Abstract
Abnormal feeding behavior is one of the main symptoms of Prader-Willi syndrome (PWS). By studying a PWS mouse mutant line, which carries a paternally inherited deletion of the small nucleolar RNA 116 (Snord116), we observed significant changes in working-for-food behavioral responses at various timescales. In particular, we report that PWS mutant mice show a significant delay compared to wild-type littermate controls in responding to both hour-scale and seconds-to-minutes-scale time intervals. This timing shift in mutant mice is associated with better performance in the working-for-food task, and results in better decision making in these mutant mice. The results of our study reveal a novel aspect of the organization of feeding behavior, and advance the understanding of the interplay between the metabolic functions and cognitive mechanisms of PWS.
Keywords: Prader-Willi, genomic imprinting, food anticipatory activity, behavioral timing
PRADER-WILLI syndrome (PWS) is a rare neurodevelopmental disorder that is caused by genomic imprinting defects within the small nuclear ribonucleoprotein N (SNPRN) cluster of the human chromosome region 15q11-13. PWS is characterized by developmental delays, feeding problems, hyperphagia, behavioral disorders, and sleep-wake disturbances (Peters 2014; Tucci 2016). In preclinical investigations, mouse models with different deletions at the orthologous locus on mouse chromosome 7C have shown the main features of the human syndrome, such as growth retardation (Rozhdestvensky et al. 2016), hyperphagia (Davies et al. 2015), and sleep abnormalities (Lassi et al. 2016). Small nucleolar RNA 116 (Snord116, also called MBII-85), a paternally expressed noncoding gene that modifies other small nuclear RNAs (snoRNAs), is considered one of the key players in PWS (Peters 2014). Expression studies of the snoRNAs MBI-36, MBII-85 (Snord116), MBII-52 (Snord115), and MBII-13 show that these snoRNAs are either exclusively or prevalently expressed in the brain (Cavaillé et al. 2000). Yet, it has been demonstrated that MBII-52, MBII-85, and MBII-13 are imprinted in the mouse brain. However, no analysis of the imprinting status of Snord116 in different areas of the brain has yet been conducted. Mice that maternally inherit the Snord116 deletion are phenotypically similar to wild type (Skryabin et al. 2007); thus, the paternal inheritance of the mutation is the critical heterozygous pattern to be investigated. However, several studies have indicated that the investigation of homozygous mutants may also be needed to fully understand the role of Snord116 (Wolf et al. 2008; Qi et al. 2016). Biallelic deletion of Snord116 in mice produces classical developmental phenotypes associated with reduced body weight and growth; however, as mutant mice enter adulthood, hyperphagia is observed, and the balance between energy intake and expenditure is altered (Qi et al. 2016). The regulatory processes involved in maintaining body weight homeostasis through daily food intake and expenditure include a proper balance between thermoregulation and physiological states (Overton and Williams 2004; Florant and Healy 2012), and between behavior and cognitive processes (Chen et al. 2016; Higgs 2016). Recently, we studied the link between thermogenesis and sleep in PWScrm+/p− mutant mice, reporting a significant increase in the circadian variations of body temperature that disrupt sleep in mutants (Lassi et al. 2016).The latter study suggests that Snord116 is involved in sleep–wake regulation, and in the daily control of body temperature. Thermogenesis plays an important role in adjusting metabolism both during sleep, when energy expenditure is reduced owing to resting, and during wakefulness, when the individual is searching for food and engaging in physical activity (Schmidt 2014). Food intake is governed by at least two important motivations, the need for calories, and the hedonic value of food (Challet and Mendoza 2010). However, food intake is also structured by fundamental cognitive/behavioral processes, such as clock-dependent mechanisms (Mistlberger 2011). By studying the PWSICdel mouse line, Davies et al. (2015) reported that hyperphagia in these PWS mutant mice is due to a constant need for calories, and that the related behavior is not due to an increased hedonic value of food intake. In particular, the PWSICdel mice presented similar behavioral licking responses compared to control mice toward palatable food. That study concluded that the hyperphagic trait in mutant mice is due mainly to one of two motivational systems; it remains to be understood whether the specific need for calories influences behavioral or cognitive processes in mutants as they access food during the day. Therefore, here, we explored fundamental aspects of the behavioral and cognitive systems associated with food-intake (i.e., working-for-food behavioral strategies) in the mouse mutant model PWScrm+/p− (Skryabin et al. 2007), which lacks Snord116. We focused our investigation on two temporally determined behaviors across different timescales. The first timing behavior is known as “food anticipatory activity” (FAA); FAA occurs at the scale of hours (Mistlberger 2011), and is important, for example, when searching for food (foraging behavior) as it becomes important in estimating the delay between meals during the day. The second timing behavior is called interval timing, and refers to an ability—one that is widely shared across species—to release behavioral responses on a time scale of seconds to minutes (Buhusi and Meck 2005; Merchant et al. 2013; Tucci et al. 2014b). Short-interval behavioral responses are fundamental for attention, for decision making, and for learning and memory processes. We report here that Snord116 is paternally expressedmaternally silenced in specific areas of the mouse brain. The brain areas in which Snord116 is imprinted regulate timing and feeding, among other functions. Interestingly, we show that PWScrm+/p− mutant mice present altered temporal responses in timing tasks, which might be a indirect effect of the mutation on timing behaviors across timescales. Indeed, we discuss whether timing abnormalities may be a direct, or an indirect, consequence of hyperphagia.
Materials and Methods
Animal husbandry
PWScrm+/p− mutant mice carry a deletion of the Prader-Willi Syndrome critical region (PWScr), which includes Snord116 and IPW exons A1/A2, B, and C (Skryabin et al. 2007). At the Istituto Italiano di Tecnologia (IIT), all mice were bred and maintained through paternal inheritance on a C57BL/6J background. During the home-cage studies, animals were housed singly, and body weights were monitored daily to ensure the welfare of the animal. Male mice, 8–9 months old, were tested because the phenotype is reported to be more evident with age, both in patients (Sinnema et al. 2011) and in mice with a deletion of Snord116 (Ding et al. 2005, 2008, 2010). All animal procedures were approved by the ethical national committee in Italy, for IIT Genova. The study followed ARRIVE guidelines (http://www.nc3rs.org.uk/arrive-guidelines).
