Abstract
Inferring and understanding changes in effective population size over time is a major challenge for population genetics. Here we investigate some theoretical properties of random-mating populations with varying size over time. In particular, we present an exact solution to compute the population size as a function of time, , based on distributions of coalescent times of samples of any size. This result reduces the problem of population size inference to a problem of estimating coalescent time distributions. To illustrate the analytic results, we design a heuristic method using a tree-inference algorithm and investigate simulated and empirical population-genetic data. We investigate the effects of a range of conditions associated with empirical data, for instance number of loci, sample size, mutation rate, and cryptic recombination. We show that our approach performs well with genomic data (≥ 10,000 loci) and that increasing the sample size from 2 to 10 greatly improves the inference of whereas further increase in sample size results in modest improvements, even under a scenario of exponential growth. We also investigate the impact of recombination and characterize the potential biases in inference of . The approach can handle large sample sizes and the computations are fast. We apply our method to human genomes from four populations and reconstruct population size profiles that are coherent with previous finds, including the Out-of-Africa bottleneck. Additionally, we uncover a potential difference in population size between African and non-African populations as early as 400 KYA. In summary, we provide an analytic relationship between distributions of coalescent times and , which can be incorporated into powerful approaches for inferring past population sizes from population-genomic data.
Keywords: effective population size, coalescent time, human evolution
NATURAL populations vary in size over time, sometimes drastically, like the bottleneck caused by the domestication of the dog (Lindblad-Toh et al. 2005) or the explosive growth of human populations in the past 2000 years (Cohen 1995). Inferring population size as a function of time has many applications, for instance, better understanding of major ecological or historical events’ impact on humans such as glacial periods (Lahr and Foley 2001; Palkopoulou et al. 2013), agricultural shifts or technological advances (Boserup 1981), and colonization of new areas (Ramachandran et al. 2005; Jakobsson et al. 2008). Knowledge about the demographic history is also important for studies of natural selection to avoid spurious finds (Nielsen 2005; Li et al. 2012).
Estimating past effective population size has gained considerable interest in recent years, in particular with the development of methods such as the Bayesian skyline plots implemented in BEAST (Drummond et al. 2012); see Ho and Shapiro (2011) for a review of this school of methods. More recently, methods based on the sequentially Markovian coalescent [SMC and its refined version SMC′ (McVean and Cardin 2005; Marjoram and Wall 2006)], such as PSMC (Pairwise Sequentially Markovian Coalescent, Li and Durbin 2011), MSMC (Multiple Sequential Markovian Coalescent, Schiffels and Durbin 2013), DiCal (Demographic Inference using Composite Approximate Likelihood, Sheehan et al. 2013), and Bayesian approaches (Palacios et al. 2015), have advanced our ability to infer past population sizes. The former type of methods can use relatively large sample sizes, but can handle only modest numbers of loci, and these methods have often been used for analyzing mitochondrial DNA. The latter group of methods can handle genome-wide data and explicitly model recombination, using a Markovian assumption for neighboring gene genealogies (McVean and Cardin 2005). PSMC works with a single (diploid) individual, which leads to simple underlying tree topologies without requiring phase information. However, the inference power is limited, in particular in the recent past, as most coalescences in a sample of size 2 are not expected to occur in that period (Li and Durbin 2011). MSMC and DiCal extend this approach by using information from multiple samples. MSMC focuses on the first coalescence event in the sample at each locus and ignores the remaining coalescence events. The algorithm can deal with genome-wide data in a computationally efficient way. DiCal, on the other hand, uses all coalescent events in the gene genealogies to provide estimates of the population size, assuming a Markov property between sites as well (Sheehan et al. 2013). The algorithm quickly becomes computationally intensive as the sample size increases and analyzing genome-wide data are challenging. Palacios et al. (2015) develop an interesting Bayesian nonparametric approach building on the SMC′ model and assuming known gene genealogies, which shows promising accuracy for inferring relatively simple past population sizes using a moderate number of loci.
There are two important steps for most of these types of approaches: the inference of the underlying gene genealogies and the inference of population size as a function of time from the inferred genealogies. In this article we introduce the Population Size Coalescent-times-based Estimator (Popsicle), an analytic method for solving the second part of the problem. We derive the relationship between the population size as a function of time, , and the coalescent time distributions by inverting the relationship of the coalescent time distributions and population size that was derived by Polanski et al. (2003), where they expressed the distribution of coalescent times as linear combinations of a family of functions that we describe below. The theoretical correspondence between the distributions of coalescent times and the population size over time implies a reduction of the full inference problem of population size from sequence data to an inference problem of inferring gene genealogies from sequence data. This result represents a theoretical advancement that can dramatically simplify the computation of for many existing and future approaches to infer past population sizes from empirical population-genetic data.
In this article, we first present the core theoretical result: the exact correspondence between the set of distributions of coalescent times for samples of any size and the population size as a function of time. We then provide an illustration of the performance of the theoretical result on simulated gene genealogies, including several assessments of how different factors (number of loci, sample size, presence of recombination) can affect the performance of our approach. Finally, we illustrate how our theoretical result could be used to estimate population size over time from sequence data (simulated and experimental). Since this latter part necessitates a method to infer gene genealogies from sequence data, we provide a simple algorithm to perform this particular task, based on the UPGMA (Unweighted Pair Group Method with Arithmetic Mean) algorithm and properties of the mutation process for the coalescent.
Model and Methods
Distributions of coalescent times and
Under the constant population size model, the waiting times between coalescent events are independent exponentially distributed random variables. In particular, the time during which there are exactly k lineages follows an exponential distribution with rate generations. When population size varies as a function of time (), the waiting times to coalescence are no longer independent of each other. Specifically, for , depends on all the previous coalescent times from to (see, e.g., Wakeley 2009 for an extensive description of the coalescent).
In this article, we derive a relationship between and the distributions of the cumulative coalescent times, which we denote by V. More specifically, for ,
| (1) |
The variables represent the sum of times from the present to each coalescent event. Because we use only the cumulative coalescent times and not the individual times , we refer to the times for as coalescent times, omitting the term cumulative for convenience. For example, the random variable represents the time to the most recent common ancestor (). All coalescent times are expressed in generations. We denote by the density function of .
Polanski et al. (2003) derived the density function of coalescent times under varying population size as linear combinations of a set of functions , where
| (2) |
Similar functions have previously been used in a context of varying population size (Griffiths and Tavare 1994). For , the relationship between the density function and is
| (3) |
with
| (4) |
We also define the integral of with respect to t as
| (5) |
From Equations 2 and 5 we can derive that
| (6) |
The principle of our method is to use the distributions of the coalescent times to get to the functions. In other words, we invert the result of Polanski et al. (2003).
