Abstract
Ypt/Rab GTPases, key regulators of intracellular trafficking pathways, are activated by guanine-nucleotide exchange factors (GEFs). Here, we identify a novel GEF complex, TRAPP IV, which regulates Ypt1-mediated autophagy. In the yeast Saccharomyces cerevisiae, Ypt1 GTPase is required for the initiation of secretion and autophagy, suggesting that it regulates these two distinct pathways. However, whether these pathways are coordinated by Ypt1 and by what mechanism is still unknown. TRAPP is a conserved modular complex that acts as a Ypt/Rab GEF. Two different TRAPP complexes, TRAPP I and the Trs85-containing TRAPP III, activate Ypt1 in the secretory and autophagic pathways, respectively. Importantly, whereas TRAPP I depletion copies Ypt1 deficiency in secretion, depletion of TRAPP III does not fully copy the autophagy phenotypes of autophagy-specific ypt1 mutations. If GEFs are required for Ypt/Rab function, this discrepancy implies the existence of an additional GEF that activates Ypt1 in autophagy. Trs33, a nonessential TRAPP subunit, was assigned to TRAPP I without functional evidence. We show that in the absence of Trs85, Trs33 is required for Ypt1-mediated autophagy and for the recruitment of core-TRAPP and Ypt1 to the preautophagosomal structure, which marks the onset of autophagy. In addition, Trs33 and Trs85 assemble into distinct TRAPP complexes, and we term the Trs33-containing autophagy-specific complex TRAPP IV. Because TRAPP I is required for Ypt1-mediated secretion, and either TRAPP III or TRAPP IV is required for Ypt1-mediated autophagy, we propose that pathway-specific GEFs activate Ypt1 in secretion and autophagy.
Keywords: macro-autophagy, Ypt1, Ypt/Rabs, TRAPP complex, TRAPP III, GEF, Trs33, TrappC6A, TrappC6B
IN autophagy, cargo destined for degradation is engulfed by the double-membrane autophagosomes (APs), and is shuttled to the lysosome. Depending on the cargo and the growth conditions, autophagy can be generic or selective (Nair and Klionsky 2005; Nakatogawa et al. 2009). All autophagy pathways start with the formation of the preautophagosomal structure (PAS), which is comprised of the autophagy-specific proteins Atgs and membrane (Weidberg et al. 2011). Like all other intracellular trafficking pathways, autophagy is regulated by the conserved Ypt/Rab GTPases (Ao et al. 2014). When stimulated by guanine-nucleotide exchange factors (GEFs), Ypt/Rabs bind their downstream effectors, which include intracellular trafficking machinery components, like motors and tethers (Segev 2001). Recently, a role for Ypt/Rabs in coordination of intracellular trafficking steps and pathways has been proposed (Segev 2011; Lipatova et al. 2015).
In yeast, three Ypts regulate the different steps of autophagy: Ypt1 is required for the beginning of autophagy, PAS formation (Lipatova et al. 2012), while Vps21 and Ypt7 play a role in later steps that lead to the fusion of APs with the vacuole (the yeast lysosome) (Wang et al. 2002; Chen et al. 2014). Ypt31/32 were also implicated in autophagy, but the step is not clear (Zou et al. 2013). The established role of Ypt1 is the regulation of ER-to-Golgi transport (Segev 1991), and that of Vps21 and Ypt7 is in endocytosis (Schimmoller and Riezman 1993; Singer-Kruger et al. 1994). Interestingly, whereas both Vps21 and Ypt7 function in autophagy and endocytosis in the context of the same GEF-GTPase-effector modules (Wang et al. 2002; Chen et al. 2014), Ypt1 does not. Instead, two different TRAPP complexes, TRAPP I and TRAPP III, stimulate Ypt1 in the secretory and autophagy pathways, respectively (Lipatova et al. 2015). Likewise, in secretion and autophagy Ypt1 interacts with different effectors; e.g., Atg11 is an autophagy-specific effector of Ypt1 (Lipatova et al. 2012).
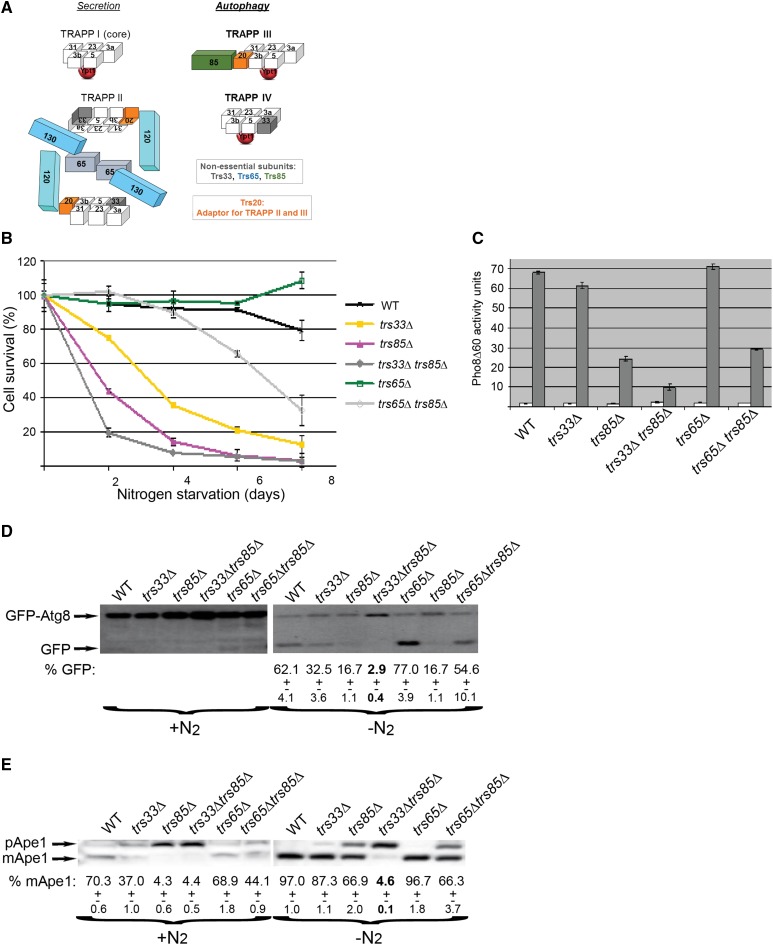
Currently, three TRAPP complexes are known: I, II, and III, and their multiple subunits are conserved from yeast to human cells (Kim et al. 2016). TRAPP I contains four core essential subunits, and TRAPP III contains the nonessential Trs85 in addition to core-TRAPP and the Trs20 adaptor. Both TRAPP I and TRAPP III act as Ypt1 GEFs (Morozova et al. 2006; Cai et al. 2008; Lynch-Day et al. 2010). TRAPP II contains two essential large subunits, Trs120 and Trs130, the nonessential subunits Trs65 and Trs33, the Trs20 adaptor, and core-TRAPP. TRAPP II localizes to trans-Golgi, and, while still controversial, acts as a GEF for Ypt31/32 (Kim et al. 2016). A role for a third nonessential subunit, Trs33, was shown in the assembly of TRAPP II only in the absence of Trs65 (Tokarev et al. 2009) (Figure 1A).