Genotyping
PCR analyses of genomic DNA from tail biopsies were performed. Deletion of PWScr resulted in a PCR product of 300 bp, which was absent in the WT genotype. To genotype mice, PCR analysis of genomic DNA from tail biopsies were performed using the primer pair PWScrF1PWScrR2 5′-AGAATCGCTTGAACCCAGGA and 5′-GAGAAGCCCTGTAACATGTCA, respectively PCR cycling conditions were as follows: 94° for 2 min, 35 cycles of 94° for 30 sec, 55° for 30 sec, 72° for 30 sec, followed by a final extension of 72° for 9 min.
Gene expression analysis by quantitative real-time PCR
PWScrm+/p+ and PWScrm+/p− littermates (all males) were killed with CO2 followed by decapitation. Brain sections and liver, as negative controls, were dissected and snap-frozen in liquid nitrogen. Total RNA was extracted using Qiazol (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. RNA samples were quantified with an ND1000 Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). Reverse transcription of 1–0.1 g of RNA was performed using ImProm-II(TM) Reverse Transcriptase (Promega, Milan, Italy) according to the manufacturer’s instructions. RT-qPCR was performed on a Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) using QuantiFast SYBR Green PCR Kit (Qiagen, Hilden, Germany) and following these conditions: 5 min at 95°, 40 cycles of denaturation at 95° for 10 sec, an annealing step at 60° for 30 sec, and extension step at 70° for 1 min. Each sample was run to obtain average Ct values according to the manufacturer’s specifications. The primers used were the following:
SNORD116 Forward TGGATCTATGATGATTCCCAG
SNORD116 Reverse TGGACCTCAGTTCCG ATGAG
Gapdh Forward GAACATCATCCCTGCATCCA
Gapdh Reverse CCAGTGAGCTTCCCGTTCA
Samples were normalized against the housekeeping gene Gapdh. Expression levels relative to this housekeeping gene was determined by the calculation of . The data were expressed as 2−ΔΔct where is the difference of each sample compared with WT-FC levels.
Working-for-food cognitive tasks
Adult mice were tested in a home-cage system that we have recently described (Maggi et al. 2014). Each cage was equipped with a COWE (Cognition and Welfare, TSE Systems, Germany). A COWE is a 3-hopper operant wall enabled to detect nose-poking activity via infrared beams, and contains LEDs for luminous stimulation. The COWE was remotely controlled via a computer (TSE Systems-OBS software). All mice were housed under a 12:12 light:dark cycle with lights on from 7 am until 7 pm. Food pellets were available ad libitum, or restricted to particular time windows according to the protocol (see below). All food pellets were delivered when the mouse self-initiated the trial according to the experimental protocol. Water was available ad libitum.
Restricted food protocol
Cohorts of 6 PWScrm+/p+, 6 PWScrm+/p−, and 5 PWScrm+/p− male mice were tested for food entrainment by making pellets available only for 6 hr (from 12 am to 6 pm) in constant dark conditions. Mice had to nose poke in the central hopper, and then in the lateral one, to obtain a food pellet. As mice learned the time of the day when food was available, they exhibited FAA. We have assessed FAA by calculating nose poking activity in mice throughout the 18 hr when food was unavailable. Nose poking activity was normalized to the maximum value of the poking activity during the 18 hr, collapsed into 15-min bins, when food was unavailable. In addition, we calculated the number of nose pokes and food intake in the time window when food was available.
Changing point analysis
Time series of nose poking activity were extracted by collapsing to time bins of 6 min, then normalizing to the maximum value, and averaging over 24 hr. For each subject, a changing point detection was performed following the bootstrap procedure described in Taylor (2000) excluding hours of food availability (from 12 am to 6 pm). Briefly, to avoid outliers, we analyzed ranked values instead of the actual activity. Because each time series X = consists of N = 180 time points, we have given a rank of N to the highest value of the series, N − 1 to the second largest and so on. Let us call the series of ranks R = . From this series, we computed a CUMSUM chart, defined as:
| (1) |
Where is the average value of the series R.
The CUMSUM chart increases if the time series is above its own average for a prolonged period, and decreases if the time series is below its average. An estimator of change could be:
| (2) |
where , and where .
We implemented a bootstrap procedure to test the hypothesis of no changes as follows:
Generate a sample of N values, = .
Compute the CUMSUM chart from this series, and extract the estimator.
Repeat points 1 and 2 a number of times (we repeated it times) to obtain an empirical distribution of the estimator under the hypothesis that no change in the series occurred.
If the estimator from the original CUMSUM chart, , exceeds the 95% percentile of the bootstrap sample distribution, we reject the hypothesis that no change has occurred.
If a change is detected, an estimator of the time where the change has occurred can be obtained by minimizing the mean square error:
| (3) |
where and .
To find multiple changing points, we split the time series in two, and , and repeat the procedure for the series. We split the data and repeated the procedure until significant changes were detected. Once all changing points were extracted, we estimated the 5th and 95th percentile of the distribution of time series in each of the intervals between consecutive changing points. All the procedures were implemented in Python, using the packages scipy, numpy, and matplotlib.
Timing tasks
Pretraining:
We subjected mice to cognitive tasks that involved time perception in the seconds-to-minutes range. All tasks were preceded by a 3-day pretraining phase, in which mice were trained to associate a light signal in the hopper with the delivery of a dustless precision pellet (20 mg, BioServ). During the pretraining, the animals obtained food from the feeder by initiating a trial with a nose poke in the central hopper, thereby triggering 2 sec of illumination. When the light was switched off, the mouse could retrieve the food pellet by poking its nose into one of the lateral hoppers within 30 sec. The inter trial interval (ITI) began after the release of the pellet, or at the end of the time limit; it lasted 30 sec plus a random interval based on a geometric distribution with a mean of 60 sec.
Fixed interval (FI)–peak interval (PI) procedure:
Six PWScrm+/p+, six PWScrm+/p−, and five PWScrm−/p− male mice were tested in two FI–PI schedules at 10 and 30-sec fixed intervals. For this procedure, one of the lateral hoppers was blocked. Mice could initiate the trial by nose-poking in the central hopper, which activates a LED for 2 sec. Then, nose-poking in the lateral hopper no sooner than 10 sec (T = 10 sec), and no later than 30 sec (T-off = 30 sec) after lights went off, caused the delivery of the pellet. For the training schedule on FI = 30 sec, we used T = 30 sec, and T-off = 60 sec; 20% of unreinforced probe trials (Lp) were randomly intermixed in the PI procedure. The 10-sec training was conducted for 10 days after pretraining, and the 30-sec training for the following 10 days.