Theorem. Given a sample of size n,
with
Corollary.
| (7) |
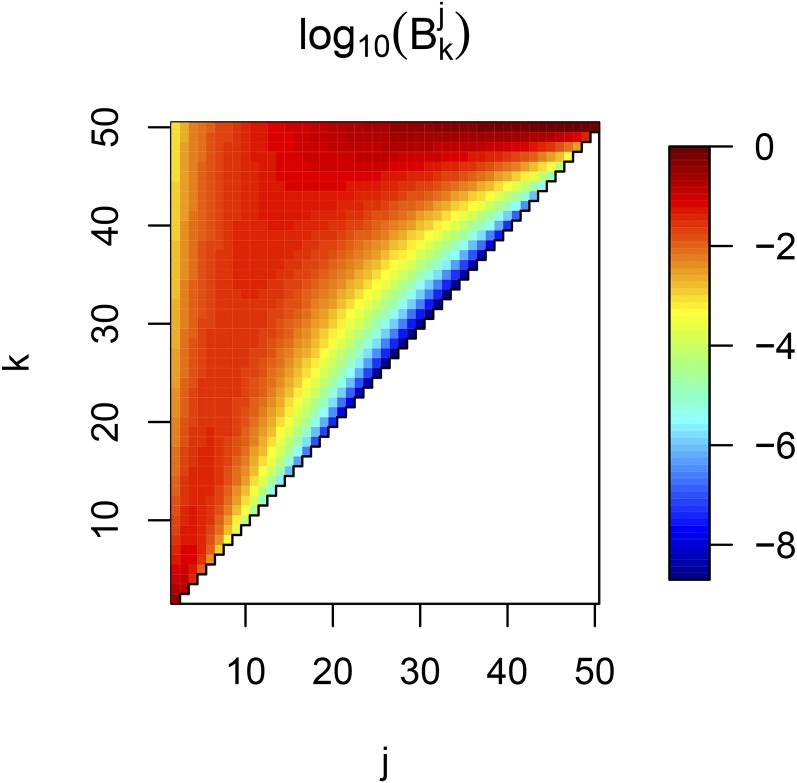
The proof of the Theorem is given in the Appendix. This Theorem implies that for any time t generations in the past, and can be obtained using the distributions of coalescent times. From each (and its integral ), the function can be obtained using Equation 6. In contrast to the coefficients (Equation 4) that can become very large as n increases and are of alternate signs (Polanski et al. 2003), the coefficients introduced in the Theorem are all positive and take values between 0 and 1 (Figure 1). Thus, our formula is not constrained by numerical limitations and can be used for very large sample sizes.
Figure 1.
Heatmap of the values of for , as function of k and j. The white area represents the region where
Finite number of observed gene genealogies: adaptation of the theorem to time intervals, the “Popsicle”
The Theorem states that the population size can be computed at any time in the past, provided that we know all the distributions of coalescent times for any time in the past. However, this knowledge would require us to observe the genealogies of an infinite number of independent loci evolving under the same N function over time. In practice, genomes are finite so we have access to only a finite number of loci to estimate the coalescent time distributions. We use empirical distribution functions to estimate the cumulative distribution functions of the coalescent times as these estimators have good statistical properties: They are unbiased and asymptotically consistent (Van der Vaart 2000).
Because of the finite number of loci, time is discretized into intervals and within each interval is estimated by its harmonic mean, as the harmonic mean of N has a simple relationship to the functions:
| (8) |
Definition (Popsicle). Given a sample of size n haploid individuals evolving in a random-mating population of variable size N over time and given a number j between 2 and n, we define the Popsicle of N over a time interval to be
with being the empirical distribution function of the cumulative coalescent time variable at time t.
In the rest of this article, we set (in Equations 7 and 8), as it incorporates information from all coalescent time distributions and performs well even for very recent times (see Supplemental Material, File S1, Figure S1, Figure S2, Figure S3, and Figure S4).
Quantifying the accuracy of the method
Let us consider a time discretization and define average relative difference (ARD) and average relative error (ARE) as
| (9) |
and
| (10) |
where is the estimate of the harmonic mean of N during the time interval as defined in Equation 8 with and replaced by its estimate , and is the value for the true harmonic mean of N for the corresponding interval.
Algorithm for inferring gene genealogies from polymorphism data
We apply a simple two-step algorithm to infer gene genealogies from polymorphism data. In the first step, we reconstruct the genealogy for each locus, using the UPGMA algorithm and the matrix of pairwise differences. We convert the branch lengths from a timescale in mutations to a timescale in generations, using the mutation rate per locus, which is considered known. Because of the discrete behavior of mutations, we do not have resolution for time intervals generations, with being the total mutation rate of each locus. We discretize the time space into equal intervals of size , starting at 0, and estimate the harmonic mean of N for each interval, using the method. This strategy gives a first estimate of . In the second step, we refine our reconstruction by using the profile computed in the first step. More precisely, we use the pairwise differences between haploid individuals/gene copies to estimate the time to the most recent common ancestor of each pair of (haploid) individuals. From this computation, we construct a distance matrix on which we apply UPGMA to reconstruct the genealogy. We compute the coalescent times between the pairs of (haploid) individuals using a Gamma distribution, following the idea that if mutations are Poisson distributed onto the coalescent tree of a given pair of (haploid) individuals, and if the height of the tree is exponentially distributed with rate [which is the case under the constant model of ], then the height of the tree T, conditional on the number of pairwise differences S between the two individuals, is Gamma distributed with shape and with rate (Tavaré et al. 1997):
| (11) |
We use the first step to compute as the harmonic mean of the inferred N from the present to the time interval corresponding to the number of observed differences between the two individuals.
Application to human data
Data preparation:
We use high-coverage sequencing data from the 1000 Genomes Project, publicly available at ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data/. The data are downloaded as VCF formatted files, from which we retain variant positions passing the filters set up by the 1000 Genomes Project, replacing the filtered-out positions by missing genotypes. We retain only the trios and within each population existing in the sample, we phase the individuals (Browning and Browning 2007) under the trios file input option, but retain only the parents after phasing, as a sample of unrelated individuals. The phasing also imputes missing genotypes. We extract sequences corresponding to regions of supposedly no/low recombination as indicated by a recombination rate of 0 in the Decode genetic map (Kong et al. 2002). The description of how those regions were ascertained is given below. We use the following population data: individuals of European ancestry from Utah (CEU), sample size of 64; southern Han Chinese individuals, China (CHS), sample size of 56; Peruvian individuals from Lima, Peru (PEL), sample size of 58; and Yoruba individuals from Ibadan, Nigeria (YRI), sample size of 38.
Genetic map and no/low recombination regions:
We use the Decode genetic map, which has been obtained by tracking >2000 meioses in Islandic lineages (Kong et al. 2002). The map is downloaded from the Table tool on the UCSC genome browser website Genome Bioinformatics Group of UC Santa Cruz (2013). We extract regions that have a recombination rate of 0. There are 22,321 such regions, of varying lengths (Figure S10), with the most common length being 10 kb (6457 regions) and mean length being ∼48 kb. An alternative would be to use the HapMap recombination maps (which can be population specific), but since they are obtained using linkage disequilibrium (LD) information, which in turn is directly linked to demography and N, we focus on the Decode map. In particular, regions of high LD can be suggestive of a low local recombination rate or a short gene genealogy of the sample used for computing LD or both. So, by extracting regions of low “recombination rate” in LD-based genetic maps, one might enrich the chosen regions in short gene genealogies, leading to inference of a smaller population size. We see this effect when applying Popsicle to regions extracted using HapMapCEU with a total recombination threshold of cM per region, although the difference of the inference is relatively small between the different genetic maps (Figure S11).
