Abstract
The mechanisms cells use to maintain genetic fidelity via DNA repair and the accuracy of these processes have garnered interest from scientists engaged in basic research to clinicians seeking improved treatment for cancer patients. Despite the continued advances, many details of DNA repair are still incompletely understood. In addition, the inherent complexity of DNA repair processes, even at the most fundamental level, makes it a challenging topic. This primer is meant to assist both educators and students in using a recent paper, “Promotion of homologous recombination by SWS-1 in complex with RAD-51 paralogs in Caenorhabditis elegans,” to understand mechanisms of DNA repair. The goals of this primer are to highlight and clarify several key techniques utilized, with special emphasis on the clustered, regularly interspaced, short palindromic repeats technique and the ways in which it has revolutionized genetics research, as well as to provide questions for deeper in-class discussion.
Keywords: Caenorhabditis elegans, RAD51, double-strand breaks (DSBs), homologous recombination, meiosis
IN all organisms, cells will invariably encounter mutagens that have the potential to alter DNA sequence by either directly or indirectly altering nucleotides. On one hand, changes in DNA sequence provide genetic variation, the raw material for evolution. Yet quite often, the accumulation of accidental lesions results in genetic instability, which manifests as cell death, senescence, or disease (Aguilera and Gómez-González 2008).
Depending on the source of the mutagenic agent, DNA damage can be classified into two categories: endogenous (e.g., errors occurring as a result of normal metabolism) or exogenous (resulting from radiation exposure, carcinogens, or paradoxically, certain drugs used to treat cancer). Fortunately, the presence of mutagens is counteracted by the existence of several robust DNA repair pathways, which ensure that the vast majority of damage that our cells encounter will not cause a permanent change in DNA sequence. At the same time, however, cellular repair machinery is not 100% efficient; thus mutations can persist. As accumulation of mutations contributes directly to aging and diseases such as cancer, there is widespread interest in understanding how these repair pathways function at the cellular level.
Among the most egregious forms of damage is the double-strand break (DSB) in which phosphodiester bonds of both strands of DNA are broken, thereby eliminating the ability of the complementary strand to serve as an intact repair template. There are numerous instigators of DSBs, including ionizing radiation (i.e., from X rays or other medical devices), oxidative stress, and exposure to certain chemicals that interfere with DNA replication. As lesions formed by unrepaired DSBs have the capacity to cause chromosome fragmentation, which is lethal to cells, multiple repair pathways have evolved solely to handle this type of damage. Of these, the two most well studied are nonhomologous end joining (NHEJ) and homologous recombination (HR). NHEJ can be thought of as applying a Band-Aid to a wound. This repair provides direct ligation of DNA fragments to keep the chromosome intact, yet due to its haphazard nature, NHEJ occasionally alters DNA sequence. These changes are analogous to scar tissue, which may prove problematic in the future, depending on its location. In contrast, HR is a multistep process that requires considerably more effort for the cell. Damage repaired by HR is analogous to a wound meticulously cared for and stitched back together by a team of emergency room physicians. Just as the the medical team uses their expertise to follow a clear plan that greatly minimizes chance scarring, HR utilizes a homologous template as its instruction manual to prevent mutations. As a result, HR is considerably more effective at preserving genome integrity and is generally viewed as “error-free,” yet the cost is greater in terms of effort required. While versions of both pathways exist in species ranging from bacteria to humans, the preference for repair pathway selection is a function of the cellular and organismal context in which the DSB occurs (such as the stage of the cell cycle) as well as the organism (Le Guen et al. 2015).
As DNA repair pathways are highly conserved, one way to advance our understanding of these processes is to use model organisms in which genetic manipulation and analysis are easily accomplished. Using certain species of bacteria, fungi, fruit flies, nematodes, fish, mice, or plants, geneticists can address fundamental questions regarding DNA repair. The evolutionary relationship among genes has enabled further analysis of repair pathways in multiple species, including humans. Researchers in the J. L. Yanowitz and K. A. Bernstein laboratories, both at the University of Pittsburgh, study DNA repair using model organisms. In their recently published study, McClendon et al. (2016) describe the role of a newly characterized Caenorhabditis elegans gene called sws-1 that they find plays a role in HR during both mitosis and meiosis. Using a combination of several established and more recently described cellular, genetic, and molecular techniques, the discoveries of McClendon et al. (2016) have direct implications for understanding how mutations in the human versions of sws-1 and related genes can predispose individuals to cancer and hinder fertility.
Background
Why study DNA damage and repair in the germ line?
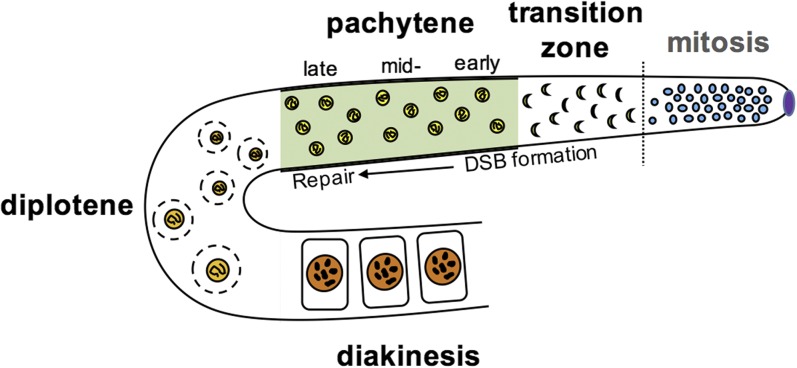
McClendon et al. (2016) studied the effects of DNA damage in germ cells that are collectively referred to as the germ line. Unlike somatic cells, which give rise to all of our “body” cells, germ cells are unique as they are the only cell type to undergo both mitosis and meiosis and therefore directly contribute to future generations by forming gametes (egg or sperm). In a stepwise series of events, the germ cells first divide mitotically, dramatically increasing in number. Each cell then undergoes one round of premeiotic S phase, during which all chromosomes replicate. Subsequently, two rounds of meiotic cellular division result in random segregation of maternal and paternal chromosomes to form haploid gametes (Figure 1). For researchers studying mechanisms of DNA repair, the germ line provides a unique opportunity: it is a confined pool of undifferentiated cells in which the effects of damage can be assessed during both mitosis and meiosis.
