Abstract
Functional annotations have been shown to improve both the discovery power and fine-mapping accuracy in genome-wide association studies. However, the optimal strategy to incorporate the large number of existing annotations is still not clear. In this study, we propose a Bayesian framework to incorporate functional annotations in a systematic manner. We compute the maximum a posteriori solution and use cross validation to find the optimal penalty parameters. By extending our previous fine-mapping method CAVIARBF into this framework, we require only summary statistics as input. We also derived an exact calculation of Bayes factors using summary statistics for quantitative traits, which is necessary when a large proportion of trait variance is explained by the variants of interest, such as in fine mapping expression quantitative trait loci (eQTL). We compared the proposed method with PAINTOR using different strategies to combine annotations. Simulation results show that the proposed method achieves the best accuracy in identifying causal variants among the different strategies and methods compared. We also find that for annotations with moderate effects from a large annotation pool, screening annotations individually and then combining the top annotations can produce overly optimistic results. We applied these methods on two real data sets: a meta-analysis result of lipid traits and a cis-eQTL study of normal prostate tissues. For the eQTL data, incorporating annotations significantly increased the number of potential causal variants with high probabilities.
Keywords: Bayesian fine mapping, annotations, summary statistics, causal variants
A large amount of annotation information of genomic elements has been generated, such as the Encyclopedia of DNA Elements (The ENCODE Project Consortium 2012). Regulatory elements, such as those marked by DNase I hypersensitive sites (DHSs), have been shown to be enriched with associations and explain a large proportion of heritability (Maurano et al. 2012; Gusev et al. 2014). Incorporation of these annotations into statistical association analyses can improve both the power in genome-wide association study (GWAS) discovery (Pickrell 2014) and the accuracy in fine mapping underlying causal variants (Quintana and Conti 2013; Kichaev et al. 2014; Wen et al. 2015). However, the number of annotations is often very large and many of them may not be informative for the underlying causal genetic variants. Currently, there is no systematic and effective way to incorporate a large number of annotations in association analyses, which is crucial to the full use of the available information. In practice, usually only a small number of annotations are considered (Quintana and Conti 2013; Wen et al. 2015). A recent proposed algorithm, PAINTOR, suggests a one-by-one test of each annotation selecting the top 4–5 for inclusion. This has potential to waste the information from the remaining annotations, which may together provide significant predictive power of the causal status. Another approach (Pickrell 2014) considered a large number of annotations, however, it has two limitations. First, it assumes the maximal number of causal variants in each genomic region locus is one, which may lead to suboptimal results (Hormozdiari et al. 2014; Kichaev et al. 2014; Chen et al. 2015). Second, the process of annotation selection may not fully explore the available annotation information. In this study, we propose a Bayesian framework to incorporate a large number of annotations simultaneously. Moreover, we extend our previous fine-mapping method, CAVIARBF, under this framework, which enables the use of summary statistics. Simulation results show that our proposed method achieves better or similar performance compared to PAINTOR and fgwas under a variety of different annotation selection strategies.
Methods
A general Bayesian hierarchical model to incorporate functional annotations
We first consider one region or locus for fine mapping and later extend it to multiple loci. For the locus of interest, suppose there are m variants and n individuals. We assume that only a subset of the m variants is causal. Denote the phenotypes by and genotypes by . The phenotypes can be quantitative traits or binary traits. The effect size of each variant is denoted by , which depends on the causal states, denoted by , where the elements of vector c are either 1, indicating the corresponding variant is causal, or 0 for noncausal variants. For the th variant, if , is assumed to be normally distributed with a mean of zero and variance specified by (Servin and Stephens 2007; Guan and Stephens 2008). For quantitative traits, Y and also depend on a random variable τ that specifies their variances, for which we assign a noninformative prior (Servin and Stephens 2007; Guan and Stephens 2008). If , then = 0. We assume the causal states represented by c depend on the annotations of the variants. For example, we can model the relationship between the annotation and the probability of being causal using a logistic model. Suppose there are d annotations for each variant and the annotation matrix is denoted by and denotes the ith row. Denote the annotation effects on the causal states by , where T means transpose, is the intercept, and is a vector of annotation effects. We assume a normal prior distribution on the annotation effects with alternate distributions considered later. The full model can be written as follows, assuming quantitative traits and a logistic model linking annotations to causal states:
We assign noninformative priors on μ, τ, and . Specifically, , , and , where is the Gamma distribution with the shape parameter and the inverse scale parameter , and we let
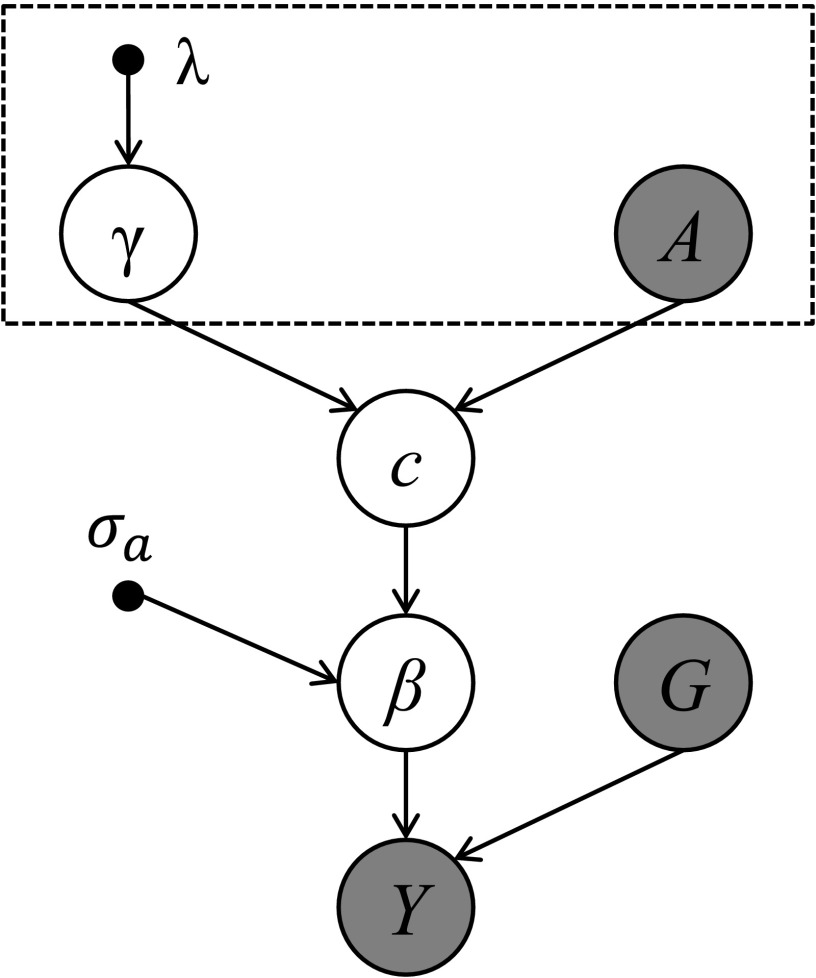
The above probabilistic model forms a hierarchical model, or directed graphical model (Bishop 2006), as shown in Figure 1. In Figure 1, the nodes are random variables, where open circles represent unobserved/hidden random variables, shaded circles represent observed random variables, small solid circles represent fixed parameters, and the arrows show the dependencies between two random variables. The arrow leaves from the node on which the receiving node depends, for example, the causal state c depends on the annotation A and the annotation effects γ. Variables with noninformative priors are not shown in Figure 1. When using summary statistics, we need only to replace Y with the Z-test statistic, and replace G with the correlation matrix ∑, which will be described later. Compared to the previous model in CAVIARBF (Chen et al. 2015), this model has additional levels to incorporate annotations, enclosed by the box in Figure 1. Compared to PAINTOR (Kichaev et al. 2014) and the practical inference procedure of FM-QTL (Wen et al. 2015), this model has an extra level specifying the prior of the annotation effects γ, which turns out to be critical and enables the incorporation of a large number of annotations simultaneously into the model. For now we assume λ is a known fixed value, and later we will discuss ways to select the optimal parameter.