Comparison with PSMC and MSMC:
Since its publication (Li and Durbin 2011), the PSMC method has been widely used to estimate past population size over time in a number of organisms. Thus, it is important to assess how our reconstruction method compares to the results of PSMC as well as to the more recent iteration of this approach, MSMC (Schiffels and Durbin 2013). We use the sequences of the parents, with missing genotypes imputed, and cut the sequences into regions of 100 bp, identical to the approach in the original article. If no pairwise difference is observed within a region between the pairs of alleles at the 100 bp, the region is considered homozygote. If at least one pairwise difference is observed, the region is considered heterozygote. PSMC and MSMC are developed as a hidden Markov model, where the hidden states are the coalescent times of each region, while the observed states are the heterozygosity of the regions. It models recombination in the transition probabilities from one region to its neighbor. Intuitively, if a locus has many heterozygote regions, its underlying coalescent time is going to be inferred as large, whereas if a locus contains mostly homozygote regions, the coalescent time is inferred as small. Chromosomes are given as independent sequences and only autosomes are used. For running PSMC, we use the same time intervals as the human study in the original PSMC article. MSMC was used with the default time discretization, which is believed to be adapted for human data. Nondefault parameters for MSMC were fixed recombination rate and a recombination to mutation ratio of 0.88.
Application of Popsicle:
We apply Popsicle to the 22,321 low-recombining regions for the four populations, under two different settings: In the first setting, we reconstruct an effective population size profile for every individual and average the results across all individuals from the same population (we refer to that setting as “Popsicle 1”); in the second setting, we use Popsicle on subsamples of size 5 and compute the average of the obtained estimates within each population (we refer to that setting as “Popsicle 5”). We use the two-step procedure described above. Because PSMC also infers the local gene genealogies when performing its MCMC computations, we also extract the local gene genealogies from PSMC’s decoding (option -d of the program) and apply Popsicle 1 to them. The results seem highly unstable, casting doubt on the reliability of the inferred local gene genealogies from PSMC (see Figure S12).
Data and code availability:
Simulated data can be regenerated using the commands given in File S1. Data from the 1000 Genomes Project are available on the ftp server ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/. Code for the computations is available at: jakobssonlab.iob.uu.se/popsicle/.
Results
Evaluation of Popsicle on simulated gene genealogies
Four different demographic scenarios:
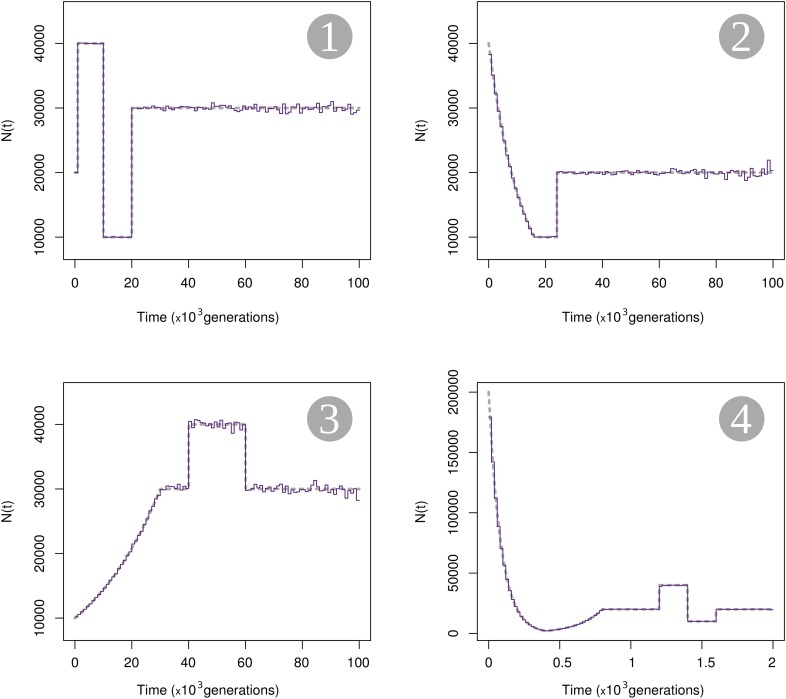
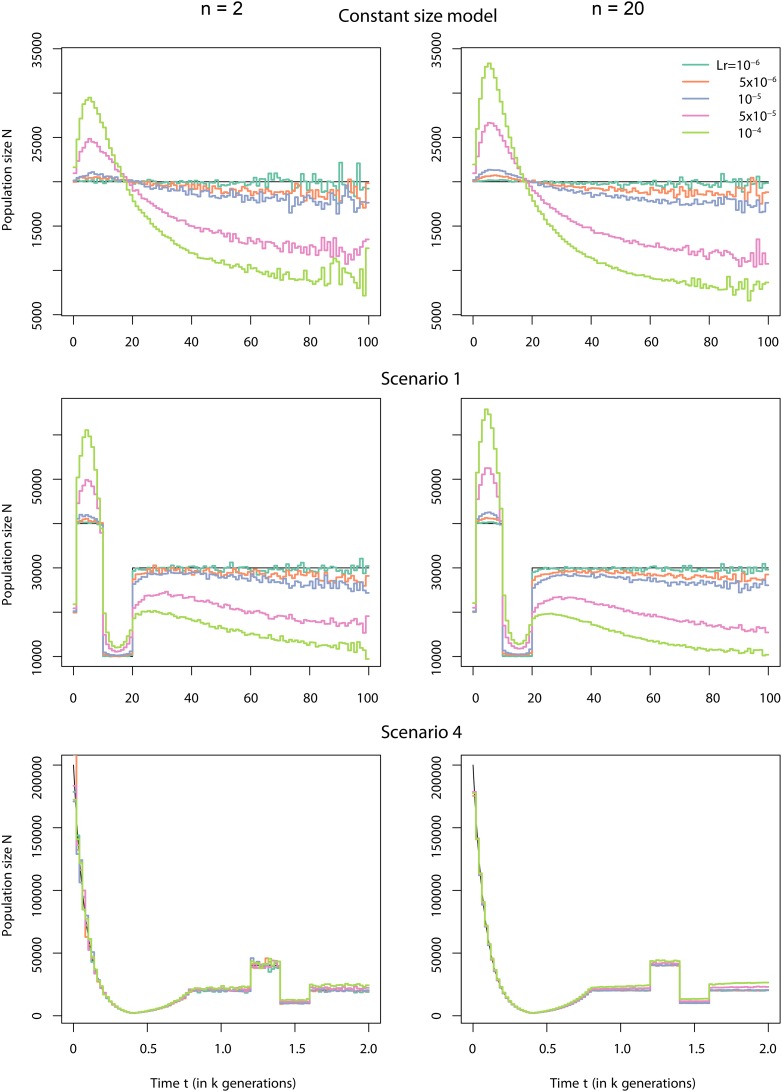
To evaluate the inference of , we used the software ms (Hudson 2002) to simulate samples under different population models with varying population size. We investigated four demographic scenarios illustrated in Figure 2. The first three scenarios describe demographic models that span between the present and 100,000 generations in the past and that include various periods of constant population size, instantaneous changes, and exponential growth or decline. In contrast to scenarios 1–3, scenario 4 describes complicated changes in size that occur in the recent past, within the last 2000 generations. Detailed descriptions of each scenario and the ms commands for the simulations are given in File S1, Table S1, Table S2, Table S3, and Table S4. In each studied scenario, we simulated 1,000,000 independent gene genealogies of 20 haploid gene copies (note that we will investigate the effect of number of loci and hence reduce that number for certain cases; see below). We assume that the true gene genealogies are known and omit any inference of genealogies from polymorphism data at this stage. The genealogies were used to estimate coalescent time distributions and in turn reconstruct the population size profile, using Popsicle. We discretized time into 100 equally long intervals (1000 generations in each interval for scenarios 1–3 and 20 generations in each interval for scenario 4).
Figure 2.