Figure 1.
Sites of mitotic and meiotic recombination in C. elegans. The gonads of C. elegans are a syncitium of nuclei, which is organized spatiotemporally. At the distal most region known as “distal tip” (purple) germ-line nuclei divide mitotically, before entering meiosis I (indicated by dotted line). During mitosis, HR typically occurs in response to stalled replication forks, whereas in meiosis, it is a normal consequence of programmed DSBs occurring in the first stages of prophase I. In the transition zone (leptotene/zygotene), DSBs are formed and chromosomes pair, and in C. elegans, this stage is visually distinguishable as the nuclei become C-shaped. In pachytene, chromosomes synapse, and DSBs are repaired to form at least one crossover between homologs. In diplotene through diakinesis, the synaptonemal complex is disassembled, and the paired chromosomes condense to form bivalents that will be packaged into each oocyte (for an extensive review of chromosome dynamics during prophase I in worms, see Woglar and Jantsch 2014).
Paradoxically, the generation of DNA damage in the meiotic germ line is required as are the mechanisms that activate the repair of this damage. During early prophase of meiosis I, induction of programmed DSBs allows chromosomes to pair and form bridges between them called chiasmata, a step that is essential in forming gametes with the correct chromosome complement (Keeney et al. 2014). Therefore, in order for meiosis to occur without incident, all chromosomes are routinely subjected to programmed DSBs, and as a result of this, appropriate DNA repair machinery must be actively engaged.
If DNA damage is a consequence of a normal cell process such as meiosis, why study repair in the first place? In certain cases, DNA damage may go unrepaired due to genetic predisposition or environmental insults; in these situations, genomic integrity is compromised and disease is a common result. When unrepaired damage occurs in germ cells, for example, this is especially hazardous as it can result in miscarriages, infertility, predisposition to cancer, and developmental disorders including Klinefelter, Turner, and Down syndromes (Hassold and Hunt 2001). Likewise, for mitotically dividing cells of the germ line, unrepaired DNA damage can also cause infertility as well as problems similar to that of somatic cells, such as tumor formation and cancer. It may be no surprise then, that mutants that lack DNA repair machinery altogether are unable to produce offspring (Handel and Schimenti 2010).
C. elegans as a model for understanding DNA repair
Several genetic models are used for DNA repair experiments, one of which is the roundworm C. elegans. As an adult, C. elegans is a 1-mm-long, transparent nematode capable of self-fertilization, with each hermaphrodite capable of producing ∼300 offspring during its 3- to 4-day life cycle, and up to 1000 offspring when mated with males. Both mutants and genetic tools to assess knockdown of gene expression are also readily available and inexpensive (see Corsi et al. 2015).
Mechanisms of DNA repair in C. elegans are frequently studied within the gonads of adult worms, which function as reservoirs of dividing mitotic and meiotic germ cells. In C. elegans, the gonads are U-shaped structures and are unique in that they form a syncytium, meaning there is no cell membrane separating the germ cell nuclei until the end of prophase I when the nuclei are packaged into oocytes. Their germ line is also spatiotemporally organized, similar to a conveyer belt system at a manufacturing plant (Figure 1). At the most distal end, germ cells divide mitotically until transitioning to a program of meiotic division. This spatiotemporal organization is especially advantageous for researchers, as both mitotically and meiotically dividing cells can be observed easily in live organisms as well as in fixed samples dissected out onto a slide. As a result, scientists can readily study the effects of genetic mutations and/or chemical perturbation on various aspects of germline development. Moreover, any defects affecting the integrity of chromosomes during this process can be directly assessed.
Ins and outs of HR
All sets of chromosomes (excluding the heterogametic sex chromosomes, such as the X–Y pair in human males) are composed of two homologs, one from the mother and one from the father. Homologs each consist of the same set and order of genes, but can vary in nucleotide sequence to produce different alleles. During DNA replication, all homologs are copied, forming pairs of identical sister chromatids corresponding to each homolog. The term homologous recombination refers to the exchange of chromosome segments between either the two identical sister chromatids or the two homologs, as occurs during several contexts such as meiosis, some types of DSB repair, and the formation of antibodies (Jasin and Rothstein 2013).
The role of HR was first described in the germ line, where it permits homologous regions of chromosomes to be exchanged during crossover, resulting in a shuffling of alleles to form new combinations (Hunter 2015). Given its fundamental role in ensuring successful chromosome segregation and the production of viable gametes, it is therefore not surprising that the genetics of HR are perhaps most frequently studied within the context of meiosis.
As HR is the primary repair pathway active during meiosis, it is important to understand not only the basic steps of HR but also the cellular context in which it operates in these cells. Prior to the start of the first meiotic prophase, all chromosomes will have been replicated and the resulting sister chromatids become bound together by ring-like structures known as cohesins (McNicoll et al. 2013). In the leptotene/zygotene substage of prophase I, all chromosomes are subject to programmed genomewide DSBs which, in C. elegans, occur simultaneously with homolog pairing. To facilitate recombination, each homolog seeks out its partner in a process known as synapsis. Synapsis involves the assembly of a proteinaceous structure called the synaptonemal complex that bridges homologs and serves as a temporary scaffold to facilitate recombination through alignment of the broken DNA strands (Cahoon and Hawley 2016).
While there are multiple ways to repair a DSB using HR, to generate chiasmata and the accurate segregation of homologs at metaphase I, these exchanges must occur between homologs, not sister chromatids. This form of HR, interhomolog (IH) repair, fulfills the minimum requirement of one crossover per homolog pair for meiosis to proceed (Hong et al. 2013). Alternatively, as the number of DSBs generally far exceeds the number of crossovers, the sister chromatid may also be used as a repair template. In this scenario, the form of HR used is referred to as intersister (IS) repair. Although IS repair cannot generate crossovers, its contributions to meiosis have been well documented (Goldfarb and Lichten 2010; Pradillo and Santos 2011). Furthermore, for mitotically dividing cells (as occurs in the premeiotic region of the germ line and in some somatic cells), IS repair is favorable, as it decreases the risk of errors such as genome rearrangements or loss of heterozygosity (Moynahan and Jasin 2010).
