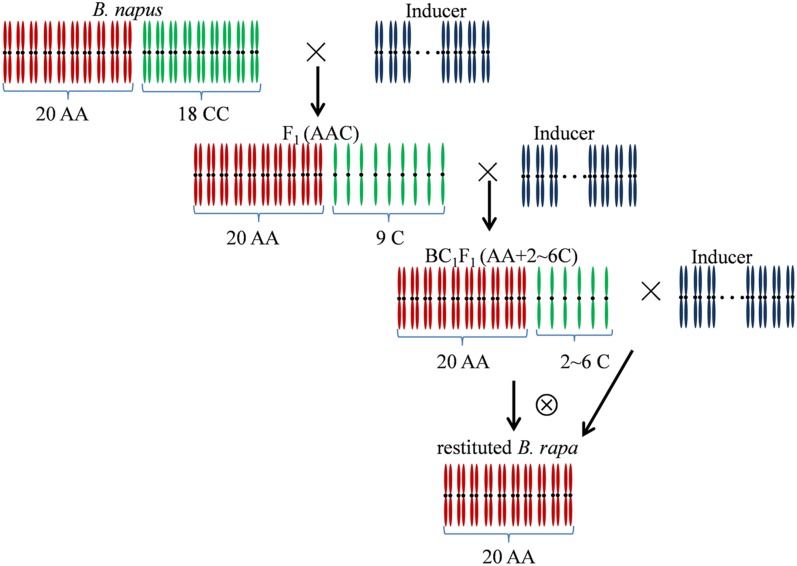
To investigate the cytological process of extracting the A subgenome from natural allotetraploid B. napus (2n = 38, AACC), the elite cultivar “Zhongshuang 11” was pollinated by another crucifer C. abyssinica (2n = 90) to induce the preferential elimination of the C-subgenome chromosomes (Figure 1). Out of 92 seeds harvested from ∼10,000 flowers pollinated, only 36 seeds germinated and produced F1 plants that showed a phenotype quite similar to the female parent. Among 36 F1 plants, 32 were found to keep the same chromosome number as the B. napus parent and were not used for further experiment. The remaining four F1 plants were identified to have 2n = 29 and were pollinated again by C. abyssinica, and 33 backcross (BC1) plantlets were obtained from more than >2000 crosses by embryo rescue on MS agar medium (Murashige and Skoog 1962) without hormones. These plantlets were propagated by subculturing the young buds on MS medium with 1.5 mg/liter−1 6-benzyl aminopurine (6-BA), and 0.25 mg/liter−1 α-naphthalenacetic acid (NAA), to produce enough plants for study. Among BC1 plants, 30 had 2n = 30–44 and showed the variations in the phenotype. The other three BC1 plants (2n = 22, 23, and 26) gave the B. rapa-type morphology. After the plant with 2n = 22 was self-pollinated with artificial assistance, the plants with 2n = 20 (AA) were derived and designated as RBR ZS11, the restituted B. rapa from Zhongshuang 11.
Figure 1.
Crossing and cytological process of A-subgenome extraction and B. rapa restitution from B. napus induced by intertribal hybridizations. The inclusion of the complete A subgenome in F1 hybrid and backcrossing progenies was traced and shown by cytological observations (see Figure 2).
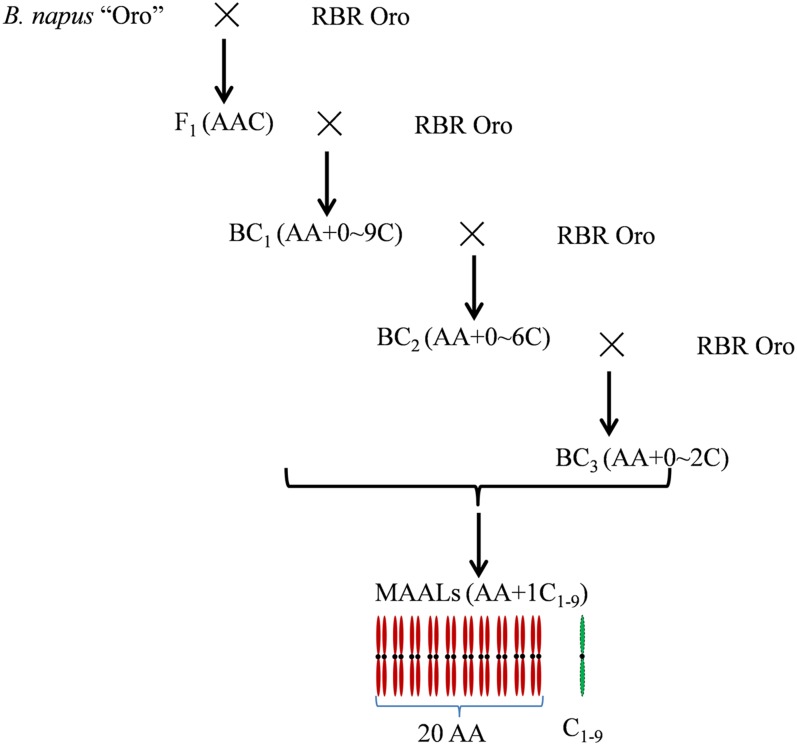
Previously, the diploid ancestor B. rapa (2n = 20, genome AA) was extracted from the allotetraploid B. napus cv. Oro (2n = 38, AACC), after being repeatedly pollinated by another crucifer I. indigotica (2n = 14, II) (Tu et al. 2010). After the restituted B. rapa genotype (RBR Oro) was crossed with Oro as female, the triploid hybrid (2n = 29, AAC) was readily obtained and pollinated by RBR Oro to generate BC1 plants (2n = 20–29). Those BC1 plants with different C-subgenome chromosomes were pollinated again by RBR Oro to produce BC2 plants (2n = 20–26), and several BC2 plants (2n = 22) that were found to carry chromosome C4 were pollinated further by RBR Oro. Both BC2 and BC3 plants (2n = 20–22) were screened to establish MAALs for different C-subgenome chromosomes (2n = 21, AA + 1C1–9), by both cytological and molecular selections (Figure 3). The cytological numerical designations of the nine C-subgenome chromosomes followed the internationally adopted system (Wang et al. 2011a; Xiong and Pires 2011; Chalhoub et al. 2014).
Figure 3.
Development of MAALs and dissection of the C subgenome in the background of RBR.
Because the growth of RBR Oro and the hybrid progenies was severely impeded under humid conditions during the winter season in the experimental field in Wuhan, these plants were grown in pots in an unheated glasshouse before flowering and later transferred to the field to produce seeds during the spring season.
Morphology, cytology, and pollen viability
Morphological characteristics of the complete set of MAALs were documented and compared to those of Oro and RBR Oro. To determine the chromosome numbers of the hybrids between Oro and RBR Oro and backcrossing progenies of successive generations, the ovaries from young flower buds were collected and treated with 2 mM 8-hydroxyquinoline for 3 hr at ∼20° and then fixed in Carnoy solution (3:1 ethanol:glacial acetic acid, v/v) and stored at −20° for use. For meiotic analyses, young flower buds from fully blooming plants were collected and fixed in Carnoy solution for 12 hr and then transferred to fresh liquid three times and stored at −20°. Cytological observation was followed according to the method of Li et al. (1995). The percentage of stainable pollen grains stained with 1% acetocarmine of >500 pollens per MAAL line was calculated to determine pollen viability.
Probe labeling, genomic in situ hybridization, and FISH analysis
The plasmid DNA of BAC BoB014O06 specific for the C genome of B. oleracea (provided by Susan J. Armstrong, University of Birmingham, Birmingham, UK) and genomic DNA of C. abyssinica were labeled with biotin-11-deoxycytidine triphosphate by random priming using the BioPrime DNA Labeling System kit (Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol. The genomic DNA of B. napus Zhongshuang 11 was fragmented in boiling water and used as a block. To identify specific C-subgenome chromosomes, four repetitive DNA sequences: 5S ribosomal DNA (rDNA), 45S rDNA, CentBr1, and CentBr2 were labeled with digoxigenin-11-deoxyuridine triphosphate (Roche, Basel, Switzerland) by random priming using the BioPrime Array CGH Genomic Labeling System kit according to the manufacturer’s protocol. Multiple target FISHs with these probes were performed to identify certain MAALs according to the work of Xiong and Pires (2011). Slide preparations were carried out mainly according to the methods of Zhong et al. (1996) and the procedures of BAC-FISH analyses following the procedure of Cui et al. (2012) with slight modification to reduce the washing time to 8 min in 0.1 × SSC with 20% deionized formamide at 40°. Before a second round of FISH, slides were washed for 10 min in 2 × SSC with 50% deionized formamide at 42° to clear former hybridized probe.
