Abstract
Introns occasionally remain in mature messenger RNAs (mRNAs) due to splicing errors and the translated, aberrant proteins that result represent a metabolic cost and may have other deleterious consequences. The nonsense-mediated decay (NMD) pathway degrades aberrant mRNAs, which it recognizes by the presence of an in-frame premature termination codon (PTC). We investigated whether selection has shaped the location of PTCs in introns to reduce waste and facilitate NMD. We found across seven model organisms, that in both first and last introns, PTCs occur earlier in introns than expected by chance, suggesting that selection favors earlier position. This pattern is more pronounced in species with larger effective population sizes. The pattern does not hold for last introns in the two mammal species, however, perhaps because in these species NMD is not initiated from 3′-terminal introns. We conclude that there is compelling evidence that the location of PTCs is shaped by selection for reduced waste and efficient degradation of aberrant mRNAs.
Keywords: premature termination codon, nonsense-mediated decay, translation, splicing errors, intron definition
IT is clear that selection can play a major role in shaping genome architecture (Lynch 2007). Identifying selection is especially straightforward for exons in protein coding genes, where tests based on silent and replacement site substitutions can be employed. However, in noncoding regions, the role of selection in shaping nucleotide content is less easily investigated because of the difficulty identifying expected patterns. In introns, length, phase, and frequency of occurrence have been studied as a product of the processes of selection and drift (Castillo-Davis et al. 2002; Lynch 2002; Whitney and Garland 2010; Kelkar and Ochman 2012) but, apart from sequence-based splicing signals and GC content (Mount 1982; Deutsch and Long 1999; Amit et al. 2012; Farlow et al. 2012), few studies have examined the nucleotide composition of introns (Lim and Burge 2001; Halligan et al. 2004; Andolfatto 2005; Ressayre et al. 2015). In this study, we investigate the role of selection in determining the position of premature termination codons (PTCs) within introns.
During post-transcriptional processing, splicing errors can result in introns being present in mature messenger RNAs (mRNAs) (Gilbert 1978). Translation of such mRNAs results in the production of proteins that are aberrant in amino acid sequence and usually shortened. A shorter protein results from the presence of in-frame, PTCs within the unspliced intron, or in a downstream exon due to a frameshift. Aberrant proteins may reduce fitness because they have an activity that is damaging to the cell, and/or because they represent wasted resources, particularly amino acids and sequestered ribosomes (Drummond and Wilke 2009). For these reasons, we hypothesize that selection favors both efficient splicing and mechanisms that minimize the effects of splicing errors.
There is strong evidence for selection acting on both the efficiency of splicing and on the effects of splicing errors. Splicing site consensus sequences are highly conserved and the surrounding nucleotides exhibit weak secondary structure, indicating they are constrained by selection for efficient splicing (Sheth et al. 2006; Zafrir and Tuller 2015). In addition, transcripts containing large introns are more dependent on 5′- and 3′-splicing context to define exons, while splice context in shorter introns appears to be more lax (Jaillon et al. 2008; Farlow et al. 2012).
The nonsense-mediated decay (NMD) pathway is present in all eukaryotes and induces rapid decay of mature mRNAs possessing PTCs, minimizing the effects of splicing errors (Jaillon et al. 2008; Ramani et al. 2009; Wen and Brogna 2010; Drechsel et al. 2013). NMD is expected to select for the presence of an in-frame PTC in an intron so that, if unspliced, the intron will rapidly cause mRNA decay. In addition, there is evidence that the number of PTCs can be selected. For example, in Paramecium tetraurelia, PTCs are more common in introns whose length is a multiple of 3 (Jaillon et al. 2008).
In this study, our goal is to test for evidence of selection on PTCs. We hypothesize that PTCs are selected to occur early in introns to promptly initiate NMD and reduce both the time taken for a ribosome to translate an aberrant mRNA containing unspliced introns and the length of the resulting aberrant proteins. Since selection is caused by the deleterious effects of having aberrantly spliced mRNAs, we expect our predictions to more likely hold for genes that are expressed at high levels, because they may produce more aberrant mRNAs, and introns with fewer than ∼250 nucleotides, where splicing errors more commonly lead to intron retention (Lim and Burge 2001; De Conti et al. 2013).
To test whether there is evidence that selection has acted to minimize the deleterious effects of failure to remove an intron from an mRNA by modifying the location of PTCs, we utilized data from seven model organisms: Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Homo sapiens, Mus musculus, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. These species have well-annotated genomes, reliable splicing information, and expression data, and thus represented all of the species with the necessary data to test our predictions. Our analysis focused on a single intron in each gene so that each intron-containing gene contributed equally to the data set. We chose the first intron because it exists in all intron-containing genes and the effect of splicing failure on the aberrant mRNA is not altered by splicing of other introns. For a few analyses, in species containing genes with more than one intron, we also analyzed PTC position in the last intron because NMD is not initiated from the last intron in mammals (Maquat 2005). If NMD is driving selection on PTC position, we do not expect to see evidence for selection on PTC position in the last intron in mammals. We determined whether first PTCs happen to occur earlier than expected and whether there was an effect of level of expression or intron length on the patterns observed.
If incorrect splicing is generally not very common (Wilhelm et al. 2008; Drummond and Wilke 2009; fox-Walsh and Hertel 2009) and NMD is reasonably effective, then the fitness costs of incorrectly spliced mRNAs may be quite small at each locus. Thus, selection on PTC position in an intron caused by the presence of aberrant mRNAs may be quite weak. We thus expected our predictions to be less robust in species with smaller effective population sizes, where genetic drift can overwhelm weak selection, as has been observed for other genomic features (Lynch and Conery 2003).
Materials and Methods
Data collection
Genome data were collected for A. thaliana, C. elegans, D. melanogaster, H. sapiens, M. musculus, S. cerevisiae, and S. pombe as GenBank files from the National Center for Biotechnology Information (NCBI) (Caenorhabditis elegans Sequencing Consortium 1998; Lin et al. 1999; Mayer et al. 1999; Adams et al. 2000; Erfle et al. 2000; Tabata et al. 2000; Theologis et al. 2000; Lander et al. 2001; Wood et al. 2002; Engel et al. 2014). Genomes were parsed using Feature Extract to collect all protein-encoding genes with annotated introns (Wernersson 2005) (Table 1). Within species, files were sorted by gene name to remove duplicate genes and genes with alternative splicing, because sequence data for the first intron, terminal intron, and 3′-UTRs of these genes were often identical. Our goal was to have each intron-containing gene contribute once to the data set. The effective population sizes for each species were obtained from previously published reports and are based on estimates from nuclear synonymous sites (Schoen and Brown 1991; Chen and Li 2001; Wright et al. 2002; Sivasundar and Hey 2003; Cutter 2006; Shapiro et al. 2007; Wright and Andolfatto 2008; Skelly et al. 2009; Brown et al. 2011; Phifer-Rixey et al. 2012; Behringer and Hall 2016). Expression data were collected for each organism from previously published data, and genes lacking expression data were removed from the study [A. thaliana (Carviel et al. 2009); C. elegans, Michael Smith Genome Sciences Centre (http://www.bcgsc.ca) D. melanogaster, FlyAtlas (http://www.flyatlas.org) (Chintapalli et al. 2007); H. sapiens (Dezso et al. 2009); S. cerevisiae (Pelechano et al. 2010); and S. pombe (Tanizawa et al. 2010)]. Remaining genes (Table 1) were then used for analysis. For most analyses, only the first intron was examined in each gene.