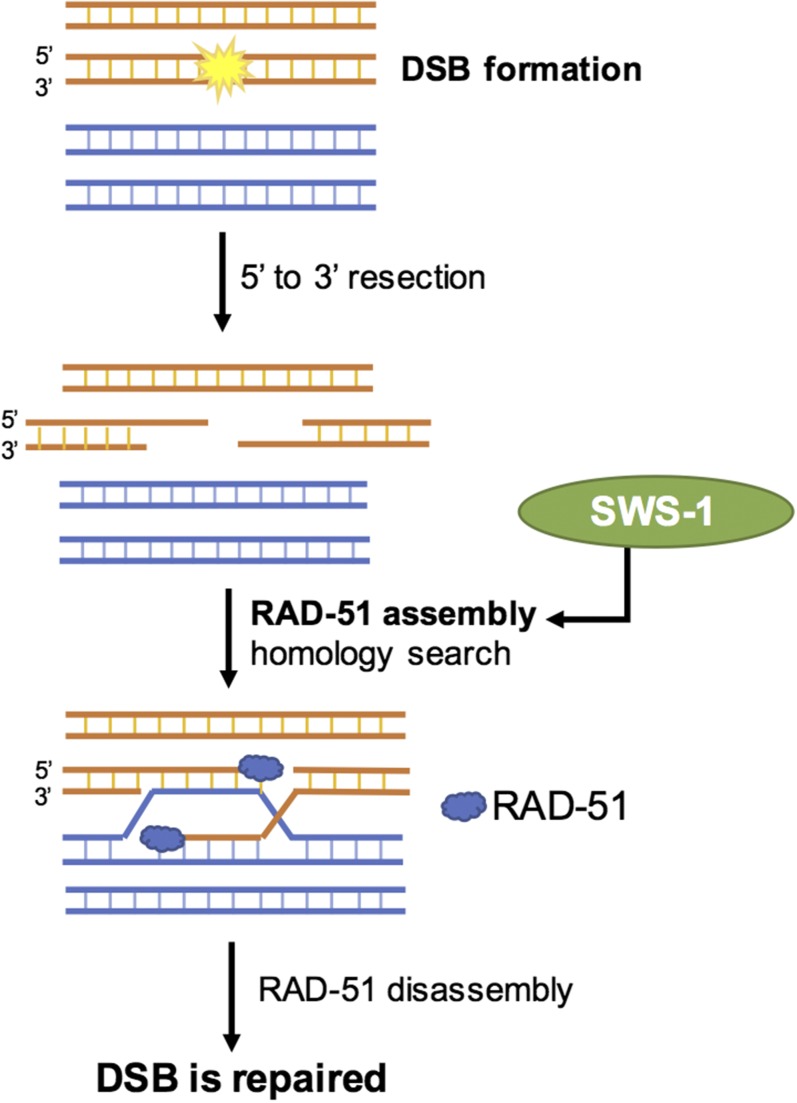
Given its widespread importance in the repair of DSBs, the individual steps of HR have been examined extensively (Kowalczykowski 2015). The major players are well conserved among species, as is the sequence of events that transpires in response to the initiation of DSBs. While daunting to consider all components involved, the basic steps are relatively straightforward. To simplify, in response to a DSB, the cell must first decide which repair pathway to utilize. Recall that there are several options, although HR is the only one considered error-free and is also the only one that can form crossovers. If the chosen pathway is HR, the first step is that the broken ends of DNA are resected, meaning that the nucleotides at the 5′ end of the break are enzymatically removed, leaving two opposing 3′ single-stranded overhangs (Figure 2). The overhangs then become coated with a protein called replication protein A (RPA), which is in turn exchanged for the recombinase RAD-51. Loading of RAD-51 onto the 3′ overhangs is essential for subsequent steps of HR, which include identification of homologous sequences in the repair template (either the homolog or sister chromatid) as well as subsequent strand invasion into the homologous DNA source. RAD-51 is therefore a key mediator in all forms of HR, and unsurprisingly, its absence is lethal to all organisms (Godin et al. 2016).
Figure 2.
DSBs are a substrate for homologous recombination. DSB formation is followed by a 59–39 resection of the DNA, yielding 39 single-stranded DNA (ssDNA) overhangs, after which RAD-51 is loaded, promoting homolog search and repair of the DSB as either a crossover or noncrossover. SWS-1 is predicted to assist in either the loading or stabilization of RAD-51 onto 39 overhangs. (For a detailed review of these processes in mitosis and meiosis, see Kohl and Sekelsky 2013).
Understanding homologs and paralogs
The physiological importance of HR-related genes is underscored by their evolutionary history. Phylogenetic analysis reveals similar HR genes in diverse species ranging from bacteria to humans (Jasin and Rothstein 2013). Of particular importance to HR are the homologs and paralogs of the Rad51 gene.
Not to be confused with the homologs (chromosomes) that pair during meiosis, homologs in the evolutionary sense are genes that share a single, ancestral DNA sequence. A related term, paralogs, refers to homologous genes that arise from a duplication event within a single genome. Paralogs often possess similar molecular roles due to their shared DNA sequences, but can develop new functions in the process of gene duplication or through subsequent mutation. Paralogs have the additional potential to function together in the same pathways or as complexes. In this regard, the existence of paralogs provides a form of genetic redundancy that can be considered an evolutionary advantage. Organisms such as the single-celled budding yeast possess an especially high number of paralogs ascribed as a form of “genetic robustness” (Kafri et al. 2005; Li et al. 2010).