Images from FISH were taken using a computer-assisted fluorescence microscope with a CCD camera (Axio Scope A1, Zeiss, Oberkochen, Germany). Photographs were manipulated by Adobe Photoshop 7.0 software to adjust contrast and brightness and change the background to black.
Data statistics
The significance of data statistics, including t test, Pearson’s correlation coefficient as well as χ2 test, was determined by the data analysis function of Microsoft Excel and the R project.
Amplified fragment length polymorphism analysis
Amplified fragment length polymorphism (AFLP) analysis was performed for the hybrids, RBR ZS11, and parents according to the protocol of Vos et al. (1995). A total of 50 ng DNA per sample was digested by the restriction endonucleases EcoRI and MseI (Thermo Fisher Scientific, Waltham, MA), then ligated with EcoRI and MseI adapters. Two steps of PCR (preselective PCR and selective PCR) were performed to amplify the ligation production. In total, 57 primers were selected and used for AFLP fingerprint analysis.
Design of C-subgenome chromosome-specific gene markers and PCR amplification
Based on the recently released genome data of B. napus (Chalhoub et al. 2014), 117 genes, anchored in one of nine C-subgenome chromosomes and including obvious sequence variations compared to their respective A subgenome homeologs, were chosen for primer design. Primers were designed by Oligo 7 software based on those sequences with variations and then blasted to the B. napus genome. Those primers with more than one potential binding site were discarded. Young leaves were collected to extract DNA according to the CTAB method. PCR reactions (10 μl) contained 1× Taq buffer, 2 mM MgCl2, 2.5 mM deoxy-ribonucleoside triphosphatesans, 5 μM forward and reverse primer, 0.35 units Taq DNA polymerase, and 50 ng genomic DNA. DNA fragments were amplified using an initial 5-min denaturation at 94° followed by 30 cycles (94° for 45 sec, 53° −57° for 30 sec, and 72° for 45 sec), and a final 10-min elongation step at 72°. Finally, PCR products were checked by 1% agarose gels.
Supplemental Material, File S1 contains Table S1, Table S2, Table S3, and the supplemental data about the detailed description of morphological characteristics of MAALs. Table S1 contains detailed information of C-subgenome chromosome-specific gene markers. Table S2 contains the number of plants used to determine the transmission rates per MAAL. Table S3 contains anchored scaffolds determined by the set of MAALs and those anchored specific gene markers.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Extraction of the A subgenome from B. napus by inducing the elimination of C-subgenome chromosomes
Although the A subgenome was extracted from the allotetraploid B. napus (2n = 38, AACC) after the C-genome chromosomes were successively and completely eliminated by pollination with another crucifer I. indigotica (2n = 14) (Tu et al. 2010), the cytological process related to the biased loss and the pervasiveness of this method needed to be elucidated in detail. Herein, B. napus cultivar Zhongshuang 11, the first genotype (Figure 2, A and B) that was selected for genome sequencing in China, was extensively pollinated by the crucifer C. abyssinica (2n = 90), which flowered at a similar time to B. napus and had a longer flowering duration compared with I. indigotica. Four F1 hybrids from ∼10,000 pollinations were identified to have 2n = 29, but showed the B. napus-type morphology (Figure 2, C and D). They contained 20 chromosomes from A-subgenome and 9 C-subgenome chromosomes as revealed by FISH with C-subgenome-specific probe (Figure 2C), but no chromosomes and chromosomal fragments from C. abyssinica were detected by genomic in situ hybridization (GISH) analyses (Figure S1, A1–A3). Additionally, FISH with C-subgenome-specific probe was also performed to check the specificity of this probe in root-tip cells of C. abyssinica (Figure S1, C1–C3). Further investigations with gene markers specific for individual C-subgenome chromosomes confirmed that these 9 C-subgenome chromosomes covered all 9 chromosomes of the C-subgenome (data not shown), indicating their chromosome component of AAC with the loss of one copy of the C subgenome from the complement of B. napus. Its chromosomes in pollen mother cells (PMCs) showed two kinds of pairing configurations, 1III + 9II + 8I (64.4%, 58/90) and 10II + 9I (35.6%, 32/90). After these hybrids (AAC) were pollinated again by C. abyssinica to induce more loss of C-subgenome chromosomes, three BC1 plants exhibited a B. rapa-type phenotype and had lower chromosome number (2n = 22, 23, and 26), including 20 A-subgenome chromosomes and additional C-subgenome chromosomes. Among the self-pollination progenies from one BC1 plant with 2n = 22 (Figure 2, E and F), >100 plants with 20 A-subgenome chromosomes appeared and gave the B. rapa-like phenotype (Figure 2, G and H), which were the restituted B. rapa from Zhongshuang 11 (RBR ZS11). In PMCs of RBR ZS11, normal chromosomes pairing as 10 bivalents and 10:10 segregation were performed. Then the flowers had well-developed anthers full of highly stainable pollen grains (90.0 ± 2.3%), which was comparable to donor B. napus (92.8 ± 2.3%) (t test, P > 0.05). However, their seed sets by selfing pollination were quite low (1.92 seeds/pod), but highly enhanced by bud pollination (6.59 seeds/pod), indicating a barrier of selfing seed setting or the regain of self-incompatibility for B. rapa. By AFLP analysis with 57 pairs of primers, the four hybrids and RBR ZS11 lost 11.33–14.76% and 34.51% of the B. napus-specific fragments, respectively, and contained 2.56–3.48% and 1.64% of the C. abyssinica-specific fragments (Table 1). Additionally, 6.24–7.48% and 13.41% of fragments novel for two parents were present in the hybrids and RBR ZS11, respectively.
Figure 2.
Chromosome complement and phenotype of plants from B. napus to RBR. Blue indicates DAPI staining of chromosomes and red indicates labeled BAC BoB014O06 probe specifically for the C genome (Bar, 10 μm). The phenotype of young plants is shown. Bar, 10 cm. (A and B) One ovary cell (2n = 38) with 20 A-subgenome chromosomes (blue) and 18 C-subgenome chromosomes (red) (A) from B. napus cv. Zhongshuang 11 (B). (C and D) One ovary cell (2n = 29) with nine C-subgenome chromosomes (red) and 20 A-subgenome chromosomes (blue) (C) from F1 hybrid (D). No signals for the C. abyssinica genomic DNA probe (green) are detected. (E and F) One ovary cell (2n = 22) with 20 A-subgenome chromosomes (blue) and 2 C-subgenome chromosomes (red) (E) from one BC1F1 plant (F). (G and H) One ovary cell (2n = 20) with no red signals on all chromosomes (G) from RBR (H).
Table 1. AFLP analysis of intertribal hybrids and RBR from B. napus Zhongshuang 11 × C. abyssinica cross.
| Plant code | Specific bands for C. abyssinica (%) | Novel bands for two parents (%) | Deleted bands for B. napus (%) |
|---|---|---|---|
| F-1a | 25 (2.56) | 65 (6.76) | 109 (11.33) |
| F-2a | 27 (2.76) | 61 (6.34) | 132 (13.72) |
| F-3a | 34 (3.48) | 72 (7.48) | 142 (14.76) |
| F-4a | 30 (3.07) | 60 (6.24) | 124 (12.89) |
| Average | 29 (2.97) | 64.5 (6.71) | 126.75 (13.18) |
| RBR ZS 11 | 16 (1.64) | 129 (13.41) | 332 (34.51) |
Intertribal hybrids.
Taken together, the present and previous results (Tu et al. 2010) manifested the feasibility to extract the complete A subgenome and restitute the ancestral B. rapa genotype from natural B. napus through inducing successive loss of C-subgenome chromosomes, after two or three rounds of pollinations by other crucifers of different genera/tribes (Figure 1). The completeness of the extracted A subgenome was shown not only by the karyotype (2n = 20) and normal meiotic division of RBR Oro and RBR ZS11, but also by the construction of a linkage map including 10 linkage groups using the F2 population from the cross between RBR Oro and the sequenced B. rapa genotype “Chiifu” (Zhang Dawei and Li zaiyun, unpublished data).