Figure 1.
Deletion of TRS33, but not TRS65, exacerbates the autophagy phenotypes of trs85∆ mutant cells. (A) Diagram showing TRAPP complex composition. Three TRAPP complexes act as GEFs for Ypt1: I, III, and IV. We propose that TRAPP I is composed of core subunits, TRAPP III contains core TRAPP, the Trs20 adaptor, and the nonessential subunit Trs85 (Kim et al. 2016), and based on results presented here, TRAPP IV contains core TRAPP and the nonessential subunit Trs33. TRAPP II contains core TRAPP, Trs20, Trs120, Trs130, and the nonessential subunits Trs65 and Trs33 (in the absence of Trs65). Diagram and positioning of Ypt1 in TRAPP I and III are based on published structures (Cai et al. 2008; Tan et al. 2013); numbers stand for TrsN, except for Bet3 and Bet5. (B–E) Generic (B–D) and selective (E) autophagy phenotypes were determined in the following strains: wild type (WT), trs33∆, trs85∆, trs33∆ trs85∆, trs65∆, and trs65∆ trs85∆ mutant cells. (B) Cell survival during nitrogen starvation. Cells were shifted from rich (YPD) medium to medium without nitrogen and the percent of viable cells at indicated time points (x-axis) is shown as percent of viable cells at time 0 (y-axis). Right: strain legend. (C) Pho8∆60 alkaline phosphatase assay. Lysates were prepared from cells grown in YPD medium (open bars) and after 4 hr of nitrogen starvation (shaded bars), and ALP activity in the lysates was determined. Pho8∆60 activity units represent nmol nitrophenol/mg protein. (D) GFP-Atg8 processing. Strains transformed with a plasmid expressing GFP-Atg8 were grown to midlog phase in rich medium and shifted to medium without nitrogen for 4 hr. Processing of GFP-Atg8 was determined by immuno-blot analysis using anti-GFP antibodies (percent GFP is shown at the bottom). All strains exhibit similar Atg8 processing under nonstarvation conditions (left). Under nitrogen starvation (right), >60% of the GFP-Atg8 protein in wild-type and trs65∆ mutant cells is processed to the GFP size, whereas this processing is defective in cells deleted for TRS33 and/or TRS85. (E) Ape1 processing during normal growth and under nitrogen starvation. Cells were grown to midlog phase in rich medium and shifted to medium without nitrogen for 4 hr. Processing of Ape1 was determined by immuno-blot analysis using anti-Ape1 antibodies (percent mApe1 is shown at the bottom). In wild-type and trs65∆ mutant cells, ∼70% of the Ape1 protein (pApe1) is processed into the mature form (mApe1) under nonstarvation conditions, whereas >95% is processed under starvation. Cells deleted for TRS33 and/or TRS85 exhibit an Ape1 processing defect under starvation and nonstarvation conditions. Error bars and ± represent SD; results shown in this figure are representative of at least two independent experiments.
Ypt1 is essential for autophagy based on the fact that the autophagy phenotypes of autophagy-specific ypt1 mutations are as severe as those of core-atg deletions (Lynch-Day et al. 2010; Lipatova et al. 2012). In contrast, whereas Trs85 plays a role in autophagy, it is not essential for this process (Lipatova et al. 2012). The question that drove this research project is why Ypt1 is essential to autophagy whereas its autophagy-specific GEF is not. Here, we show that Trs33 plays a role in autophagy, and together with Trs85 is required for Ypt1-mediated PAS formation. Based on results presented here, we propose the existence of a new TRAPP complex, the Trs33-containing TRAPP IV, which together with TRAPP III activates Ypt1 in the onset of autophagy. Because all players are conserved from yeast to human cells, we propose that the human homologs of Trs33, TrappC6A and B, regulate Rab1-mediated autophagy.
Materials and Methods
Strains, plasmids, and reagents
Strains used in this paper are summarized in Supplemental Material, Table S1. Plasmids used in this study are summarized in Table S2. All chemical reagents were purchased from Fisher Scientific (Hampton, NH), except for the following: Nitrogen bases were purchased from US Biological (Swampscott, MA); ProtoGel for Western blots from National Diagnostics (Atlanta, GA); Bacto peptone and Bacto agar from BD Difco (Franklin Lakes, NJ); salmon testes DNA, amino acids, p-nitrophenyl phosphate, and protease inhibitors from Sigma (St. Louis, MO); glutathione Sepharose 4B beads from Amersham Biosciences (Little Chalfont, UK); glass beads from BioSpec Products (Bartlesville, OK); EDTA-free protease inhibitor mixture from Roche Diagnostics (Indianapolis, IN); restriction enzymes and buffers from New England Biolabs (Ipswich, MA).
Antibodies used in this study included mouse monoclonal anti-GFP (Roche Diagnostics), rabbit anti-GST (Molecular Probes, Eugene, OR), rabbit anti-Ape1 (a kind gift from Dr. Ohsumi), affinity-purified rabbit anti-Ypt1 (Segev et al. 1988), rabbit anti-G6PDH (Sigma), goat anti-rabbit HRP and goat anti-mouse HRP (GE Healthcare), and TexasRed dye-conjugated goat anti-rabbit (Jackson ImmunoResearch).
Yeast culture conditions and viability analysis
Medium preparation and yeast culture growth for nitrogen starvation shift experiments were done as described (Segev and Botstein 1987).
Protein level analyses
To determine levels of GST- or GFP-tagged proteins in yeast lysates, exponentially growing cell cultures (7 × OD600) were spun down, resuspended in 100 µl of Laemmli buffer, boiled, vortexed with glass beads, and subjected to Western blot analysis using appropriate antibodies. Preparation of protein lysates for Ape1 and GFP-Atg8 processing analyses was done as described (Cheong and Klionsky 2008). ImageJ was used for quantification of protein bands.
Autophagy assays
Cell survival, Atg8-GFP processing, and Ape1 processing assays were done as previously described (Lipatova et al. 2012). Alkaline phosphatase activity assay of Pho8∆60 was done as previously described (Abeliovich et al. 2003).
GST pull-downs from yeast extracts
Yeast culture growth for pull-down experiments and purification of GST-tagged proteins was done as previously described (Morozova et al. 2006).