Given the essential role of HR in numerous cellular contexts such as genome maintenance and meiotic recombination, the discovery of Rad51 paralogs was not entirely surprising. Further demonstrating their importance, mutations in Rad-51 paralogs cause several pathologies in humans such as an increased risk of cancer (Suwaki et al. 2011). The majority of studies of Rad51 paralog function have been conducted in the budding yeast, Saccharomyces cerevisiae. In this organism, two Rad51 paralog complexes have been identified, one of which is called the Shu complex. Similar to Rad51 itself, the paralogs whose protein products comprise Shu act in the early steps of HR, where they aid in assembly of the RAD-51 filament onto the broken strand of DNA.
Tools, Techniques, and Results
CRISPR: Generating the sws-1 mutant
McClendon et al. (2016) investigated the role of the C. elegans gene sws-1, a homolog of the yeast gene and Shu complex component Shu2, and they probed its genetic and molecular interactions with the Rad51 paralogs rfs-1 and rip-1. They began with the goal of isolating a sws-1 mutant strain.
To determine the function of a previously uncharacterized gene, the approach most commonly used by geneticists is to characterize its mutant phenotype. Model genetic organisms such as C. elegans provide a powerful tool for reverse genetic screens, in which a gene is selectively targeted for mutation and the resulting phenotype is used to infer the normal function of the gene. Commonly used strategies include targeted RNA-mediated knockdown (Gunsalus and Piano 2005), a genome-editing technique involving transposons called Mos1 mutagenesis (Robert and Bessereau 2007), and most recently, a tool known as the clustered, regularly interspaced, short palindromic repeats (CRISPR)–CRISPR-associated protein 9 (Cas9) system, which has revolutionized genetics research and is now an indispensable tool in basic biology through clinical research (Dickinson and Goldstein 2016).
CRISPR serves as part of an adaptive immune response to resist viral infection, present in the majority of archaea and many bacteria (Barrangou et al. 2007; Horvath and Barrangou 2010; Mojica and Rodriguez-Valera 2016), providing sequence-specific memory against pathogens. Upon subsequent exposure, this nucleic acid memory allows the host to direct a robust attack against pathogen genomes to eliminate the pathogen (Rath et al. 2015). In response to viral infection, the host organisms will integrate foreign genetic material, referred to as spacers, between 24- and 48-bp repeats within the CRISPR locus. In the event of a recurring viral attack, these spacer sequences are subsequently transcribed to form specialized RNAs known as a long pre-CRISPR RNA (pre-crRNA) and trans-activating CRISPR RNA (tracrRNA) (Karvelis et al. 2013). Formation of the pre-crRNA:tracrRNA complex is mediated by another essential component, the CRISPR-associated protein, Cas. Along with the host RNase III (an enzyme that degrades RNA), this specialized complex cleaves the repeat sequences to form mature crRNA. In turn, the mature crRNA guides Cas, via crRNA complementarity, to the invading viral nucleic acid sequence, where it is cut by the Cas protein.
The ability of this system to target defined regions of a genome quickly spawned interest among the scientific community as a tool for genetic manipulation. An important finding was that the Cas/crRNA complex targets only specific sequences containing what is known as a protospacer adjacent motif (PAM), a particular three-base sequence on the target DNA molecule. In prokaryotes, the purpose of the PAM is to prevent unintended cleavage of their own genome (Marraffini and Sontheimer 2010). To exploit the CRISPR system as a genetic tool for modifying a sequence of interest, a target sequence must fit the following criteria: (1) it is unique in the host genome and (2) it must be immediately upstream of a PAM (Briner and Barrangou 2016). The most extensively researched and most ubiquitously used Cas protein for genome editing is Cas9, hence the editing tool is often referred to as “CRISPR–Cas9.” The CRISPR/Cas9 system can deliver site- and sequence-specific mutations both effectively and cheaply. Notably, it was the pioneering work of the research group led by Jennifer Doudna that first demonstrated how to manipulate the CRISPR–Cas9 system to easily and efficiently alter the DNA of any organism (Jinek et al. 2014; O’Connell et al. 2014). In addition to being especially useful for scientists, the implications of her discovery for the treatment of human disease quickly brought her recognition among the medical community and the general public (Pollack 2015).
In the laboratories of J. L. Yanowitz and K. A. Bernstein, CRISPR was used to generate the sws-1(ea12) allele, specifically a nonsense mutant that does not encode functional SWS-1 protein. To design the needed RNA sequences, they used bioinformatic tools to generate crRNA guides. Once amplified (using PCR) and inserted into a plasmid, the purpose of these guides is to provide the complementary sequence, which specifies the site of Cas9 action (Briner et al. 2016). In this case, the authors chose guides that would target the sws-1 reading frame, meaning the area spanning the start and stop codons within the gene. Using recombinant DNA technology, these guides were cloned into a specialized plasmid vector containing the Cas9 gene sequence. Plasmid DNA was purified from bacterial colonies and microinjected into the germ line of young adult worms, and transgenic worms (meaning those containing the foreign DNA) were isolated.
Along with the plasmid containing the sws-1 guides and Cas9 sequence, the injection mix also contained guides corresponding to a gene called dpy-10. F1 offspring of the injected worms that are successfully transformed, will express a “rolling” phenotype due to a twisted cuticle conferred by the presence of a new dpy-10 mutation (Higgins and Hirsh 1977) and one-quarter of the F2 generation should be homozygous for the dpy-10 mutation. At this stage, rolling worms were selected, which were presumed to have also taken up the construct containing Cas9 and the crRNA guides engineered to disrupt the genetic sequence of sws-1. To verify the presence of the sws-1 mutation, the “roller worms” were lysed and their DNA was isolated for PCR amplification of the Cas9 target site. This step was followed by DNA sequencing to positively identify candidates containing the desired sws-1 mutation. To eliminate the dpy-10 mutation, the strain was backcrossed to wild-type worms, selecting just for sws-1.
To test whether the mutation eliminated sws-1 gene products, McClendon et al. measured the abundance of sws-1 mRNA in their mutant strain. They found a fivefold reduction compared to wild-type controls, which supported their conclusion that the allele was incapable of producing normal levels of SWS-1 protein. Armed with their key reagent in hand (the sws-1 mutant worms), they proceeded to determine the role of sws-1 in DNA repair.