Switch test:
A different cohort of six PWScrm+/p+, six PWScrm+/p−, and five PWScrm−/p− inbred male mice was subjected to the Switch test as described in (Balci et al. 2009). In this task, the mouse, after initiating the trial, had to make a decision within 30 sec after the light switched off. It had to decide whether to nose-poke in the left hopper, associated with short light signals (3 sec), or in the right hopper, associated with long light signals (9 sec), to receive a pellet. The probabilities for the short (PS) and long (PL) trials were equal (PS = PL). When the animal thought that the short-latency had elapsed, it decided to leave the short location and moved (switched) to the long location. The reward was obtained only if the first nose poke of the animal occurred at the correct location. Indeed, if it switched too early or too late, the animal received no payoff. Mice were subjected to the switch test for 10 days after the pretraining.
Food intake and nose poking analyses:
The analyses of the number of pellets eaten were performed while performing the FI–PI procedure. Mice were weighed systematically at the same time of day.
We counted the pellets eaten by each animal during 24 hr. In both cases, we normalized food intake to body weight 0.75. We calculated the average food intake for the PWScrm+/p− and PWScrm−/p− mice and littermate controls.
For the FI–PI procedure, the normalized distribution of the STARTs and STOPs of the nose pokes in probe trials was computed as in Church et al. (1994). The first 2 days of testing were not considered, so that only established behavior was evaluated. We obtained individual raster plots. By averaging all trials, we obtained a peak curve that is distributed around the target time for reward. We compared the average of all subjective peak curves, for each genotype, for each FI–PI schedule. In the Switch task, the latency to switch, and accuracy, were analyzed only for long trials. As in the previous test, the first 2 days were not included in the analysis. The distributions of switch latencies were fitted with a mixed Gaussian function as described in Maggi et al. (2014). All parameters were estimated using the maximum likelihood estimation method. We represented the empirical distribution of the switch latency values, and estimated the optimal target-switch latency according to the subjective timing uncertainty (Balci et al. 2009). The latency to switch from the short to the long location in time was used as a predictor of their temporal strategy (conceptualized as an accuracy measure), while timing uncertainty was estimated from the coefficient of variation (CV) of the cumulative distribution functions (CDFs) of switch latencies. The error rate was assessed by indexing it per hour, defined as , where is the number of incorrect trials (hour ), and is the number of total trials per hour. All parameters were analyzed comparing light and dark phases.
Open field test:
We assessed locomotor and anxiety-like behavior within an open field arena. Animals were tested in a square gray arena, 44 cm 44 cm, and the central area was 8 cm 8 cm. We placed each mouse in the periphery of the arena, and allowed the animal to explore the apparatus freely for 15 min. The apparatus was cleaned between animals. Mice tend to explore the entire arena over the trial, which allows for assessment of their general locomotor activity. Additionally, mice display thigmotaxis, i.e., they remain close to the wall and prefer to explore the periphery rather than the center of the arena, which is the most anxiogenic part. Using a video-tracking system, we recorded the amount of time spent in different areas, the distance traveled, and the speed of movement, which are measures that can be used as indicators of both motor activity and anxiety.
Rotarod:
We tested the mice with a rotarod (TSE, Homburg, Germany) to assess their motor coordination and balance, as previously described (Mandillo et al. 2008). Mice were placed on a rotating rod, and we recorded the latency to fall for each animal. We first ran a training session in which the rod was rotating at the constant speed of 4 rpm. All mice were able to keep their balance at 4 rpm for at least 60 sec, and were then tested on an accelerating rod (i.e., from 4 to 40 rpm for 300 sec). The mice were tested in the accelerating mode three consecutive times. The intertrial interval lasted at least 15 min. The latency to fall from the rod was determined automatically, but, in the event of a passive rotation, i.e., when a mouse held on to the rod while rotating, the timer was stopped manually. A maximum of three mice were tested at once.
Statistical analyses:
We used unpaired two-tailed t-tests for comparison of the circadian period and the changing point between genotypes. We used a two-way mixed ANOVA (Genotype Days) for the food intake of the ad libitum experiment, and two-way mixed ANOVAs (Genotype Time) to analyze food intake, NP activity, error rate, and switch latency.