Additions of C-subgenome chromosomes to RBR
To dissect the C subgenome of natural allotetraploid B. napus, B. napus cv. Oro was pollinated by RBR Oro (Tu et al. 2010), and the hybrid seeds were easily produced (Figure 3). The F1 hybrid plants were confirmed to be triploid (2n = 29, AAC) and their chromosome pairings were as follows: 1III + 9II + 8I (65.06%), 10II + 9I (26.51%), 2III + 8II + 7I (6.02%), and 1IV + 8II + 9I (2.41%). Then, more than 20 hybrids were pollinated again by RBR Oro to generate BC1 plants. Out of 201 BC1 plants (2n = 20–29) with chromosome numbers determined in the young ovary cells (Table 2), 13 (6.5%) had 2n = 20 for euploid B. rapa, 12 (6.0%) had 2n = 21 for putative monosomic alien addition lines (MAALs), and the remaining kept the higher chromosome number (Table 2). Unfortunately, the plants of these putative MAALs grew weakly and failed to produce seeds due to high sensitivity to some diseases in the field, and only one matured and gave the seeds after pollination by RBR Oro, which was later confirmed to carry chromosome C8 (AA + C8). Nine plants had 2n = 28 and three plants still kept one whole set of C genome (2n = 29, AAC), and the remaining plants had 2n = 22–27 (Table 2). Then about 90 BC1 plants with 2n = 21–27 were pollinated by RBR Oro to generate BC2 progenies. Among 445 BC2 plants observed (2n = 20–26) (Table 2), 237 (53.3%) were of RBR Oro-type (2n = 20) and 117 (26.3%) were MAALs with one of nine C-subgenome chromosomes except C4. Out of the other BC2 plants with two or more C-genome chromosomes, several double MAALS (DMAALs) harboring C4 and another C-subgenome chromosome were pollinated again by RBR Oro, and MAALs with C4 were selected among BC3 progenies from two DMAALs (C4 + C3 and C4 + C7) (Figure 3). Because of severe low selfing seed sets for most additional lines, all MAALs were preserved by backcrossing to RBR Oro.
Table 2. Distributions of chromosome numbers among BC1 and BC2 plants from the hybrids (2n = 29, AAC) between Oro and RBR Oro backcrossed to RBR Oro.
| Generations | Chromosome nos. (2n) | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | ||
| BC1 | 13 | 12 | 29 | 30 | 26 | 27 | 28 | 24 | 9 | 3 | 201 |
| BC2 | 237 | 117 | 49 | 24 | 8 | 9 | 1 | 0 | — | — | 445 |
Specific gene markers of individual C-subgenome chromosomes
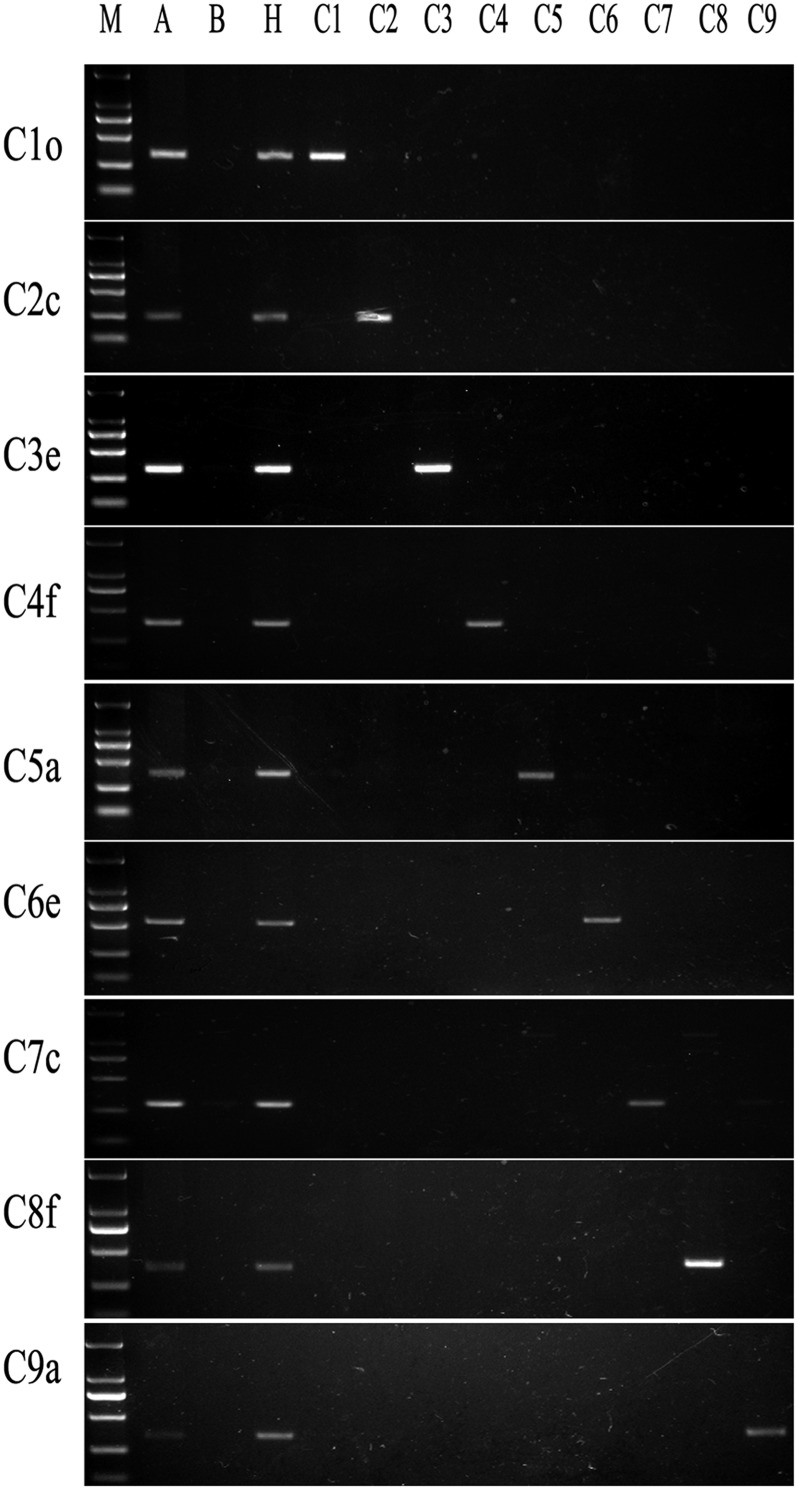
Genome sequences of B. napus genotype “Darmor” (Chalhoub et al. 2014) gave references for chromosome-specific genes of the C subgenome to identify the complete set of MAALs. Totally, 117 specific genes or gene sequences distributed on nine C-subgenome chromosomes were selected to design primers to determine their identities in each MAAL. Among 117 gene markers, 49 pairs of primers (Table S1) gave rise to specific fragments of the C subgenome after being screened in four B. napus cultivars (Oro, Zhongshuang 11, Huashuang 3, and Zhongyou 821), four B. rapa genotypes (RBR Oro, RBR ZS11, and Chiifu, Baiyou 1), and four BC1 plants carrying different numbers of C-subgenome chromosomes (2n = 21, 23, 24, and 26). The remaining primers generated either no amplified products or no polymorphic products among these screened materials, probably attributed to improper designed primers and the difference of genomic backgrounds between Darmor and Oro. The number of specific markers for each C-subgenome chromosome was four to seven, which were located on two arms of one assembled chromosome in sequencing. Those markers along every C-subgenome chromosome presented perfectly consistent results, when used to identify and classify the complete set of MAALs designated as AAC1–AAC9 (Figure 4).
Figure 4 .
C-subgenome chromosome-specific gene markers for the complete set of MAALs. A, B. napus cv. Oro; B, RBR Oro; H, hybrid of Oro and RBR Oro; and C1–C9, nine MAALS. The sizes of marker ladder from top to bottom are 2000, 1000, 750, 500, 250, and 100 bp, respectively.
Additionally, one plant with 2n = 20 also presented all the amplified products of specific primers for chromosome C1, suggesting the monosomic substitution line of B. rapa, which was confirmed by the FISH analysis with C-genome-specific BAC probe. One plant with 2n = 21 showed the amplified products of all C1-specific markers and some C8-specific markers, which included one intact additional C1 chromosome and one A-subgenome chromosome with one segment translocated from C8. These two plants were not further investigated, for not being MAALs.