Immunofluorescence: Monitoring DSB repair
McClendon et al. hypothesized that SWS-1 is involved in HR repair through the regulation of RAD-51 nucleoprotein filament dynamics. As described earlier, RAD-51 is a recombinase that loads onto the resected 3′ overhangs at the sites of DSBs and replication forks and stimulates recombination between either homologs or sister chromatids (Figure 2). Consequently, RAD-51 foci, meaning sites where this protein is most concentrated, are detected on chromosomes undergoing HR repair. As a reminder, this can occur as a consequence of sporadic DNA damage or during normal meiotic division. To demonstrate a role for SWS-1 in HR repair, McClendon et al. used immunofluorescence to monitor RAD-51 foci in sws-1 mutants and wild-type controls in both meiotic nuclei and in mitotic nuclei of worms exposed to the DNA damaging agent camptothecin (CPT).
Immunofluorescence allows one to assess protein levels, determine colocalization with other proteins, and to visualize specific proteins within the context of the intact cellular structure. The location of proteins within discrete cellular compartments, for example, the nucleus or the cytosol, can provide clues to their function. Immunofluorescence makes use of antibodies, or immunoglobulins, that bind strongly and specifically to proteins of invading viruses and bacteria. Researchers use fluorescently tagged antibodies that bind to proteins of interest within intact cells for visualization under fluorescence microscopes. The binding function of immunoglobulins is conferred by a variable region. A separate constant region has an amino acid sequence that is conserved within a species. The antibody that recognizes the cellular target is called a primary antibody and is typically generated in a small animal such as a rabbit or mouse. In the studies performed by McClendon et al., a rabbit primary antibody was used to localize and quantify RAD-51 foci.
Prior to adding the antibody, worms were dissected onto slides, and their gonads were fixed in paraformaldehyde, which forms chemical cross-links between proteins, thereby preserving the normal structure of the cell. The specimens were then subjected to freeze cracking, which disrupts the exterior surface of the specimen to permit subsequent antibody labeling. RAD-51 in the gonads was labeled with the primary antibody, followed by several washes to remove unbound antibody. Then, secondary antibodies were added that have a variable region that binds to the constant region of the primary antibody. Secondary antibodies for immunofluorescence are chemically linked to a fluorophore, that when stimulated with light of a specific wavelength, emits light of a longer wavelength. In this study, the secondary antibody used to label the rabbit anti-RAD-51 primary was anti-rabbit 568, indicating that the fluorophore emits light at 568 nm. A guinea pig antibody specific for XND-1 in combination with anti-guinea pig 633 was used as a positive control as it stains all nuclei within the gonad.
After washing to remove unbound secondary antibodies, the compound DAPI was added. Unlike an antibody, DAPI binds to A- to T-rich regions of DNA, and it emits blue light when stimulated with an ultraviolet light source. DAPI staining alone enabled the researchers to identify defects in the structural integrity of chromosomes in the mutants they examined, whereas in combination with anti-RAD-51, it allowed them to identify and count nuclei in which RAD-51 dynamics were altered in sws-1 germ lines.
In the sws-1; helq-1 double mutant as well as each of the corresponding single mutants, they examined DAPI staining in diakinesis, the last stage of meiotic prophase I when homologs are attached by chiasmata and are highly condensed in preparation for metaphase (Figure 1). The rationale for this experiment was that helq-1 encodes a protein involved in HR repair whose absence confers synergistic lethality with either rfs-1, rip-1, or sws-1 (Ward et al. 2010; Taylor et al. 2015; McClendon et al. 2016). By examining chromosomes in the the sws-1; helq-1 double mutant, the researchers could test the hypothesis that these genes play overlapping roles in HR repair. Combining the techniques of DAPI staining along with use of the RAD-51 antibody, they also examined nuclei in earlier stages of prophase I and discovered that RAD-51 kinetics were altered in sws-1 mutants. Unlike wild-type germ lines, wherein RAD-51 foci disappear by late pachytene (Figure 1), sws-1 germ lines had persisting RAD-51 foci, indicative of a defect in DSB repair. Further, in response to CPT-induced damage (which exclusively affects mitotic nuclei), RAD-51 staining was also disrupted in sws-1 germ lines, suggesting that DSB repair is compromised in premeiotic nuclei lacking functional SWS-1. As they hypothesized, these observations support a model wherein the normal role of SWS-1 is to modulate HR repair in the germ line.
Genotoxins: Assessing the damage response
Throughout our lifetime, we are exposed to numerous agents that can wreak havoc on our cells, including radiation, chemicals, viruses, and certain types of bacteria. The term genotoxin refers to a damaging agent that can lead to mutations in our DNA. In C. elegans, there are well-established assays used to quantify an organism’s response to genotoxins, as measured by elevated germ cell death, sterility, or progeny inviability (Kim and Colaiácovo 2015).
Given the strong genetic and cellular evidence implicating SWS-1 in HR repair, the researchers next decided to monitor the ability of C. elegans to handle genotoxic damage in the presence or absence of the sws-1 mutation. Specifically, they tested the effects of a subset of genotoxins that generate genetic lesions which, under normal circumstances, would result in the recruitment of HR repair machinery. The logic is that if the animals normally utilize HR to repair damage, elimination of SWS-1, a presumed regulator of HR repair, would be predicted to impair HR capability in mitotically and/or meiotically dividing nuclei. A failure to repair damage would therefore be expected to manifest in decreased viability of sws-1 offspring. Indeed, this is what was found: when treated with each of the genotoxins, the researchers observed a statistically significant decrease in the survival of sws-1 offspring vs. those of wild-type controls exposed to the same chemicals.
Given that each of the genotoxins used in this study affect different substrates, comparing the effect of each agent on progeny viability provides further clues into the mechanism by which SWS-1 responds to damage. For example, in their assay, the drug CPT induced the most lethality in sws-1 mutants. CPT is known to disrupt DNA replication by inducing breaks in DNA that directly interfere with the replication fork. In normal cases, stalled replication forks caused by CPT typically become substrates for HR repair (Sakasai and Iwabuchi 2016).