Microscopy
Live-cell microscopy was done as follows: Wild-type and mutant cells carrying constructs for expression of GFP-, YFP-, yEGFP-, RFP-, or mCherry-tagged protein(s) were grown to midlog phase in appropriate selective media. Fluorescent microscopy was performed using a deconvolution Axioscope microscope (Carl Zeiss) with FITC and TexasRed filter sets. Immuno-fluorescence microscopy using affinity-purified anti-Ypt1 antibodies was done as previously described (Segev et al. 1988). Colocalization was quantified by counting puncta that do or do not overlap on a single plane. For statistical analyses we used Student’s t-test.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
We have noticed that the autophagy phenotypes of the autophagy-specific ypt1-1 mutation are more severe than those of trs85∆ mutant cells (Lipatova et al. 2012). While it was possible that Ypt1 can function in autophagy without being activated by an autophagy-specific GEF, we hypothesized that there is an additional GEF that can activate Ypt1 in autophagy. One candidate was Trs33, which was originally identified as a TRAPP I/II subunit using pull-down experiments (Sacher et al. 2001). However, unlike other TRAPP I subunits, Trs33 is not essential for viability (Sacher et al. 2000) or for the Ypt1-GEF activity of TRAPP I (Kim et al. 2006). Interestingly, a negative genetic interaction was reported in high-throughput screens between TRS33 and TRS85. Specifically, trs33∆ trs85∆ double deletion cells exhibit slower growth than each of the single deletions (Schuldiner et al. 2005; Costanzo et al. 2010). Because ypt1-1 mutant cells also exhibit slower growth than wild-type cells (Segev and Botstein 1987), we decided to explore a possible role for Trs33 in autophagy.
Trs33 is required for autophagy in trs85∆ mutant cells
The effect of TRS33 deletion in wild-type and trs85∆ mutant cells on autophagy was determined. Generic autophagy was determined by survival, delivery of the cytosolic protein Pho8∆60 to the vacuole, and processing of GFP-Atg8 and Ape1 under nitrogen starvation. The selective autophagy cytosol-to-vacuole (CVT) pathway was determined by processing of Ape1 under normal growth conditions (Lipatova et al. 2012). The trs33∆ mutation resulted in mild generic autophagy phenotypes under nitrogen starvation, i.e., viability, alkaline phosphatase assay, processing of GFP-Atg8, and processing of Ape1. This single deletion also caused a defect in selective autophagy, based on Ape1 processing under normal growth conditions. The observed autophagy phenotypes of trs33∆ are less severe than those of trs85∆. However, deletion of TRS33 in trs85∆ mutant cells resulted in severe generic and selective autophagy phenotypes (Figure 1, B–E), more severe than those of either single deletion and similar to those reported for the ypt1-1 mutation (Lipatova et al. 2012).
We have previously reported that Trs33 is important for TRAPP II assembly in the absence of the TRAPP II nonessential subunit Trs65 (Tokarev et al. 2009), and that mutation in the TRAPP II-specific subunit Trs130, trs130ts, can cause defects in autophagy at the nonpermissive temperature (Zou et al. 2013). Therefore, we wished to determine whether the effect of the trs33∆ mutation on autophagy is due to a defect in TRAPP II function. First, deletion of TRS65 in wild-type cells does not cause generic or selective autophagy defects in WT cells. Second, deletion of TRS65 in trs85∆ mutant cells does not cause more severe phenotypes than those of trs85∆ mutant cells (Figure 1, B–E). Moreover, we have previously shown that while the trs130tsatg11∆ double mutation results in a more severe autophagy phenotype than that of atg11∆ (Zou et al. 2013), the trs85∆ atg11∆ double mutation does not (Lipatova et al. 2012). Similar to trs85∆, the trs33∆ atg11∆ double mutation also does not cause a more severe autophagy phenotype than that of the atg11∆ deletion (Figure S1), suggesting that Trs33 functions in autophagy in the context of the Ypt1-Atg11 module while TRAPP II does not. Together, these results indicate that the effects of trs33∆ on autophagy are not due to its role in TRAPP II assembly. In agreement with this conclusion we show below that Trs33 and Trs85, but not Trs65, are required for bringing TRAPP to PAS, and that Ypt31 does not suppress the autophagy defect of trs33∆, while it can suppress that of trs130ts mutant cells (Zou et al. 2013).
Trs33 functions in autophagy through Ypt1
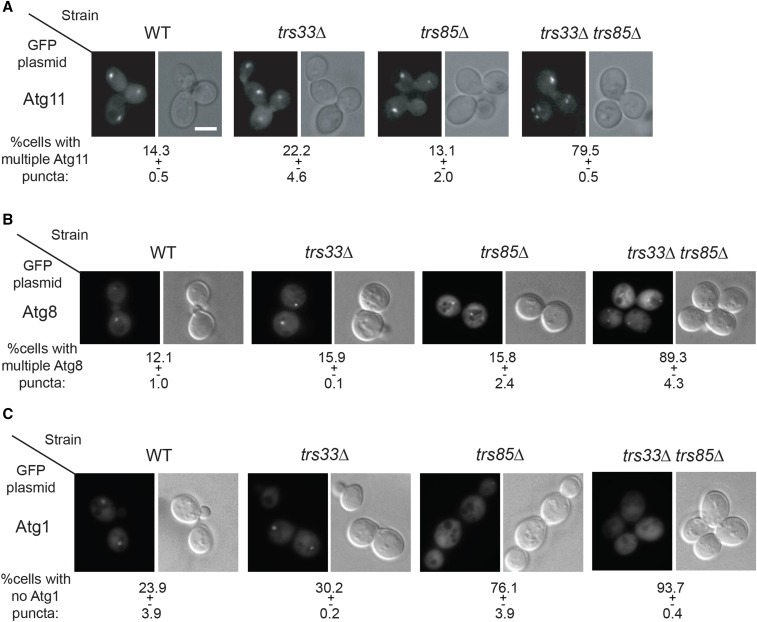
Ypt1 regulates autophagy (Lynch-Day et al. 2010; Lipatova et al. 2012) and a Trs33-containing TRAPP complex can act as its GEF in vitro (Kim et al. 2006). Therefore, we tested whether the role of Trs33 in autophagy is through Ypt1. First, we have previously shown that Ypt1 plays a role in the onset of autophagy: the formation of PAS (Lipatova et al. 2012). Specifically, whereas in >80% of wild-type cells, the core-Atg components – Atg11, Atg8, and Atg1 – localize to a single dot per cell that represents PAS or AP, PAS is not formed in >70% of ypt1-1 mutant cells; i.e., Atg11 and Atg8 show multiple dots and Atg1 is diffuse (even though their levels do not change) (Lipatova et al. 2012). Here, the effect of deletion of TRS33 in wild-type and trs85∆ mutant cells on PAS formation was determined by following the cellular distribution of Atg11, Atg8, and Atg1. While the proportion of cells with multiple Atg11 or Atg8 dots is not significantly higher in trs33∆ and trs85∆ single mutant cells when compared to wild-type cells, ∼75% of trs85∆, but not trs33∆, mutant cells exhibit diffuse Atg1 staining (Figure 2). This is in agreement with the more severe autophagy phenotypes of trs85∆ than those of trs33∆ mutant cells. Importantly, the trs33∆ trs85∆ double mutant cells exhibit more severe defects in Atg11, Atg8, and Atg1 distribution than the single deletions (Figure 2), similar to those of ypt1-1 (Lipatova et al. 2012). As in the ypt1-1 mutant cells, the changes in the distribution of PAS components were not caused by changes in GFP-Atg protein levels (Figure S2). These results support the idea that, like Ypt1, either a Trs33- or Trs85-containing TRAPP complex is required for PAS formation.
Figure 2.