Integrity of C-subgenome chromosomes in MAALs
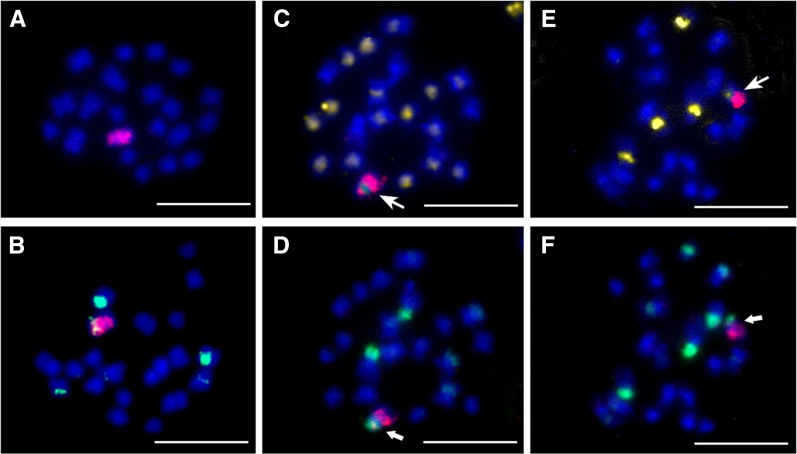
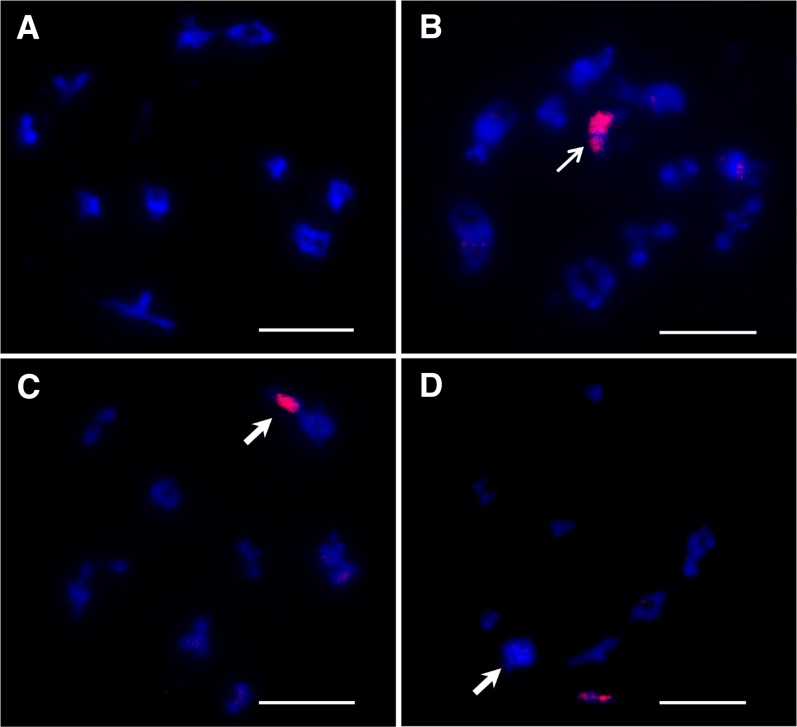
To confirm the integrity of C-subgenome chromosomes in MAALs, FISH analyses with BAC BoB014O06 as probe (Cui et al. 2012; Zhu et al. 2015) were performed on several selected plants from each MAAL (Figure 5A). It was manifested that all plants observed carried one complete C-genome chromosome, which was in line with the results from molecular makers above. Then, according to the method of Xiong and Pires (2011), a dual FISH with BAC BoB014O06 and 5S rDNA as probes was applied to MAALs with C4 (Figure 5B). Two continuous rounds of multiple target FISH with repeated sequences CentBr1, CentBr2, 45S rDNA, and BAC BoB014O06 as probes were applied to MAALs with C7 (Figure 5, C and D) and C8 (Figure 5, E and F), to prove the reliability of these specific gene markers. In addition, all or some of the five probes above were applied to detect the other MAALs and showed consistent results of molecular markers (Table 3). Taken together, the results from these cytological and gene-specific markers provided robust evidence for the complete set of MAALs with each C-subgenome chromosome in the background of the A subgenome extracted from the same natural B. napus, i.e., the in situ dissection of the C subgenome.
Figure 5.
BAC-FISH analyses of mitotic cells in MAALs. Blue indicates DAPI staining and red indicates labeled BAC BoB014O06 probe. (A) One mitotic cell for MAAL with 20 A-subgenome chromosomes (blue) and one C-subgenome chromosome (red). (B) One mitotic metaphase of AAC4 in which the chromosome C4 (red) harbors 5S rDNA locus (green). (C–F) Identification of AAC7 and AAC8 by applying two rounds of multiple target FISH. (C and D) One mitotic cell of AAC7. The chromosome C7 (red) gives the signals from CentBr1 probe (yellow, big arrow) in the first round FISH (C) and from 45S rDNA probe (green, solid arrow) in the second round FISH (D). (E and F) One mitotic cell of AAC8. The alien chromosome (red) is labeled with CentBr2 (yellow, big arrow) (E) and 45S rDNA probe (green, solid arrow) (F), respectively. Bar, 10 μm.
Table 3. Cytological markers for the complete set of MAALs.
| MAALs | Probes and signals | |||
|---|---|---|---|---|
| AAC1 | BAC BoB014O06 | CentBr1 | CentBr2 | 5S rDNA |
| AAC2 | BAC BoB014O06 | CentBr1 | CentBr2 | 45S rDNA |
| AAC3 | BAC BoB014O06 | CentBr1 | CentBr2 | 45S rDNA |
| AAC4 | BAC BoB014O06 | 5S rDNA | ||
| AAC5 | BAC BoB014O06 | CentBr1 | CentBr2 | 45S rDNA |
| AAC6 | BAC BoB014O06 | CentBr1 | CentBr2 | 45S rDNA |
| AAC7 | BAC BoB014O06 | CentBr1 | 45S rDNA | |
| AAC8 | BAC BoB014O06 | CentBr1 | CentBr2 | 45S rDNA |
| AAC9 | BAC BoB014O06 | CentBr1 | 45S rDNA | |
Boldface type denotes that FISH signal with this probe is detected in given MAAL and italics denote no signal for this probe.
Chromosome pairing of MAALs
By FISH with the probe BAC BoB014O06 specific for the C genome, the chromosome pairings for each MAAL were observed in 97–123 PMCs at diakinesis, respectively (Table 4). Totally, the majority of PMCs (68.91–87.13%) had the pairing configuration with 10IIAA + 1IC (Figure 6B) and the minority (10.89–26.05%) showed the pairing with 9IIAA + 1IIIAAC (Figure 6C). Additionally, in a small part of PMCs (0.81–6.03%) from all MAALs, one autosyndetic quadrivalent was formed by A-subgenome chromosomes, resulting in the pairing with 8IIAA + 1IC + 1IVAAAA (Figure 6D). But the quadrivalent was not observed in RBR Oro (Figure 6A). AAC1 and AAC2 showed the two highest percentages of PMCs (26.05% and 24.14%) with the allosyndetic trivalent, likely due to the high homeology between A1 and C1 and A2 and C2. They also had the highest percentages of PMCs (6.03% and 5.04%) with one A-subgenome quadrivalent. Notably, the percentage of PMCs (10.89%) with allosyndetic trivalent was lowest in AAC9. AAC4 also showed comparably low frequency of allosyndetic trivalent (14.42%) (χ2 test, P > 0.05). The other MAALs had 17.48–23.89% PMCs with 9IIAA + 1IIIAAC and 0.81–3.09% PMCs with 8IIAA + 1IC + 1IVAAAA (Table 4). The average pairing per cell for each MAAL is summarized in Table 4.