Figure 1.
Hierarchical models incorporating functional annotations. The box indicates the added modeling block related to annotations. Shaded ○ indicates observed data. A, the annotation matrix of variants; c, the causal configuration vector of variants; G, full genotype matrix; Y, phenotype vector; β, effect size vector of genotypes on the phenotype; , the parameter specifying the prior distribution of β; γ, the effect size vector of annotations on the causality state; λ, the parameter specifying the prior distribution of γ.
Exact Bayes factors using summary statistics for quantitative traits
We previously showed (Chen et al. 2015) that the fine-mapping method CAVIAR (Hormozdiari et al. 2014) is approximately equivalent to BIMBAM (Servin and Stephens 2007; Guan and Stephens 2008). Based on this equivalence, we developed an approximate Bayesian framework for fine mapping multiple causal variants in a locus using only summary statistics. When we assume at most one causal variant in a locus, it reduces to the approximate Bayes factor (ABF) derived by Wakefield (2009), and it produces exactly the same Bayes factor as in fgwas (Pickrell 2014) when the variance of the effect size is estimated (see Appendix). The approximation depends on a key assumption that the explained variance by each model is very small compared to the total variance of the phenotype. This assumption is reasonable for most GWAS hit regions due to the small effect sizes. However, for fine mapping expression quantitative trait loci (eQTL), this assumption may not hold very well because it is not uncommon that the gene expression trait is moderately or highly correlated with certain variants close to the gene. Fortunately, we found that for quantitative traits the exact Bayes factors using a D2 prior (Servin and Stephens 2007) can still be calculated using only the marginal test statistics and correlation matrix. We assume a linear model as follows:
where , denotes the n×n identity matrix, and n is the number of individuals. is the coded genotype matrix from putative causal variants. The D2 prior assumes that β has a prior normal distribution of , where is a diagonal matrix and β and are independent. The jth marginal test statistic is calculated from the model
where is the jth column of . Denote the correlation matrix by . Let be the column vector where the jth component is , and let S be a diagonal matrix where the jth entry is the unbiased sample variance of . The exact Bayes factor can be calculated as follows:
The detailed proof is shown in the Appendix. Besides the exact calculation, to make it robust to different magnitudes of effect sizes, we also added the option in CAVIARBF to set multiple prior distributions for β using different variance values and to calculate the averaged Bayes factor as in BIMBAM (Servin and Stephens 2007; Guan and Stephens 2008).
Maximum a posteriori estimate using expectation maximization
In general, techniques such as Markov chain Monte Carlo (MCMC) can be used for inference based on the model in Figure 1. However, this can be time consuming. Instead, we calculate a point estimate of the annotation effects γ, the maximum a posteriori (MAP) estimate, and use it for all other inferences. We may sacrifice some accuracy by using the MAP estimate to represent the full posterior distribution of γ, but we gain computational speed. Given Y, G, A, and specified λ, , the posterior probability of γ is
Therefore,
where is a vector with elements , and is the norm of a vector. The term constant represents all other terms which are not a function of γ.
To maximize with respect to γ, we only need to maximize
Because , L is simplified to
This also shows the well-known correspondence between a Bayesian model using a Gaussian prior and a penalized likelihood model with l2/ridge penalty.
To maximize L, notice that the first part of the equation is a likelihood function with missing data, or hidden random variables. Therefore we can use the expectation-maximization (EM) algorithm to obtain the maximum likelihood estimate (MLE) of γ. For example, the causal status configuration c can be considered missing. Even though the effect size parameter β is also unobserved, it can be integrated out analytically for quantitative traits or using approximations for binary traits (Servin and Stephens 2007; Guan and Stephens 2008; Chen et al. 2015). Compared to the EM method for MLE, the MAP estimate requires only a small change at the maximization (M) step. We first show how to use the EM method for MLE when λ is set to 0 (Kichaev et al. 2014; Wen et al. 2015). If c were observed, the full-data likelihood would be , where we have used the dependence relationship in Figure 1. Therefore,
Denote by the conditional distribution of c given the observed data Y, G, A and the estimate at iteration t. In the expectation (E) step, we calculate the expectation of the log likelihood of full data on this conditional distribution as
The first term is not a function of γ, so it is irrelevant to the M step where we maximize this expectation with respect to γ. Therefore, we only need to focus on the second term. Assume given A, γ, the probability of each variant being causal is independent from each other. In implementation, we use a logistic regression model which satisfies this assumption. Then we have:
And because
we have
In the M step, we update γ by . If we use a logistic model that has , is the ith component of , and then the objective function is exactly the form of the log-likelihood function in a logistic regression model. The only difference is that the observed data, , , are not ones or zeros, but in the range of [0, 1]. Therefore for each iteration, the updated γ can be solved by fitting a logistic model treating as the response and A as the data matrix. This was also used in Wen et al. (2015). Because our aim is to find the MAP estimate instead of the MLE, the M step changes to:
| (1) |
The objective function to maximize has the same form of a penalized log likelihood for logistic regression, with the response being . In our implementation, we use the R package glmnet (Friedman et al. 2010) to solve the penalized logistic regression model.
For each iteration, needs to be recalculated. It is actually the posterior inclusion probability (PIP) for variant i, and can be calculated based on existing methods. Specifically, we first calculate the prior probability of each SNP being causal given the annotations and the estimated annotation effect size using . Then, any method that can calculate can be used. For example, given the prior probability of each SNP being causal, the original CAVIARBF can output the PIPs for each variant using summary statistics (Chen et al. 2015). It needs to be calculated for each iteration because the prior probability of each variant being causal changes depending on the current estimate .
Extension to LASSO and elastic net penalties
In Equation 1, we can change the penalty to other penalties, for example, (e.g., LASSO) penalty or elastic net (ENET), which uses both and penalties. These penalties correspond to different prior distributions of . Specifically, denote each component of by . If we assume a Laplace prior, , and are independent of each other, then for the MAP estimate of γ, the objective function to maximize becomes
where the penalty is . For ENET penalty, the corresponding objective function is
where α is a parameter that determines the mixture of and penalties. The corresponding prior is , assuming the is independent. Since only the penalty term changes, the same EM method can be applied except that in the M step, a different penalized model is solved. In our implementation, the R package glmnet is used to fit all three different penalized models.
Using summary statistics
The same framework can be applied if we have only the summary statistics. Denote the marginal test statistics for individual variants as , of which each component approximately follows a normal distribution. Denote the correlation matrix among variants as . As shown in Chen et al. (2015), in the fine-mapping setting, using (Y, G) is approximately equivalent to using (Z, Σ). Therefore when summary statistics are used, simply replace G with Σ, and Y with Z. The posterior probability of γ is
To calculate the MAP estimate of γ, similarly as using (Y, G), we take the limit , and maximize the following penalized likelihood
which assumes an penalty. A similar EM method can be used to obtain the MAP estimate. Specifically, for the M step, we only need to replace with , which can be calculated using CAVIARBF. Using the penalty or ENET penalty is also straightforward.