Table 1. Species used in this study.
| Species | Assembly | Ne (× 103) | Total genes | Genes analyzed |
|---|---|---|---|---|
| A. thaliana | TAIR10 | 40 | 33,583 | 10,141 |
| C. elegans | WBcel215 | 80 | 21,187 | 10,869 |
| H. sapiens | GRCh27.p10 | 90a | 37,150 | 6075 |
| M. musculus | GRCm38.p2 | 120 | 34,293 | 8373 |
| D. melanogaster | Release 5 | 1150 | 15,581 | 5260 |
| S. cerevisiae | R64-1-1 | 8530 | 6352 | 161 |
| S. pombe | ASM294v2 | 8800 | 5883 | 652 |
Included are species, genome assemblies, effective population size estimates, total genes including nonprotein coding genes in each assembly, and number of protein-coding genes meeting the criteria used in the study. Effective population sizes were obtained from the primary literature (Schoen and Brown 1991; Chen and Li 2001; Wright et al. 2002; Sivasundar and Hey 2003; Cutter 2006; Shapiro et al. 2007; Wright and Andolfatto 2008; Skelly et al. 2009; Brown et al. 2011; Phifer-Rixey et al. 2012). Species are listed in order of increasing effective population size.
Ancestral estimate used since analysis relies upon genome-wide data.
The first intron was chosen for several reasons. First, examining a single intron per gene prevents some genes from contributing more to patterns than others (pseudoreplication). Second, many genes contain only a single intron, especially in S. cerevisiae where 229 of the 241 intron-containing genes have only one intron. Third, if a first intron is retained, the effect on the sequence of the message can be reliably determined. This is not true for later introns if multiple splicing errors can occur in the same message, which will be particularly likely because errors may not be independent (Hossain et al. 2011). For example, with multiple introns in a gene, aberrant splicing might retain one, or more than one in a message. When a first intron is retained in an aberrant mRNA, the ribosome will translate the first exon, then proceed into the first intron, and then into the second exon, regardless of presence/absence of downstream introns. However, a prediction cannot be reliably made for downstream introns because failure to splice an upstream intron will have consequences both on whether a ribosome will reach a downstream intron (an in-frame, premature termination codon will end translation early) and what the reading frame will be when it gets there. Fourth, practically, performing such an analysis would be difficult because of the need to determine the correct reading frame for each successive intron when previous introns have been spliced correctly, across genes that vary substantially in the number of introns. Given the available data sets, we are not sure exactly how we would program this analysis. Reading frame was not a problem for the first and last introns. For the first intron, determining frame is straightforward since there are no prior introns. For the last intron, frame was determined by working backward from the stop codon. Fifth, it is not clear to us the appropriate way to compare across genes with a few vs. many introns, or those that vary in the proportion of introns that are short vs. long (see next section), which is important for intron definition (De Conti et al. 2013).
We did perform some analyses using last introns in an attempt to test the hypothesis that the source of the selection on PTC position was the NMD pathway. In mammals, retention of the final intron does not trigger NMD and so we predicted that PTC position would be early in invertebrate last introns but not in mammalian last introns.
Identification of first PTCs, and other nonsense codon positions
Using the annotation data for each species, we determined the length of the first intron within the coding sequence for each gene. We then indexed the position of the first 5′-splice site, the position of the first PTC (frame 0), and the first intronic nonsense codon (NC) in the +1 and +2 reading frames for each gene using custom Perl scripts. Throughout our study, we distinguish in-frame (frame 0) intronic termination codons, PTCs, from out-of-frame (frame +1 and +2) intronic NCs (TAA, TAG, or TGA), as the latter are unable to cause translation termination when an intron is unspliced.) Comparing the position of the PTCs to that expected by randomizing intron base composition did not alter the results qualitatively. However, it is clear that introns vary in base composition along their length (Supplemental Material, Figure S5), which could mislead conclusions concerning PTC position, and so we utilized out-of-frame NCs to control for base composition effect. The distances between the 5′-splice site and both PTCs and intronic NCs, were determined for each gene and then assigned to 30-nucleotide bins (equivalent to the coding sequence for 10 amino acids). Differences between the in-frame and out-of-frame distributions were determined using the Wilcoxon signed-rank test. We also identified the last introns for A. thaliana, C. elegans, H. sapiens, M. musculus, D. melanogaster, and S. pombe genes that contained at least two introns within the coding sequence. This was done by reversing the annotation and sequences of all genes using custom Perl scripts and counting backward to determine phase and extract the final introns and exons. Once extracted, the final introns and exons were returned to the proper orientation, and PTC position was identified as before. We manually checked that the predicted and actual PTC positions were identical in several genes with different phases of their last introns to make sure the scripts worked as expected.
In addition, we compared the position of the PTC to the length of the first intron and last intron, as well as the position of the PTC to the level of gene expression. Intron length is strongly associated with intron-definition (for introns with fewer than ∼250 nucleotides) vs. exon-definition (for introns with more than ∼250 nucleotides) splicing (De Conti et al. 2013). When a splicing error occurs, intron-definition splicing generally results in the retention of an intron, whereas exon-definition splicing generally results in exon skipping. Introns that undergo intron-definition splicing are expected to have earlier PTCs because the intron is retained in the aberrant message. We thus divided introns into two groups: short introns of ≤250 nucleotides and long introns of >250 nucleotides in length to determine the effects of intron- vs. exon-definition splicing behavior. S. pombe and S. cerevisiae were excluded from the intron length analysis since both species have only intron-definition splicing (Brown et al. 1992; Romfo et al. 2000). Gene expression was also divided into two groups: highly expressed genes, consisting of the top 100 expressed genes within a species, similar to Castillo-Davis et al. (2002), and medium/low expressed genes, consisting of all other genes. We tried other cut-offs for gene expression, including the upper 10% and upper quartile, but results were unaffected (data not shown). Positions of PTCs and frame +1 and +2 intronic NCs were compared in short and long introns and high and low expressed genes using the Mann–Whitney U-test.
Data and reagent availability
All data were acquired from the NCBI and public repositories. Custom Perl scripts written to parse the data are available at: https://github.com/behrimg/Scripts/tree/master/Hall_Projects/Introns.
Results
In seven model species, we analyzed protein-coding genes that met the following four criteria: they (1) contained at least one intron, (2) could be matched to published expression data, (3) were not reported to have alternative splice forms, and (4) were not duplicates according to annotation data. For the seven species examined, there were between 161 (S. cerevisiae) and 10,869 genes (C. elegans) that met these criteria (Table 1).