Table 4. Chromosome pairing, transmission rate of additional chromosomes, and pollen fertility for MAALs.
| Chromosome pairings | Transmission rate ♀ (%) | Transmission rate ♂ (%) | Pollen viability (%) | ||||
|---|---|---|---|---|---|---|---|
| MAALs | Total cells | IC (%) | IIIAAC (%) | IVAAAA (%) | |||
| AAC1 | 119 | 82 (68.91d) | 31 (26.05a) | 6 (5.04a) | 33.0a | 13.3a | 93.1 ± 1.3B |
| AAC2 | 116 | 81 (69.83cd) | 28 (24.14a) | 7 (6.03a) | 27.5ab | 8.6a | 96.2 ± 1.1A |
| AAC3 | 108 | 81 (75.00bcd) | 24 (22.22a) | 3 (2.78ab) | 14.5c | 3.2b | 80.4 ± 0.6D |
| AAC4 | 104 | 85 (81.73ab) | 15 (14.42b) | 4 (3.85a) | 8.3d | 8.8a | 71.1 ± 2.3E |
| AAC5 | 113 | 84 (74.34bcd) | 27 (23.89a) | 2 (1.77ab) | 19.4bc | 3.3b | 86.2 ± 1.8C |
| AAC6 | 97 | 76 (78.35bc) | 18 (18.56ab) | 3 (3.09ab) | 26.3ab | 6.7ab | 86.0 ± 2.1C |
| AAC7 | 103 | 82 (79.61b) | 19 (18.45ab) | 2 (1.94ab) | 20.5bc | 6.0ab | 93.4 ± 0.8B |
| AAC8 | 123 | 96 (78.05bcd) | 26 (21.14a) | 1 (0.81b) | 27.4ab | 7.5a | 89.1 ± 3.1BC |
| AAC9 | 101 | 88 (87.13a) | 11 (10.89b) | 2 (1.98ab) | 7.8d | 0.0c | 69.0 ± 0.5E |
| Average | — | 76.99 | 19.97 | 3.03 | 20.5 | 7.2 | |
Shared letters (a–d) within each association type denote that the values are insignificantly different (χ2 test, P < 0.05). Average of male transmission rate excludes the data from AAC9. Groups A–E detected as significantly different by t test, P < 0.05.
Figure 6.
BAC-FISH analyses of meiotic chromosome pairings in MAALs. (A) One PMC at diakinesis of RBR Oro with 10 bivalents of the A subgenome (10IIA). (B) One diakinesis PMC of AAC1 with 10IIA and one IC (arrow). (C) One diakinesis PMC of AAC2 with 9IIA and one allosyndetic trivalent (IIIAAC; solid arrow). (D) One PMC at diakinesis of AAC2 with 8IIA, one IC (red signal) and one autosyndetic quadrivalent (IVAAAA; big arrow). Bar, 5 μm.
Transmission rate of additional chromosomes through male and female gametes
To assess the transmission rates of additional C-subgenome chromosomes, 95–131 (from two plants of MAAL per chromosome except for AAC9 with only one plant obtained in BC2) and 66–93 (from one plant of MAAL per chromosome) plants from reciprocal crosses between each MAAL and RBR Oro were obtained to analyze the female and male transmission rates, respectively (Table S2). Then four gene markers distributed along both arms per C-subgenome chromosome were selected to determine its existence in the progeny plants. For female transmission of each C-subgenome chromosome in MAALs, the rates varied from 7.9% for AAC9 to 33.0% for AAC1 (Table 4), averaging 20.5%. For male transmission (except AAC9), the rates were from 3.2% for AAC3 to 13.3% for AAC1 (Table 4), averaging 7.2%, much lower than the female transmission rate. The male transmission of C9 was not detected, likely due to the lowest pollen fertility of AAC9 (69.0 ± 0.5%) and the limited number of progenies observed. The C1 chromosome showed the highest transmission rates by both male (13.3%) and female gametes (33.0%). In AAC4 and AAC9, the transmission rates (8.3 and 7.8%) for female were significantly lower than those of the others (χ2 test, P < 0.05). In AAC4, the transmission rate for male (8.8%) was slightly higher than that for female (8.3%). Interestingly, the female transmission rate was highly correlated with pollen viability of MAALs (R2 = 0.81, P = 8 × 10−4), but the correlation between male transmission rate and pollen viability was not as significant (R2 = 0.29, P > 0.05), implying the weaker competitiveness of aneuploidy male gametes in the process of fertilization. In several AAC2 plants and progenies, two markers, C2b and C2c, were missing, indicating the addition of the incomplete C2 chromosome and its inheritance to progeny.
Anchoring undetermined scaffolds of B. napus by MAALs
As ∼13% of scaffolds remained undetermined to certain chromosomes in the sequenced genome of B. napus (Chalhoub et al. 2014), this complete set of MAALs would help to anchor the elusive C-subgenome scaffolds to specific chromosomes. Therefore, 22 nonanchored C-subgenome scaffolds, ranging from 0.049 to 1.231 Mb and totally covering >10 Mb as well as assembling 1097 predicted genes, were selected to be anchored to C-subgenome chromosomes, based on presence/absence of amplified products of designed primers in nine MAALs. In total, 108 gene markers, corresponding to 108 anchored genes assembled to these scaffolds, were picked out. DNA extracted from three plants per genotype was mixed to prepare a pool. After PCR amplification, 45 gene markers from all 22 scaffolds (Table S3) were uniquely amplified in a single MAAL, resulting in the successful anchoring of these scaffolds to eight chromosomes of the C subgenome except C1.
Morphology of RBR and MAALs
As the exact progenitors of B. napus cv. Oro were unknown, RBR Oro would reflect largely the phenotype of the B. rapa ancestor several thousand years ago. While Oro, the first cultivar with low content of erucic acid in the world, which was selected from the local variety “Liho” in Germany, was a spring oilseed type, RBR Oro showed a flowering habit similar to the winter type of B. rapa (Guo et al. 2014). When planted in October in Wuhan, the young plants produced the procumbent leaves (Figure 7) with trichomes and many accessory buds under the low temperature, not observed in Oro (Figure S2). Similar phenotypes were also observed in RBR ZS11. The leaves and whole plants of RBR were slightly green, not so deep as those of their parental B. napus, and the phenotype and growth habitat were different from those B. rapa genotypes in our hands. The plants of RBR Oro flowered and matured earlier than Oro, and showed much lower seed yield. It also had weak growth and was more susceptible to diseases during and after flowering in the humid and high temperature environment, resulting in quick withering. In particular, it showed a barrier of selfing seeds, probably due to self-incompatibility, for its normal anthers were full of pollen grains with high stainability (89.2 ± 4.1%), while the seed set by self-pollination was quite low (1.32 seeds/pod) but very high (9.48 seeds/pod) by bud pollination.
Figure 7.
Morphology of donor B. napus (Oro), RBR Oro, and MAALs at seedling stage. The seedling and young plant in winter for RBR Oro are given (RBR2). C1–C9 mark nine MAALs. Bar, 15 cm.
All MAAL plants generally showed the morphology or architecture biased to RBR Oro, but expressed some peculiar features likely associated with each additional C-subgenome chromosome. Attractively, only AAC2 produced the plants with dark green leaves and stems covered with much waxen powder, as Oro did, while the other MAALs gave the plants with slight green leaves and stems. In comparison with RBR Oro, MAAL plants gave rise to decreased vigor, reduced stature and branch number, as well as delayed flowering time except for AAC6 (Figure S2A), probably because of deleterious impact of aneuploidy. Some particular features of RBR Oro were presented by all or some of the MAALs, such as the hindered growth and sensitivity to diseases, barrier of selfing seed sets, as well as procumbent leaves in winter, but AAC2 had a good selfing seed set in higher temperature and AAC9 did not have procumbent leaves in winter. MAALs were distinguishable by their detectable differences in the morphology of leaves from seedlings (Figure 7) and flowers (Figure S2B). Interestingly, six MAALs (except AAC3, AAC4, and AAC9) had high pollen fertility comparable to Oro and RBR Oro (t test, P < 0.05, Table 4), contrasting the much reduced fertility in the other B. rapa–B. oleracea MAALs (Heneen et al. 2012).
There existed some detectable differences in morphological characteristics among these MAALs, which are described separately in File S1 and summarized in Table 4.