Estimation of N based on simulated gene genealogies. Four scenarios of variable population sizes are used to generate 1,000,000 independent loci in each scenario, for a sample of size 20 (10 diploid individuals). Time is divided into 100 regular intervals and estimates of the harmonic mean of N (purple solid lines) for all intervals are plotted. The true values of N over time are indicated by gray dashed lines. Figure S4 further explore the uncertainty of the estimate of N based on finite loci.
The harmonic mean estimates are very close to the true size in all four scenarios, with better accuracy in the recent past than in the distant past (Figure 2). The division of time into 100 intervals is arbitrary and dividing time using the true breakpoints of the scenarios leads to an almost perfect fit for the time periods where the population size is constant, whereas dividing time more finely in the periods of variable size improves the estimation, as long as there are enough coalescent times occurring within the interval to get a good estimate of the cumulative distribution function (results not shown). The estimation is very accurate in periods of small population size, especially when it is followed by an expansion. Estimates of are more variable around the true value when population size was larger in the past (scenario 3). These observations can be understood intuitively by the fact that will be better estimated in time periods of small N as the coalescence rate is proportional to the inverse of N. The resolution of the reconstruction method for N is also accurate in the recent past, even for drastic or rapid changes in size over a couple of hundred generations (scenario 4). In summary, with a finite but sufficiently large number of loci to estimate the cumulative distributions of coalescent times, we can accurately reconstruct the global shape of the population size over time, from very recent times to far into the past.
Effect of sample size:
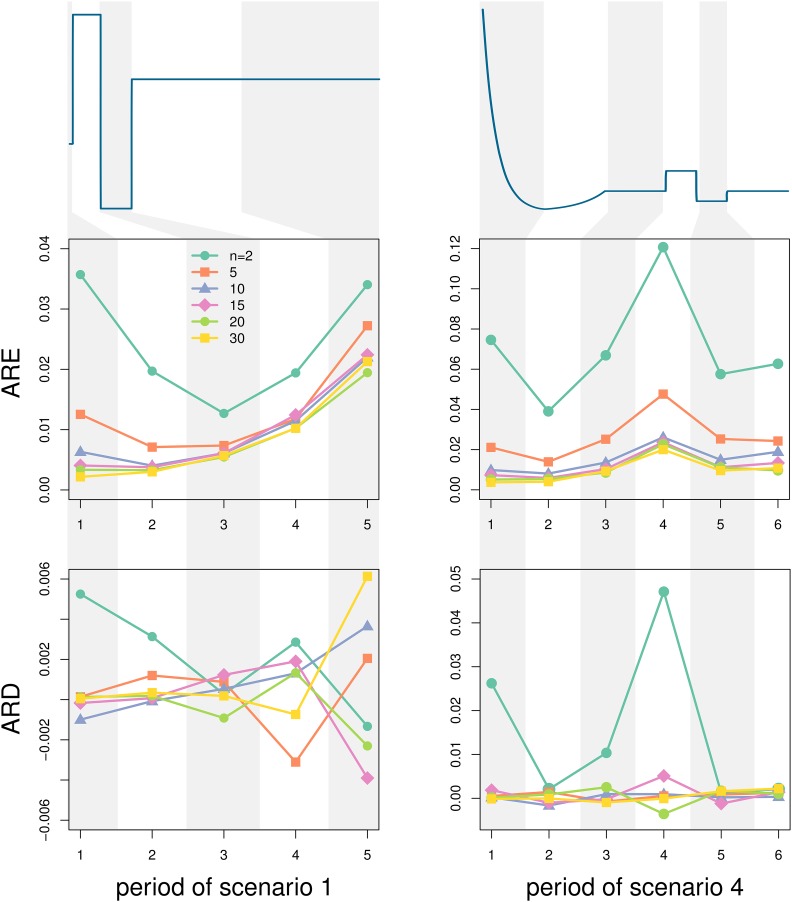
We tested the accuracy of our method for different sample sizes. To be able to quantify the performance in reconstructing the population size over time, we introduced two statistics: the ARD and the ARE. The former quantifies a systematic deviation from the true value of the population size, while the latter quantifies the error of the estimation (see Model and Methods for the computation of ARD and ARE). We used scenarios 1 and 4, from which we simulated 1,000,000 independent gene genealogies with sample sizes taken from the values . Each scenario was divided into large periods, to be able to discriminate the effect of sample size in the reconstruction between recent and old time periods and between periods of large and small population sizes. Scenario 1 was divided into five periods, while scenario 4 was divided into six periods (Figure 3, Table 1). Within each period, we discretized the time into 100 equally long intervals and assessed the reconstruction with ARD and ARE (Figure 3).
Figure 3.
Effect of sample size. We divide scenario 1 (left panel) and scenario 4 (right panel) into smaller periods of time where we assess the average relative error and the average relative difference on the N estimates compared to the true values of N, as functions of the sample size used for the estimation. A total of 1,000,000 loci were simulated for each scenario and each sample size. The original scaling for the x- and y-axes of both scenarios can be found in Figure 2.
Table 1. Division of scenarios 1 and 4.
| Scenario | ||
|---|---|---|
| Period | 1 | 4 |
| 1 | [0–1,000] | [0–400] |
| 2 | [1,000–10,000] | [400–800] |
| 3 | [10,000–20,000] | [800–1200] |
| 4 | [20,000–60,000] | [1,200–1,400] |
| 5 | [60,000–100,000] | [1,400–1,600] |
| 6 | — | [1,600–2,000] |
Time intervals are given in generations.
In general, scenario 1 is predicted more accurately than scenario 4, with an average relative error ranging between 0.2% and 3.6% compared to a range of 0.4–12.1% for scenario 4. There is little bias in the reconstruction of the two scenarios, except maybe for sample of size 2 in scenario 4, where there may be an upward bias of some 5% in period 4. In both scenarios and in all periods, the accuracy of the estimates is improved by increasing the sample size. The improvement is substantial when increasing the sample size from 2 to 10 and increasing the sample size further results in only modest improvements. Note the relatively higher error for the instantaneous population expansion of scenario 4 (period 4), irrespective of sample size, suggesting that a large population size for a brief period of time is difficult to infer. Accurate estimates of N for such periods require a greater number of loci to obtain resolution on par with time periods with smaller N, as the number of coalescences is reduced for periods of large N. This effect is investigated further in the next section.