The statistical tests were performed with MATLAB, Python, and GraphPad Prim 5.0. For all tests, P-values were considered significant to different levels: *, **,and ***
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Snord116 is imprinted in adult time-keeping brain tissues
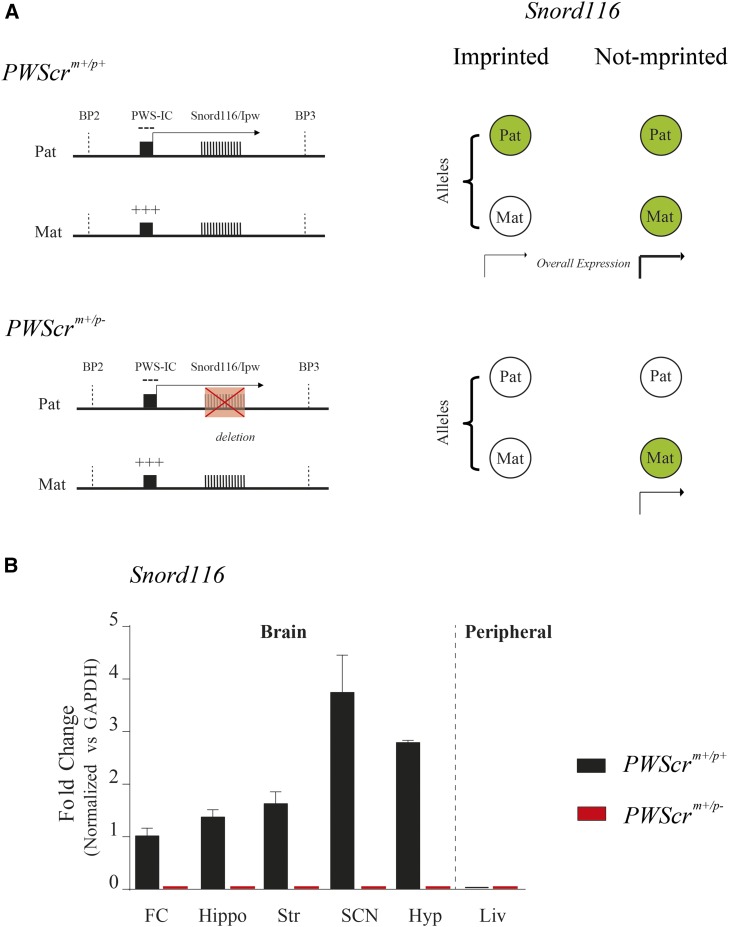
To study whether Snord116 is subject to imprinting control in adult brain tissues, we quantified its level of transcription in a series of brain tissues (frontal cortex, hippocampus, striatum, SCN, and hypothalamus) in adult heterozygous mice carrying the paternally inherited allele. The model of imprinted expression in this case assumes that heterozygous mutants result in a homozygous suppression because the maternal allele is already silenced (Figure 1A). Remarkably, in all brain tissues we examined, we observed a total absence of Snord116 in PWScrm+/p− heterozygous mutant mice; in contrast, Snord116 was generally expressed in PWScrm+/p+ wild-type animals (Figure 1B). This result indicates that the maternal allele in mutant adults is silenced, hence imprinted. Interestingly, our results imply that Snord116 is monoallelically expressed in the wild-type suprachiasmatic nucleus (SCN) of the hypothalamus, the hypothalamus, and the striatum, which are important regulatory centers for the organization of the long-interval circadian clock, food anticipatory behaviors (at hourly scale), and short-interval (seconds-to-minutes) behavioral responses (Merchant et al. 2013), respectively. The lack of expression of Snord116 in both wild-type and mutant mice in the liver is in agreement with the report that this snoRNA is expressed only in the brain in mice (Skryabin et al. 2007).
Figure 1.
Snord116 imprinting in adult brain tissues. (A) Representation (not to scale) of the mouse Snord116 region at chromosome 7 between two breakpoints (BP2 and BP3). The PWScrm+/p+ wild-type (upper panel) and the PWScrm+/p− mutant (lower panel) regions are shown. The expression of the imprinted genes in this region is controlled by the PWS imprinting center (PWS-IC), which includes a CpG island that is differentially methylated. While the maternal (Mat) allele is methylated (+++), then repressed, the paternal (Pat) allele is unmethylated (−), then expressed. The red crossed box, on the PWScrm+/p− paternal mutant allele, represents the deleted region. The right panel summarizes a model used to distinguish whether Snord116 expression depends on an imprinted or unimprinted control. The allelic combination for both conditions is represented as green filled circles (expressed) or empty circles (not expressed). Briefly, an imprinted regulation is signified by a condition in which a wild-type allele expresses, while mutants do not express at all. (B) Relative Snord116 expression analysis by RT-PCR in PWScrm+/p+ and PWScrm+/p− in mouse brain tissues. Graphs are presented as the meansSEM (n = 3). The results are normalized vs. GAPDH mRNA levels, and compared to wild type FC Snord116 expression. Liver was used as a negative control. FC, frontal cortex; Hippo, hippocampus; Str, striatum; Hyp, hypothalamus; SNC, suprachiasmatic nucleus of the hypothalamus; Liv, liver.
Working-for-food: PWScrm+/p− and PWScrm−/p− mutants show delayed food anticipatory activity compared to controls
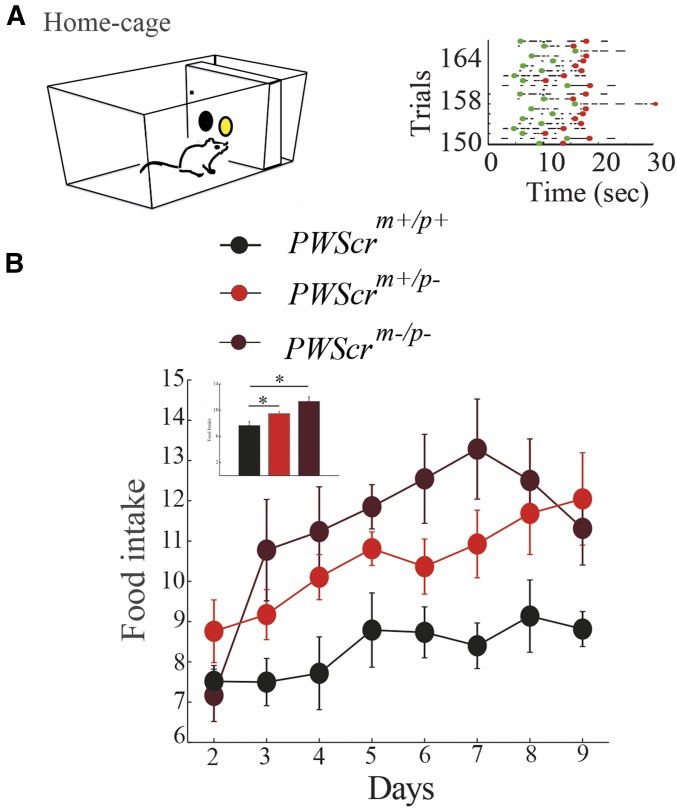
All behavioral tests were conducted in automated home-cages as previously described (Maggi et al. 2014). Mice self-initiated each trial during the 24-hr cycle by nose-poking in ad hoc operant walls mounted in the cage (see Materials and Methods). This experimental setup allowed us to continuously monitor both the behavioral activity (i.e., nose-poking) and the amount of food pellets eaten (food intake) during the day (Figure 2A). The overall food intake of PWScrm+/p− and PWScrm−/p− mutants was significantly higher than that of wild-type littermates (Figure 2B). This hyperphagic behavior echoes the severe overeating phenotype of PWS patients, confirming that the imprinting defect in mice is associated with an increase in food intake sought.
Figure 2.