Multi-loci extension
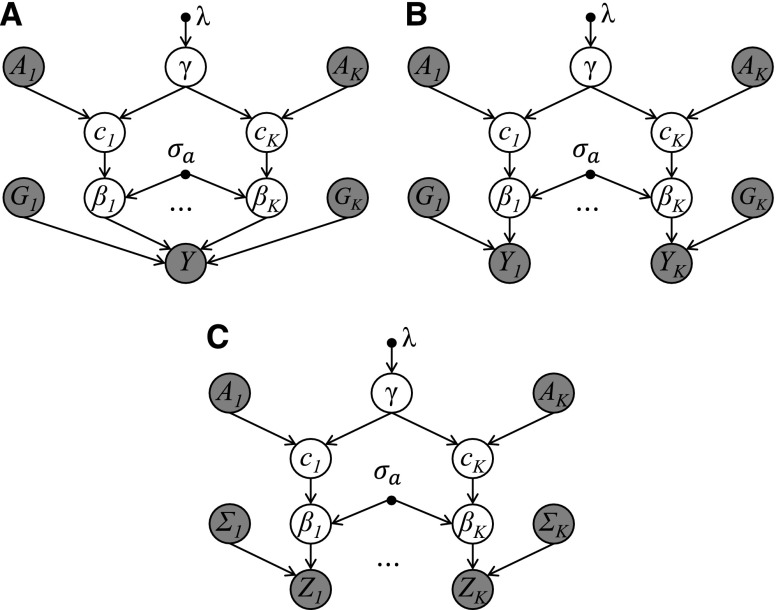
When there are multiple loci of interest, a joint model aggregating annotation information across all loci may be more powerful, as proposed in Kichaev et al. (2014) and Wen et al. (2015). We consider two scenarios here. For the first scenario, there is a single trait for all loci. This corresponds to fine mapping multiple GWAS hit regions for a single trait. For the second scenario, each locus has its own independent trait. For example, in fine mapping of eQTL, each locus has its own targeted gene expression trait. Figure 2 shows the hierarchical model of K loci incorporating annotation effects with subscripts denoting loci indices. All loci share the same annotation effect γ, which further depends on the parameter λ. Figure 2A corresponds to the model for fine mapping of multiple GWAS hit regions for a single trait and Figure 2B corresponds to the model for fine mapping of eQTLs, where are gene expression traits and are variants usually in a cis region of the corresponding genes. We assume independence among given γ and the corresponding . The two models in Figure 2, A and B, require both the phenotype and individual genotype data. Figure 2C shows the model using only summary statistics, which can be applied for both scenarios. It assumes that the marginal test statistics are independent from each other given γ and the corresponding . For fine mapping GWAS hit regions for a single trait, this assumption is usually reasonable if the genotypes of GWAS hit regions are independent, e.g., they are not too close to each other, or on different chromosomes. For the eQTL model, the independence assumption of implies the independence assumption of . If we define , and , and for the eQTL model, then Figure 2, A and B, can be collapsed to Figure 1, and Figure 2C can be collapsed to Figure 1 with Y and G replaced by Z and Σ, respectively.
Figure 2.
Hierarchical models of K loci incorporating functional annotations. All K loci share the same annotation effects. Shaded ○ indicates observed data. The definitions of symbols are the same as in Figure 1 except the extra subscript indicating the locus index. There are three different models depending on the input data: (A) full genotype and a single phenotype, e.g., a GWAS data set; (B) full genotype and multiple phenotypes for each locus, e.g., a eQTL data set; (C) marginal test statistics and correlations among variants for each locus, which can be derived from either a GWAS data set or an eQTL data set.
The same EM method can be used for the MAP estimate of . For example, in case of using summary statistics, the likelihood term in the penalized likelihood to maximize is
The only change in the EM method is that now depends on observed data from all loci. However, we can show that , where j is the locus where variant i is in, as follows:
In the above, we have assumed that are independent from each other given γ. This means the PIPs can be calculated based on data from each locus, which reduces the computational cost. Similarly, for the eQTL model, we can use to compute the PIPs.
Penalty parameters selection using Akaike information criterion, Bayesian information criterion, and cross validation
In the above, we assumed parameter λ is a known fixed value. For real data analysis we need to make it data adaptive for better model fitting. To select the best parameter, we fit the model using different penalty parameters λ and choose the best model based on several criteria, including the Akaike information criterion (AIC), the Bayesian information criterion (BIC), and cross validation (CV). Here the use of AIC and BIC are more heuristic than mathematically strict, however, they may provide easy-to-calculate ways to compare different models. For AIC, we use the formula
where loglik is the log likelihood of the observed data using the MAP estimate of γ, which is the first term of the penalized likelihood . is the effective number of parameters, which will be explained soon. For BIC, we use the formula
where is the number of variants. We follow the calculation of effective number of parameters/d.f. in Park and Hastie (2008) and Tibshirani and Taylor (2012). Specifically, for the penalty,
where is the trace function on a matrix, is a diagonal matrix with the diagonal entries being λ except the first one being 0. W is a diagonal matrix with diagonal entry , where is the final fitted PIPs. For the penalty,
where is a submatrix of A, with columns selected corresponding to nonzero values in γ. For ENET penalty, based on results derived by Park and Hastie (2008) and Tibshirani and Taylor (2012), we use
Compared to AIC and BIC, CV is more time consuming but usually achieves better performance. When we have the genotype- and phenotype-level data, i.e., Y, G are available, we can partition the individuals into subsets, and do CV over different values of λ (and α for ENET penalty). When we only have summary statistics, we can partition the loci into subsets and do CV, as used in Pickrell (2014). The partition based on loci assumes independence among loci. For CV using partitioned loci, we calculate the MAP estimate of γ using the training data, and then use it to calculate the log likelihood of the testing data. The best parameter is chosen based on the maximum of likelihood on the testing data, i.e., the maximum of the sum of the log likelihood of all the testing data.
Significance of individual annotations and estimating marginal effect sizes
Because the above framework incorporates all annotations into the model, whether an annotation is included in the final model and the estimated effect sizes depend on all the available annotations. For screening a large number of annotations, it is common practice to first assess their marginal association significance and effect size. The top ranked annotations may provide functional insight into the underlying mechanism, and can be further combined in the final model (Pickrell 2014). This may produce overly optimistic results and needs validation on external independent data sets. To perform this type of screening, we use the likelihood-based method by setting λ to 0; therefore, no parameter selection is needed and it can be computed quite quickly. To test the significance of individual annotations, we choose the likelihood ratio test of nested models, similarly as in PAINTOR (Kichaev et al. 2014). Denote the log likelihood as for the null model without any annotation. Denote the log likelihood of adding a single annotation in the model as . Then, under the null hypothesis that the annotation is not associated with the causal states, approximately follows a distribution. To verify the null distribution, we took the eQTL data (see later eQTL data analysis) and simulated 105 independent annotations from the standard normal distribution. Supplemental Material, Figure S1 shows the Q–Q plot under the null hypothesis, where the observed and theoretical quantiles match well.