Location of in-frame PTCs
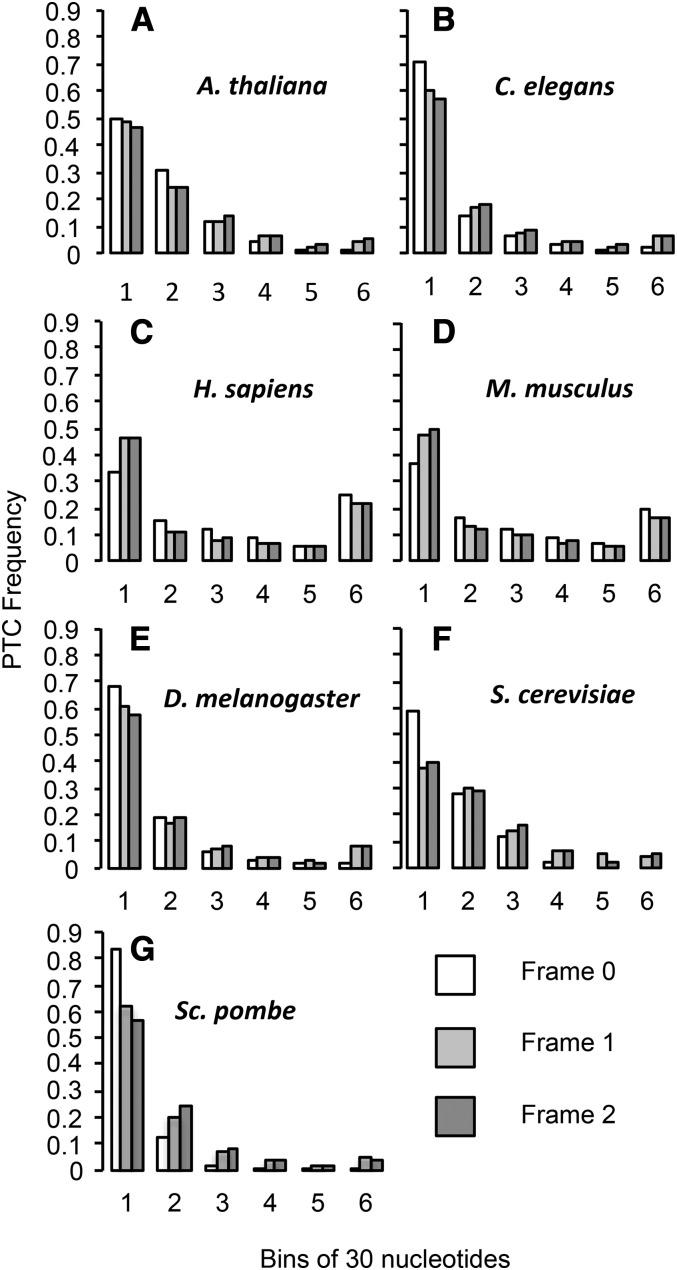
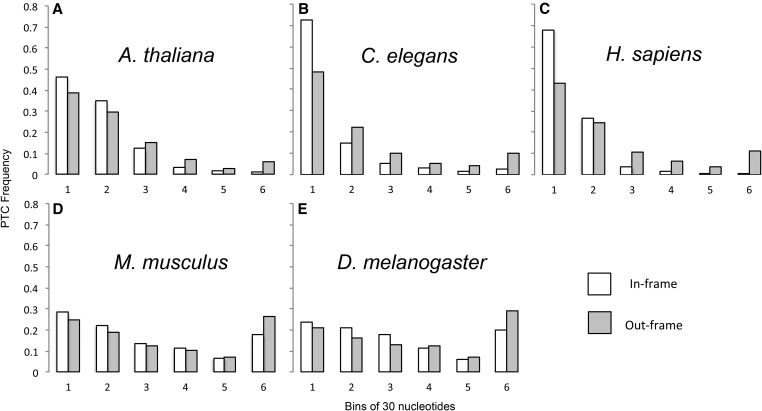
The position of the first PTC, and the first intronic frame +1 or +2 NC, relative to the 5′-splice site was determined for the first intron of each gene. Since out-of-frame NCs do not terminate translation, they should not be visible to selection and their position thus controls for the pattern of nucleotide composition within introns. We determined whether the first PTCs occur earlier than the first out-of-frame NCs. For all nonmammal species, first PTCs appear from 1 to 23 bases earlier in the intron than first out-of frame NCs, which is a highly significant difference in four of five comparisons (Wilcoxon sign-rank test: A. thaliana, P = 1.38 × 10−9; C. elegans, P < 2.20 × 10−16; D. melanogaster, P = 0.784; S. pombe, P < 2.20 × 10−16; and S. cerevisiae, P ≈ 6.23 × 10−7). In contrast, PTC position in mammals is 19–24 bases later than first out-of-frame NCs, which is again highly significant (Wilcoxon sign-rank test: H. sapiens, P = 1.59 × 10−15; M. musculus, P < 2.20 × 10−16) (Table S1, Figure 1).
Figure 1.
First PTCs occur significantly earlier than expected in the first intron for all nonmammals and significantly later for mammals. Observed distances in base pairs between the 5′-splice site and first PTCs (white bar) and out-of-frame NCs (gray bars). Distances are separated into six bins, each of which is 30 nucleotides in length, except for bin 6, which contains all distances longer than 150 nucleotides. A–G are in order of increasing effective population size (Table 1).
In S. cerevisiae, there are data that identify the genes that are especially NMD sensitive, obtained through analysis of NMD knockout strains (Sayani et al. 2008). Such NMD-sensitive genes are expected to have especially early PTCs. Across all 161 (intron containing) genes in our data set, PTCs are ∼1.5 times more likely to occur in the first 30 nucleotides of the first intron than are frame +1 and +2 NCs across all genes. In the 27 genes that are especially NMD sensitive, the excess is ∼1.9-fold (Figure S1), which is the expected pattern, although it is not significant (P = 0.09, Fisher’s exact test comparing number of PTCs in bin 1 for NMD-sensitive and NMD-insensitive genes), likely because the number of genes is quite small.
In mammals, entry into the NMD pathway differs from nonmammals (see Discussion), and PTCs in the last intron are unable to trigger NMD. To determine whether the position of the first PTCs is affected by recruitment of the NMD pathway, we performed the same position analysis in terminal introns. S. cerevisiae has only nine genes containing multiple introns and was thus not included in this analysis. While for S. pombe there is no significant difference between in-frame PTCs and out-of-frame NCs, in the remaining species NCs in frame 1 occur earlier in the first 30 nucleotides than PTCs and frame 2 NCs (Wilcoxon sign-rank test: A. thaliana, P < 2.20 × 10−16; C. elegans, P < 2.20 × 10−16; H. sapiens, P < 2.20 × 10−16; M. musculus, P < 2.20 × 10−16; D. melanogaster, P = 0.003; and S. pombe, P = 0.051) (Figure S2, Table S2).