Deletion of TRS33 in trs85∆ mutant cells causes a severe defect in PAS formation. The distribution patterns of PAS components, Atg11, Atg8, and Atg1, were compared in WT, trs33∆, trs85∆, and trs33∆ trs85∆ mutant strains. Cells were transformed with a plasmid expressing from the alcohol dehydrogenase (ADH1) promoter GFP-Atg11 (A), GFP-Atg8 (B), or GFP-Atg1 (C), grown under normal conditions, and analyzed using live-cell microscopy. Whereas in WT cells these PAS markers localize to a single dot per cell, in mutant cells Atg11 and Atg8 show multiple puncta, and Atg1 is diffuse. Shown for each strain, GFP (left) and DIC (right). Bottom: Percent cells with multiple Atg11 and Atg8 puncta (A and B) or with no Atg1 puncta (C). The differences between trs33∆ and WT are not statistically significant (P-value = 0.1 for GFP-Atg11, GFP-Atg8, and GFP-Atg1); for GFP-Atg1 the differences between trs85∆ and WT and between trs85∆ and trs33∆ trs85∆ are statistically significant (P-values are 0.0005 and 0.015, respectively). At least 45 cells visualized for each strain. ± represents SD. Results shown in this figure are representative of at least two independent experiments. ± represents SD. Bar, 5 μm.
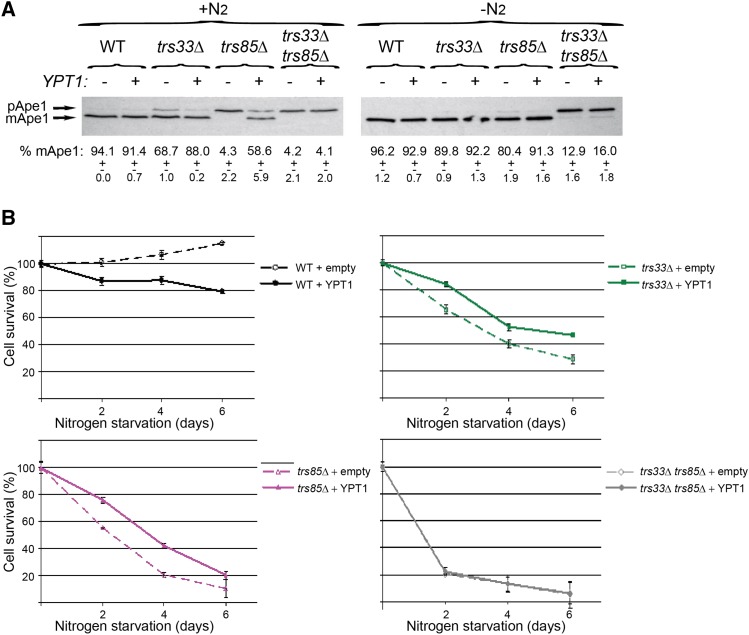
Second, we expect that overexpression of the Ypt substrate of a GEF might suppress the phenotypes of partial depletion of its GEF. For example, we have previously shown that overexpression of Ypt31, but not Ypt1, can suppress the autophagy phenotype of trs130ts (Zou et al. 2013). This result is in agreement with our view that TRAPP II acts as a Ypt31/32 GEF (Morozova et al. 2006). We have also shown that overexpression of Ypt1, but not Ypt31, can suppress the Ape1 processing defect of trs85∆ mutant cells (Lipatova et al. 2012), which is in agreement with the role of Trs85-containing TRAPP III as a GEF for Ypt1 (Lynch-Day et al. 2010). Here, we show that overexpression of Ypt1, but not Ypt31, can suppress the mild Ape1 processing defect of trs33∆ mutant cells (Figure 3A and Figure S3). In addition, overexpression of Ypt1 can partially suppress the growth defects of trs33∆ and trs85∆ mutant cells under nitrogen starvation (Figure 3B). In contrast, overexpression of Ypt1 cannot suppress the severe Ape1 and growth phenotypes of trs33∆ trs85∆ double mutant cells (Figure 3 and Figure S3). These results indicate that, like Trs85, Trs33 functions in autophagy through activation of Ypt1, and that Ypt1 activation by an autophagy-specific GEF is required for its function in autophagy.
Figure 3.
Overexpression of Ypt1 can suppress the autophagy phenotypes of trs33∆ and trs85∆, but trs33∆ trs85∆, mutant cells. WT, trs33∆, trs85∆, and trs33∆ trs85∆ mutant cells were transformed with a 2 μ plasmid for overexpression of Ypt1 (+), or empty plasmid as a negative control (−). Cells were tested for: (A) Ape1 processing under normal growth (left) and nitrogen starvation (right), and (B) viability under nitrogen starvation, as described for Figure 1. Results shown in this figure are representative of at least two independent experiments. Error bars and ± represent SD.
Trs33 and Trs85 bring core-TRAPP and Ypt1 to PAS
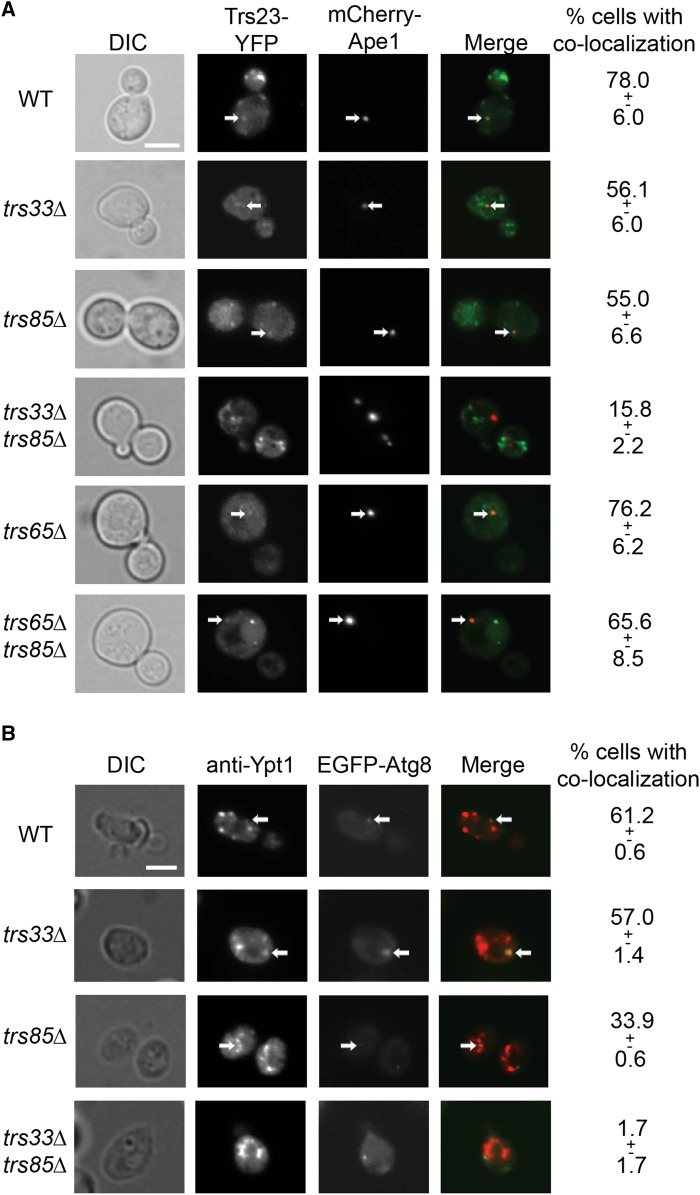
If Trs33 and Trs85 act in the context of TRAPP to activate Ypt1, which in turn mediates PAS formation, we expect that in their absence, TRAPP and Ypt1 do not colocalize with PAS markers. The colocalization of the tagged core-TRAPP subunit Trs23 with the PAS marker mCherry-Ape1 was determined by live-cell microscopy in wild-type, trs33∆, trs85∆, and trs33∆ trs85∆ mutant cells. Whereas Trs23 colocalizes with Ape1 in ∼80% of wild-type cells, colocalization was observed in only ∼15% of the trs33∆ trs85∆ double mutant cells. The single deletions, trs33∆ and trs85∆, result in a partial defect (∼55%, Figure 4A). In contrast, deletion of the TRAPP II subunit Trs65 does not affect Trs23 localization to PAS, and does not exacerbate the trs85∆ phenotype (Figure 4A). These results suggest that together Trs33 and Trs85 are required for recruitment of core-TRAPP to PAS, and support the idea that Trs33 does not function through TRAPP II.