Food intake in PWScrm+/p−, PWScrm+/p− and PWScrm−/p− mutants. (A) Representation of the working-for-food behavior in automated home cages. (B) Food intake of PWScrm+/p+ (n = 6), PWScrm+/p− (n = 6), and PWScrm−/p− (n = 5) was defined as cumulative food intake across days of the experiment. The means ± SEM are shown. Significant differences are indicated as follows:
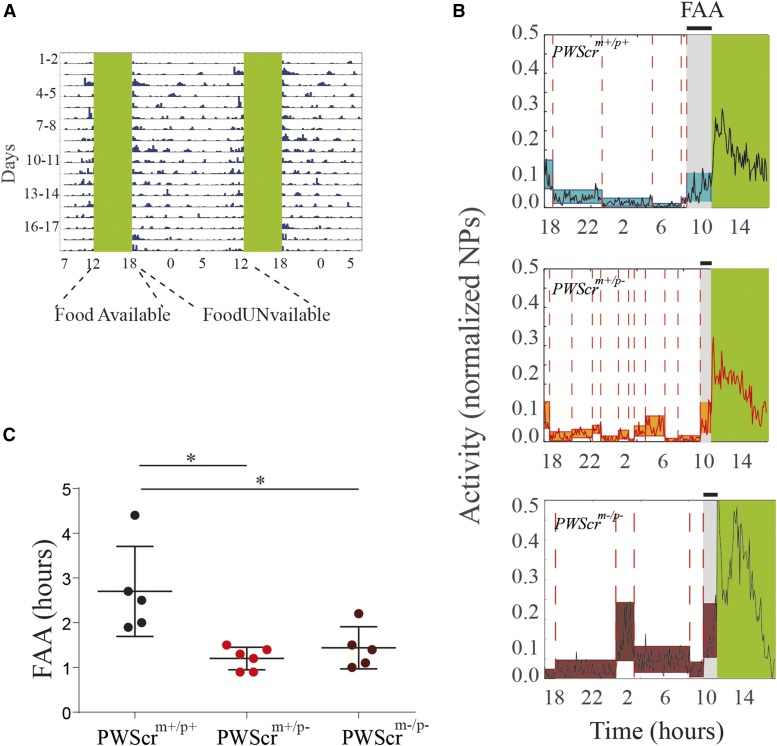
Food intake is strongly modulated by the endogenous circadian clock; it is entrained by both light-entrainment and food-entrainment oscillators (Mistlberger 2011). In a different experiment, we subjected all mice to a timed food schedule in constant darkness (see Materials and Methods and Figure 3A). In the absence of light cues, animals relied only on food timing to entrain their biological clock, and to develop FAA before each meal. Using a changing point algorithm (see Materials and Methods and Figure 3B), we identified the time at which each individual mouse changed its activity to anticipate the meal, and we observed significant differences in the development of FAA according to the genotype. In Figure 3C, we show the time when mice start their FAA before the meal. PWScrm+/p− and PWScrm−/p− mutants significantly postpone their anticipation compared to controls.
Figure 3.
FAA in PWScrm+/p+, PWScrm+/p−, and PWScrm+/p− mutants. (A) Representative double-plotted actograms for nose poke (NP) activity in the food restriction protocol in constant darkness. Mouse activity is collapsed into 15-min time bins, and is represented by blue bars over the 18 hr of food unavailability. Green regions indicate the time when mice had food available. (B) Examples (i.e., a PWScrm+/p+, a PWScrm+/p−, and a PWScrm−/p− mouse) of nose poking and changing points in FAA. We plotted the nose-poke activity time trace as a black continuous line. The colored rectangles represent the percentile of each time trace split, and the changing points are marked by a dashed red line. The gray area represents the FAA time interval. (C) Boxplots of the mean latency time to FAA are plotted for PWScrm+/p+ (n = 6), PWScrm+/p− (n = 6), and PWScrm−/p− (n = 5); preceding food availability, expressed as number of hours as identified by the changing point algorithm. Graphs are presented as the mean SEM. Significant differences are indicated as follows:
To check the general motor activity of the mutants compared to wild types, we subjected cohorts of mutants (PWScrm−/p− and PWScrm−/p− mice) and littermate controls to a classical “open field test” (Mandillo et al. 2008), and the “rotarod test” (Mandillo et al. 2008). We observed no differences between mutants and controls in either tests (Supplemental Material, Figure S1).
Timing: PWScrm+/p− and PWScrm−/p− mice start later and stop earlier
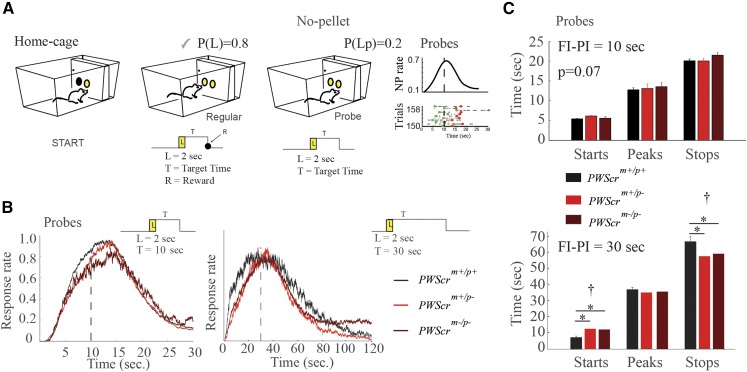
The observation of imprinted gene regulation in a brain area (i.e., the striatum) that is central for short-interval timing mechanisms, along with a defect in FAA at hour scale, motivated us to study seconds-to-minutes timing behaviors in mutants. Cohorts of adult mice were trained in automated home-cages to perform a task called the “peak procedure task” (see Materials and Methods and Figure 4A). Briefly, the task measures the ability of the animal to perceive the duration of a short-interval before obtaining a reward. Mice were trained either with a 10-sec fixed schedule, or with a longer 30-sec fixed schedule to test the scale-invariance of this timing behavior across different temporal intervals (Maggi et al. 2014). Our experimental setup included a small percentage (20%) of trials that went unrewarded (probes) to observe the distribution of behavioral responses around the target (expected) time.
Figure 4.