PTCs remain early when accounting for splice site consensus
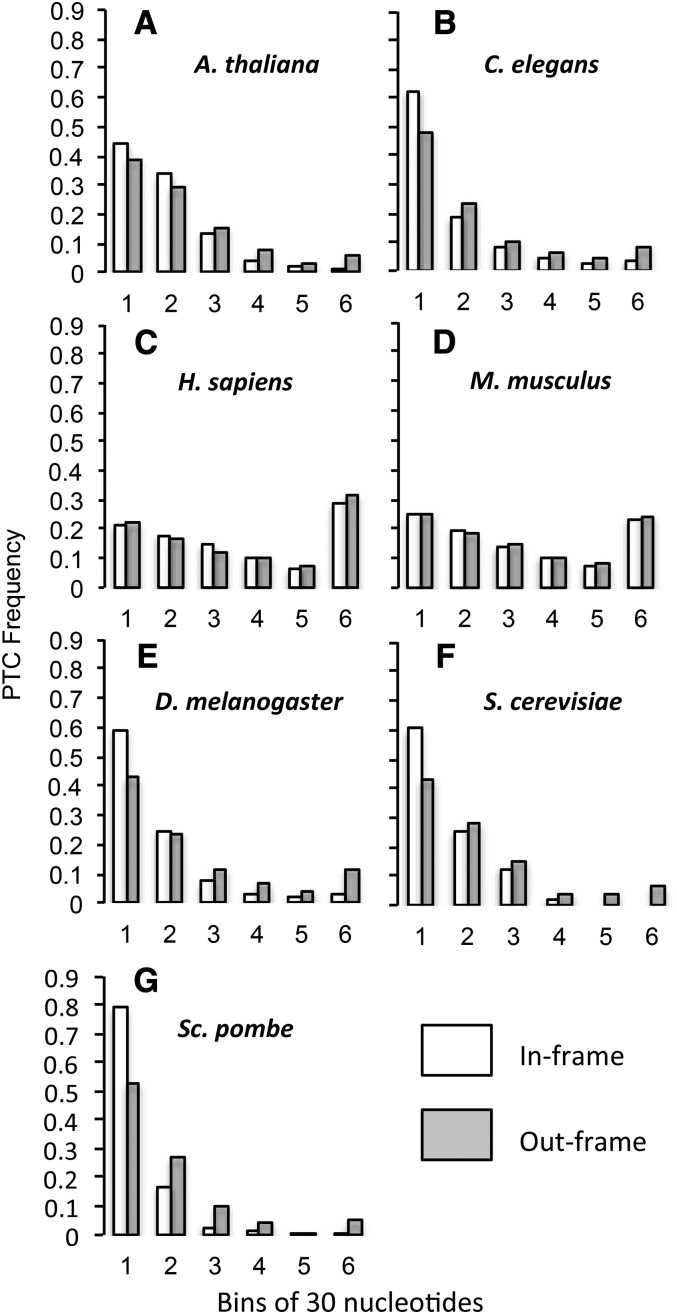
It is possible that the conserved splice consensus at the 5′ end of the intron affects PTC position. Except for S. cerevisiae, all of the species examined contain a stop-codon-like trinucleotide (TGA or TAA) within the 5′-splice consensus GTRAGT. Phase 2 introns, which are those in which the first base of the intron begins, if unspliced, in the third position of a codon, would have a PTC introduced by the 5′-splice consensus sequence. Phase 1 (unspliced first intron base is second position in a codon) and phase 0 (unspliced first intron base is first position in a codon) introns would both have an out-of-frame NC in the splice consensus sequence. If a majority of introns were phase 2, this would make PTCs appear relatively earlier. We thus repeated our analysis, discounting phase/frame combinations that introduce splice site PTCs and out-of-frame NCs. To do this, we reduced the data set by examining the position of PTCs in phase 0 and phase 1 introns and compared them to the position of out-of-frame NCs in phase 0, frame +2 and phase 1, frame +1 introns, respectively, and found that the pattern of earlier PTC position relative to out-of-frame NC position in first introns was now significant across all nonmammals (A. thaliana, P < 2.20 × 10−16; C. elegans, P < 2.20 × 10−16; D. melanogaster, P < 2.20 × 10−16; and S. pombe, P < 2.20 × 10−16) (Figure 2, Table S3). In mammals, the position of the first PTC was no longer significantly later than the first out-of-frame NC and was actually significantly earlier in humans (Wilcoxon sign-rank test: H. sapiens, P = 0.003; and M. musculus, P = 0.270; Bonferroni correction: P ≤ 0.008) (Figure 2, Table S3).
Figure 2.
After removing the effect of the splice site, the first PTC still occurs significantly earlier than expected in the first intron for nonmammals, while no difference was observed in mammals. Observed distances in base pairs between the 5′-splice site and first PTCs (white bar) and out-of-frame NCs (gray bars). Distances are separated into six bins, each of which is 30 nucleotides in length, except for bin 6, which contains all distances longer than 150 nucleotides. A–G are in order of increasing effective population size (Table 1).
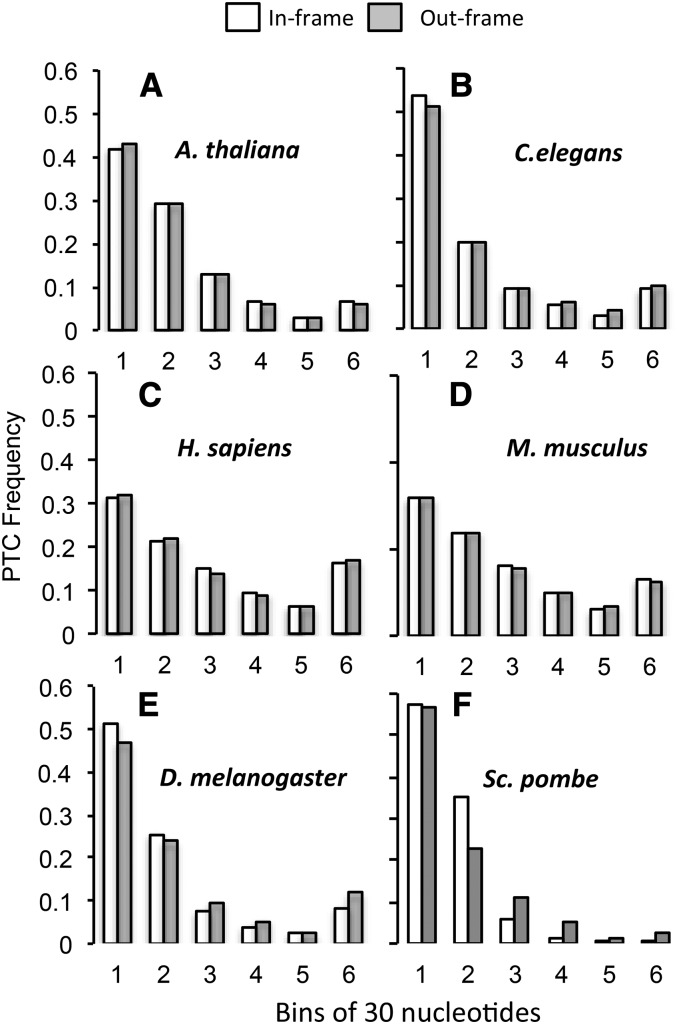
In last introns, after controlling for stop codons introduced by the splice site consensus sequence, PTCs occurred significantly earlier in D. melanogaster, and were not different from NCs in A. thaliana, C. elegans, H. sapiens, M. musculus, or S. Pombe. (Wilcoxon sign-rank test: A. thaliana P = 0.036; C. elegans, P = 0.013; H. sapiens, P = 0.518; M. musculus, P = 0.938; D. melanogaster, P = 1.15 × 10−6; and S. pombe, P = 0.244; Bonferroni correction: P ≤ 0.008) (Figure 3, Table S4).