Yeast hybrid assays: Uncovering interactions between SWS-1 and RAD-51 paralogs
sws-1 mutants exhibit reduced survival and a high incidence of male progeny, (the him phenotype), which results from X chromosome nondisjunction (Hodgkin et al. 1979). Both of these defects can arise from HR repair defects. Importantly, these characteristics have also been shown for the RAD-51 paralog mutants, rfs-1 and rip-1 (Taylor et al. 2015), leading McClendon et al. to hypothesize that sws-1 and these RAD-51 paralogs act in the same pathway. To investigate this possibility, the authors generated double (e.g., rfs-1; sws-1 and rip-1; sws-1) and triple (e.g., rfs-1; rip-1; sws-1) mutants and assayed each mutant combination for the him phenotype and lethality, both of which result when HR repair is compromised. The goal of this experiment was to test whether the genes normally function in one pathway or different pathways. If these genes are involved in different pathways, one would expect an additive effect in these mutants compared to the single mutants. However, no significant difference in lethality was observed between the triple and single mutants, leading to the conclusion that SWS-1, RFS-1, and RIP-1 likely participate in a common pathway.
The central hypothesis of McClendon and colleagues' study is that SWS-1 is a functional homolog of yeast Shu2 given their shared protein structure, the SWIM domain. Previous studies have shown that Shu2 physically interacts with Rad51 paralogs to form a complex that participates in HR (Godin et al. 2016). Given its role in mediating interactions between yeast and human paralogs of these proteins, the SWIM domain was therefore a likely candidate for promoting similar interactions in C. elegans. Additionally, McClendon et al. tested the role of the Walker B protein motif, which participates in ATP binding and is a conserved feature of the Rad51 paralogs (Wiese et al. 2006). Based on these data and their finding that SWS-1 and the C. elegans RAD-51 paralogs likely participate in the same pathway, McClendon et al. next performed yeast-2-hybrid (Y2H) and yeast-3-hybrid (Y3H), techniques that allowed them to probe physical interactions between SWS-1, RFS-1, and RIP-1 and to define the protein domains involved.
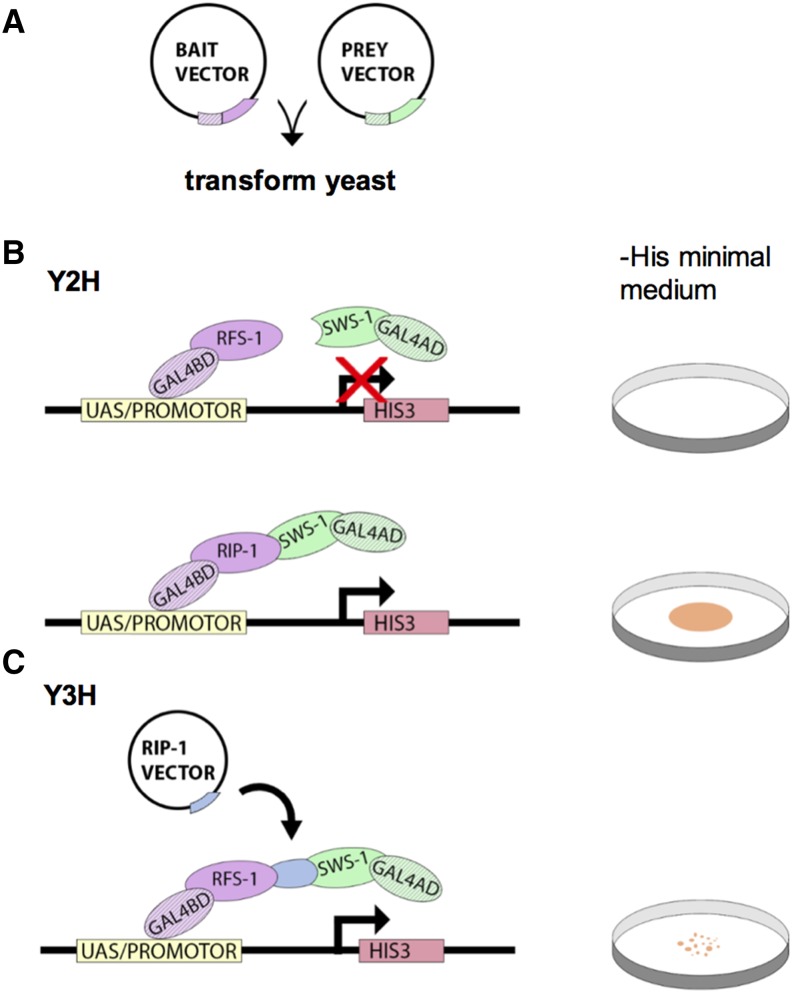
Y2H and Y3H studies are used to assay for protein–protein interactions (Figure 3). The assay makes use of a characteristic of eukaryotic transcription factors in which functionally distinct domains participate in DNA recognition [binding domain (BD)] or activation of transcription [activating domain (AD)]. For transcription to occur, these domains must be in close proximity, but physical contact is not required. In Y2H assays, the BD and AD of the Gal4 transcription factor are encoded by different plasmids that are introduced into yeast cells by transformation. A bait protein is fused to the BD and a prey protein is fused to the AD. If the bait and prey proteins interact, the BD and AD domains are brought close enough together to initiate transcription of a downstream reporter gene.
Figure 3.
Yeast hybrid experiments detect protein–protein interactions. (A) In yeast hybrid screens, the budding yeast S. cerevisiae are cotransformed with a bait vector and a prey vector, which encode a protein of interest fused to the BD or the AD of the Gal4 transcription factor, respectively. (B) The yeast strain is typically auxotrophic, meaning that it is unable to synthesize an essential nutrient, in this case the amino acid histidine. Transformed yeast plated on a medium lacking this nutrient (i.e., −His minimal medium; see Petri dish schematic on right) will not grow unless interactions between bait and prey proteins bring the BD and AD in close enough proximity to form a functional Gal4 transcription factor. This allows for transcription of HIS3, an enzyme involved in histidine biosynthesis. Thus, growth on −His medium serves as a marker for bait/prey interactions. Various combinations of bait and prey vectors can be used to effectively assess a multitude of protein–protein interactions. From these Y2H assays, the authors determined that SWS-1 interacts with RIP-1 but not RFS-1. (C) Y3H assays are similar, with the addition of a vector encoding a third gene of interest (RIP-1 vector). The transcription product of this gene may interact with both the bait and prey, bridging the interaction between the BD and AD. In this study, RIP was shown to bridge RFS-1 and SWS-1. Growth of the yeast was less robust (as indicated by Petri dish schematic on right), suggesting that the three-protein complex was less stable than the RIP-1/SWS-1 complex.