Figure 4.
Trs33 and Trs85 are required for the localization of core-TRAPP and Ypt1 to PAS. (A) The core-TRAPP subunit Trs23 was tagged on the chromosome with YFP in the indicated strains (left column). Cells were transformed with a plasmid expressing the PAS marker mCherry-Ape1, and the colocalization of Trs23 and Ape1 was determined using live-cell microscopy. From top to bottom: WT, trs33∆, trs85∆, trs33∆ trs85∆, trs65∆, and trs65∆ trs85∆. Shown from left to right: DIC, green, red, merge, and % cells with colocalization. The differences between WT and the single deletions trs33∆ and trs85∆ are statistically significant (P-values = 0.05). (B) Cells were transformed with a plasmid expressing the PAS marker EGFP-Atg8, and the localization of Ypt1 was determined by immuno-fluorescence microscopy. From top to bottom: WT, trs33∆, trs85∆, and trs33∆ trs85∆. Shown from left to right: DIC, Ypt1, Atg8, merge, and % cells with colocalization. While the difference between WT and trs33∆ is not statistically significant (P-value = 0.1), the difference between WT and trs85∆ is (P-value = 0.0005). Arrows point to colocalizing puncta. At least 35 cells visualized for each strain. Results shown in this figure are representative of at least two independent experiments. ± represents SD. Bar, 5 μm.
The colocalization of Ypt1 with the PAS marker yEGFP-Atg8 was determined in wild-type, trs33∆, trs85∆, and trs33∆ trs85∆ mutant cells, using immuno-fluorescence microscopy. Whereas colocalization of Ypt1 and Atg8 was observed in ∼60% of wild-type cells, no colocalization was observed in the trs33∆ trs85∆ double mutant cells. Deletion of TRS33 did not have an effect, while localization of Ypt1 to PAS was partially impaired in trs85∆ single mutant cells (Figure 4B). These results are in agreement with the severity of the autophagy phenotypes of the single and double mutant cells shown above.
Trs33 and Trs85 localize to PAS independently
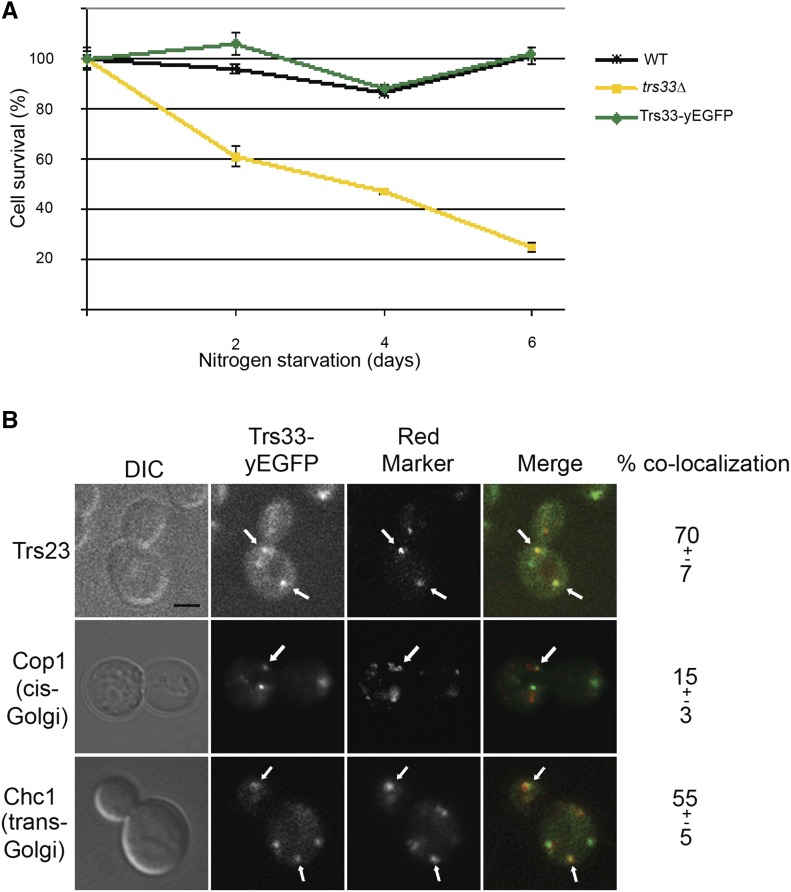
The aforementioned evidence suggests that either Trs33 or Trs85 is required for bringing core-TRAPP and Ypt1 to PAS in the onset of autophagy. Therefore, we expect that Trs33 and Trs85 can be recruited to PAS independently of each other. To test this idea, the colocalization of fluorescently tagged Trs33 or Trs85 with a PAS marker was compared in wild type and in cells deleted for the other subunit. Trs33 tagged on the chromosome with yEGFP is functional based on the observation that, unlike trs33∆, it does not cause a growth defect under nitrogen starvation (Figure 5A). Live-cell microscopy shows about four Trs33-yEGFP puncta/cell slice, of which 70% colocalizes with the core-TRAPP subunit Trs23, 55% colocalizes with a trans-Golgi marker (Chc1), and only 15% colocalizes with a cis-Golgi marker (Cop1) (Figure 5B). These results indicate that Trs33-yEGFP can incorporate into the TRAPP complex, and agree with our previous observation that Trs33 can function in the context of TRAPP II in the late Golgi (Tokarev et al. 2009). While Trs85-GFP was reported to colocalize with the Golgi marker Sec7 (especially under starvation) and not with a PAS marker (Shirahama-Noda et al. 2013), we have previously shown using bimolecular fluorescence complementation analysis that Trs85-Ypt1 interaction puncta do not colocalize with any exocytic compartment, including the Golgi (Lipatova et al. 2012). Thus, it is still unclear whether Trs33 and Trs85 colocalize with each other on secretory compartments.
Figure 5.