Figure 3.
After removing the effect of splice site on PTC and NC frequencies, in-frame PTCs occur significantly earlier than expected in the last intron for C. elegans and D. melanogaster and later than expected for A. thaliana. Observed distances in base pairs between the 5′-splice site and first PTCs (white bar) and out-of-frame NCs (gray bars). Distances are separated into six bins, each of which is 30 nucleotides in length, except for bin 6, which contains all distances longer than 150 nucleotides. A–F are in order of increasing effective population size (Table 1).
PTC position is affected by intron length but not gene expression
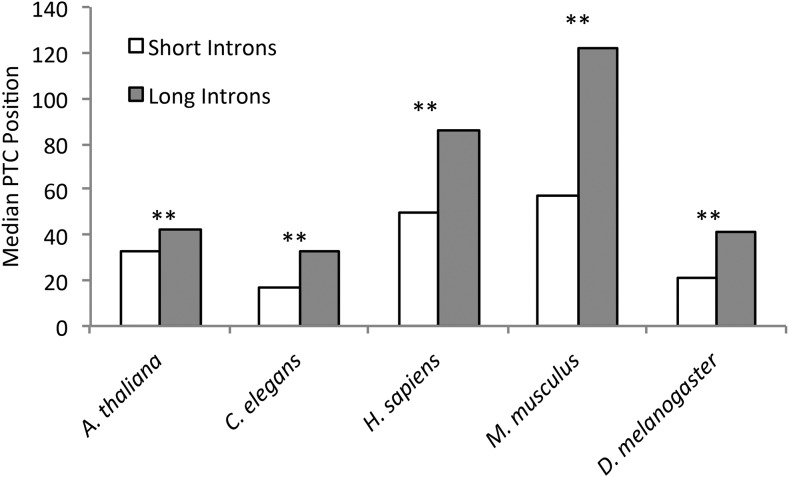
In species with both intron-definition and exon-definition splicing, mis-splicing of introns shorter than ∼250 nucleotides results in intron retention, while mis-splicing of introns longer than ∼250 nucleotides results in exon skipping (De Conti et al. 2013), which should lead to earlier PTCs in short introns if position is under selection. When we examined PTC position as a function of intron length, we found that PTCs occur closer to the 5′-splice site in first introns that are shorter than 250 nucleotides (short introns) (Figure 4) (Wilcoxon sign-rank test: A. thaliana, P < 2.20 × 10−16; C. elegans, P < 2.20 × 10−16; H. sapiens, P = 2.09 × 10−8; M. musculus, P = 0.007; and D. melanogaster, P < 2.20 × 10−16). S. cerevisiae and S. pombe were omitted from the intron length analysis because all of their introns utilize intron-definition splicing (Brown et al. 1992; Romfo et al. 2000).
Figure 4.
PTCs occur earlier in short introns. Median position of the first PTC in first introns, measured in nucleotides downstream of the 5′-splice site, for short and long introns. Introns ≤250 nucleotides are considered short introns and generally result in intron retention when mis-spliced. Introns >250 nucleotides are considered long introns and generally result in exon skipping when mis-spliced. Bonferroni corrected ** P-value <0.01.
To determine whether the cause of early PTC position in short introns was due to NMD, we repeated this analysis in last introns. For C. elegans and D. melanogaster, PTCs in short introns were significantly earlier, and in A. thaliana, M. musculus, and H. sapiens, PTCs in short introns occurred no earlier or later than out-of-frame PTCs (Figure S4, Wilcoxon sign-rank test: A. thaliana, P = 0.595; C. elegans, P = 0.001; H. sapiens, P = 0.129; M. musculus, P = 0.191; and D. melanogaster, P = 9.41 × 10−10; Bonferroni correction P ≤ 0.01).
Further, previous work has shown that in C. elegans and humans, highly expressed genes tend to have shorter introns (Castillo-Davis et al. 2002). We find a similar pattern in D. melanogaster, M. musculus, but no clear relationship between intron size and expression in A. thaliana. When analyzing the effect of gene expression on PTC position, we found PTC position did not differ in highly expressed genes in any of the species (Figure S3; Mann–Whitney U-test: A. thaliana, P = 0.756; C. elegans, P = 0.253; H. sapiens, P = 0.486; M. musculus, P = 0.680; D. melanogaster, P = 0.016; S. pombe, P = 0.819; Bonferroni correction P ≤ 0.008).
Correlations with effective population size
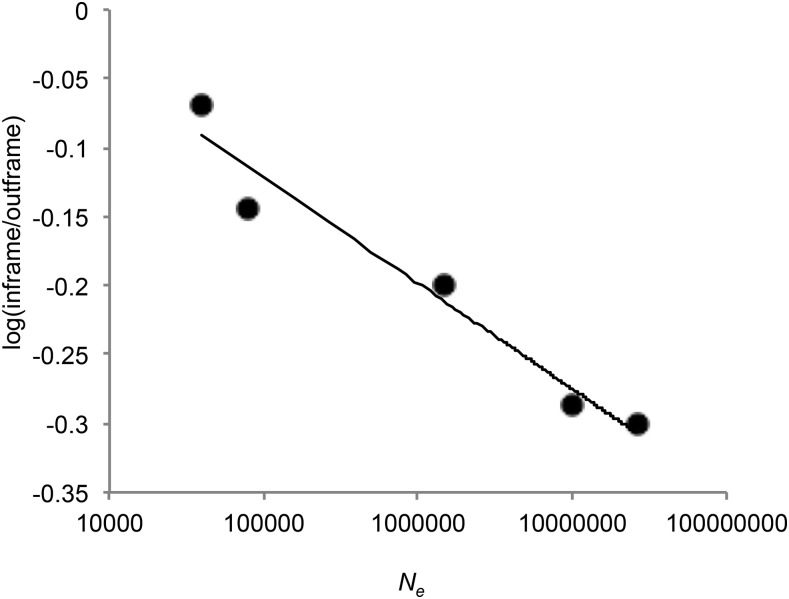
Several genomic patterns are often more apparent in species with larger effective population sizes, where selection is more effective (Lynch and Conery 2003). For the subset of introns ≤250 nucleotides in all species, the logarithm of the ratio of median PTC nucleotide position to median first NC nucleotide position is negatively correlated with the effective population size after the effect of splice site is removed (d.f. = 6, Spearman’s r = −0.893, P = 0.007; Figure 5). Thus, for species that show early position of the first PTC, those with larger effective population sizes have earlier PTCs in their first intron. In terminal introns, the correlation is also negative, although not significant (d.f. = 2, Spearman’s r = −0.257, P = 0.623), perhaps because there are only three species that have earlier PTCs in last introns.