Methods settings
We mainly compared the performance of CAVIARBF with PAINTOR, which can also incorporate functional annotations and showed very competitive performance over other methods (Kichaev et al. 2014). Because PAINTOR does not provide a direct way to simultaneously incorporate all annotations, several different strategies of selecting informative annotations were tested. We also compared our method with fgwas, assuming one causal variant and focusing on the different annotation selection scheme, where the Bayes factors are almost the same (they are exactly the same under certain settings). We implemented the forward-backward annotation selection proposed in Pickrell (2014) for both fgwas and CAVIARBF for the comparison. For the forward-backward annotation selection, we defined the significance for individual annotations using the P-value threshold 0.05. This may result in false inclusion of irrelevant annotations at the first step simply due to randomness given the large number of annotations. The later backward exclusion might reduce this effect. However, we still see obvious inflation of PIP and decreased performance under the null annotations using this search scheme. Because FM-QTL only considers as few annotations as PAINTOR, it is not compared here. All tested analysis methods are described in Table 1.
Table 1. Different methods compared in this study.
| Name format | Examples | Annotation usage | Explanation |
|---|---|---|---|
| <method>_non | CAVIARBF_non, PAINTOR_non, fgwas_non | No | Fit the intercept using the traditional logistic regression in the EM method. For fgwas_non, no annotation is used |
| CAVIARBF_glm | CAVIARBF_glm | Yes | Fit all annotations using the traditional logistic regression in the EM method |
| CAVIARBF_<penalty>_<criterion> | CAVIARBF_L2_CV, CAVIARBF_L1_CV, CAVIARBF_ENET_CV, CAVIARBF_ENET_AIC, CAVIARBF_ENET_BIC | Yes | Use penalized models. L2, penalty; L1, penalty; ENET, elastic net penalty; <criterion> can be AIC, BIC, and CV |
| CAVIARBF_fb_CV | CAVIARBF_fb_CV | Yes | Assume one causal variant and use the forward-backward annotation selection proposed in fgwas |
| <method>_top<k> | PAINTOR_top40 | Yes | First, annotations are ranked by their individual increase in likelihood compared to the model without any annotation. Then the top k annotations are used |
| <method>_top<k>t | PAINTOR_top10t, CAVIARBF_top10t | Yes | All annotations are ranked as in top<k>. Then we sequentially select the annotations with additional two thresholds: (1) the absolute correlations between already selected annotations and the annotation to select are less than a threshold, 0.2 in our simulations; (2) the individual P-value of the annotation to select against the null model without any annotation is less than a threshold, 0.05 in our simulations. This method may not be able to select exactly k annotations due to the additional constraints |
| <method>_step<k>t | PAINTOR_step5t, PAINTOR_step10t | Yes | The difference between step<k>t and top<k>t is that before selecting a new annotation to include, all remaining candidate annotations are ranked based on their individual contributions in likelihood to the model with all current selected annotations. Specifically, let be the log likelihood from the model using the current selected annotations and be the log likelihood from the model using the current selected annotations plus candidate annotation i. Then the difference is the contribution of adding annotation i. It starts with the candidate annotation with the largest contribution and checks the threshold in the same way as in top<k>t. Once an annotation is selected, all the remaining candidate annotations will be evaluated based on the increase of likelihood for the next selection. This method may not be able to select exactly k annotations due to the additional constraints |
| fgwas | fgwas | Yes | fgwas with annotation and the forward-backward annotation selection |
Without any specific change of settings, here is the default setting for CAVIARBF: We used fivefold CV, when CV is used for penalty parameter selection. We set the maximal number of causal variants in each locus to 3, and the parameter For CAVIARBF_ENET_CV, the grid search set for λ and α was set to for simulated data sets. For real data analysis, λ was searched from for a finer grid search. For CAVIARBF_L2 _CV and CAVIARBF_L1 _CV, the search set for λ was the same as that in CAVIARBF_ENET_CV. PAINTOR version 2.1 was used in all simulations and we set the maximal number of causal variants to 3. For lipid data analysis, we assumed the genotype effect sizes follow a normal distribution on the standardized genotypes (mean, 0; variance, 1), to be consistent with the assumption in PAINTOR (Chen et al. 2015). But for the eQTL analysis, we assumed the genotype effect size follows a normal distribution on the original scale of genotypes, i.e., the counts of the specified allele.
Data simulation and evaluation
We first simulated a large number of annotations associated with each variant and some of them were correlated. Specifically, 10 groups of annotations were simulated. Each group had 10 correlated annotations, and annotations between groups were independent. Annotations in a group were simulated from a multivariate normal distribution with the pairwise correlation coefficient of 0.4. Then, the probability of being causal for each variant was simulated based on the annotations using a logistic model. Specifically, , where A is the annotation matrix including the intercept and all m variants, γ is the annotation effect size vector, is the ith component of . Due to different annotation effects reported (Carbonetto and Stephens 2013; Kichaev et al. 2014; Wen et al. 2015), we considered two different settings for the annotation coefficient γ: a small number of annotations with large effects, and a large number of annotations with moderate effects. For the first setting we chose 5 out of 100 annotations with an annotation coefficient/effect size of log5. For the second setting we chose 40 out of 100 annotations with an annotation coefficient of log1.5. The rest of the annotations had a coefficient of zero except the intercept. To ensure the number of causal variants in each locus was not too large, we set the intercept in γ to , where is the number of variants in each locus, 35 for multiple loci data simulation. We only kept samples where the maximal number of causal variants in each locus is no more than 5, because having >5 causal variants in a locus may not be very realistic, and we used three maximal causal variants in all simulations and would not like the true underlying model to deviate from our assumption too much. For fine-mapping analysis, similarly to the simulation in Kichaev et al. (2014), we only focused on the loci with at least one causal variant. Therefore, the number of loci used for analysis was often smaller than the initial simulated loci. We simulated two different numbers of initial loci: 20 and 100. The average numbers of loci with 1, 2, 3, 4, and 5 causal variants across 100 data sets were summarized in Table S1.
Next we simulated the genotypes for multiple loci. We used HAPGEN (Su et al. 2011) to simulate genotypes, using the CEU population from the 1000 Genomes Project (Abecasis et al. 2012) as the reference panel. We first randomly sampled multiple loci on chromosome 8 and made sure the distance between loci was no less than 1 cM. For each locus, we randomly selected a region of common SNPs (minor allele frequency >0.01) and simulated n = 10,000 individuals. The genotypes were additively coded as 0, 1, and 2. Quantitative traits were simulated based on the genotypes and specified causal variants using a linear model. Let be the sampled genotype matrix of causal variants in a locus, and the corresponding coefficients. Suppose is centered with zero mean in each column, then the sample variance of the genotype component of this locus is . We uniformly sampled a value between 1 and 2 and set it to the sample variance and obtained a solution of β, which made sure the variance explained by each locus was similar. The quantitative trait vector for n individuals was sampled from , where r is the total number of loci and and are the causal genotype matrices and coefficients, respectively. We set according to the total proportion of variance explained (PVE) by all loci. Specifically, we used the following definition of PVE (Guan and Stephens 2011; Zhou et al. 2013):
where . Function is the sample variance, i.e., , where x is a vector with each component denoted by , and is the mean of all values in x. Therefore . We set for 100 initial loci and for 20 initial loci. Note that the way we simulated genotype effects and the traits were different from that in Chen et al. (2015). Here we did not enforce the noncentrality parameters of causal variants to be in a certain range, which may not be easy to control when simulating a large number of loci. Similar as in Kichaev et al. (2014), we also did not require a significance threshold P-value of 5 × 10−8 for each locus.
One interesting question is whether we gain much by incorporating annotation information if we only have one GWAS hit, which is not uncommon in real data. To address this question, we also simulated 100 data sets with only one locus. We set and The simulation was the same as above except the following: When simulating 40 informative annotations with the effect size log1.5, the intercept was set to , which ensured the number of causal variants was not too large. The total numbers of data sets with 1, 2, 3, 4, and 5 causal variants across 100 data sets were 31, 28, 21, 16, and 4, respectively, for five informative annotations. The distribution of causal variants was similar for 40 informative annotations.