Trs33-yEGFP is functional and localizes to the Golgi. Trs33 was tagged with yEGFP at its C-terminus on the chromosome. (A) Trs33-yEGFP is functional autophagy. The survival of these cells under nitrogen starvation was compared with those of WT and trs33∆ mutant cells. Whereas trs33∆ mutant cells exhibit an autophagy phenotype, cells expressing Trs33-yEGFP behave like WT. Error bars represent SD. (B) Trs33-yEGFP colocalizes with a core-TRAPP subunit and Golgi markers. Trs33-yEGFP was tagged in cells also expressing RFP-tagged the core-TRAPP subunit Trs23 (top), the cis-Golgi marker Cop1, or the trans-Golgi marker Chc1. The colocalization of Trs33 and the red markers was determined using live-cell microscopy. Sown from left to right: DIC, green, red, merge, and % dots of Trs33-GFP that colocalize with the red marker. Arrows point to colocalizing puncta. At least 43 cells visualized for each strain. Results shown in this figure are representative of two independent experiments. Bar 2 μm.
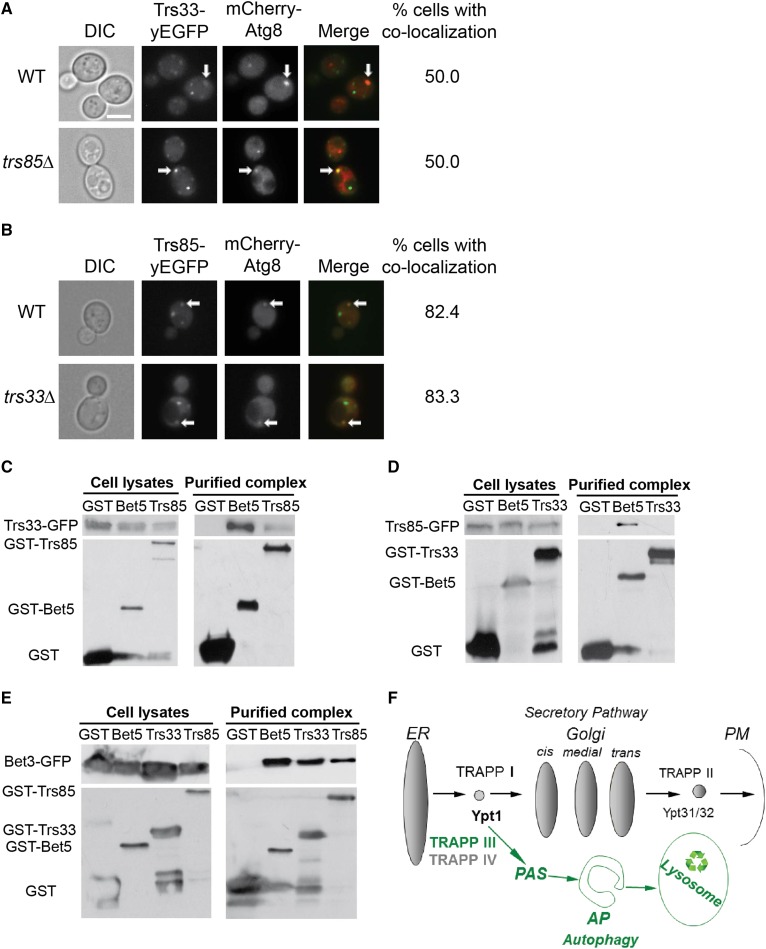
In addition to its localization to the Golgi, in about 50% of wild-type cells that contain PAS marked with mCherry-Atg8, Trs33-yEGFP localizes to PAS (Figure 6A), and deletion of TRS85 does not affect this localization. Functional Trs85-yEGFP (Lipatova et al. 2012) localizes to PAS in >80% of cells that contain PAS, and deletion of TRS33 does not affect this localization either (Figure 6B). Thus, the localization of Trs33 and Trs85 to PAS is not dependent on each other.
Figure 6.
Trs33 and Trs85 localize to PAS independently from each other. (A) The localization of Trs33 to PAS is not affected by deletion of TRS85. Trs33 was tagged on the chromosome with yEGFP in WT (top) and trs85∆ mutant cells (bottom). The cells were transformed with a plasmid for expression of mCherry-Atg8. Shown from left to right: DIC, green, red, merge, and % cells with colocalization. (B) The localization of Trs85 to PAS is not affected by deletion of TRS33. Trs85 was tagged on the chromosome with yEGFP in WT and trs85∆ mutant cells. The cells were transformed with a plasmid for expression of mCherry-Atg8. Shown from left to right: DIC, green, red, merge, and % cells with colocalization. (A and B) Arrows point to colocalizing puncta; at least 40 cells visualized for each strain. Bar, 5 μm. (C–E) Trs85 and Trs33 are present on two separate TRAPP complexes. Cells expressing GFP-tagged Trs33 (C), Trs85 (D), or Bet3 (E) were transformed with a plasmid for expression of GST, GST-Bet5, GST-Trs85, or GST-Trs33. The coprecipitation of the GFP-tagged proteins (right) with the GST-tagged proteins from yeast cell lysates (left) was determined. Trs33-GFP coprecipitates with GST-Bet5, but not with GST or GST-Trs85 (C); Trs85-GFP coprecipitates with Bet5, but not with GST or Trs33 (D); Bet3-GFP coprecipitates with GST-Bet5, GST-Trs85, and GST-Trs33, but not with GST (E). Results shown in A–E are representative of at least two independent experiments. (F) Model for TRAPP complexes function in the secretory pathway and autophagy. Whereas TRAPP I and II regulate secretion (Sacher et al. 2001), TRAPP III and IV regulate autophagy [(Lynch-Day et al. 2010), and results presented here]. See text for discussion.
The ability of Trs33 and Trs85 to localize to PAS independently suggests that they can assemble into separate TRAPP complexes. To test this idea, we used a GST pull-down assay. Trs33, Trs85, or the core-TRAPP subunit Bet3 were tagged on the chromosome with GFP in wild-type cells. The cells were transformed with a plasmid that expresses GST, GST-Bet5 (core-TRAPP), GST-Trs33, or GST-Trs85. The coprecipitation of GFP-tagged proteins with the GST-tagged protein (or GST as a negative control) was determined using immuno-blot analysis. The core-TRAPP subunit Bet3-GFP coprecipitated with GST-tagged Bet5, Trs33, and Trs85, but not GST. This result shows that all the GST-tagged proteins, including Trs33 and Trs85, can pull down a core-TRAPP subunit. In contrast, Trs33-GFP coprecipitated with GST-Bet5, while very little coprecipitates with GST-Trs85 or GST. Likewise, Trs85-GFP coprecipitated with GST-Bet5, but not with GST-Trs33 or GST (Figure 6, C–E). These results show that both Tss33-GFP and Trs85-GFP can precipitate with TRAPP (GST-Bet5). However, while GST-tagged Trs33 and Trs85 can pull down a core-TRAPP subunit (Bet3-GFP), they cannot pull down each other. These results support the idea that Trs33 and Trs85 form separate TRAPP complexes.