Figure 5.
Median position of in-frame PTCs correlates with effective population size. Data points represent log ratios of median in-frame PTC distance and out-of-frame PTC distance for introns ≤250 nucleotides once splice site context is removed from the 5′-splice site. (d.f. = 4, Spearman’s r = −0.893, P = 0.007). Organisms in order of increasing Ne (i.e., points going left to right): A. thaliana, C. elegans, H. sapiens, M. musculus, D. melanogaster, S. cerevisiae, and S. pombe.
Discussion
We find evidence for selection acting on PTC position within introns, supporting our hypothesis that selection favors early PTC position for prompt entry into the NMD pathway leading to efficient termination of translation. The pattern remains after removing the possibly confounding effect of splice site consensus. Evidence for early PTC position was also reported in a recent paper that examined four species of ascomycete fungi, including the two in this study (Zafrir et al. 2016). While both studies reach a similar conclusion regarding PTC position, the Zafrir et al. (2016) study showed a statistically less significant difference than we report. Zafrir et al. (2016) used a randomization procedure for generating the expected PTC position, which we abandoned as inappropriate because of variation in nucleotide content along the first intron (see Materials and Methods and Figure S5).
We initially examined the first intron across seven species since this intron is present in all intron-containing genes. We found that in the five nonmammalian species, PTCs occur earlier than expected, regardless of whether the possibly confounding effects of splice site are removed. In the two mammals, however, PTCs appear significantly later than expected. However, when the effect of splice site is removed, the distribution of mammalian PTCs is not significantly different from the distribution of intronic out-of-frame NCs. We hypothesize that mammals do not show the same pattern of early PTCs in first introns observed in nonmammals because they mainly contain long introns. In fact, 94.3 and 95.1% of human and mouse introns are long, compared to 0–41.4% in the other five species.
We found that long introns show less evidence for selection favoring early PTC position. Long introns differ from short introns because they usually use an exon-definition splicing strategy, where splicing errors result in exon skipping (Berget 1995), and there is thus no opportunity for selection to act on PTC position in a retained intron. Short introns, in contrast, use an intron-definition splicing strategy, where splicing errors result in intron inclusion (Talerico and Berget 1994). Therefore, error in splicing shorter introns is more likely to result in the aberrant mRNA containing an intron, which can then be translated. Under our hypothesis, selection on PTC position is mediated by intron translation, making PTC position more likely to be early in short introns, which is what we find across all species. This observation supports our conjecture for why mammals do not show early PTC position when averaged across all of their introns because most of their introns are long. Mammals do have earlier PTC position in short but not in long introns (Figure 6).
Figure 6.
The first PTC occurs significantly earlier than expected in the first intron for all organisms after accounting for retained introns and splice site effects. Observed distances in base pairs between the 5′-splice site and first PTCs (white bar) and out-of-frame NCs (gray bars). Distances are separated into six bins, each of which is 30 nucleotides in length, except for bin 6, which contains all distances longer than 150 nucleotides. A–E are in order of increasing effective population size (Table 1).
Gene expression appears to have no effect on PTC position. This is an unexpected finding if the cost of translation is an important factor causing PTCs to be early because highly expressed genes might be expected to produce higher numbers of aberrant mRNAs. One explanation for highly expressed genes not exhibiting earlier PTCs is that they experience stronger selection for higher splicing efficiency compared to genes expressed at lower levels. If splicing efficiency is higher, then a lower percentage of transcripts will be aberrant at highly expressed genes, such that the number of aberrant transcripts might not differ as a function of gene expression. If the number of aberrant transcripts does not differ with expression, then the opportunity for selection on PTCs in introns for highly expressed genes is no greater than for lowly expressed genes (Wilhelm et al. 2008).
In contrast to our finding, the Zafrir et al. (2016) study found a significant and substantial effect of gene expression in both S. cerevisiae and S. pombe. In S. cerevisiae (and S. pombe), they examined the 60 (500) highest and 60 (500) lowest expressed intron-containing genes. They found that the average PTC position in the first intron of highly expressed genes is earlier than 99.9 and 99.5% of the randomized introns in S. cerevisiae and S. pombe, respectively. In contrast, in lowly expressed genes, the average PTC position is in the middle of their randomized intron distribution. It is not obvious to us what is driving the different findings between the two studies. They used protein abundance as a proxy for expression level, while we used RNA expression, but that difference seems unlikely to alter the ranking of genes with respect to gene expression enough to make a difference. They also use randomized introns to generate the expected PTC position (see above), though it is not clear why that might disproportionately affect highly expressed genes. Given the different findings between the two studies, coupled with similar findings in the other species that we analyzed, we feel it is most conservative to conclude that there is not a significant effect of gene expression on PTC location in these species.
We also examined PTC position in last introns for genes containing two or more introns in the six species with genes containing multiple introns. We analyzed PTC position in last introns because in mammals, PTCs in last introns do not trigger NMD, and therefore PTC position is not expected to be as early in mammals if entry into the NMD pathway is a primary factor favoring early position (Maquat 2005). Two of the three nonmammals, C. elegans and D. melanogaster, exhibit earlier PTC position in last introns, although the pattern is weaker, i.e., PTC position is not as early as in first introns. However, A. thaliana has PTCs that occur slightly, but significantly later than expected in its last introns. Additionally, in mammals, where NMD is not induced in last introns, PTCs do not occur earlier than expected. To determine whether this pattern is affected by intron length as it was in first introns, we analyzed the effect of intron length on PTC position in the last intron. In A. thaliana, C. elegans, and D. melanogaster we find short introns have earlier intronic PTCs. Mammals show a contrasting pattern, however, where short introns have PTCs that are significantly later in the last intron.
PTCs may be not as early in last introns because selection is weaker. One possibility is that the deleterious effect on the encoded protein of incorrectly splicing the last intron may be less than for the first intron, perhaps because only a relatively small number of amino acids at the carboxyl end of the protein are altered. Another possibility may be caused by the connection between polyadenylation of mature mRNA and the removal of the last intron (Nesic and Maquat 1994; Proudfoot 2011). If the last intron is not spliced correctly, then the mRNA may not be polyadenylated and therefore not exported from the nucleus for translation. If the aberrant mRNA is not translated, then there is no selective pressure for an earlier PTC.
In summary, our results suggest that selection may favor early translation termination in the incorrectly spliced mRNAs that retain the first intron in all examined species and the last intron in nonmammals. We postulate that selection favors timely termination of translation in aberrantly spliced mRNAs. The muting of the pattern in last introns suggests that selection is less effective in terminal introns. When our predictions do hold, they are more pronounced in species with larger effective population sizes (Figure 5). Because the frequency of aberrant splicing varies, the strength of selection acting on unspliced transcripts is likely weak, which explains the strong sensitivity to effective population size (Wilhelm et al. 2008; Drummond and Wilke 2009; Fox-Walsh and Hertel 2009).