Effect of the number of loci:
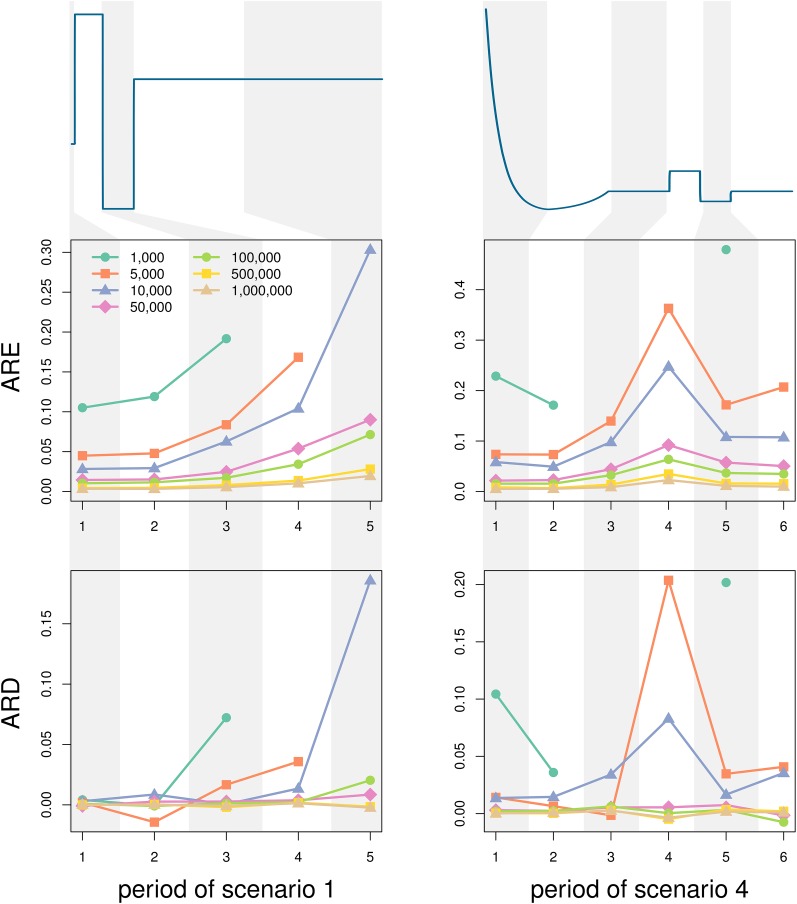
With the full knowledge of the density functions , we could potentially compute N at any time in the past. However, in practice, the distributions can be estimated only where observations are made, and hence we are limited to the time ranges where reasonable estimates of the distributions can be computed because we have enough observations. For that reason, the more loci there are, the more coalescent times can be observed within a time interval and the better the estimate of the cumulative distributions. Here we investigate the robustness of Popsicle to varying the number of loci, by simulating genealogies of samples of size 20 under scenarios 1 and 4. We compare the effect of the number of loci for different periods in the past, as described in Table 1, divide each period into 100 regular intervals on which we estimate the harmonic mean of N, and measure the accuracy within each period with ARD and ARE.
The accuracy of the N estimates in all periods for both scenarios increases with increasing number of loci (Figure 4). Generally, for these investigated (and human realistic) scenarios, the ARE and the ARD from the true values are low for cases with 50,000 loci (ARE , ARD ) and still moderate for 10,000 loci (ARE , ARD ). For smaller numbers of loci, errors can reach ≥40%. For scenario 1 and 1000 loci, no coalescence occurred during periods 4 and 5 in any of the simulations, making the inference impossible for these periods. Similarly, there were no coalescence events in period 5 of scenario 1 with 5000 loci, as well as in periods 4 and 6 in scenario 4 with 1000 loci. This illustrates the greater difficulty of accurate N reconstruction for older time periods (in particular, if the period is preceded by a severe bottleneck) and periods of large population size, both subject to low probabilities of coalescences, to occur. Thus, depending on the history of the population and how far back in time N is of interest, the required number of loci will vary. Subsampling from some particular number of loci might give an idea of whether a particular number of loci is enough for a good estimation of .
Figure 4.
Effect of the number of loci. We divide scenario 1 (left panel) and scenario 4 (right panel) into smaller periods of time where we assess the average relative error (ARE) and the average relative difference (ARD) on the N estimates compared to the true values of N, as functions of the number of loci used for the estimation. The sample size for all simulations is 20. The original scaling for the x- and y-axes of both scenarios can be found in Figure 2.
Effect of recombination:
We explore the robustness of the reconstruction if recombination occurs in the loci, but when each locus is treated as nonrecombining. This case is equivalent to considering the entire sequence fragment as nonrecombining and having a single underlying genealogy, represented by an average tree, instead of considering the multiple underlying genealogies within the (recombining) locus. We investigate the effect of ignoring recombination for samples of size 2 and for samples of size 20, for different levels of recombination within each simulated locus. For a sample of size 2, the average tree is simply the weighted mean of the trees of the nonrecombining segments, with the weight being the relative length of each nonrecombining segment compared to the total segment length. For the case of 20 gene copies, we build the “average tree” by applying a UPGMA algorithm to the weighted average matrix of pairwise time to coalescence between all pairs of haploid individuals. We use scenarios 1 and 4, as well as the constant-size model, to study the robustness of the method to different levels of recombination. We tested five levels of recombination within the locus: , , , , and . Assuming a recombination rate of per site per generation, which is around the estimated average for the human recombination rate, these five levels represent loci of length 80, 400, 800, 4000 and 8000 bp.
Cryptic recombination can lead to inference of spurious changes in population size, even under the simple model of constant population size (Figure 5), although the effect is limited to a factor of at most ∼2 in the investigated cases. For instance, scenarios 1 and 4 that are relatively realistic for, e.g., humans show low to moderate bias due to cryptic recombination, even for the cases of high levels of recombination (or long fragments). Overall, the effect of cryptic recombination appears to be indifferent to sample size. For the case of constant size, we can provide some intuition on the effects of cryptic recombination. We note that genealogies inferred from recombining loci are weighted averages of the underlying genealogies of the nonrecombining fragments of the loci and therefore tend to be more star-like as well as of intermediate size. Estimating one single gene genealogy from such a mosaic of correlated gene genealogies will have an impact on the distributions of coalescent times (see Figure S5). Star-like gene genealogies are typically associated with rapid and recent expansions, which is what the inferred shows in the case of constant population size and high level of cryptic recombination.
Figure 5.
Effect of omitting recombination. Shown is a comparison between reconstructed using gene genealogies computed as a weighted average of the gene genealogies obtained from ms and true (black lines) under three different demographic scenarios. We generated 1,000,000 independent loci for two different sample sizes, 2 and 20 haploid gene copies, and for five different levels of recombination within each locus. The three different demographic scenarios were the constant-size model (top), scenario 1 (middle), and scenario 4 (bottom). The different cryptic recombination rates for each locus (in morgans) are indicated by different colors and the values of the recombination of the segments are given in the key.
Toward solving the full inference problem
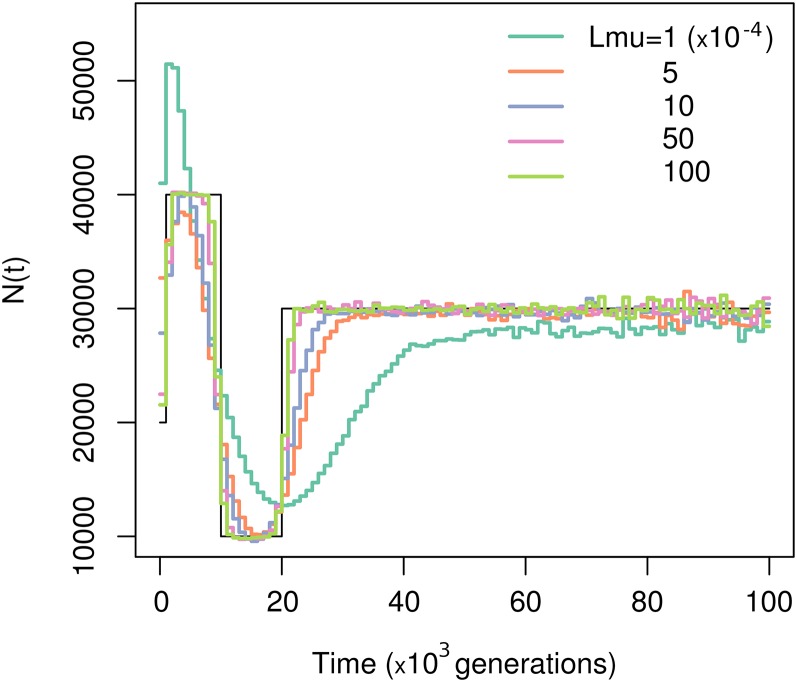
Popsicle is designed to infer effective population size over time from samples of gene genealogies obtained under the demographic model studied and not directly from observed sequence data. However, to provide an illustration and one example of a solution to integrate Popsicle into a full inference method that would take sequence data as input, we outline one heuristic approach here. This approach builds local gene genealogies from sequence data and applies Popsicle to the distributions of coalescent times obtained using inferred gene genealogies. Our aim is notably to apply this full-resolution method on human sequences, to be able to compare these population size profiles to previously published results. Hence, we have to find a way to obtain the gene genealogies from sequence data. One way could be to use ARGWeaver (Rasmussen et al. 2014), as is done in Palacios et al. (2015). However, Palacios et al. (2015) found a systematic bias in their reconstruction of effective population size over time profiles when using ARGWeaver to infer gene genealogies, even for rather simple demographic scenarios. Here, we develop a simple two-step algorithm based on UPGMA and properties of the coalescent to infer gene genealogies from sequences, as it seemed to perform well on simulated nonrecombining sequences. A detailed outline of the algorithm is described in Model and Methods. Inferring gene genealogies from sequences is a challenging problem, especially for recombining sequences, and we note that our algorithm is merely a heuristic solution to the problem that performs well.
