By the early 1920s, the existence of mutations was well established, but how they could be generated remained a topic of lively speculation. One interesting case was the Drosophila Bar mutation (Tice 1914). While normal flies have round eyes, the X-linked mutation Bar (B) caused the eyes to be small and slit-like in males and homozygous females; female heterozygotes had kidney bean-shaped eyes (Figure 1A). Intriguingly, the Bar mutation was somewhat unstable: it tended to revert to wild-type spontaneously (May 1917). In 1919, Zeleny reported that females homozygous for Bar had progeny with round eyes at a frequency of ∼1/1000 (Zeleny 1919). What could be going on? Why was this mutation so unstable? And what were the more extreme “ultra-Bar” progeny of Bar females, whose eyes were even more severely reduced than Bar mutants’, and which appeared at a frequency similar to that of the apparent revertants?
Figure 1.
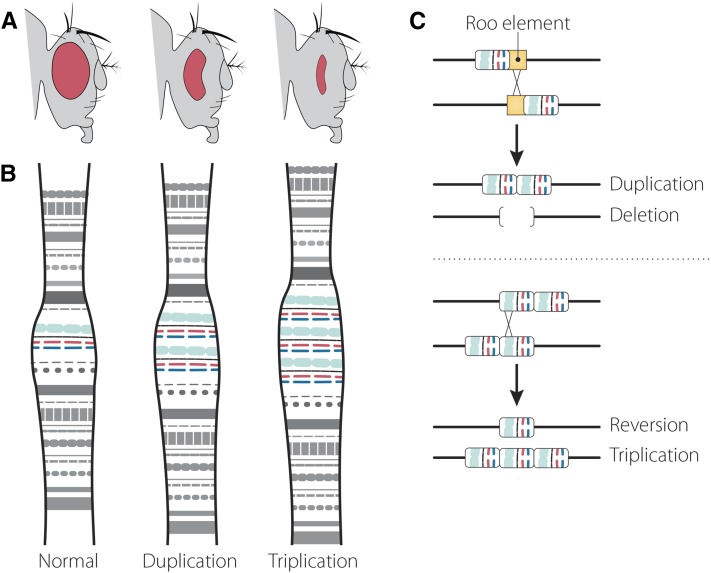
Bar alleles and their phenotypes. (A) Wild-type Drosophila have round eyes. Flies homozygous (or hemizygous) for the Bar mutation have thin, slit-like eyes. Flies homozygous (or hemizygous) for the double-Bar mutation have even smaller eyes. (B) Schematic of the Bar region of polytene chromosomes. The Bar mutation is a tandem duplication and double-Bar a tandem triplication of the region. (C) The Bar mutation arose by unequal crossing over between two Roo transposable elements (yellow), resulting in a tandem duplication. Reversion and triplication alleles arose from the Bar mutant by unequal crossover between duplicates that had aligned out of register.
Having figured out as an undergraduate how to map gene positions using three-point crosses, Alfred Sturtevant turned his talents to exploring the Bar mutation. He realized that he could apply similar logic to test the hypothesis that revertant Bar alleles reflected a structural change in the gene or chromosome caused by recombination; one simply had to examine the progeny of flies carrying Bar alleles flanked by other markers. In a 1923 Science paper (Sturtevant and Morgan 1923), he and his mentor T. H. Morgan reported that females heterozygous for Bar and a Bar allele flanked by forked (f) and fused (fu) alleles gave round-eyed (Bar-revertant) progeny that carried only one of the two flanking mutations. Females heterozygous for one Bar allele flanked only by f and another Bar allele flanked only by fu gave revertant progeny that were either wild-type for both flanking markers or carried both flanking mutations. These two results suggested that the revertants arose from crossovers within or very near to the Bar allele.
In his 1925 GENETICS paper (Sturtevant 1925), Sturtevant proved that unequal crossing over was responsible for generating not only the revertants but also the reciprocal crossover products: the new, stronger ultra-Bar alleles (which he renamed double-Bar). He showed that unequal recombination between two Bar alleles could generate non-Bar and double-Bar alleles at roughly equal frequencies, suggesting that the Bar mutant-allele was comprised of two units that could be separated by recombination. In this model, the wild-type condition was a single unit at Bar, and the Bar mutation was a duplication. If one of the two Bar units on one homolog paired out-of-register with a unit on the other homolog, a crossover could generate one “revertant” product with a single unit, and one double-Bar product that contained three units (Figure 1C). This was a remarkable insight that would take nearly 90 years to confirm at the molecular level (Miller et al. 2016b). The crossovers between Bar alleles only occurred in females; in a screen of the progeny from over 10,000 males, Sturtevant recovered no evidence of intra-allelic crossovers.