Nature of selection on PTCs
If first PTCs are selected to occur earlier, as our results suggest, then there must be one or more benefits to early, efficient termination of aberrant mRNAs containing introns. There are several possible costs that might cause selection to favor earlier PTC position. These include: (1) The opportunity cost of not creating enough functional proteins because ribosomes are translating aberrant mRNAs encoding nonfunctional protein products; (2) the energetic cost of synthesizing aberrant proteins; (3) the energetic cost of breaking down abnormal proteins; and (4) the functional cost of creating proteins that are deleterious to the cell. All of these costs would be reduced by more expeditious induction of NMD since aberrant mRNAs would be more rapidly degraded. In addition, earlier ribosome release would reduce both the opportunity cost and the elongation and break down energetic costs of the aberrant proteins, which would select for earlier position of first PTCs.
Of these four possibilities, we suspect that the opportunity cost of ribosomes translating nonfunctional products and the clean up costs of the abnormal proteins may be the most important contributors to early PTC position within introns. The cost to decay an aberrant protein is directly proportional to the amount of aberrant protein produced, and the occupation of aberrant mRNAs by ribosomes, producing nonfunctional protein products, represents an opportunity cost to the organism (Drummond and Wilke 2009). Ribosomes translating aberrant mRNAs are unavailable for production of other functional protein products. Selection for efficient translation termination as early as possible in the incorrectly spliced message would minimize the amount of aberrant protein the cell has to decay, as well as the duration that the aberrant transcript occupies the ribosome.
Elongation and sequestering of limited amino acid resources in aberrant product has been hypothesized to be a major energetic cost (Stoebel et al. 2008). However, studies in Escherichia coli show that supplementing amino acids does not change the cost of protein expression, which suggests that amino acid limitation is therefore not likely a major selection pressure on PTC position (Stoebel et al. 2008).
Finally, while it is clear that creating aberrant proteins that are toxic will be deleterious in terms of fitness, and this is likely the primary selection pressure associated with the evolution of the NMD pathway itself, we suggest that it is less likely to be an explanation for the position of PTCs. The reason is that an earlier PTC position only moderately shortens the aberrant protein, which may generally have little or no effect on its toxicity.
Future directions
The data suggest that the position of PTCs may be shaped by selection. Data from additional species is clearly needed to confirm the patterns, especially in species with large effective population sizes where the patterns are expected to be more robust. In addition, the fact that patterns are more apparent for introns that are particularly sensitive to NMD in S. cerevisiae suggests that distinguishing these two classes of genes in other species would be useful for elucidating the predicted patterns. While studies have been conducted in A. thaliana, C. elegans, D. melanogaster, and H. sapiens identifying transcripts that are upregulated in NMD-deficient cells, these studies generally rely on gene expression and do not indicate specifically introns that are particularly sensitive to NMD (Mendell et al. 2004; Metzstein and Krasnow 2006; Ramani et al. 2009; Drechsel et al. 2013).
One variable that we have not addressed is the frequency with which aberrant mRNAs are produced. The expectation is that genes that routinely produce aberrant mRNAs would experience selection on PTC position and termination efficiency more often. For example, in the extreme case in which a gene is never aberrantly spliced, it would never experience selection on PTCs. Tantalizing data that can be interpreted as supporting this prediction comes from S. cerevisiae in which highly NMD-sensitive introns exhibit earlier first PTC positions than introns less sensitive to NMD (Figure S1). If NMD sensitivity is related to the probability of aberrant splicing, such that commonly retained introns are more likely to be NMD sensitive, then NMD-sensitive introns will also be those with earlier PTCs. Testing the prediction that introns with less efficient splicing will show greater PTC position effects is challenging. The problem is that steady-state levels of aberrant mRNAs, such as would be measured in a standard RNA-sequencing experiment, are affected by both the production of and the degradation of the aberrant mRNAs (Pelechano et al. 2010) For this reason, it would be necessary to measure aberrant mRNA production directly, which is a more challenging, and expensive, undertaking.
Acknowledgments
We would like to thank the Associate Editor and two anonymous reviewers for very helpful comments on a previous versions of this manuscript. We would also like to thank K. Dyer, J. Mrazek, and J. Wares for their helpful insight throughout the course of this project. This work was supported by the National Institutes of Health grants R01GM097415 and T32 GM007103.
Footnotes
Communicating editor: C. D. Jones
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.189894/-/DC1.
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Amit M., Donyo M., Hollander D., Goren A., Kim E., et al. , 2012. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Reports 1: 543–556. [DOI] [PubMed] [Google Scholar]
- Andolfatto P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Behringer M. G., Hall D. W., 2016. Genome-wide estimates of mutation rates and spectrum in Schizosaccharomyces pombe indicate CpG sites are highly mutagenic despite the absence of DNA methylation. G3 (Bethesda) 6: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S. M., 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270: 2411–2414. [DOI] [PubMed] [Google Scholar]
- Brown J. D., Plumpton M., Beggs J. D., 1992. The genetics of nuclear pre-mRNA splicing: a complex story, pp. 35–46 in Molecular Biology of Saccharomyces. Springer-Verlag, The Netherlands. [DOI] [PubMed] [Google Scholar]
- Brown W. R., Liti G., Rosa C., James S., Roberts I., et al. , 2011. A geographically diverse collection of Schizosaccharomyces pombe isolates shows limited phenotypic variation but extensive karyotypic diversity. G3 (Bethesda) 1: 615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caenorhabditis elegans Sequencing Consortium , 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2018. [DOI] [PubMed] [Google Scholar]
- Carviel J. L., Al-Daoud F., Neumann M., Mohammad A., Provart N. J. et al, 2009. Forward and reverse genetics to identify genes involved in the age‐related resistance response in Arabidopsis thaliana. Mol. Plant Pathol. 10: 621–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis C. I., Mekhedov S. L., Hartl D. L., Koonin E. V., Kondrashov F. A., 2002. Selection for short introns in highly expressed genes. Nat. Genet. 31: 415–418. [DOI] [PubMed] [Google Scholar]
- Chen F.-C., Li W.-H., 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68: 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Cutter A. D., 2006. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics 172: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Conti L., Baralle M., Buratti E., 2013. Exon and intron definition in pre‐mRNA splicing. Wiley Interdiscip. Rev. RNA 4: 49–60. [DOI] [PubMed] [Google Scholar]
- Deutsch M., Long M., 1999. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 27: 3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezso Z., Nikolsky Y., Nikolskaya T., Miller J., Cherba D., et al. , 2009. Identifying disease-specific genes based on their topological significance in protein networks. BMC Syst. Biol. 3: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G., Kahles A., Kesarwani A. K., Stauffer E., Behr J., et al. , 2013. Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. The Plant Cell Online 25: 3726–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D. A., Wilke C. O., 2009. The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 10: 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S. R., Dietrich F. S., Fisk D. G., Binkley G., Balakrishnan R., et al. , 2014. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda) 4: 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erfle H., Ventzki R., Voss H., Rechmann S., Benes V., et al. , 2000. Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana. Nature 408: 820–822. [DOI] [PubMed] [Google Scholar]