We evaluate our ability to reconstruct the population size over time, using Popsicle together with our algorithm to infer gene genealogies. In particular, we study the impact of the mutation rate on the reconstruction, to get a sense of how large the mutation rate needs to be to obtain reasonable results. We present the results for samples of size 20, simulated with values of taken from , for 1,000,000 nonrecombining loci and under scenarios 1 and 4 (Figure 6 and Figure S7). For reference, with a mutation rate of /bp per generation, the range of values corresponds to loci of 8, 40, 80, 400, and 800 kb, respectively. With a mutation rate of , we can already uncover a good estimate of the population size profile. Unsurprisingly, the more mutations there are, the better the estimates of times to coalescence and the more accurate the reconstruction is. This fact is particularly important for recent times where enough mutations are required to accumulate to infer the very recent population sizes (Figure S6).
Figure 6.
Effect of estimating gene genealogies from polymorphism data. Shown is reconstruction of from distributions of coalescent times computed from gene genealogies inferred from polymorphism data. We used a sample size of 20 and 1,000,000 independent loci, evolving under scenario 1. The mutation rate per locus is indicated by the color of the line and the key gives the mutation rates.
Application to human sequence data
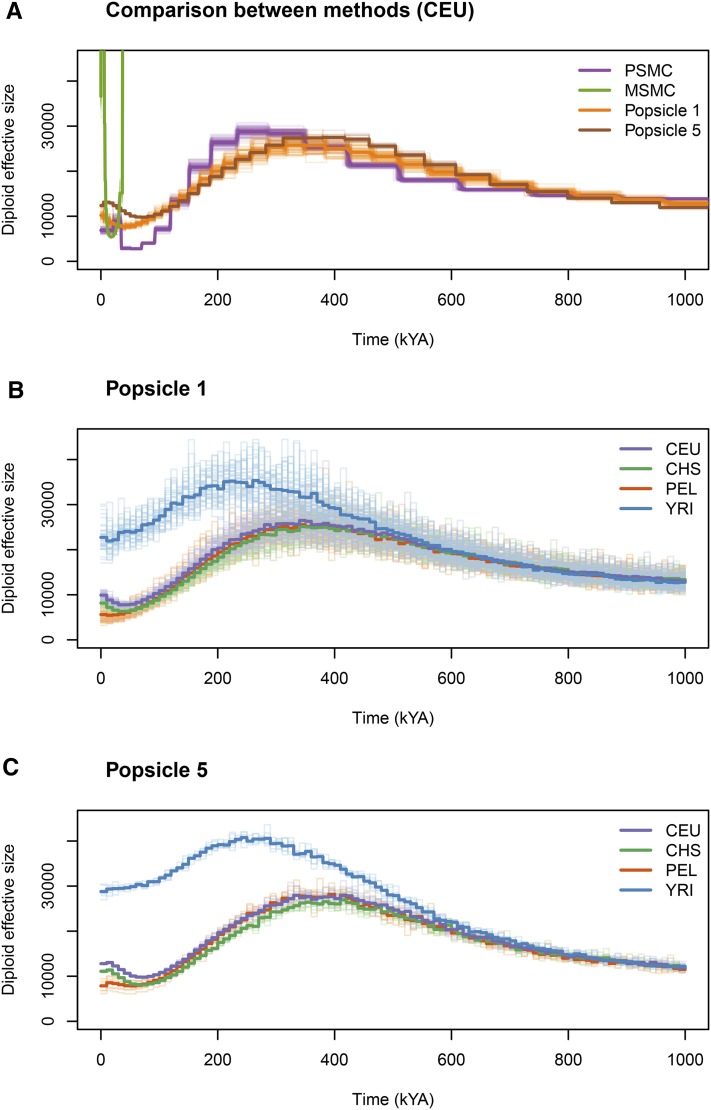
We apply the developed heuristic algorithm of gene-genealogies inference followed by Popsicle to empirical sequence data. The effect of recombination can be mitigated by considering only regions of the genome with low or no recombination, provided that we have access to a good genetic map. Following this principle, we applied Popsicle to human genome sequence data from the 1000 Genomes Project (Complete Genomics high-coverage samples from the Complete Genomics Data from 1000 Genomes Public Repository 2013), for Yoruba individuals from Nigeria, for American individuals of European ancestry from Utah, for Han Chinese individuals from southern China, and for Peruvian individuals. We extracted regions of no recombination according to the Decode recombination map (Kong et al. 2002) (see Model and Methods for a description of the data preparation). For comparison, we also use PSMC (Li and Durbin 2011) and MSMC (Schiffels and Durbin 2013) to infer profiles from the data. We inferred profiles for the four populations in two ways: (a) using single individuals (as PSMC does) and averaging across single individuals (denoted Popsicle 1) and (b) using five individuals from the population (denoted Popsicle 5). From simulations, we have observed that >10 haploid sequences result in only a minor improvement of the inference in population size (see Figure 3).
Overall, the Popsicle profiles of effective population size in the last 1 MY for every population largely resemble the vague knowledge about past human population sizes as well as the profiles inferred by, e.g., PSMC (Figure 7A). In contrast, the profile reconstructed by MSMC is very different from that of PSMC and Popsicle. As MSMC traces only the first coalescent event between any pair of the 10 chromosomes in the data, it provides estimates of the population size only for the last 50,000 years or so. A comparison on the log scale between the three methods applied to CEU data is provided in Figure S7. Results of MSMC and PSMC across populations are given in Figure S8 and Figure S9, respectively.
Figure 7.
Comparison of inference among different methods. (A) Comparison of profiles inferred using PSMC, MSMC, Popsicle 1, and Popsicle 5. (B) Inferred profiles for four populations, CEU, CHS, PEL, and YRI based on Popsicle 1. (C) Inferred profiles for four populations, CEU, CHS, PEL, and YRI based on Popsicle 5. The timescale is computed assuming a mutation rate of and a generation time of 25 years.