Discussion
After B. napus was pollinated by several crucifers out of the Brassiceae tribe, including Orychophragmus violaceus (Cheng et al. 2002; Hua and Li 2006), Capsella bursa-pastoris (Chen et al. 2007), Lesquerella fendleri (Du et al. 2008), I. indigotica (Tu et al. 2010) and C. abyssinica (present study), only nonclassical hybrids were identified and contained the variable chromosome numbers (2n = 19–38) from B. napus and few alien chromosomes or DNA sequences. The chromosomes from the pollinators were likely eliminated mostly during certain stages of embryo or hybrid plant development, because of their distant relationships with B. napus. The production of B. napus euploid plants (2n = 38) might originate from the natural chromosome doubling after the alien elimination, from parthenogenesis of unreduced gametes, or from occasional pollen contamination. Of relevance for the extraction of the A subgenome, the C-subgenome chromosomes were also lost to different extents in individual hybrids, together with those from pollinators that functioned as inducers (Figure 1). Specially, the hybrids with 2n = 29 were recurrently obtained in all crosses with these pollinators, and only two representative pairings (1III + 9II + 8I and 10II + 9I) showed that 10 duplicated and 9 individual chromosomes were maintained (Cheng et al. 2002; Chen et al. 2007; Tu et al. 2010; present study). The A-subgenome origin of 10 paired chromosomes and C-subgenome origin of unpaired ones were speculated (Chen et al. 2007) but confirmed by the derivation of the restituted B. rapa from the hybrids after two more rounds of the induced chromosome eliminations (Tu et al. 2010; present study). FISH and molecular analyses of the hybrids (2n = 29) in this study showed the AAC complement with the whole copy of the C subgenome. The completeness of the A subgenome was retained in the progenies of generations and the continuous loss of C-subgenome chromosomes was realized after further induction, finally leading to the production of RBR (Figure 1 and Figure 2).
The introgression of alien DNA elements into the extracted A subgenome probably occurred at a low rate, for such sequences specific for pollinators were detected at low frequencies in the hybrids (2n = 29) with Oro pollinated by O. violaceus (Cheng et al. 2002) and I. indigotica (Tu et al. 2010). Subsequently, with the loss of all C-subgenome chromosomes and the alien individual chromosome or segments in the hybrids, the alien introgression would decrease further, i.e., from 3.48% in the hybrid to 1.64% in RBR ZS11 (Table 1). However, the A–C homeologous pairing (1III + 9II + 8I, other than 10II + 9II) in the hybrids (2n = 29) could probably move certain segments of the C-subgenome chromosomes to the specific A-subgenome chromosome, as the trivalent occurred in ∼60% PMCs (Cheng et al. 2002; Tu et al. 2010; present study). The exchange rate should be much reduced as more C-subgenome chromosomes were eliminated. Possibly, the transposon activation during the extraction process should cause more genomic changes of RBR (Table 1). Similarly, the preferential loss of C-subgenome chromosomes and different stabilities of three subgenomes (B > A > C) were also observed in the synthesized Brassica allohexaploids (2n = 54, AABBCC), after being pollinated by O. violaceus (Ge et al. 2009) or after self-pollinations of successive generations (Zhou et al. 2016). The distinct behaviors of these genomes were possibly attributable to their differences in size, inherent cytology, and homeology (Zhou et al. 2016).
In another scheme to extract AABB component from allohexaploid bread wheat and restitute the ancestral species, the hybridization between the extant tetraploid wheat (AtAtBtBt, the superscripts denoting the origin of the subgenomes) (Triticum turgidum subsp. durum) and hexaploid wheat (AaAaBaBaDaDa) (T. aestivum) was carried out, followed by the nine cycles of backcrossing of the hybrids (AatAatBatBatDa) to the hexaploid wheat to recover the genome (AaAaBaBa) of the restituted wheat (Kerber 1964). The presence of the Ph1 locus on the 5B chromosome, which prevented homeologous exchanges, likely contributed to the high karyotype stability of the restituted wheat (Zhang et al. 2014). Theoretically, the extracted genome should be >99.8% identical to the AaAaBaBa subgenomes of its bread wheat donor after the ninth backcrossing (Zhang et al. 2014). Although the extracted AABB genomes would largely represent those in bread wheat, the minor part of the genomic composition of the extant tetraploid still retained could make them diverge from the original situation to certain extents. So our extraction of the A subgenome from natural B. napus by inducing the preferential loss of the C-subgenome chromosomes had advantages for keeping its real constituent, and inversely for in situ dissecting of the C subgenome of the same B. napus donor. Otherwise, by following the backcrossing scheme of wheat (Kerber 1964), natural B. napus (AnAnCnCn) with progenitors unknown was crossed to extant B. rapa (ArAr) and the resultant hybrid (AnArCn) with the two A subgenomes of different origins was backcrossed as female four times to B. rapa, which resulted in the derivation of the diploid AA hybrid containing the lower proportion of the An subgenome but much higher proportion of the Ar subgenome than expected, together with some Cn-subgenome introgressions (Pelé et al. 2016). The heterogeneity of these two A subgenomes should change the genetic effects of the individual C-subgenome chromosome in established MAALs, which hindered the precise determination of genes and their performance, and specifically the study of genome interplay.
The morphological features in diploid hybrids and derived allotetraploids were of plurality, and ranged from intermediate to parental to transgressive (Soltis et al. 2014). The phenotype of RBR should reflect the proximal image of the actual B. rapa parent, in consideration of the short history of its hybridization with B. oleracea ∼7500 yr ago (Chalhoub et al. 2014). Although RBR Oro resembled morphologically a winter type of European B. rapa, for Oro originated from Europe, it presented obvious genetic divergence from the B. rapa types from the Old World or East Asia, as revealed by SSR markers (Guo et al. 2014). The causes for the divergence of the A subgenome in B. napus were different from those of natural B. rapa types, which evolved as independent taxa. As extensively observed, the resynthesized B. napus at initial generations showed the changes at genomic, transcriptomic, and proteomic levels (Song et al. 1995; Albertin et al. 2006; Gaeta et al. 2007; Xiong et al. 2011; Cui et al. 2013). Furthermore, due to the close relationship between A and C subgenomes and the lack of genetic mechanism suppressing the homeologous pairing (Cui et al. 2012), high rates of homeologous exchanges involving large segments to single SNPs occurred in natural B. napus (Chalhoub et al. 2014). As B. napus cv. Oro was normal, the defects in growth vigor and resistance to biotic and abiotic stresses shown by RBR Oro suggested that some genes of the A subgenome lost their functions, but those of the C subgenome were still active during the evolution. The normal growth and vigor of resynthesized B. napus parented by RBR (data not shown) further proved the correction of the altered functions of the AA subgenome by the CC subgenome. Similarly, the extracted tetraploid wheat (AaAaBaBa) with a stable karyotype exhibited the wide-ranging phenotypic abnormalities (Kerber 1964; Zhang et al. 2014), and many (rather than a few) genes were likely affected in its genome. But the resynthesized allohexaploid wheat parented by extracted tetraploid wheat fully restored normal phenotypes and high fertility, which also demonstrated that the DD subgenome compensated for the compromised functionality of the BBAA subgenomes of bread wheat (Kerber 1964; Zhang et al. 2014).
The whole exterior of the synthetic and natural B. napus was more biased to the parent B. oleracea, particularly by expressing its trait of the deeply green leaves covered with a thicker layer of waxen powder (Cui et al. 2012; Heneen et al. 2012). So B. napus was called B. oleracea-type rapeseed in China, and replaced the native B. rapa and B. juncea for its higher seed yield and stronger resistance to biotic and abiotic stresses. The better resistance of B. napus was largely contributed by B. oleracea. The weak vigor and strong susceptibility to diseases of RBR also suggested that the functional partitioning of the resistance was enhanced for the C subgenome but attenuated for the A subgenome in B. napus. But the biased expression toward only one parental genome (i.e., the genomic asymmetry) at different levels in B. napus was not so obvious as in wheat (Feldman et al. 2012) and upland cotton (Paterson et al. 2012). The characteristic B. oleracea-type exterior similarity between AAC2 and Oro indicated that the C2 chromosome harbored the gene(s) for the synthesis of waxen powder. The distinct phenotypes given by the different MAALs suggested the genetic effects of the gene(s) located on the extra C-subgenome chromosomes in the background of the A subgenome (Figure 7 and Figure S2), and some traits not observed in B. napus were likely suppressed after genome merger. For example, the trichomes appeared on the leaves of young plants for RBR Oro, but Oro never produces the leaves with the trichomes. As B. oleracea did not show this characteristic, the gene(s) from B. rapa was silenced in B. napus. Only MAAL (AAC7) produced the densely hairy leaves, implying that each C-subgenome chromosome except C7 completely or partly suppressed the expression of this trait.