Interval timing in PWScrm+/p+, PWScrm+/p−, and PWScrm−/p− mutants (A) Cartoon of the PI task. The mouse can initiate the trial with a nose poke (NP) in the central hopper (light on for 2 sec) and, to obtain a food pellet, it had to NP in the lateral hopper, after lights off, no sooner than 10 or 30 sec ( sec or sec), and no later than 30 or 60 sec, according to the experimental paradigm (see Materials and Methods). Regular and probe trials are shown. The right panel also shows a representative raster plot, and the distribution of start and stop times of NPs in probe trials. (B) Distribution of the response (NP) rate in probe trials ( sec on the left panel, and sec on the right panel). (C) STARTs and STOPs for the PWScrm+/p+ (n = 6), PWScrm+/p− (n = 6), and PWScrm−/p− (n = 5) are shown for the two 10-sec and 30-sec FI–PI schedules. Graphs are presented as the mean SEM. The cross notation mark above the asterisks in the bars represent the one-way ANOVA between groups, while the asterisks represent post hoc significance () corrected by Holm’s correction.
From the distribution of the responses during probes, we observed that the onset of activity in PWScrm+/p− and PWScrm−/p− mice occurs later, although the peak response at the target time is similar to wild-type mice (Figure 4B). This delay to the target time was observed both in the 10 and 30-sec protocols in both heterozygous and homozygous mutants compared to wild-type littermate control animals (Figure 4B). Furthermore, in the 30-sec delay, the postpeak response also changes in mutants. In particular, an accurate analysis of the STARTs and STOPs (Maggi et al. 2014) of the behavioral activity demonstrates that PWScrm+/p− and PWScrm−/p− mice start later, and stop earlier, compared to control mice (Figure 4C). Genotype-specific differences at 10 and 30-sec interval timing suggest that scale invariance of timing is maintained in mutants.
Moreover, we compared the slope of the response rate before the target time in normally rewarded trials during the first hour of food availability, when animals were hungry, with the rate observed during the last hour of food availability, when animals were fed. We found no differences in timing between the two time windows (Figure S2). The way the latter assessment was designed excludes the possibility that changes in timing occur as an effect of satiety, and this result reinforces the idea that the change in timing is due to genotype differences.
Decision making: PWScrm+/p− and PWScrm−/p− mice show altered risk assessment
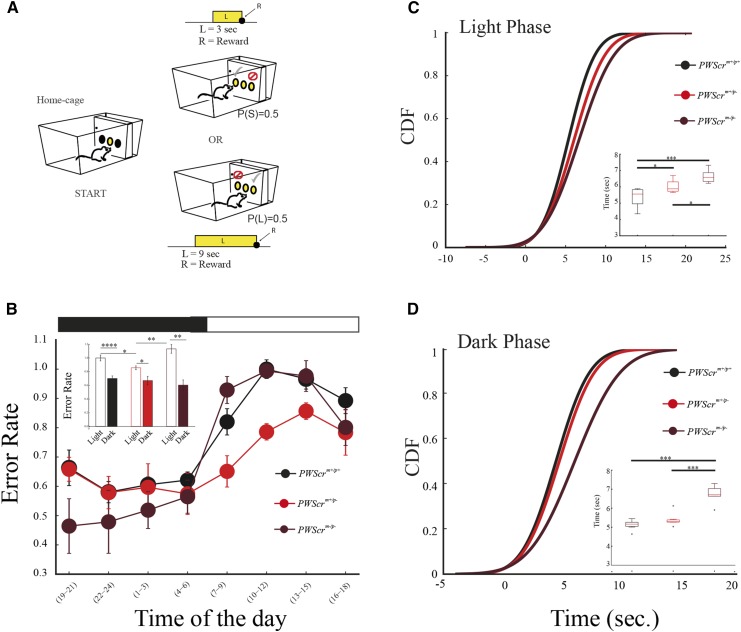
Timing is a fundamental cognitive function useful in circumstances in which a payoff depends on our promptness in making a decision (Maggi et al. 2014). Moreover, the precision in timing an event influences the assessment of the risk we are willing to take in delaying or anticipating a response. Because the PWS heterozygous and homozygous mutants displayed peculiar behavior in delaying the mode of responding around a target time, we wanted to explore whether this timing trait has consequences in risk assessment in mice by testing their temporal decision-making capabilities. We subjected different cohorts of mutant and wild-type littermate control mice to a behavioral test called the “switch task” (Balci et al. 2009). In this task, mice learned to discriminate between a short (3 sec) and a long (9 sec) light signal (Figure 5A). Each light signal was associated with a location in the cage in which to obtain a reward pellet if the response after the end of the signal was correct. Therefore, the obvious behavior for an animal, as soon as the trial started, was to move toward the location associated with the short-signal, but to change (switch) location when the signal was perceived as long.
Figure 5.
Switch task in PWScrm+/p+, PWScrm+/p−, and PWScrm+/p− mutants (A) Cartoon of the switch task. From left to right: a mouse nose pokes in the central hopper and activates a light signal. The mouse has to NP in the left hopper after 3 sec (short trial) or switch to the right hopper, and NP after 9 sec (long trial) according to the duration of light on no later than 30 sec after light off. L, light; P(S), probability of short trial; P(L), probability of long trial. (B) The error rate in the switch test, according to genotype, in the light and dark phases. Inset, histograms of total average errors for all groups of mice in light vs. dark. (C) The cumulative distribution function (CDF) of the switch latency values for PWScrm+/p+ (n = 6), PWScrm+/p− (n = 6), and PWScrm−/p− (n = 5) in dark and light phases. Inset, boxplot of the mean latencies for all genotypes during dark and light phases. All graphs are presented as the means SEM. Significant differences are indicated as follows:
When we assessed the general performance of all mice in the test, we observed that PWScrm+/p− heterozygous mice had a lower error rate in the light phase compared to control and homozygous mutants (Figure 5B). Moreover, during the light phase, both heterozygous and homozygous mutants switch locations later than control animals (Figure 5C), while only PWScrm−/p− homozygous mutants showed a significant delay during the dark phase (Figure 5D).
Discussion
In this study, we report a significant role of the Snord116 imprinted gene in working-for-food behaviors. We describe how an imprinted regulation of Snord116 normally occurs in specific brain sites in mice, reinforcing and refining previous evidence of imprinted expression throughout the entire brain (Cavaillé et al. 2000). The specific brain areas we investigated in our study are involved in the organization of many behaviors, including timing behaviors.