Popsicle reveals a steady but slow increase in effective population size starting around 1 MYA, reaching a maximum between 200 and 500 KYA, followed by a sharper decline and a recovery during the last 100 KY for European and East Asian populations. However, prior to 1 MYA, the population size inferred by PSMC is higher than the population size inferred by Popsicle (Figure S7). In addition, Popsicle infers a less sharp decline in population size than PSMC does, for all four populations, and infers a population size history markedly different for Yoruba compared to the three other non-African populations (Figure 7, B and C) whereas the Yoruban population follows the non-African populations rather closely in the PSMC results (Figure S9). Popsicle results suggest a somewhat larger ancestral population for Yoruba than the ancestral population size of the three non-African populations, which could be interpreted as deep and long-lasting population structure within Africa between 400 and 100 KYA. Note, however, that the nonrecombining regions have been chosen using the Decode recombination map, a genetic map formed by tracking >2000 meioses in Islandic lineages. Recombination patterns and hotspots in particular are believed to be variable across populations (Myers et al. 2005; Baudat et al. 2010), and thus the nonrecombining regions selected using the Decode map might be in fact recombining in Yoruba, resulting in a bias of the population size estimates (see Figure 5). Recombination maps for Yoruba have been computed (Frazer et al. 2007), but because they have been inferred using properties of linkage disequilibrium that itself depends on demography, they would not be ideal to use for selecting regions of low/no recombination. A future pedigree- or sperm-typing-based recombination map for the Yoruba would help in understanding the differently inferred profiles for African and non-African populations.
Popsicle 1 and Popsicle 5 give similar effective population size profiles (Figure 7, B and C) but the times of the major features in Popsicle 5 are shifted to older times compared to Popsicle 1. Whereas Popsicle 1 suggests a bottleneck in non-African populations that reaches its strongest effect between 30 and 40 KYA, Popsicle 5 places the bottleneck between 70 and 80 KYA, which is more in line with the estimates of timing of the founder effects due to a dispersal out of Africa (Scally and Durbin 2012). In neither Popsicle nor PSMC do we see the superexponential increase in size that has occurred in all populations since the spread of agriculture (Keinan and Clark 2012), but we possibly do in the MSMC results (Figure S8). It is possible that for Popsicle and PSMC too few loci are included for a reliable inference in the recent times, or too few individuals, or that the mutation rate per locus is too low to observe a dramatic expansion in population size (as most terminal branches will be very short in genealogies from models of rapid recent expansion). Keinan and Clark (2012) suggest that observing enough rare variants is necessary to infer the exponential growth that human populations have been going through in the past thousands of years.
The resolution of Popsicle can be better than that of PSMC, as Popsicle does not constrain the coalescent times into a finite (and usually rather small) set of values like PSMC does. In principle, any time discretization for computing the harmonic mean of the effective population size over time can be used, although in practice we need to make sure that there are enough coalescences within each time interval to get reliable estimates of the effective size. Popsicle is also markedly faster than PSMC, not only because it uses a moderate number of nonrecombining regions, but also because of the closed-form relationship between population size and coalescent time distributions. Most of the computational time is spent on inferring the gene genealogies (which takes <20 min for the 22,321 loci in the data application). Once the gene genealogies are computed, the application of the Theorem for reconstructing the population size takes a few seconds. Finally, Popsicle accommodates samples of any size, which should lead to more reliable results, especially in the recent times, provided that the phasing of the genomes is accurate.
Applying Popsicle to extracted regions of limited recombination should not bias the results in principle. Regardless of the molecular reason explaining the low rate of recombination in the region (for instance, limited access for crossovers or conservation constraints due to functional importance of the region), the fact that there is one local gene genealogy for the entire region is what matters for the method to work. However, for applications to empirical data, variation in the local mutation rate, due to purifying selection for example, will affect the reconstruction of the gene genealogy by changing the estimates of the branch lengths for different loci. This could potentially cause bias in the reconstructed Popsicle profiles, as all gene genealogies are inferred using one mutation rate. Using a mutation map obtained from the study of de novo mutations in trios or pedigrees could alleviate this issue and infer the local gene genealogies from genetic data, using a specific mutation rate for each region.
Discussion
The major implication of our main result is to reduce the problem of reconstruction from polymorphism data to a problem of gene-genealogy inference. If local gene genealogies in the genome can be inferred accurately from observed polymorphism data, then our Theorem can be used to estimate with great accuracy as well. Currently, however, local gene-genealogy inference remains a challenge. First, most genomes do not consist of large sets of independent nonrecombining loci, but rather of sets of recombining chromosomes. Each chromosome can be seen as a linear structure of successive nonrecombining loci whose underlying genealogies are correlated with one another. This correlation decays with distance between loci due to recombination. Also, in a given sample, the exact positions on the chromosome of the recombination events, and hence the breakpoints between the nonrecombining bits of DNA, are unknown. Fully recovering the genealogies along the chromosome means reconstructing the ancestral recombination graph from polymorphism data and this is a challenging problem (Griffiths and Marjoram 1996; McVean and Cardin 2005; Parida et al. 2008; Rasmussen et al. 2014; Zheng et al. 2014). We noted based on simulations that a low to moderate level of cryptic (unaccounted) recombination leads to accurate estimates of , but the bias increases with greater levels of cryptic recombination.
The problem of inferring gene genealogies can also be challenged by a lack of mutation events to accurately estimate coalescent times. For some species, there might not be enough mutation events to be able to infer the local gene genealogies of nonrecombining segments. In humans for example, the ratio between the mutation rate per site and per generation and the recombination rate per site and per generation is likely close to 1 (or 2, depending on assumptions on mutation rate; for the pedigree-based mutation rate or the divergence-based mutation rate, see, e.g., Scally and Durbin 2012). Hence, on average, for each mutation observed locally in a sample, there is also a recombination breakpoint nearby. A targeted approach, where only low-recombining regions of sufficient length are considered, could yield better results and we have shown that such a strategy can provide profiles that are similar to estimates based on approaches that specifically model recombination. These challenges are inherent to the problem of estimating local gene genealogies from sequence data. There have been interesting developments in this area (see, e.g., Rasmussen et al. 2014), and we look forward to the further methodological improvements to infer the ancestral recombination graph.
To gauge some intuition of usefulness of Popsicle for human genome data, we can make a computation of the number of regions that can be recruited for analysis. Assume a genome of 3 billion bp, a mutation rate of 1.25 /bp and generation, a recombination rate of 1.25 /bp and generation, and an effective population size of 10,000 diploid individuals. Assume further that the genome is organized into recombination “hotspots” and “cold regions,” where the former account for 99% of the recombination events and the cold regions have a 100 times lower recombination rate compared to the genome average. Assuming an average cold region extends for 40 kbp (compare with Figure S10), the average recombination rate in such a locus is 5 (orange line in Figure 5) and the average number of pairwise mutations would be 20. Hence, the genome would consist of 75,000 genome regions of length 40 kbp that contain abundant polymorphism data to obtain a good estimate of gene genealogies. This rough computation illustrates that at least the human genome harbors favorable properties that Popsicle can utilize.
We present a novel method for inferring population size over time, a problem that has recently gained much interest due to the availability of genome sequence data. By analytically solving the relationship between and the distribution of coalescent times, we have connected to the problem of inferring the ancestral recombination graph from polymorphism data, which remains a challenge in population genetics. We show that, using a moderate number of loci and a simple algorithm for genealogy inference, our method Popsicle was able to recover the general pattern of population size as a function of time with high resolution and using modest computational time, properties that will be useful for future large-scale studies of many full genomes.