We simulated 100 data sets for each scenario: two settings for annotation effect sizes and two settings for initial number of loci. To compare different methods, we calculated the proportion of causal variants included among 100 data sets when selecting the top SNPs ranked by PIPs.
We also simulated binary annotation with 100 initial loci when comparing CAVIARBF with fgwas. We first simulated the continuous annotations as described above and then dichotomized them into binary annotations using a random threshold. Everything else was the same as simulating continuous annotations. We also simulated scenarios under the null where no annotation was informative. The data were simulated in the same way as that with binary annotations except that the annotation effect size was zero.
Lipid data
The lipid data provides the meta-analysis results on four phenotypes: total cholesterol (TC), low-density lipoprotein cholesterol (LDL), high-density lipoprotein cholesterol (HDL), and triglycerides (TG) (Teslovich et al. 2010). To include as many SNPs as possible in the analysis, we downloaded the imputed results using ImpG (Pasaniuc et al. 2014) from http://bogdan.bioinformatics.ucla.edu/software/impg/. SNPs with potential ambiguous allele coding, i.e., A/T or C/G, and SNPs with a meta-analysis sample size of <80,000 were removed before applying ImpG. We extracted the hit regions using a 100-kb window, the same sized region as used in the previous analysis (Kichaev et al. 2014), with the reported lead SNPs (Teslovich et al. 2010) in the center. We excluded several hit regions where SNPs had high correlations (>0.9 calculated using EUR populations from the 1000 Genomes Project) but showed large differences in Z statistics. In the end, we had 30 hit regions for LDL, 41 for HDL, 45 for TC, and 26 for TG.
Annotations used for lipid data
We downloaded the same annotation used in a previous study (Pickrell 2014) from https://github.com/joepickrell/1000-genomes. In brief, there are 451 annotations, where 450 are binary and only 1 is quantitative [distance to the nearest transcription start site (TSS)]. The annotations are mainly maps of DHSs in a wide range of primary cell types and cell lines. It also includes the predicted genome segmentations from six ENCODE cell lines (Hoffman et al. 2013). We normalized the quantitative annotation to have a mean of 0 and a variance of 1.
eQTL data
In the first stage, we identified all potential genes related to prostate cancer (PrCa)-risk SNPs for each risk interval. Specifically, from 146 reported PrCa-risk SNPs, we included a total of 6324 risk and in-linkage-disequilibrium (LD) tag SNPs (r2 > 0.5) in 100 unique risk intervals. After quality control and normalization, both RNA sequencing (RNA-seq) data of normal prostate tissue and genotype data were obtained from 471 individuals with European ancestry. Samples were genotyped using Illumina Infinium 2.5 M bead arrays and then imputed using SHAPTIT and IMPUTE2 with reference files from the 1000 Genomes Phase I integrated variant set. We analyzed all SNPs in the risk intervals for association with surrounding gene transcripts. In total there were 127,276 SNP-gene pairs with 51 out of 100 risk intervals demonstrating a Bonferroni-significant eQTL signal (P-value < 1.96 × 10–7) and these were associated with 88 genes. More details on the data collection and analysis can be found in Thibodeau et al. (2015).
In the second stage, to fully identify all cis-eQTLs for these 88 target genes, we chose all candidate SNPs within 1.1 Mb of each gene’s TSS or transcription end site positions. Because some candidate SNPs overlapped in multiple target genes, these violated our assumption of independence among loci. As a proof of principle, we selected 41 target genes with nonoverlapping candidate SNPs in this analysis. The phenotype of each target gene is the residual from regressing normalized gene expression level on covariates: the histologic characteristics, percent lymphocytes, percent epithelium, and 14 expression principal components. To reduce computational burden, we first filtered candidate SNPs using a P-value threshold of 1 × 10−4 and kept a maximum of 250 SNPs for each target gene/locus.
Annotations used for eQTL data
We collected 76 annotation tracks that may be related to cis-eQTLs in normal prostate tissue. These include predicted chromatin state using ChromHMM (Ernst and Kellis 2012) in breast and prostate normal and cancer cells (Taberlay et al. 2014), DHSs from the normal and cancer prostate cells from the ENCODE project (The ENCODE Project Consortium 2012), histone modifications such as histone acetylation, histone methylation (He et al. 2010; Wang et al. 2011; Bert et al. 2013; Hazelett et al. 2014), and transcription factor bindings (Tan et al. 2012; The ENCODE Project Consortium 2012; Sharma et al. 2013; Hazelett et al. 2014; Takayama et al. 2014). All annotations were aligned to human genome version 19 coordinates. For each category of the predicted chromatin state from ChromHMM, we created a binary annotation where 1 indicates the SNP is in that category and 0 otherwise. Other annotations are quantitative. For simplicity and proof of principle, we set the missing annotation values to 0. More advanced annotation imputation (Ernst and Kellis 2015) may achieve higher imputation accuracy. We normalized all annotations to have mean 0 and variance 1.
Data availability
The detailed annotation list and Gene Expression Omnibus track numbers are shown in File S2. The summary statistics and annotation matrix used of the eQTL data are available upon request. Software is available at https://bitbucket.org/Wenan/caviarbf.
Results
Proportion of causal variants identified by different methods
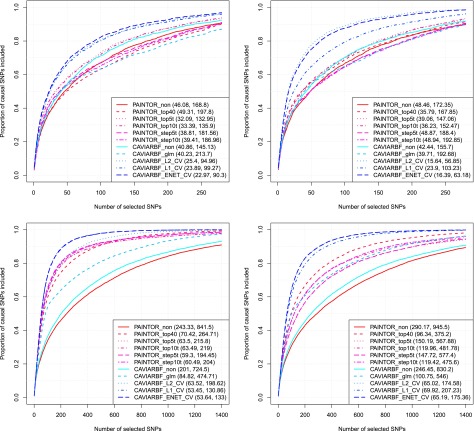
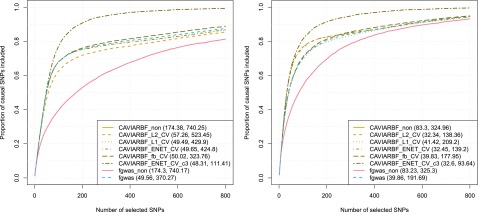
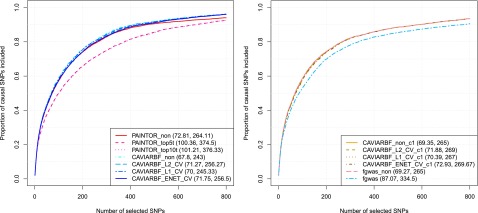
Results for all analyses are shown in Figure 3. The top panels correspond to 20 initial loci and the bottom panels correspond to 100 initial loci. The left column corresponds to five informative annotations with large effects, and the right column corresponds to 40 informative annotations with moderate effects. CAVIARBF applied using the ENET penalty with CV (CAVIARBF_ENET_CV) is the most robust and among the best methods across all scenarios. When the number of informative annotations is small and their effects are large, CAVIARBF using the ENET penalty usually has similar performance as that using the penalty. On the other hand, when the number of informative annotations is large and their effects are moderate, CAVIARBF using the ENET penalty usually has similar performance as that using penalty. This is consistent with the characteristics of these penalties (Tibshirani 1996; Zou and Hastie 2005). As expected, for both CAVIARBF and PAINTOR, including annotations increases the probability of identifying the causal variants. CAVIARBF_glm, which uses only the MLE without any penalty term when incorporating the annotations, has much lower accuracy compared to all penalized models, sometimes even lower than CAVIARBF_non (Figure 3, top two panels). This demonstrates the overfitting problem when fitting a large number of annotations without regularization. The same problem can be observed for PAINTOR. For example, PAINTOR including the top 40 annotations (PAINTOR_top40) has worse performance than PAINTOR applied with no annotation (PAINTOR_non) in the top left panel. This shows the importance of using penalties in the model when incorporating a large number of annotations. There is no single best annotation selection method for PAINTOR; the performance depends on the number of loci and the number of informative annotations. PAINTOR_top5t and PAINTOR_top10t appear to be good options in general, even though when the number of informative annotations and loci are large (bottom right panel), simply choosing the top 40 annotations (PAINTOR_top40) achieves the best result for PAINTOR. The advantage of CAVIARBF_ENET_CV over PAINTOR is larger when the number of informative annotations is large. This illustrates the benefit of incorporating all annotations with regularization instead of only using the top annotations. When no annotation is used, CAVIARBF has slightly better performance than PAINTOR, although the magnitude of differences varies among different scenarios.