Discussion
Two autophagy-specific TRAPPs
Although Trs85 plays a role in autophagy, it is not essential for this process. Results presented here show that in addition to Trs85, another TRAPP subunit, Trs33, is important for activation of Ypt1 in autophagy. While trs33∆ mutant cells exhibit mild autophagy phenotypes, trs33∆ trs85∆ double mutant cells exhibit more severe autophagy defects than either deletion alone, similar to defects observed in ypt1-1 mutant cells. In addition, like Ypt1 and Trs85, Trs33 plays a role in the onset of autophagy, namely PAS assembly. Moreover, like Trs85, Trs33 functions in the context of TRAPP to activate Ypt1, because either Trs33 or Trs85 is required for efficient recruitment of core-TRAPP and Ypt1 to PAS. The ability of overexpressed Ypt1 to suppress the autophagy defects of trs33∆ and trs85∆ mutant cells supports this idea. Finally, Trs33 and Trs85 can localize to PAS independently of each other, and form biochemically distinct TRAPP complexes. Therefore, we conclude that two TRAPP complexes can activate Ypt1 in autophagy: Trs85-containing TRAPP III and Trs33-containing TRAPP IV.
Trs33 is currently considered a TRAPP I/II subunit based on coprecipitation with core-TRAPP (Sacher et al. 2001). However, there is no evidence for a role for Trs33 with TRAPP I in secretion, and allocation to a specific TRAPP complex solely based on coprecipitation can be misleading because coprecipitation of TRAPP subunits is dependent on the purification conditions (Choi et al. 2011; Brunet et al. 2012). For example, based on coprecipitation with core-TRAPP, Trs85 was also considered to be in TRAPP I/II (Sacher et al. 2001). However, when a role for this subunit was shown in autophagy (Meiling-Wesse et al. 2005; Nazarko et al. 2005), the TRAPP complex containing Trs85 was termed TRAPP III (Lynch-Day et al. 2010). Likewise, based on a role for Trs33 in autophagy, distinct from that of Trs85, we term the Trs33-containing complex TRAPP IV (Figure 1A). The EM structure of TRAPP III was reported to contain core-TRAPP and Trs20 (Tan et al. 2013), and the latter is required for the assembly of Trs85 with TRAPP (Taussig et al. 2014). We suggest that TRAPP IV also contains core-TRAPP subunits based on coprecipitation with Bet3 and Bet5 (Figure 6, C and E) and colocalization with Trs23 (Figure 5B). However, we propose that Trs20 is not required for the assembly and the function of TRAPP IV in autophagy. First, Trs33 associates with TRAPP in the absence of Trs20 (Kim et al. 2006). Second, the autophagy phenotypes of trs20 mutations are similar (not more severe) to those of the trs85∆ mutation (Brunet et al. 2013; Taussig et al. 2014), supporting the idea that Trs20 does not function with TRAPP IV in autophagy.
TRAPP III and IV could colocalize on PAS or APs based on the observation that Trs85 and Trs33 colocalize with Atg8 in ∼80 and 50% of the cells, respectively (Figure 6, A and B). However, whereas the other Trs85 puncta colocalize with the membrane protein Atg9 (Lipatova et al. 2012), Trs33 colocalizes with cis- and trans-Golgi markers (Figure 5B). This different cellular distribution might reflect a distinct mechanism of recruitment of the two complexes to the autophagy pathway.
We have previously shown that Trs33 is required for formation of the Ypt31/32 GEF TRAPP II in the absence of another nonessential TRAPP subunit, Trs65 (Tokarev et al. 2009). The colocalization of Trs33-yEGFP mostly with a trans-Golgi marker agrees with this idea (Figure 5B). In addition, a role for TRAPP II in autophagy has been suggested based on autophagy defects of trs130ts mutant cells at their nonpermissive temperature (Zou et al. 2013). We show that the role of Trs33 in autophagy is not connected to TRAPP II or Ypt31/32 based on the following evidence: First, deletion of the TRAPP II-specific Trs65 subunit in wild-type or trs85∆ mutant cells does not exacerbate their autophagy phenotypes. Second, while the autophagy defects of trs130ts mutant cells can be suppressed by Ypt31 and not Ypt1 (Zou et al. 2013), those of trs33∆ mutant cells can be suppressed by overexpression of Ypt1 and not Ypt31 (shown here). Finally, the autophagy defects of the double mutant atg11∆ trs130ts are more severe than those of either single mutation, suggesting that Atg11 and the Trs130-containing TRAPP II function in parallel pathways (Zou et al. 2013). In contrast, deletion of TRS33 does not exacerbate the autophagy phenotypes of the atg11∆ mutation (shown here), indicating that like Trs85 (Lipatova et al. 2012), Trs33 functions in the context of the same Ypt1 GTPase module as Atg11.
The existence of TRAPP III and IV raises the question of why two GEFs exist for the activation of a single Ypt, Ypt1, in a single process — PAS formation. One possibility is that in the absence of TRAPP III, which is the major Ypt1 GEF in generic autophagy as judged by the severity of the autophagy phenotypes, TRAPP IV compensates for its absence. The alternative is based on the breadth of the autophagy pathways, which are all dependent on PAS formation. Thus, while TRAPP III and IV compensate for each other in autophagy pathways that were tested, they might be specific for yet untested ones.
Pathway-specific Ypt/Rab GEFs
Based on previous data and results presented here, we conclude that pathway-specific GEFs enable Ypt/Rab-dependent regulation of two distinct pathways. Thus, Ypt1-dependent initiation of the secretory and autophagy pathways is regulated by TRAPP complexes: TRAPP I is required for Ypt1-mediated ER-to-Golgi transport, whereas TRAPP III or IV is required for Ypt1-mediated PAS assembly (Figure 6F). While the requirement of TRAPP I for Ypt1-dependent ER-to-Golgi transport is established (Sacher et al. 2001), the requirement of a GEF for Ypt1-mediated autophagy was not clear, because depletion of TRAPP III resulted in less severe autophagy defects than depletion of Ypt1. Here we show that TRAPP III or IV is required for Ypt1-mediated PAS assembly. Moreover, we show that overexpression of Ypt1 can suppress the autophagy defects of trs33∆ and trs85∆ single mutant cells, but not those of the double mutant. This finding indicates that Ypt1 needs a GEF to mediate autophagy. Thus, in cells deleted for one GEF, this function is provided by the remaining GEF, while in cells depleted for both GEFs, autophagy is completely blocked. One possible explanation for this dependency of Ypt1-mediated autophagy on a GEF is that at least one autophagy-specific GEF, TRAPP III or TRAPP IV, is essential for the recruitment of Ypt1 from the secretory pathway to autophagy.
We have proposed that Ypt/Rab GTPases coordinate shuttling of cargo from a single compartment to different destinations (Lipatova et al. 2015). For example, Ypt1, which regulates delivery of cargo from the ER to the secretory pathway and autophagy (Lipatova et al. 2013), is a candidate for coordination of these two pathways, and pathway-specific GEFs are candidates for enabling such coordination.