To further test his hypothesis that the alleles at Bar encompassed varying numbers of linked units, Sturtevant used an allele named infraBar that gave a weaker phenotype. Using the f and fu flanking markers, he showed that recombination in a Bar/infraBar female could occasionally generate round-eyed revertants or Bar-infraBar chromosomes (analogous to how Bar/Bar females could occasionally generate round revertants or double-Bar chromosomes). Interestingly, the Bar and infraBar units in double-mutant chromosomes could be in either order (Bar-infraBar or infraBar-Bar) and would retain this order. Therefore, Sturtevant’s work showed that Bar alleles were comprised of units that could be added together or separated by recombination. All of this was achieved without sequencing, PCR, or knowing that Tice’s original Bar mutation—still in use in hundreds of fly labs today—arose from unequal crossing over between two Roo elements (Figure 1C) (Tsubota et al. 1989), let alone the knowledge that the Bar gene encodes homeodomain transcription factors (Higashijima et al. 1992) important for neural and sense organ development. About 10 years after Sturtevant’s paper, cytological studies by Bridges (1936), in the context of interpretations by Muller (1936), confirmed that the Bar mutation was indeed a tandem duplication (Figure 1B).
Sturtevant’s 1925 study was huge (> 100,000 flies) and carefully controlled for genetic background and temperature, factors he noted affected the severity of Bar phenotypes. Though he recognized that unequal crossing over is not the only mechanism for generating mutations, Sturtevant’s discovery was extremely influential. His findings are highly relevant to many areas of current study. Recent work has shown that unequal crossing over among repeats in tandem units analogous to Bar occurs de novo in about 1% of Drosophila meioses (Miller et al. 2016a). This mechanism also underlies the expansion of gene families like Hox genes, key events in the evolution of different body plans (Holland 2015). The influence of gene copy number on phenotype is now evident in many other cases, including for copy number variants in humans (Zhang et al. 2009). Indeed, a study looking at de novo duplications and triplications in Charcot-Marie Tooth disease identified a similar phenomenon: individuals with a triplication at the disease locus have a stronger phenotype than those with a duplication (Liu et al. 2014). Finally, Sturtevant made a very important finding concerning the effects of gene position/context on phenotype: the phenotypic severity of Bar alleles depended on the relative cis/trans position of their repeat units. Measurements of eye-facet number showed that the phenotype of a female with four Bar-locus units depends on how they are distributed across chromosomes: a female with double-Bar (three units) on one X chromosome that is wild-type (one unit) on the other X has a more severe Bar phenotype than a female who has four Bar-locus units arranged as Bar/Bar (two units on each X chromosome). Sturtevant proposed that this could reflect a “different balance of modifying genes in the (duplicated) section of the chromosome;” we would now phrase this as creation of new enhancers. Alternatively, he suggested, the difference could reflect “localized regions of activity” inside what we now know to be the nucleus. Both ideas foreshadowed molecular findings to be made decades later (Gonzalez-Sandoval and Gasser 2016), phenomena that remain under intense study nearly a century after Sturtevant crossed his first bar-eyed flies.
Acknowledgments
Many thanks to Scott Hawley for insightful comments, and to Angela Miller for drawing the figure.
Footnotes
Communicating editor: C. Gelling
ORIGINAL CITATION
Alfred H. Sturtevant
GENETICS March 1, 1925 10: 117–147
Photo caption: Photo of Alfred Sturtevant, 1922, from History of the Marine Biological Laboratory (http://hpsrepository.asu.edu/handle/10776/2527). Licensed as Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported. http://creativecommons.org/licenses/by-nc-sa/3.0/.