Acknowledgments
We thank Martin Lascoux and Michael G. B. Blum for helpful comments on the manuscript. This work was supported by grants to M.J. from the Knut and Alice Wallenberg foundation, the Swedish Research Council (no. 642-2013-8019), and the Göran Gustavsson foundation.
Appendix: Derivation of the
The relationship between the density function of the cumulative coalescent times and the family of functions can be written in matrix form. We define as the vector of density functions of cumulative coalescent times , as the vector , and the upper triangular matrix as . Then from Equation 3, from Polanski et al. (2003) we have
To prove that the defined in the Theorem can invert the relationship between and , we show that the matrix B defined by is the inverse matrix of A. We define . Our aim is to prove that C is in fact the identity matrix. First, we know that C is an upper triangular matrix, as both A and B are upper triangular matrices. To prove that C is the identity matrix, we cover four separate cases: for , for , for , and finally . For the computation of the two first cases, we need to introduce a notation:
| (A1) |
We know from partial fraction decomposition that
| (A2) |
We compute the coefficients , for
| (A3) |
In the above calculation, we go from line 3 to line 4 by using Equation A1. Then on the next line we exchange the two sums and by noticing that the terms under the k-indexed sum are not dependent on k, we obtain line 6. On line 6, we can notice that the factor after is exactly the same as in line 3, thus is equal to . Since , only can satisfy . We go on by computing our second case: the coefficients for
| (A4) |
Similarly to the computation of above, the only way to satisfy for is to have Now, the remaining coefficients to be computed are the diagonal coefficients. For ,
| (A5) |
Finally,
| (A6) |
All the above computed coefficients prove that the matrix C is the identity matrix, and hence B is the inverse matrix of A, which demonstrates the Theorem.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.185058/-/DC1.
Communicating editor: J. Wakeley
Literature Cited
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., et al. , 2010. Prdm9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boserup E., 1981. Population and Technological Change: A Study of Long-Term Trends. University of Chicago Press, Chicago. [Google Scholar]
- Browning S. R., Browning B. L., 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81: 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. E., 1995. How many people can the earth support? Science 35: 18–23. [Google Scholar]
- Complete Genomics Data from 1000 Genomes Public Repository, 2013 File location for Complete Genomics high coverage data. Available at: ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/complete_genomics_indices/20130820.cg_data.index. Accessed: September 30, 2013.
- Drummond A. J., Suchard M. A., Xie D., Rambaut A., 2012. Bayesian phylogenetics with beauti and the beast 1.7. Mol. Biol. Evol. 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K. A., Ballinger D. G., Cox D. R., Hinds D. A., Stuve L. L., et al. , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome Bioinformatics Group of UC Santa Cruz, 2013 Genome Browser Table Tool. Available at: http://genome-euro.ucsc.edu/cgi-bin/hgTables?hgsid=208520476_rAHGRV4HFcmGAOng8Gp4ETNO9vYF. Accessed: October 12, 2016.
- Griffiths R. C., Tavare S., 1994. Sampling theory for neutral alleles in a varying environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 344: 403–410. [DOI] [PubMed] [Google Scholar]
- Griffiths R. C., Marjoram P., 1996. Ancestral inference from samples of dna sequences with recombination. J. Comput. Biol. 3: 479–502. [DOI] [PubMed] [Google Scholar]
- Ho S. Y. W., Shapiro B., 2011. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol. Ecol. Resour. 11: 423–434. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Jakobsson M., Scholz S. W., Scheet P., Gibbs J. R., VanLiere J. M., et al. , 2008. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451: 998–1003. [DOI] [PubMed] [Google Scholar]
- Keinan A., Clark A. G., 2012. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science 336: 740–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Gudbjartsson D. F., Sainz J., Jonsdottir G. M., Gudjonsson S. A., et al. , 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Lahr M. M., Foley R., 2001. Genes, fossils and behaviour: When and where do they fit, pp. 13–48 in Genes, Fossils, and Behaviour: An Integrated Approach to Human Evolution, Vol. 310, edited by Donnelly P., Foley R. IOS Press, Amsterdam. [Google Scholar]
- Li H., Durbin R., 2011. Inference of human population history from individual whole-genome sequences. Nature 475: 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li H., Jakobsson M., Li S., Sjodin P., et al. , 2012. Joint analysis of demography and selection in population genetics: Where do we stand and where could we go? Mol. Ecol. 21: 28–44. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C. M., Mikkelsen T. S., Karlsson E. K., Jaffe D. B., et al. , 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438: 803–819. [DOI] [PubMed] [Google Scholar]
- Marjoram P., Wall J. D., 2006. Fast “coalescent” simulation. BMC Genet. 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A. T., Cardin N. J., 2005. Approximating the coalescent with recombination. Phil. Trans. R. Soc. Lond. B Biol. Sci. 360: 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P., 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310: 321–324. [DOI] [PubMed] [Google Scholar]
- Nielsen R., 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39: 197–218. [DOI] [PubMed] [Google Scholar]
- Palacios J. A., Wakeley J., Ramachandran S., 2015. Bayesian nonparametric inference of population size changes from sequential genealogies. Genetics 201: 281–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palkopoulou E., Dalén L., Lister A. M., Vartanyan S., Sablin M., et al. , 2013. Holarctic genetic structure and range dynamics in the woolly mammoth. Proc. Biol. Sci. 280: 20131910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida L., Melé M., Calafell F., Bertranpetit J., Genographic Consortium , 2008. Estimating the ancestral recombinations graph (arg) as compatible networks of SNP patterns. J. Comput. Biol. 15: 1133–1153. [DOI] [PubMed] [Google Scholar]
- Polanski A., Bobrowski A., Kimmel M., 2003. A note on distributions of times to coalescence, under time-dependent population size. Theor. Popul. Biol. 63: 33–40. [DOI] [PubMed] [Google Scholar]
- Ramachandran S., Deshpande O., Roseman C. C., Rosenberg N. A., Feldman M. W., et al. , 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. USA 102: 15942–15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M. D., Hubisz M. J., Gronau I., Siepel A., 2014. Genome-wide inference of ancestral recombination graphs. PLoS Genet. 10: e1004342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A., Durbin R., 2012. Revising the human mutation rate: implications for understanding human evolution. Nat. Rev. Genet. 13: 745–753. [DOI] [PubMed] [Google Scholar]
- Schiffels S., Durbin R., 2013. Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46: 919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan S., Harris K., Song Y. S., 2013. Estimating variable effective population sizes from multiple genomes: a sequentially Markov conditional sampling distribution approach. Genetics 194: 647–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S., Balding D. J., Griffiths R. C., Donnelly P., 1997. Inferring coalescence times from DNA sequence data. Genetics 145: 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Vaart A. W., 2000. Asymptotic Statistics, Vol. 3 Cambridge University press, Cambridge, UK. [Google Scholar]
- Wakeley J., 2009. Coalescent Theory: An Introduction Roberts and Company Publishers, Greenwood Village, CO. [Google Scholar]
- Zheng C., Kuhner M. K., Thompson E. A., 2014. Bayesian inference of local trees along chromosomes by the sequential Markov coalescent. J. Mol. Evol. 78: 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Simulated data can be regenerated using the commands given in File S1. Data from the 1000 Genomes Project are available on the ftp server ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/. Code for the computations is available at: jakobssonlab.iob.uu.se/popsicle/.