The cytological diploidization, i.e., the inhibition of pairing to homeologous chromosomes, was fundamental for the evolution success of the allopolyploid species. The cytological process in mostly studied allohexaploid wheat was realized through two independent but complementary systems: the genetic control of pairing (Ph1 gene) and the physical divergence of the homeologous chromosomes (Feldman and Levy 2012). Owing to the close relatedness between A and C subgenomes in B. napus, homeologous pairings and exchanges were frequently detected in synthetic and natural B. napus (Nicolas et al. 2007; Szadkowski et al. 2010, 2011; Cui et al. 2012; Chalhoub et al. 2014). Specially, whole-chromosome aneuploidy and structural alterations occurred in its progenies of several generations and were biased to those chromosomes with extensive homeology (A1/C1 and A2/C2) (Xiong et al. 2011), likely resulting from their nondiploidized meiotic pairing (Cui et al. 2012). Accordingly, the two MAALs (AAC1 and AAC2) showed the higher rates of allosyndetic trivalents as well as higher transmission rate of the added C-subgenome chromosomes by both male and female gametes (Table 4). Although diploid-like meiosis in Brassica allopolyploids was proposed to be genetically regulated (Prakash 2009), the major QTL PrBn (pairing regulator in B. napus) found so far to suppress the homeologous pairing functioned only in its haploids (Jenczewski et al. 2003). The locus was mapped on linkage group C9, and two minor QTL on C1 and C6 (Liu et al. 2006), while its molecular function and mechanism of action remained elusive (Nicolas et al. 2009). The lowest frequency of allosyndetic trivalents was observed in our AAC9, potentially owing to the locus. The occurrence of one A-subgenome quadrivalent in all MAALs not in RBR Oro hinted that the extra unpaired C-subgenome chromosomes could promote the crossovers between A-subgenome chromosomes. These data recalled the “interchromosomal effect,” whereby crossover rates in inversion heterozygotes were sometimes elevated in collinear parts of the genome (Schultz and Redfield 1951). However, the corresponding ArArCo9 in MAALs of B. rapa–B. oleracea showed the highest frequency of allosyndetic trivalents (Heneen et al. 2012), implying that no suppressor-like PrBn located on C9 in B. oleracea, or no such pairing regulating system was established in newly synthesized B. napus, as the Ph1 gene in wheat also emerged, likely through allopolyploidization (Griffiths et al. 2006).
In comparison with the complete set of B. rapa var. trilocularis–B. oleracea var. alboglabra MAALs carrying the individual C-genome chromosomes (Heneen et al. 2012), the rates of PMCs with the pairing of 10 bivalents from the A subgenome and the additional univalent of the C subgenome in our MAALs were much higher (68.91–87.13%, averaging 76.99%) than those in B. rapa var. trilocularis–B. oleracea var. alboglabra MAALs (39.0–82.5%, averaging 55.8%). Furthermore, the additional C-subgenome chromosomes in all our MAALs showed only one type of homeologous pairing by forming the trivalent with two A-subgenome chromosomes. But most of C-genome chromosomes in other MAALs had more than one type of homeological pairing partner among the A-genome chromosomes, resulting in the formation of heteromorphic bivalents, trivalents, or pentavalents (Heneen et al. 2012). The less flexibility of homeologous pairing in our MAALs also showed that not only the A and C subgenomes of natural B. napus, which coevolved in the same nuclei for thousands of years, underwent more physical divergence from each other than from those of extant diploids, but also the homeologous pairing was weakened or restricted probably by some genetic control, which should develop during the evolution.
As Brassica genomes experienced a Brassiceae lineage-specific whole genome triplication, which was followed by abundant rearrangements and divergence (Lysak et al. 2005; Wang et al. 2011b; Liu et al. 2014), the A and C subgenomes in B. napus would be complex mosaics of triplicated ancestral genomic blocks. Therefore, the scaffold assignment to the correct chromosomes should be a challenge for the genome sequencing of B. napus, because ∼28% assembly scaffolds in B. oleracea (Liu et al. 2014) and ∼13% in B. napus were undetermined (Chalhoub et al. 2014). With this complete set of MAALs with each C-genome chromosome isolated, PCR amplification of specific gene markers from the C genome across MAALs was feasible for anchoring those nonanchored C-subgenome scaffolds to corresponding chromosomes. In total, 22 comparatively large scaffolds were successfully anchored to their specific C-subgenome chromosomes. However, it should be pointed out that the method of PCR amplification was laborious and time consuming, because of the huge number of undetermined scaffolds and small size for the majority of them. With aneuploidy lines, two studies employing chromosome-based sequencing were performed to decipher the genome information of allohexaploid wheat (Mayer et al. 2014), which highlighted the possibility of drawing support from the combination of current MAALs and high-throughput sequencing, such as resequencing as well as RNA-sequencing technology, to overcome barriers in genome sequencing of B. napus.
In conclusion, the distinct stabilities of two subgenomes in young allotetraploid B. napus made it feasible to first extract the more stable A subgenome and restitute the progenitor B. rapa, by inducing the biased loss of the unstable C subgenome. Subsequently, the C subgenome of the same B. napus genotype was readily dissected with each chromosome added to the extracted A subgenome. The RBR and the whole set of additions provided direct observation of the phenotypes of the progenitors after the evolution of thousands of years, and also provided unprecedent insights into structure and function of two subgenomes and their interplay in this allotetraploid. The ongoing studies of these novel materials for genome, transcriptome, DNA methylation, and proteome are expected to reveal more information about the contributions and interactions of two progenitors that produced B. napus.
Acknowledgments
We are grateful to Susan J. Armstrong (University of Birmingham, Birmingham, United Kingdom) for providing the clone BoB014O06 and Boulos Chalhoub for constructive comments. The work was supported by the National Key Research and Development Program of China (grant 2016YFD0100202) and the National Natural Science Foundation of China (grant 31471530).
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.190967/-/DC1.
Communicating editor: A. Houben
Literature Cited
- Albertin W., Balliau T., Brabant P., Chevre A. M., Eber F., et al. , 2006. Numerous and rapid nonstochastic modifications of gene products in newly synthesized Brassica napus allotetraploids. Genetics 173: 1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allender C. J., King G. J., 2010. Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley R., Spannagl M., Pfeifer M., Barker G. L., D’Amore R., et al. , 2012. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggs R. J., Wendel J. F., Doyle J. J., Soltis D. E., Soltis P. S., et al. , 2014. The legacy of diploid progenitors in allopolyploid gene expression patterns. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B., Denoeud F., Liu S., Parkin I. A., Tang H., et al. , 2014. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953. [DOI] [PubMed] [Google Scholar]
- Clausen R. E., Cameron D. R., 1944. Inheritance in Nicotiana tabacum. Xviii. Monosomic analysis. Genetics 29: 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. F., Wang H., Li Z. Y., 2007. Production and genetic analysis of partial hybrids in intertribal crosses between Brassica species (B. rapa, B. napus) and Capsella bursa-pastoris. Plant Cell Rep. 26: 1791–1800. [DOI] [PubMed] [Google Scholar]
- Cheng B.F., Seguin-Swartz G., Somers D. J., 2002. Cytogenetic and molecular characterization of intergeneric hybrids between Brassica napus and Orychophragmus violaceus. Genome 45: 110–115. [DOI] [PubMed] [Google Scholar]
- Cui C., Ge X., Gautam M., Kang L., Li Z. Y., 2012. Cytoplasmic and genomic effects on meiotic pairing in Brassica hybrids and allotetraploids from pair crosses of three cultivated diploids. Genetics 191: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C., Ge X., Zhou Y., Li M., Li Z. Y., 2013. Cytoplasmic and genomic effects on non-meiosis-driven genetic changes in Brassica hybrids and allotetraploids from pairwise crosses of three cultivated diploids. PLoS One 8: e65078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Flagel L. E., Paterson A. H., Rapp R. A., Soltis D. E., et al. , 2008. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42: 443–461. [DOI] [PubMed] [Google Scholar]