The analysis of the behavioral responses of all animals to the probes allowed us to conclude that the time perception of mutant mice (i.e., the peak of the response rate) was comparable to that of control animals. Interestingly, although mutants presented intact time perception, they use different strategies to time their behaviors. For example, when anticipating the event, both heterozygous and homozygous mutants start responding later than control animals, and, when terminating the behavior, mutants stop earlier.
The temporal switching behavior of mice during the decision-making task also suggests that, in assessing the risk of changing location within the cage, mutants used a different strategy than controls. Mutants enhance their performance by delaying their switching from one location to the other, and this behavior is evident in both mutants during the light phase, resulting in a reduction of the error rate in heterozygous mutants. In addition, the delay in switching is even longer in the homozygous mutants during the dark phase, also resulting in error rate reduction in this phase for homozygous mutants. Some differences emerged between heterozygous and homozygous mutants, suggesting that additional factors associated with the deletion might intervene in the control of specific measures. Assessing this behavior of mutant animals for optimality, we can argue that mutants have developed an optimal strategy to gather more food, regardless of whether this behavior is due to calorie need or the hedonic value of food. In our previous work in mice (Lassi et al. 2012), we reported that the lack of imprinted regulation (e.g. in the Ex1a mutant line) causes a reduction of endogenous uncertainty, and a reduction in the error rate of timed behaviors, hence an improvement in performance. Together, the results from our works advocate for an imprinting control of endogenous uncertainty in temporal behaviors. Closely constrained uncertainty is not a favorable trait for the development of many behavioral responses. For example, it might not allow for the generalizing of a learning process across different situations (e.g., responding to a dangerous signal when the environment changes). Indeed, genomic imprinting defects that improve timing performance result in defects in fear responses in mice (Lassi et al. 2012).
Generally, mice have the highest occurrence of sleep episodes during the light phase. The change in performance (i.e., error rate) between light and dark phases in all mice confirms our previous observation that the 24-hr timing performance in mice varies along light-dark cycles, and is modulated by a sleep inertia-like effect (Maggi et al. 2014). Tentatively, our behavioral results, and the expression pattern of Snord116 in brain regulatory centers for timing, suggest a direct role for this gene in timing; however, our study cannot conclude such a direct effect. Indeed, the overall behavioral differences we observed in mutants throughout the entire 24 hr of every day may not necessarily be due to a central regulation of this specific behavior. The same differences may also result from different physiological (e.g., sleep) and metabolic (e.g., caloric need, thermoregulation) traits that characterize these mutants and thereby influence cognitive performance.
The evolution of the central nervous system in many species has prioritized, through either direct or indirect adaptive mechanisms, the selection of behavioral traits that allow mice to process temporal information within a continuously changing environment (Tucci 2012). Timing behaviors occur at various timescales, but whether these processes are regulated by similar biological mechanisms is still unclear. Current investigations in seconds-to-minutes (interval timing) cognitive processes are focused on deciphering the properties of neuronal activity, and/or to map the intricate circuits that control this fundamental behavior (Buhusi and Meck 2005). Moreover, previous research has been conducted to identify direct or indirect effects on short-interval timing processes by studying timing in gene (Agostino et al. 2013; Yin and Meck 2014) and disease (Balci et al. 2009; Poryazova et al. 2013; Jones and Jahanshahi 2014; Tucci et al. 2014a) models. Our study introduces a new gene as a player in the behavioral biology of timing.
Genomic imprinting has been shown to play important roles in cognitive functions (Isles and Wilkinson 2000; Davies et al. 2007), so it is not surprising that new roles for imprinted genes in important behavioral functions continue to be revealed. Importantly, imprinted genes play central roles during prenatal brain development—a period that sets the basis for the development of the cognitive system. Therefore, a role of particular imprinted genes in the regulation of fundamental aspects of working-for-food behaviors is to be expected, particularly in the case of PWS, in which the hyperphagia may trigger the reorganization of behavioral food-seeking strategies.
In our study, the overall performance of heterozygous and homozygous mutant animals, compared to their wild-type littermate controls, demonstrated that mutants maximize the allocation of resources; they increased their food intake, enhancing their performance in working-for-food activity. The study of the FAA in mutants reinforced the conclusion that a particular delayed behavioral strategy is used by mice to time events. Short-interval timing behaviors and food anticipatory activity are ancestral evolutionary mechanisms that allow the predicting of meals, and play an important role in learning, memory and circadian biology. Interestingly, these two time-scale disparate behaviors share similarities. Indeed, it has been argued that mice, like humans, learn to time seconds-to-minutes and daily meals according to similar conditioning (Pavlovian) rules (Balsam et al. 2009; Balsam and Gallistel 2009).
Acknowledgments
We thank Jürgen Brosius and Boris Skryabin for making the PWScrm+/p− mice available. We thank Jo Peters for help and discussions on genomic imprinting.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.192286/-/DC1.