- Farlow A., Dolezal M., Hua L., Schlötterer C., 2012. The genomic signature of splicing-coupled selection differs between long and short introns. Mol. Biol. Evol. 29: 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Walsh K. L., Hertel K. J., 2009. Splice-site pairing is an intrinsically high fidelity process. Proc. Natl. Acad. Sci. USA 106: 1766–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., 1978. Why genes in pieces? Nature 271: 501. [DOI] [PubMed] [Google Scholar]
- Halligan D. L., Eyre-Walker A., Andolfatto P., Keightley P. D., 2004. Patterns of evolutionary constraints in intronic and intergenic DNA of Drosophila. Genome Res. 14: 273–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M. A., Rodriguez C. M., Johnson T. L., 2011. Key features of the two-intron Saccharomyces cerevisiae gene SUS1 contribute to its alternative splicing. Nucleic Acids Res. 39: 8612–8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Bouhouche K., Gout J.-F., Aury J.-M., Noel B., et al. , 2008. Translational control of intron splicing in eukaryotes. Nature 451: 359–362. [DOI] [PubMed] [Google Scholar]
- Kelkar Y. D., Ochman H., 2012. Causes and consequences of genome expansion in fungi. Genome Biol. Evol. 4: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer D. M., Drabenstot S. D., Buchanan K. L., Lai H., Zhu H., et al. , 2004. Introns and splicing elements of five diverse fungi. Eukaryot. Cell 3: 1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., et al. , 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lim L. P., Burge C. B., 2001. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl. Acad. Sci. USA 98: 11193–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Kaul S., Rounsley S., Shea T. P., Benito M. I., et al. , 1999. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402: 761–768. [DOI] [PubMed] [Google Scholar]
- Lynch M., 2002. Intron evolution as a population-genetic process. Proc. Natl. Acad. Sci. USA 99: 6118–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2007. The Origins of Genome Architecture. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Lynch M., Conery J. S., 2003. The origins of genome complexity. Science 302: 1401–1404. [DOI] [PubMed] [Google Scholar]
- Maquat L. E., 2005. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 118: 1773–1776. [DOI] [PubMed] [Google Scholar]
- Mayer K., Schüller C., Wambutt R., Murphy G., Volckaert G., et al. , 1999. Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402: 769–777. [DOI] [PubMed] [Google Scholar]
- Mendell J. T., Sharifi N. A., Meyers J. L., Martinez-Murillo F., Dietz H. C., 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 36: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Metzstein M. M., Krasnow M. A., 2006. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2: e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., 1982. A catalogue of splice junction sequences. Nucleic Acids Res. 10: 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic D., Maquat L. E., 1994. Upstream introns influence the efficiency of final intron removal and RNA 3′-end formation. Genes Dev. 8: 363–375. [DOI] [PubMed] [Google Scholar]
- Pelechano V., Chávez S., Pérez-Ortín J. E., 2010. A complete set of nascent transcription rates for yeast genes. PLoS One 5: e15442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phifer-Rixey M., Bonhomme F., Boursot P., Churchill G. A., Piálek J., et al. , 2012. Adaptive evolution and effective population size in wild house mice. Mol. Biol. Evol. 29: 2949–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., 2011. Ending the message: poly (A) signals then and now. Genes Dev. 25: 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A. K., Nelson A. C., Kapranov P., Bell I., Gingeras T. R., et al. , 2009. High resolution transcriptome maps for wild-type and nonsense-mediated decay-defective Caenorhabditis elegans. Genome Biol. 10: R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressayre A., Glémin S., Montalent P., Serre-Giardi L., Dillmann C., et al. , 2015. Introns structure patterns of variation in nucleotide composition in Arabidopsis thaliana and rice protein-coding genes. Genome Biol. Evol. 7: 2913–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romfo C. M., Alvarez C. J., van Heeckeren W. J., Webb C. J., Wise J. A., 2000. Evidence for splice site pairing via intron definition in Schizosaccharomyces pombe. Mol. Cell. Biol. 20: 7955–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S., Janis M., Lee C. Y., Toesca I., Chanfreau G. F., 2008. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol. Cell 31: 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen D. J., Brown A., 1991. Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants. Proc. Natl. Acad. Sci. USA 88: 4494–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Huang W., Zhang C., Hubisz M. J., Lu J., et al. , 2007. Adaptive genic evolution in the Drosophila genomes. Proc. Natl. Acad. Sci. USA 104: 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth N., Roca X., Hastings M. L., Roeder T., Krainer A. R., et al. , 2006. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 34: 3955–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasundar A., Hey J., 2003. Population genetics of Caenorhabditis elegans: the paradox of low polymorphism in a widespread species. Genetics 163: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelly D. A., Ronald J., Connelly C. F., Akey J. M., 2009. Population genomics of intron splicing in 38 Saccharomyces cerevisiae genome sequences. Genome Biol. Evol. 1: 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoebel D. M., Dean A. M., Dykhuizen D. E., 2008. The cost of expression of Escherichia coli lac operon proteins is in the process, not in the products. Genetics 178: 1653–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata S., Kaneko T., Nakamura Y., Kotani H., Kato T., et al. , 2000. Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408: 823. [DOI] [PubMed] [Google Scholar]
- Talerico M., Berget S. M., 1994. Intron definition in splicing of small Drosophila introns. Mol. Cell. Biol. 14: 3434–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H., Iwasaki O., Tanaka A., Capizzi J. R., Wickramasinghe P., et al. , 2010. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res. 38: 8164–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A., Ecker J. R., Palm C. J., Federspiel N. A., Kaul S., et al. , 2000. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 408: 816–820. [DOI] [PubMed] [Google Scholar]
- Wen J., Brogna S., 2010. Splicing‐dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO J. 29: 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R., 2005. FeatureExtract: extraction of sequence annotation made easy. Nucleic Acids Res. 33: W567–W569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney K. D., Garland T., Jr, 2010. Did genetic drift drive increases in genome complexity? PLoS Genet. 6: e1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm B. T., Marguerat S., Watt S., Schubert F., Wood V., et al. , 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243. [DOI] [PubMed] [Google Scholar]
- Wood V., Gwilliam R., Rajandream M.-A., Lyne M., Lyne R., et al. , 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Wright S. I., Andolfatto P., 2008. The impact of natural selection on the genome: emerging patterns in Drosophila and Arabidopsis. Annu. Rev. Ecol. Evol. Syst. 39: 193. [Google Scholar]
- Wright S. I., Lauga B., Charlesworth D., 2002. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 19: 1407–1420. [DOI] [PubMed] [Google Scholar]
- Zafrir Z., Tuller T., 2015. Nucleotide sequence composition adjacent to intronic splice sites improves splicing efficiency via its effect on pre-mRNA local folding in fungi. RNA 21: 1704–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrir Z., Zur H., Tuller T., 2016. Selection for reduced translation costs at the intronic 5′ end in fungi. DNA Res. 23: 377–394. [DOI] [PMC free article] [PubMed] [Google Scholar]