Literature Cited
- Bridges C. B., 1936. The Bar “gene” a duplication. Science 83(2148): 210–211. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A., Gasser S. M., 2016. On TADs and LADs: spatial control over gene expression. Trends Genet. 32(8): 485–495. [DOI] [PubMed] [Google Scholar]
- Higashijima S., Kojima T., Michiue T., Ishimaru S., Emori Y., et al. , 1992. Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 6(1): 50–60. [DOI] [PubMed] [Google Scholar]
- Holland P. W, 2015. Did homeobox gene duplications contribute to the Cambrian explosion? Zoological Lett. 1: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Gelowani V., Zhang F., Drory V. E., Ben-Shachar S., et al. , 2014. Mechanism, prevalence, and more severe neuropathy phenotype of the Charcot-Marie-Tooth type 1A triplication. Am. J. Hum. Genet. 94(3): 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May H. G., 1917. Selection for higher and lower facet numbers in the bar-eyed race of Drosophila and the appearance of reverse mutations. Biol. Bull. 33(6): 361–395. [Google Scholar]
- Miller D. E., Smith C. B., Kazemi N. Y., Cockrell A. J., Arvanitakas A. V., et al. , 2016a Whole-genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere effect. Genetics 203(1): 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. E., Cook K. R., Yeganeh Kazemi N., Smith C. B., Cockrell A. J., et al. , 2016b Rare recombination events generate sequence diversity among balancer chromosomes in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 113(10): E1352–E1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1936. Bar duplication. Science 83(2161): 528–530. [DOI] [PubMed] [Google Scholar]
- Sturtevant A. H., 1925. The effects of unequal crossing over at the Bar locus in Drosophila. Genetics 10(2): 117–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., Morgan T. H., 1923. Reverse mutation of the Bar gene correlated with crossing over. Science 57(1487): 746–747. [DOI] [PubMed] [Google Scholar]
- Tice S. C., 1914. A new sex-linked character in Drosophila. Biol. Bull. 26(4): 221–230. [Google Scholar]
- Tsubota S. I., Rosenberg D., Szostak H., Rubin D., Schedl P., 1989. The cloning of the Bar region and the B breakpoint in Drosophila melanogaster: evidence for a transposon-induced rearrangement. Genetics 122(4): 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeleny C., 1919. A change in the Bar gene of Drosophila involving further decrease in facet number and increase in dominance. J. Gen. Physiol. 2(1): 69–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gu W., Hurles M. E., Lupski J. R., 2009. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10: 451–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further readings in GENETICS
- Lewis E. B., 1995. Remembering Sturtevant. Genetics 141: 1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof K. D., 1988. Unequal crossing over then and now. Genetics 120: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Other GENETICS articles by A. H. Sturtevant
- Dobzhansky T., Sturtevant A. H., 1938. Inversions in the Chromosomes of Drosophila pseudoobscura. Genetics 23: 28–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S., Sturtevant A. H., 1932. The linkage relations of certain genes in Oenothera. Genetics 17: 393–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1917. Crossing over without Chiasmatype? Genetics 2: 301–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1920. Genetic studies on Drosophila simulans. I. Introduction. Hybrids with Drosophila melanogaster Genetics 5: 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1921a Genetic studies on Drosophila simulans. II. Sex-linked group of genes. Genetics 6: 43–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1921b Genetic studies on Drosophila simulans. III. Autosomal genes. General discussion. Genetics 6: 179–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1925. The effects of unequal crossing over at the bar locus in Drosophila. Genetics 10: 117–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1928. A further study of the so-called mutation at the bar locus of Drosophila. Genetics 13: 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1936. Preferential segregation in Triplo-IV females of Drosophila melanogaster. Genetics 21: 444–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1940. Genetic data on Drosophila affinis, with a discussion of the relationships in the subgenus Sophophora. Genetics 25: 337–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1945. A gene in Drosophila melanogaster that transforms females into males. Genetics 30: 297–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1946. On the dot chromosomes of Drosophila repleta and D. hydei. Genetics 31: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1956. A highly specific complementary lethal system in Drosophila melanogaster. Genetics 41: 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1963. “Genetics” from 1916 to 1962. Genetics 48: 7–8. [PubMed] [Google Scholar]
- Sturtevant A. H., 2001. Reminiscences of T. H. Morgan. Genetics 159: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., Beadle G. W., 1936. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics 21: 554–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., Dobzhansky T., 1936. Geographical distribution and cytology of “sex ratio” in Drosophila pseudoobscura and related species. Genetics 21: 473–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., Novitski E., 1941. The homologies of the chromosome elements in the genus Drosophila. Genetics 26: 517–541. [DOI] [PMC free article] [PubMed] [Google Scholar]