Figure 3.
Proportion of causal variants identified by different methods. The top panels correspond to 20 initial loci (∼12 causal loci) and the bottom 5panels correspond to 100 initial loci (∼65 causal loci). The left column corresponds to 5 informative annotations with large effects, and the right column corresponds to 40 informative annotations with moderate effects. The numbers shown in the parentheses correspond to the average number of SNPs required to identify 50 and 80% of the causal SNPs, respectively.
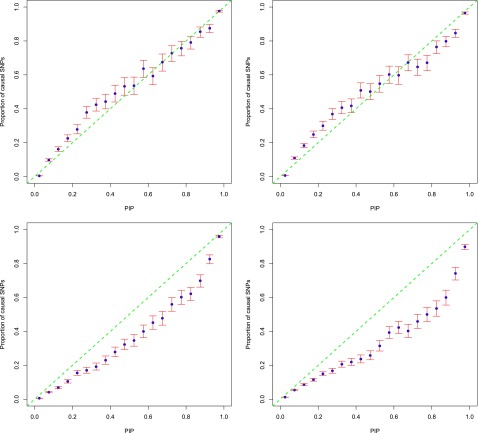
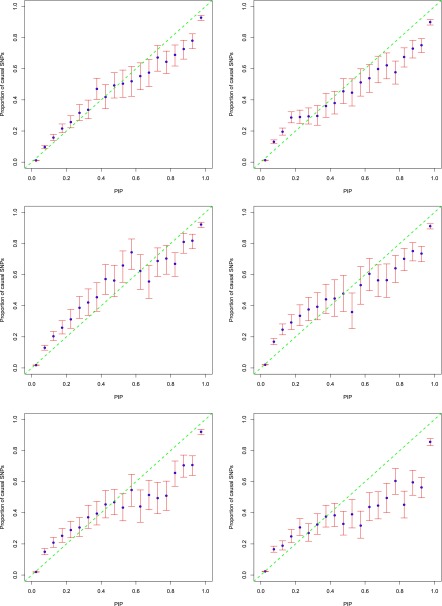
PIPs calibration
To assess the accuracy of estimated PIPs, we put SNPs into bins according to their PIPs and then compared the proportion of causal SNPs in each bin with the center PIP of that bin (Guan and Stephens 2011). Figure 4 shows the results using the data sets with 100 initial loci. Note that the initial loci include all causal and noncausal loci. The final data sets used in testing include only causal loci (Table S1). This follows the pattern of simulation used in PAINTOR (Kichaev et al. 2014). The left two panels represent the scenario of five informative annotations with an effect size of log5, the right two panels represent the scenario of 40 informative annotations with an effect size of log1.5. PIPs from CAVIARBF_ENET_CV, the top two panels with different annotation patterns, are calibrated well in general; while PIPs from PAINTOR_top5t, the bottom two panels, are overestimated. PIPs from CAVIARBF_ENET_CV show some downward bias in the middle, e.g., from 0.2 to 0.5, resulting in conservative PIPs. However, this is preferable to upward bias which results in overly optimistic PIPs. When there are a large number of annotations with moderate effects (top right), CAVIARBF_ENET_CV shows a small upward bias, e.g., from 0.7 to 0.9. PAINTOR_top5t shows consistent upward bias, which is larger in the middle and becomes smaller when PIPs are close to one. We note that the number of loci harboring at least one causal variant has an influence on the calibration and accuracy of PIPs. Figure S2 and Figure S3 show the results for 40 and 20 initial loci, corresponding to ∼25 and 12 loci on average harboring at least one causal variant (Table S1), respectively. For these small numbers of loci, even though CAVIARBF_ENET_CV always shows better calibration than PAINTOR_top5t, there exists upward bias, e.g., in the range of 0.6–0.9. Therefore, when incorporating annotations and the number of loci harboring at least one causal variant is <20, we need to interpret the PIPs with caution and consider the potential upward bias. We discuss potential sources of these biases in the Discussion section.
Figure 4.
PIP calibration with 100 initial loci (∼65 causal loci). The top two panels show results from CAVIARBF_ENET_CV, bottom two panels are from PAINTOR_top5t. Left two panels, 5 informative annotations with effect size log5; right two panels, 40 informative annotations with effect size log1.5. The x-axis shows the center of 20 bins of width 0.05. The y-axis is the proportion of causal SNPs. The blue points show the proportion of causal SNPs in each bin. The red bars show the 95% C.I. (Wilson inversion of score statistic) of the proportion assuming a binomial distribution in each bin. 100 data sets were used in each panel.
Comparison with fgwas assuming one causal variant
To make sure the Bayes factors were equivalent, we set W in fgwas to 0.01, to 0.1, and SNP variance as the weight input in CAVIARBF. A 10-fold CV was used for CAVIARBF as in fgwas. Figure 5 shows the results. When no annotation was used, CAVIARBF_non and fgwas_non had the same performance (CAVIARBF_non is overlapped and not visible). This also reflects their equivalence in Bayes factors. When there were five informative annotations with large effects (left panel), fgwas and CAVIARBF_ENET_CV showed similar results when the number of selected SNPs was small, and fgwas was slightly better when the number of selected SNPs was large. When there were 40 informative annotations with moderate effects (right panel), CAVIARBF_ENET_CV showed better performance than fgwas. CAVIARBF_fb_CV uses the same forward-backward annotation selection scheme as in fgwas. The performance was similar to fgwas in general, even though CAVIARBF_fb_CV was slightly better when the number of selected SNPs was large. The reason is not clear to us. It might be due to a different optimization, where CAVIARBF uses the EM method and fgwas uses a gradient search. In both annotation scenarios, CAVIARBF_ENET_CV_c3, assuming three maximal causal variants, showed much better performance than those assuming one causal variant. The forward-backward annotation selection was faster than CAVIARBF_ENET_CV. It might be a good option when CAVIARBF_ENET_CV takes too much time or we know that only a small portion of annotations are informative among candidate annotations.
Figure 5.
Proportion of causal variants identified by CAVIARBF and fgwas. The left panel corresponds to 5 informative annotations with large effects, and the right panel corresponds to 40 informative annotations with moderate effects. The numbers shown in the parentheses correspond to the average number of SNPs required to identify 50 and 80% of the causal SNPs, respectively. CAVIARBF_ENET_CV_c3 assumes three causal variants.