Conservation
Trs33 has two mammalian homologs, TrappC6A and TrappC6B (Kim et al. 2016). While hTrappC6A was implicated in neurodegenerative disease (Hamilton et al. 2011; Chang et al. 2015), nothing is currently known about its physiological role. The role of Ypt1 in autophagy is conserved and was shown for Rab1 (Zoppino et al. 2010). We propose that the role of Trs33 in autophagy is also conserved.
Acknowledgments
We thank X. Zhang for technical help at the beginning of the project, and J. Kim for critical reading of the manuscript. We thank Dr. Y. Ohsumi for his kind gift of anti-Ape1 antibodies and Dr. D. Klionsky for the GFP-Atg8 plasmid. This research was supported by grant GM-45444 from the National Institutes of Health (NIH) to N.S.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.194910/-/DC1.
Communicating editor: M. D. Rose
Literature Cited
- Abeliovich H., Zhang C., Dunn W. A., Jr, Shokat K. M., Klionsky D. J., 2003. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell 14: 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao X., Zou L., Wu Y., 2014. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 21: 348–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S., Noueihed B., Shahrzad N., Saint-Dic D., Hasaj B., et al. , 2012. The SMS domain of Trs23p is responsible for the in vitro appearance of the TRAPP I complex in Saccharomyces cerevisiae. Cell. Logist. 2: 28–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S., Shahrzad N., Saint-Dic D., Dutczak H., Sacher M., 2013. A trs20 mutation that mimics an SEDT-causing mutation blocks selective and non-selective autophagy: a model for TRAPP III organization. Traffic 14: 1091–1104. [DOI] [PubMed] [Google Scholar]
- Cai Y., Chin H. F., Lazarova D., Menon S., Fu C., et al. , 2008. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell 133: 1202–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. Y., Lee M. H., Lin S. R., Yang L. Y., Sun H. S., et al. , 2015. Trafficking protein particle complex 6A delta (TRAPPC6ADelta) is an extracellular plaque-forming protein in the brain. Oncotarget 6: 3578–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhou F., Zou S., Yu S., Li S., et al. , 2014. A Vps21 endocytic module regulates autophagy. Mol. Biol. Cell 25: 3166–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Klionsky D. J., 2008. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 451: 1–26. [DOI] [PubMed] [Google Scholar]
- Choi C., Davey M., Schluter C., Pandher P., Fang Y., et al. , 2011. Organization and assembly of the TRAPPII complex. Traffic 12: 715–725. [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton G., Harris S. E., Davies G., Liewald D. C., Tenesa A., et al. , 2011. Alzheimer’s disease genes are associated with measures of cognitive ageing in the lothian birth cohorts of 1921 and 1936. Int. J. Alzheimers Dis. 2011: 505984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J., Lipatova Z., Segev N., 2016. TRAPP complexes in secretion and autophagy. Front. Cell Dev. Biol. 4: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. G., Raunser S., Munger C., Wagner J., Song Y. L., et al. , 2006. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell 127: 817–830. [DOI] [PubMed] [Google Scholar]
- Lipatova Z., Belogortseva N., Zhang X. Q., Kim J., Taussig D., et al. , 2012. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc. Natl. Acad. Sci. USA 109: 6981–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z., Shah A. H., Kim J. J., Mulholland J. W., Segev N., 2013. Regulation of ER-phagy by a Ypt/Rab GTPase module. Mol. Biol. Cell 24: 3133–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z., Hain A. U., Nazarko V. Y., Segev N., 2015. Ypt/Rab GTPases: principles learned from yeast. Crit. Rev. Biochem. Mol. Biol. 50: 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day M. A., Bhandari D., Menon S., Huang J., Cai H., et al. , 2010. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 107: 7811–7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling-Wesse K., Epple U. D., Krick R., Barth H., Appelles A., et al. , 2005. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 280: 33669–33678. [DOI] [PubMed] [Google Scholar]
- Morozova N., Liang Y., Tokarev A. A., Chen S. H., Cox R., et al. , 2006. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat. Cell Biol. 8: 1263–1269. [DOI] [PubMed] [Google Scholar]
- Nair U., Klionsky D. J., 2005. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J. Biol. Chem. 280: 41785–41788. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y., 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10: 458–467. [DOI] [PubMed] [Google Scholar]
- Nazarko T. Y., Huang J., Nicaud J. M., Klionsky D. J., Sibirny A. A., 2005. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy 1: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Barrowman J., Schieltz D., Yates J. R., III, Ferro-Novick S., 2000. Identification and characterization of five new subunits of TRAPP. Eur. J. Cell Biol. 79: 71–80. [DOI] [PubMed] [Google Scholar]
- Sacher M., Barrowman J., Wang W., Horecka J., Zhang Y., et al. , 2001. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol. Cell 7: 433–442. [DOI] [PubMed] [Google Scholar]
- Schimmoller F., Riezman H., 1993. Involvement of Ypt7p, a small GTPase, in traffic from late endosome to the vacuole in yeast. J. Cell Sci. 106(Pt 3): 823–830. [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., et al. , 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123: 507–519. [DOI] [PubMed] [Google Scholar]
- Segev N., 1991. Mediation of the attachment or fusion step in vesicular transport by the GTP-binding Ypt1 protein. Science 252: 1553–1556. [DOI] [PubMed] [Google Scholar]
- Segev N., 2001. Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 13: 500–511. [DOI] [PubMed] [Google Scholar]
- Segev N., 2011. Coordination of intracellular transport steps by GTPases. Semin. Cell Dev. Biol. 22: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N., Botstein D., 1987. The ras-like yeast YPT1 gene is itself essential for growth, sporulation, and starvation response. Mol. Cell. Biol. 7: 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N., Mulholland J., Botstein D., 1988. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell 52: 915–924. [DOI] [PubMed] [Google Scholar]
- Shirahama-Noda K., Kira S., Yoshimori T., Noda T., 2013. TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J. Cell Sci. 126: 4963–4973. [DOI] [PubMed] [Google Scholar]
- Singer-Kruger B., Stenmark H., Dusterhoft A., Philippsen P., Yoo J. S., et al. , 1994. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J. Cell Biol. 125: 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D., Cai Y., Wang J., Zhang J., Menon S., et al. , 2013. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. USA 110: 19432–19437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussig D., Lipatova Z., Segev N., 2014. Trs20 is required for TRAPP III complex assembly at the PAS and its function in autophagy. Traffic 15: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarev A. A., Taussig D., Sundaram G., Lipatova Z., Liang Y., et al. , 2009. TRAPP II complex assembly requires Trs33 or Trs65. Traffic 10: 1831–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. W., Stromhaug P. E., Shima J., Klionsky D. J., 2002. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J. Biol. Chem. 277: 47917–47927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Elazar Z., 2011. Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80: 125–156. [DOI] [PubMed] [Google Scholar]
- Zoppino F. C., Militello R. D., Slavin I., Alvarez C., Colombo M. I., 2010. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 11: 1246–1261. [DOI] [PubMed] [Google Scholar]
- Zou S., Chen Y., Liu Y., Segev N., Yu S., et al. , 2013. Trs130 participates in autophagy through GTPases Ypt31/32 in Saccharomyces cerevisiae. Traffic 14: 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.