- Du X. Z., Ge X. H., Zhao Z. G., Li Z. Y., 2008. Chromosome elimination and fragment introgression and recombination producing intertribal partial hybrids from Brassica napus × Lesquerella fendleri crosses. Plant Cell Rep. 27: 261–271. [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A. A., 2012. Genome evolution due to allopolyploidization in wheat. Genetics 192: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Levy A. A., Fahima T., Korol A., 2012. Genomic asymmetry in allopolyploid plants: wheat as a model. J. Exp. Bot. 63: 5045–5059. [DOI] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., Iniguez-Luy F., Leon E., Osborn T. C., 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X. H., Wang J., Li Z. Y., 2009. Different genome-specific chromosome stabilities in synthetic Brassica allohexaploids revealed by wide crosses with Orychophragmus. Ann. Bot. (Lond.) 104: 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geleta M., Heneen W. K., Stoute A. I., Muttucumaru N., Scott R. J., et al. , 2012. Assigning Brassica microsatellite markers to the nine C-genome chromosomes using Brassica rapa var. trilocularis-B. oleracea var. alboglabra monosomic alien addition lines. Theor. Appl. Genet. 125: 455–466. [DOI] [PubMed] [Google Scholar]
- Griffiths S., Sharp R., Foote T. N., Bertin I., Wanous M., et al. , 2006. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752. [DOI] [PubMed] [Google Scholar]
- Guo Y., Chen S., Li Z. Y., Cowling W. A., 2014. Center of origin and centers of diversity in an ancient crop, Brassica rapa (Turnip Rape). J. Hered. 105: 555–565. [DOI] [PubMed] [Google Scholar]
- Heneen W. K., Geleta M., Brismar K., Xiong Z., Pires J. C., et al. , 2012. Seed colour loci, homoeology and linkage groups of the C genome chromosomes revealed in Brassica rapa-B. oleracea monosomic alien addition lines. Ann. Bot. (Lond.) 109: 1227–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y. W., Li Z. Y., 2006. Genomic in situ hybridization analysis of intergeneric hybrids between Brassica napus and Orychophragmus violaceus and production of B. napus aneuploids. Plant Breed. 125: 144–149. [Google Scholar]
- Jenczewski E., Eber F., Grimaud A., Huet S., Lucas M. O., et al. , 2003. PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164: 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber E. R., 1964. Wheat: reconstitution of the tetraploid component (AABB) of hexaploids. Science 143: 253–255. [DOI] [PubMed] [Google Scholar]
- Kihara H., 1924. Cytologische und genetische studien bei wichtigen getreidearten mit besonderer rücksicht ouf das verhalten der chromosomen und die sterilitat in den bastarden. Mem. Cell. Sci. Kyoto Imp. Univ. B1: 1–200. [Google Scholar]
- Levin D. A., 2003. The cytoplasmic factor in plant speciation. Syst. Bot. 28: 5–11. [Google Scholar]
- Li A. L., Liu D. C., Wu J., Zhao X. B., Hao M., et al. , 2014. mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26: 1878–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. Y., Liu H., Luo P., 1995. Production and cytogenetics of intergeneric hybrids between Brassica napus and Orychophragmus violaceus. Theor. Appl. Genet. 91: 131–136. [DOI] [PubMed] [Google Scholar]
- Liu S., Liu Y., Yang X., Tong C., Edwards D., et al. , 2014. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5: 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Adamczyk K., Manzanares-Dauleux M., Eber F., Lucas M. O., et al. , 2006. Mapping PrBn and other quantitative trait loci responsible for the control of homeologous chromosome pairing in oilseed rape (Brassica napus L.) haploids. Genetics 174: 1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M. A., Koch M. A., Pecinka A., Schubert I., 2005. Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K. F. X., Rogers J., Dolezel J., Pozniak C., Eversole K., et al. , 2014. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- McGrath J. M., Quiros C. F., 1990. Generation of alien chromosome addition lines from synthetic Brassica napus: morphology, cytology, fertility, and chromosome transmission. Genome 33: 374–383. [Google Scholar]
- Murashige T., Skoog F., 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nagaharu U., 1935. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7: 389–452. [Google Scholar]
- Nicolas S. D., Le Mignon G., Eber F., Coriton O., Monod H., et al. , 2007. Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175: 487–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas S. D., Leflon M., Monod H., Eber F., Coriton O., et al. , 2009. Genetic regulation of meiotic cross-overs between related genomes in Brassica napus haploids and hybrids. Plant Cell 21: 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., 2007. The evolutionary consequences of polyploidy. Cell 131: 452–462. [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Wendel J. F., Gundlach H., Guo H., Jenkins J., et al. , 2012. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492: 423–427. [DOI] [PubMed] [Google Scholar]
- Pelé A., Trotoux G., Eber F., Lodé M., Gilet M., et al. , 2016. The poor lonesome A subgenome of Brassica napus var. Darmor (AACC) may not survive without its mate. New Phytol. DOI: 10.1111/nph.14147. [DOI] [PubMed]
- Prakash S. B., 2009. Brassica and its close allies: cytogenetics and evolution. Plant Breed. Rev. 31: 21–187. [Google Scholar]
- Quiros C. F., Ochoa O., Kianian S. F., Douches D., 1987. Analysis of the Brassica oleracea genome by the generation of B. campestris-oleracea chromosome addition lines: characterization by isozymes and rDNA genes. Theor. Appl. Genet. 74: 758–766. [DOI] [PubMed] [Google Scholar]
- Ramsey J., Schemske D. W., 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Evol. Syst. 29: 467–501. [Google Scholar]
- Schultz J., Redfield H., 1951. Interchromosomal effect on crossing over in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 16: 175–197. [DOI] [PubMed] [Google Scholar]
- Sears E. R., 1954. Aneuploids of common wheat. Mo. Agric. Exp. Stn. Res. Bull. 572: 1–58. [Google Scholar]
- Soltis P. S., Soltis D. E., 2012. Polyploidy and Genome Evolution. Springer, Berlin. [Google Scholar]
- Soltis P. S., Liu X., Marchant D. B., Visger C. J., Soltis D. E., 2014. Polyploidy and novelty: Gottlieb’s legacy. Phil. Trans. R. Soc. B 369: 20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Lu P., Tang K., Osborn T. C., 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92: 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szadkowski E., Eber F., Huteau V., Lode M., Huneau C., et al. , 2010. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186: 102–112. [DOI] [PubMed] [Google Scholar]
- Szadkowski E., Eber F., Huteau V., Lode M., Coriton O., et al. , 2011. Polyploid formation pathways have an impact on genetic rearrangements in resynthesized Brassica napus. New Phytol. 191: 884–894. [DOI] [PubMed] [Google Scholar]
- Tu Y. Q., Sun J., Ge X. H., Li Z. Y., 2010. Production and genetic analysis of partial hybrids from intertribal sexual crosses between Brassica napus and Isatis indigotica and progenies. Genome 53: 146–156. [DOI] [PubMed] [Google Scholar]
- Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., et al. , 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lydiate D. J., Parkin I. A., Falentin C., Delourme R., et al. , 2011a Integration of linkage maps for the amphidiploid Brassica napus and comparative mapping with Arabidopsis and Brassica rapa. BMC Genomics 12: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., et al. , 2011b The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Xiong Z. Y., Pires J. C., 2011. Karyotype and identification of all homoeologous chromosomes of allopolyploid Brassica napus and its diploid progenitors. Genetics 187: 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z., Gaeta R. T., Pires J. C., 2011. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. W., Pan Q., Cui C., Tan C., Ge X. H., et al. , 2015a Genome-specific differential gene expressions in resynthesized Brassica allotetraploids from pair-wise crosses of three cultivated diploids revealed by RNA-seq. Front. Plant Sci. 6: 957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. K., Zhu B., Qi B., Gou X. W., Dong Y. Z., et al. , 2014. Evolution of the BBAA component of bread wheat during its history at the allohexaploid level. Plant Cell 26: 2761–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Hu Y., Jiang W., Fang L., Guan X., et al. , 2015b Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33: 531–537. [DOI] [PubMed] [Google Scholar]
- Zhong X. B., de Jong J. H., Zabel P., 1996. Preparation of tomato meiotic pachytene and mitotic metaphase chromosomes suitable for fluorescence in situ hybridization (FISH). Chromosome Res. 4: 24–28. [DOI] [PubMed] [Google Scholar]
- Zhou J. N., Chen T., Cui C., Ge X. H., Li Z. Y., 2016. Distinct subgenome stabilities in synthesized Brassica allohexaploids. Theor. Appl. Genet. 129: 1257–1271. [DOI] [PubMed] [Google Scholar]
- Zhou R. C., Moshgabadi N., Adams K. L., 2011. Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc. Natl. Acad. Sci. USA 108: 16122–16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Shao Y. J., Pan Q., Ge X. H., Li Z. Y., 2015. Genome-wide gene expression perturbation induced by loss of C2 chromosome in allotetraploid Brassica napus L. Front. Plant Sci. 6: 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.