Communicating editor: M. R. Terry
Literature Cited
- Agostino P., Cheng R.-K., Williams C., West A., Meck W., 2013. Acquisition of response thresholds for timed performance is regulated by a calcium-responsive transcription factor, carf. Genes Brain Behav. 12: 633–644. [DOI] [PubMed] [Google Scholar]
- Balci F., Day M., Rooney A., Brunner D., 2009. Disrupted temporal control in the r6/2 mouse model of Huntington’s disease. Behav. Neurosci. 123: 1353. [DOI] [PubMed] [Google Scholar]
- Balsam P. D., Gallistel C. R., 2009. Temporal maps and informativeness in associative learning. Trends Neurosci. 32: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsam P., Sanchez-Castillo H., Taylor K., Van Volkinburg H., Ward R. D., 2009. Timing and anticipation: conceptual and methodological approaches. Eur. J. Neurosci. 30: 1749–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhusi C. V., Meck W. H., 2005. What makes us tick? Functional and neural mechanisms of interval timing. Nat. Rev. Neurosci. 6: 755–765. [DOI] [PubMed] [Google Scholar]
- Cavaillé J., Buiting K., Kiefmann M., Lalande M., Brannan C. I., et al. , 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 97: 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challet E., Mendoza J., 2010. Metabolic and reward feeding synchronises the rhythmic brain. Cell Tissue Res. 341: 1–11. [DOI] [PubMed] [Google Scholar]
- Chen J., Papies E. K., Barsalou L. W., 2016. A core eating network and its modulations underlie diverse eating phenomena. Brain Cogn. DOI:10.1016/j.bandc.2016.04.004. [DOI] [PubMed] [Google Scholar]
- Church R. M., Meck W. H., Gibbon J., 1994. Application of scalar timing theory to individual trials. J. Exp. Psychol. Anim. Behav. Process. 20: 135. [DOI] [PubMed] [Google Scholar]
- Davies J. R., Humby T., Dwyer D. M., Garfield A. S., Furby H., et al. , 2015. Calorie seeking, but not hedonic response, contributes to hyperphagia in a mouse model for Prader-Willi syndrome. Eur. J. Neurosci. 42: 2105–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W., Isles A. R., Humby T., Wilkinson L. S., 2007. What are imprinted genes doing in the brain? Epigenetics 2: 201–206. [DOI] [PubMed] [Google Scholar]
- Ding F., Prints Y., Dhar M. S., Johnson D. K., Garnacho-Montero C., et al. , 2005. Lack of Pwcr1/mbii-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm. Genome 16: 424–431. [DOI] [PubMed] [Google Scholar]
- Ding F., Li H. H., Zhang S., Solomon N. M., Camper S. A., et al. , 2008. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS One 3: e1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F., Li H. H., Li J., Myers R. M., Francke U., 2010. Neonatal maternal deprivation response and developmental changes in gene expression revealed by hypothalamic gene expression profiling in mice. PLoS One 5: e9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florant G. L., Healy J. E., 2012. The regulation of food intake in mammalian hibernators: a review. J. Comp. Physiol. B 182: 451–467. [DOI] [PubMed] [Google Scholar]
- Higgs S., 2016. Cognitive processing of food rewards. Appetite 104: 10–17. [DOI] [PubMed] [Google Scholar]
- Isles A. R., Wilkinson L. S., 2000. Imprinted genes, cognition and behaviour. Trends Cogn. Sci. 4: 309–318. [DOI] [PubMed] [Google Scholar]
- Jones C. R., Jahanshahi M., 2014. Motor and perceptual timing in Parkinson’s disease, pp. 265–290 in Neurobiology of Interval Timing, edited by Merchant H., de. Lafuente V. Springer, New York. [DOI] [PubMed] [Google Scholar]
- Lassi G., Ball S. T., Maggi S., Colonna G., Nieus T., et al. , 2012. Loss of Gnas imprinting differentially affects REM/NREM sleep and cognition in mice. PLoS Genet. 8: e1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassi G., Priano L., Maggi S., Garcia-Garcia C., Balzani E., et al. , 2016. Deletion of the Snord116/Snord116 alters sleep in mice and patients with Prader-Willi syndrome. Sleep 9: 637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi S., Garbugino L., Heise I., Nieus T., Balcı F., et al. , 2014. A cross-laboratory investigation of timing endophenotypes in mouse behavior. Timing & Time Perception 2: 35–50. [Google Scholar]
- Mandillo S., Tucci V., Hölter S. M., Meziane H., Al Banchaabouchi M., et al. , 2008. Reliability, robustness, and reproducibility in mouse behavioral phenotyping: a cross-laboratory study. Physiol. Genomics 34: 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant H., Harrington D. L., Meck W. H., 2013. Neural basis of the perception and estimation of time. Annu. Rev. Neurosci. 36: 313–336. [DOI] [PubMed] [Google Scholar]
- Mistlberger R. E., 2011. Neurobiology of food anticipatory circadian rhythms. Physiol. Behav. 104: 535–545. [DOI] [PubMed] [Google Scholar]
- Overton J., Williams T., 2004. Behavioral and physiologic responses to caloric restriction in mice. Physiol. Behav. 81: 749–754. [DOI] [PubMed] [Google Scholar]
- Peters J., 2014. The role of genomic imprinting in biology and disease: an expanding view. Nat. Rev. Genet. 15: 517–530. [DOI] [PubMed] [Google Scholar]
- Poryazova R., Mensen A., Bislimi F., Huegli G., Baumann C. R., et al. , 2013. Time perception in narcolepsy in comparison to patients with Parkinson’s disease and healthy controls—an exploratory study. J. Sleep Res. 22: 625–633. [DOI] [PubMed] [Google Scholar]
- Qi Y., Purtell L., Fu M., Lee N. J., Aepler J., et al. , 2016. Snord116 is critical in the regulation of food intake and body weight. Sci. Rep. 6: 18614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky T. S., Robeck T., Galiveti C. R., Raabe C. A., Seeger B., et al. , 2016. Maternal transcription of non-protein coding RNAs from the PWS-critical region rescues growth retardation in mice. Sci. Rep. 6: 20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. H., 2014. The energy allocation function of sleep: a unifying theory of sleep, torpor, and continuous wakefulness. Neurosci. Biobehav. Rev. 47: 122–153. [DOI] [PubMed] [Google Scholar]
- Sinnema M., Einfeld S. L., Schrander-Stumpel C. T., Maaskant M. A., Boer H., et al. , 2011. Behavioral phenotype in adults with Prader-Willi syndrome. Res. Dev. Disabil. 32: 604–612. [DOI] [PubMed] [Google Scholar]
- Skryabin B. V., Gubar L. V., Seeger B., Pfeiffer J., Handel S., et al. , 2007. Deletion of the mbii-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 3: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, W. A., 2000 Change-point analysis: a powerful new tool for detecting changes. Available at: http://www. variation. com/cpa/tech/changepoint. html. Accessed June 2016.
- Tucci V., 2012. Sleep, circadian rhythms, and interval timing: evolutionary strategies to time information. Front. Integr. Neurosci. 5: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci V., 2016. Genomic imprinting: a new epigenetic perspective of sleep regulation. PLoS Genet. 12: e1006004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci V., Buhusi C. V., Gallistel R., Meck W. H., 2014a Towards an integrated understanding of the biology of timing. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20120470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucci V., Kleefstra T., Hardy A., Heise I., Maggi S., et al. , 2014b Dominant β-catenin mutations cause intellectual disability with recognizable syndromic features. J. Clin. Invest. 124: 1468–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J. B., Cheverud J. M., Roseman C., Hager R., 2008. Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet. 4: e1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin B., Meck W. H., 2014. Comparison of interval timing behaviour in mice following dorsal or ventral hippocampal lesions with mice having δ-opioid receptor gene deletion. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20120466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.