Performance under null annotations
Figure 6 (left panel) shows the performance of PAINTOR and CAVIARBF. CAVIARBF with different penalties showed similar or slightly decreased performance compared with CAVIARBF_non. CAVIARBF_ENET_CV performed similarly to CAVIARBF_non in terms of ranking causal variants when there were no informative annotations. On the other hand, the performance of PAINTOR_top5t or PAINTOR_top10t was far poorer than PAINTOR_non. Figure 6 (right panel) shows the comparison between fgwas and CAVIARBF assuming one causal variant. CAVIARBF with different penalties had similar performance, but fgwas had decreased performance compared to fgwas_non. Figure S4 shows the result of PIP calibration. Assuming three maximal causal variants, CAVIARBF_non (top left) shows good calibration, and CAVIARBF_ENET_CV (top right) has some inflated PIP estimation. The pattern is similar for CAVIARBF assuming one causal variant (middle panels). For fgwas without annotations, it shows good calibration (bottom left), however, when null annotations are included (bottom right), the PIP is more inflated than CAVIARBF. We also compared the number of SNPs with PIP >0.9. Figure S5 (top panel) shows the histogram of the difference between CAVIARBF_ENET_CV and CAVIARBF_non. Out of 100 data sets, 68 had exactly the same number between CAVIARBF_ENET_CV and CAVIARBF_non. There are only 8 out of 100 data sets where CAVIARBF_ENET_CV has more than three SNPs than CAVIARBF_non. Given that the average number of causal variants was 50, the increase of the number of SNPs >0.9 for CAVIARBF_ENET_CV under the null was either zero or very small. We also compared the number of SNPs that reached 90% of the expected causal variants (called “90% confidence set” as in Table 2 in Kichaev et al. 2014, although it is not much related to a confidence level or a credible set if the number of causal variants in each locus is more than one). Figure S5 (bottom panel) shows the histogram of the difference. Out of 100, 40 had exactly no difference. However, there were also ∼36 data sets with a reduction of SNPs >30 by CAVIARBF_ENET_CV. Therefore, the reduction of SNPs by the current implementation of CAVIARBF_ENET_CV does not provide convincing evidence of the informativeness of annotations. After examination of the difference between CAVIARBF_non and CAVIARBF_ENET_CV, we conclude this is due to falsely choosing a small penalty parameter instead of a large penalty parameter, even though the CV likelihood increased very little. Better ways to avoid this may be useful, such as choosing the largest penalty that achieves similar CV likelihood as the maximal CV likelihood.
Figure 6.
Proportion of causal variants identified when the annotation effect size is zero. The left panel shows comparison between PAINTOR and CAVIARBF assuming three maximal causal variants. The right panel shows comparison between fgwas and CAVIARBF assuming one causal variant.
Table 2. Computing time of different methods in seconds.
| PAINTOR | CAVIARBF | fgwas | |||
|---|---|---|---|---|---|
| c = 1 | |||||
| PAINTOR_non | 0.46 | CAVIARBF_non | 1.8 | fgwas_non | 0.2 |
| PAINTOR_top10t | 50 | CAVIARBF_top10t | 259 | fgwas | 454 |
| CAVIARBF_L2_CV | 1,341 | ||||
| CAVIARBF_L1_CV | 1,789 | ||||
| CAVIARBF_ENET_CV | 10,727 | ||||
| c = 3 | |||||
| PAINTOR_non | 9 | CAVIARBF_non | 10 | ||
| PAINTOR_top10t | 1151 | CAVIARBF_top10t | 621 | ||
| CAVIARBF_L2_CV | 3,126 | ||||
| CAVIARBF_L1_CV | 6,541 | ||||
| CAVIARBF_ENET_CV | 46,184 |
Comparison of AIC, BIC, and CV for penalty parameters selection
We also compared performance using different model criteria including AIC, BIC, and CV (Figure S6). The performances of CV are better than that of using AIC or BIC. In terms of computing time, CV is more computationally intensive than AIC or BIC. Even though AIC and BIC are not the best options, incorporating annotations using AIC or BIC can still achieve improved performance than that without annotations. For example, the performance of methods using AIC is consistently better than CAVIARBF_non. In most situations, the performance of methods using AIC is better than or similar to those using BIC, which are usually close to CAVIARBF_non. This may be due to too much penalization in BIC. The only exception is when the number of informative loci is 5 and the total initial loci is 100 (bottom left panel), for methods using or ENET penalties, the performance using BIC is better than that using AIC.
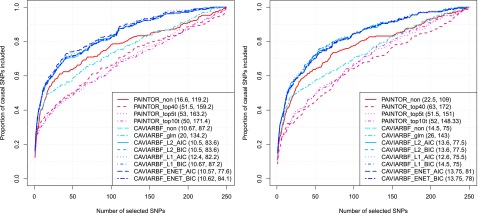
Performance when there is only one GWAS hit
Figure 7 shows the results incorporating annotations when there is only one locus of interest. Because CV partitions loci, it requires at least two loci, so only AIC or BIC can be used for parameter selection. There is not much difference between CAVIARBF_non and CAVIARBF using annotations. This is reasonable because the number of causal variants in a single locus is very small, no more than 5, even though the number of noncausal variants is large. The information from the causal/noncausal contrast is limited, which makes it hard to determine informative annotations and make use of them. It is similar to the situation where there are only a few cases but a large number of controls in a logistic regression, in which case it is difficult to estimate the effects of covariates or determine their significance. And this is exactly what happens in the computation: for each iteration of the EM method, a logistic regression is fitted with the PIPs as the outcome and annotations as the covariates. Because PAINTOR uses the MLE without regularization, simply choosing a large number of annotations, e.g., PAINTOR_top40, severely deteriorates the performance. Even with careful screening of annotations considering correlation and significance levels, e.g., PAINTOR_top10t or PAINTOR_step10t, the performance can still be much worse than that using no annotations. This also illustrates the advantage of the proposed penalized model or Bayesian framework: it is adaptive to the information available in the annotation. When there is very little information in the annotation, the penalized framework has similar results as that using no annotation. When there is enough information in the annotation, the penalized framework can use it and achieve better performance.
Figure 7.
Proportion of causal variants identified when there is only one locus. The left column corresponds to 5 informative annotations with large effects, and the right column corresponds to 40 informative annotations with moderate effects. The numbers shown in the parentheses correspond to the average number of SNPs required to identify 50 and 80% of the causal SNPs, respectively.
Fine mapping when the true correlation matrix is not available
Sometimes only the marginal Z statistics resulting from association analysis are available, for example, meta-analysis results based on marginal Z statistics of multiple studies without direct access to the underlying genotype data. If we assume there is at most one causal variant for each locus, CAVIARBF reduces to the ABF (Wakefield 2009). Under this assumption, both CAVIARBF and PAINTOR do not need the correlation matrix. Alternatively we can choose a reference population similar to the population being studied, e.g., from the 1000 Genome Project, and use the correlation calculated from the reference population as an approximation. Applying penalization/shrinkage on the correlation matrix may also improve the performance (Wen and Stephens 2010; Chen and Schaid 2014; Pasaniuc et al. 2014). Specifically in our simulation, we added a small positive value to the main diagonal of the correlation matrix calculated from the haplotypes of 379 EUR subjects in the 1000 Genome Project. We empirically set this positive value to 0.2; using other values showed similar conclusion.
To compare different options for the unknown correlation matrix, we simulated data sets as described above using 40 initial loci and set the PVE to 0.1. In total, 100 data sets were simulated for each type of annotation setting and ∼25 loci on average harboring causal variants were generated (Table S1). This number of loci matches better with that of the lipid data we will analyze later than using 20 or 100 initial loci.