In this study, the reporter gene encodes a histidine biosynthesis gene (HIS3) such that a successful bait–prey interaction allows yeast cells to grow on medium lacking histidine (−His; Figure 3). Thus, yeast containing both plasmids, whether the bait and prey protein interact or not will be able to grow on control media containing histidine, but whether those same yeast can grow in −His medium will be dependent on bait and prey proteins interacting with each other. In addition, the compound 3-amino-1,2,4-triazole (3AT), which inhibits the enzyme encoded by HIS3, was used to determine the relative stability of these protein–protein interactions. More stable bait–prey interactions will yield higher transcription of HIS3 and will show less inhibition by 3AT.
Using various combinations of SWS-1, RFS-1, and RIP-1 as bait or prey, the authors investigated whether SWS-1 directly interacts with RIP-1 and RFS-1 (McClendon et al. 2016). The authors used Y3H analysis in which SWS-1 was fused to the AD, RFS-1 was fused to the BD, and RIP-1 was expressed from a third plasmid to test whether there is an interaction between all three proteins. Additionally, Y2H was used to define the protein domains that participate in the interaction between proteins of this complex. To accomplish this, different point mutations in the SWS-1 SWIM domain were tested for their effect on protein interactions.
See questions 11–13 below to follow and interpret these experiments.
Questions for Review and Discussion
McClendon et al. (2016) note that reduced progeny viability and increased frequency of males in C. elegans, both indicative of nondisjunction, can result from HR repair defects during meiosis. How might a problem in HR lead to a nondisjunction event?
The authors used CRISPR to generate a nonsense allele of sws-1. What is a nonsense allele, and how is it possible to generate it using CRISPR technology?
In figure 1 in McClendon et al. (2016), the authors indicate the sites of the CRISPR guides with arrows. Why did they choose these locations? What can be inferred from the gel they ran before sequencing the mutation site?
Why do helq-1; rfs-1 and helq-1; rip-1 mutants exhibit synthetic lethality? Given this information, why did the authors generate the helq-1; sws-1 double mutant?
When analyzing chromosomes in the the helq-1; sws-1 double mutant (figure 2, B–D and p. 138 in McClendon et al. (2016), why did the authors choose to examine nuclei in diakinesis vs. another stage of prophase I? For what types of abnormalities were they screening? Finally, what new information was revealed by the abnormalities in the helq-1; sws-1 mutant that was not already explained by the researchers’ fertility assay?
The researchers note that the transition zone is a region of prophase I in which DSBs are repaired and RAD-51 foci first become apparent. What other meiotic event(s) would you expect to be occurring during this stage?
What was the rationale for testing for increased deletions in sws-1 mutants (figure 4 and p. 139 in McClendon et al. (2016)? Why was it important to use the double mutant dog-1; sws-1 for these experiments?
In figure 5 in McClendon et al. (2016), worms were treated with genotoxins and their survival was quantified. Why were these particular genotoxins selected? Briefly compare and contrast the function of each agent. What control was used for this experiment?
Based on the results of the CPT assay (figure 6 and p. 140 in McClendon et al. (2016), and prior RAD-51 scoring data (figure 3 and p. 139 in McClendon et al. (2016), would you expect that SWS-1 normally responds to programmed breaks, breaks induced by DSB-inducing agents, or both? Explain.
Briefly explain the purpose of adding DAPI to the dissected germ lines. Why was it necessary to stain germ lines with DAPI in addition to the RAD-51 antibody?
Identify the two types of controls used in each Y2H and Y3H assay. Why were these controls used? What is preventing some yeast strains from growing but not others?
What does the addition of 3AT to the −His medium tell us about the strength of the interaction between the various proteins in the Y2H and Y3H assays? How do the Y2H and the Y3H assays differ? What additional information do they provide? Based on the two assays, which proteins show an interaction and how strong is that interaction?
In figure 7C in McClendon et al. (2016), what is the difference between the SWS-1, SWS-1–C133S, and SWS-1–A156T strains? Which of these strains is considered a control? What is the difference between RIP-1 and RIP-1–D131A? What do all these strains tell us about the interaction between SWS-1 and RIP-1?
Using the authors’ findings and their speculations in Discussion, construct a plausible model showing how SWS-1 promotes HR repair in C. elegans. Expand upon what is shown in figure 8 in McClendon et al. (2016).
With regards to its biochemical function, what remains unknown about SWS-1? What experiments can you think of that could be performed to answer additional questions?
Acknowledgments
The authors thank Erika Rosenkranse, Elizabeth De Stasio, and Kara Bernstein for proofreading the manuscript and for helpful suggestions. P.M.C. is supported by NIH grant 1R15GM117479-01.
Footnotes
Related article in GENETICS: McClendon, T. B., M. R. Sullivan, K. A. Bernstein, and J. L. Yanowitz, 2016 Promotion of homologous recombination by SWS-1 in complex with RAD-51 paralogs in Caenorhabditis elegans. Genetics 203: 133–145.
Communicating editor: E. A. De Stasio
Literature Cited
- Aguilera A., Gómez-González B., 2008. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9(3): 204–217. [DOI] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., et al. , 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315(5819): 1709–1712. [DOI] [PubMed] [Google Scholar]
- Briner A. E., Barrangou R., 2016. Guide RNAs: a glimpse at the sequences that drive CRISPR-Cas systems. Cold Spring Harb. Protoc. 2016(7): pdb.top090902. [DOI] [PubMed] [Google Scholar]
- Briner A. E., Henriksen E. D., Barrangou R., 2016. Prediction and validation of native and engineered Cas9 guide sequences. Cold Spring Harb. Protoc. 2016(7): pdb.prot086785. [DOI] [PubMed] [Google Scholar]
- Cahoon C. K., Hawley R. S., 2016. Regulating the construction and demolition of the synaptonemal complex. Nat. Struct. Mol. Biol. 23(5): 369–377. [DOI] [PubMed] [Google Scholar]
- Corsi A. K., Wightman B., Chalfie M., 2015. A transparent window into biology: a primer on Caenorhabditis elegans. Genetics 200: 387–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Goldstein B., 2016. CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics 202: 885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin S. K., Sullivan M. R., Bernstein K. A., 2016. Novel insights into RAD51 activity and regulation during homologous recombination and DNA replication. Biochem. Cell Biol. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb T., Lichten M., 2010. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 8(10): e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus K. C., Piano F., 2005. RNAi as a tool to study cell biology: building the genome-phenome bridge. Curr. Opin. Cell Biol. 17(1): 3–8. [DOI] [PubMed] [Google Scholar]
- Handel M. A., Schimenti J. C., 2010. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11(2): 124–136. [DOI] [PubMed] [Google Scholar]
- Hassold T., Hunt P., 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2(4): 280–291. [DOI] [PubMed] [Google Scholar]
- Higgins B. J., Hirsh D., 1977. Roller mutants of the nematode Caenorhabditis elegans. Mol. Gen. Genet. 150(1): 63–72. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., Brenner S., 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Sung Y., Yu M., Lee M., Kleckner N., et al. , 2013. The logic and mechanism of homologous recombination partner choice. Mol. Cell 51(4): 440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P., Barrangou R., 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327(5962): 167–170. [DOI] [PubMed] [Google Scholar]
- Hunter N., 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7(12): pii: a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M., Rothstein R., 2013. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5(11): a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Jiang F., Taylor D. W., Sternberg S. H., Kaya E., et al. , 2014. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343(6176): 1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R., Bar-Even A., Pilpel Y., 2005. Transcription control reprogramming in genetic backup circuits. Nat. Genet. 37(3): 295–299. [DOI] [PubMed] [Google Scholar]
- Karvelis T., Gasiunas G., Miksys A., Barrangou R., Horvath P., et al. , 2013. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 10(5): 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Lange J., Mohibullah N., 2014. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet. 48: 187–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. M., Colaiácovo M. P., 2015. DNA damage sensitivity assays in Caenorhabditis elegans. Bio Protoc. 5(11): pii: e1487. [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., Sekelsky J., 2013. Meiotic and mitotic recombination in meiosis. Genetics 194: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S. C., 2015. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7(11): pii: a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guen T., Ragu S., Guirouilh-Barbat J., Lopez B. S., 2015. Role of the double-strand break repair pathway in the maintenance of genomic stability. Mol. Cell. Oncol. 2(1): e968020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yuan Z., Zhang Z., 2010. The cellular robustness by genetic redundancy in budding yeast. PLoS Genet. 6(11): e1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L. A., Sontheimer E. J., 2010. Self vs. non-self discrimination during CRISPR RNA-directed immunity. Nature 463(7280): 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClendon T. B., Sullivan M. R., Bernstein K. A., Yanowitz J. L., 2016. Promotion of homologous recombination by SWS-1 in complex with RAD-51 paralogs in Caenorhabditis elegans. Genetics 203: 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicoll F., Stevense M., Jessberger R., 2013. Cohesin in gametogenesis. Curr. Top. Dev. Biol. 102: 1–34. [DOI] [PubMed] [Google Scholar]
- Mojica F. J., Rodriguez-Valera F., 2016. The discovery of CRISPR in archaea and bacteria. FEBS J. 283(17): 3162–3169. [DOI] [PubMed] [Google Scholar]
- Moynahan M. E., Jasin M., 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11(3): 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M. R., Oakes B. L., Sternberg S. H., East-Seletsky A., Kaplan M., et al. , 2014. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516(7530): 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradillo M., Santos J. L., 2011. The template choice decision in meiosis: Is the sister important? Chromosoma 120(5): 447–454. [DOI] [PubMed] [Google Scholar]
- Pollack A. 2015. Jennifer Doudna, a Pioneer Who Helped Simplify Genome Editing. [Internet] New York Times. Available at: http://www.nytimes.com/2015/05/12/science/jennifer-doudna-crispr-cas9-genetic-engineering.html?_r=0. [Google Scholar]
- Rath D., Amlinger L., Rath A., Lundgren M., 2015. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie 117: 119–128. [DOI] [PubMed] [Google Scholar]
- Robert V., Bessereau J. L., 2007. Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 26(1): 170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakasai R., Iwabuchi K., 2016. The distinctive cellular responses to DNA strand breaks caused by a DNA topoisomerase I poison in conjunction with DNA replication and RNA transcription. Genes Genet. Syst. 90(4): 187–194. [DOI] [PubMed] [Google Scholar]
- Suwaki N., Klare K., Tarsounas M., 2011. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin. Cell Dev. Biol. 22(8): 898–905. [DOI] [PubMed] [Google Scholar]
- Taylor M. R., Špírek M., Chaurasiya K. R., Ward J. D., Carzaniga R., et al. , 2015. Rad51 paralogs remodel pre-synaptic Rad51 filaments to stimulate homologous recombination. Cell 162(2): 271–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. D., Muzzini D. M., Petalcorin M. I., Martinez-Perez E., Martin J. S., et al. , 2010. Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic doublestrand break repair. Mol. Cell 37: 259–272. [DOI] [PubMed] [Google Scholar]
- Wiese C., Hinz J. M., Tebbs R. S., Nham P. B., Urbin S. S., et al. , 2006. Disparate requirements for the Walker A and B ATPase motifs of human RAD51D in homologous recombination. Nucleic Acids Res. 34(9): 2833–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A., Jantsch V., 2014. Chromosome movement in meiosis I prophase of Caenorhabditis elegans. Chromosoma 123(1–2): 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]