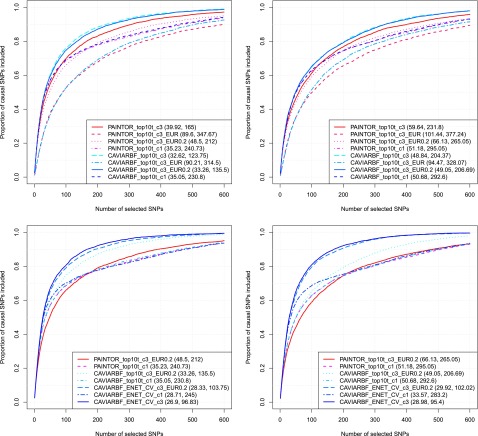
We used either one or three maximal number of causal variants, denoted by c1 or c3, respectively. Results are shown in Figure 8. The left two panels correspond to five informative annotations with an effect size of log5, and the right two panels correspond to 40 informative annotations with an effect size of log1.5. We first compare different options of PAINTOR and CAVIARBF using the top10t strategy (the top two panels). Unsurprisingly, for both CAVIARBF and PAINTOR, using the true correlation matrix (CAVIARBF_top10t_c3 and PAINTOR_top10t_c3) perform the best compared to other options. When the true correlation matrix is not available, CAVIARBF using a well-matched reference panel with shrinkage (CAVIARBF_top10t_c3_EUR0.2) achieves the best performance. Using the shrinkage on the reference correlation matrix significantly improves the performance compared to no shrinkage (CAVIARBF_top10t_c3_EUR). Even though assuming at most one causal variant is not optimal, it is better than using the reference panel without shrinkage. This can be useful if a well-matched reference panel is not available. For the same option, CAVIARBF has similar or slightly better performance than PAINTOR. Next we compare the top10t strategy with ENET using CV (the bottom two panels). CAVIARBF using the ENET to incorporate all annotations (CAVIARBF_ENET_CV_c3_EUR0.2) further improves the performance of CAVIARBF_top10t_c3_EUR0.2, and is very close to the performance assuming known correlation matrix (CAVIARBF_ENET_CV_c3). When assuming at most one causal variant, ENET using CV (CAVIARBF_ENET_CV_c1) is in general still better than the top10t strategy (CAVIARBF_top10t _c1). We noted that PAINTOR 2.1 uses a new default strategy of setting the noncentrality parameters (NCP). Switching to the old setting of NCP showed a slight improvement but did not change the general pattern of the results.
Figure 8.
Proportion of causal variants identified with and without the true correlation matrix or with reference correlation matrix. The two panels on the left correspond to 5 informative annotations with effect size log5, and the two on the right correspond to 40 informative annotations with effect size log1.5. The numbers shown in the parentheses correspond to the average number of SNPs required to identify 50 and 80% of the causal SNPs, respectively. c3 and c1 indicates that the maximal number of causal variants is 3 or 1, respectively. EUR means using the correlation matrix calculated from the EUR population in the 1000 Genomes Project and EUR0.2 means adding 0.2 to the main diagonal of the correlation matrix calculated from the EUR population.
Figure 9 shows the PIP calibration of CAVIARBF_ENET_CV_c3_EUR0.2 (two top panels), CAVIARBF _ENET_CV_c1 (two middle panels), and PAINTOR _top10t _c1 (two bottom panels). The left column corresponds to five informative annotations with effect size log5, and the right column corresponds to 40 informative annotations with an effect size of log1.5. In general, CAVIARBF_ENET_CV_c3_EUR0.2 and CAVIARBF _ENET_CV_c1 show slightly better calibration than PAINTOR _top10t _c1, especially when the annotation’s effect size is moderate (the right column). The PIP of CAVIARBF_ENET_CV_c3_EUR0.2 and CAVIARBF_ENET_CV_c1 is a little inflated when its value is >0.5. For high PIPs, e.g., PIP >0.95, PIPs of all three methods are closer to the true proportion of causal variants in the data sets.
Figure 9.
PIP calibration of CAVIARBF _ENET_CV_c3_EUR0.2 (two top panels), CAVIARBF _ENET_CV_c1 (two middle panels), and PAINTOR _top10t_c1 (two bottom panels). c3 and c1 indicates that the maximal number of causal variants is 3 or 1, respectively. EUR0.2 means adding 0.2 to the main diagonal of the correlation matrix calculated from the EUR population. The left column of panels, 5 annotations with effect size log5; the right column of panels, 40 annotations with effect size log1.5. The bins and calculation of C.I.s are the same as in Figure 4.
In summary, it is best to use the true correlation matrix when it is available. When the true correlation matrix is not available but there is a well matched reference panel, the correlation matrix from the reference panel with shrinkage achieves the best performance. To find a well-matched reference panel, we can use the continental populations from the 1000 Genomes Project, e.g., matching samples with European ancestry with the EUR population. When the (imputed) genotypes of a subsample are available, we can use correlations computed from the subsample as an approximation of that from the full data. Otherwise, assuming at most one causal variant and using ENET to incorporate all annotations seems to be the current best practice.
Estimated annotation effect sizes and bootstrap-based confidence intervals
Figure S7 shows the distribution of estimated annotation effect sizes averaged over 100 data sets for CAVIARBF_ENET_CV. For null annotations, the median of the estimate is close to zero with both positive and negative variations; for informative annotations, the estimate is more likely have the correct sign but with shrinkage of the effect size. It is clear that the estimated coefficients reflect the underlying annotation status in the averaged plot. However, in real data analysis, we need to distinguish random variation from true nonzero annotation effect size. We tried to use a bootstrap method to estimate the confidence interval of estimated annotations effect sizes. Specifically, we resampled the loci with replacement and calculated the confidence interval using the percentile method. Even though bootstrap-based confidence intervals can be problematic for penalized models (Kyung et al. 2010), we still tried to assess how helpful it could be for our model. Table S2 shows the coverage of 90% bootstrap confidence intervals based on 20 data sets and averaged over different categories of SNPs. For five informative annotations with large effects, the coverage for nonnull SNPs does not reach the specified level. One challenge is the biased (shrunken) estimation resulting from penalized models. For 40 informative annotations, the coverage for nonnull SNPs is close to the nominal. However, it can be hard to distinguish the nonnull from null annotations based on the confidence intervals. One potential reason may be the small effect sizes. Figure S8 shows the individual bootstrap confidence intervals for each SNP. In our applications in this article, we simply rank annotations based on the size of the effects (corresponding to the standardized annotations). Better ways to rank the annotations and distinguish between null and nonnull annotations will need further investigation. For example, a fully Bayesian approach might provide a valid interval (credible interval).
Computing time
Table 2 shows the computing time of each method on a simulated data set with 34 loci, each with 35 SNPs, and 100 annotations. The central processing unit is Intel(R) Xeon(R) E5-4640 2.40 GHz. For CAVIARBF with penalized models, 10-fold CV was used. The parameter search space was the same as in the simulation. Because CAVIARBF is now provided in an R package using Rcpp to interface with the C++ code, it may appear slower when the R code takes a large portion of the time. This might explain that CAVIARBF_top10t is slower than PAINTOR_top10t for one maximal causal variant but faster for three maximal causal variants. When a penalized model is applied, CAVIARBF takes much longer than fgwas, but is still practical for fine mapping. For example, assuming three maximal causal variants in each locus, CAVIARBF_ENET_CV takes ∼13 hr to finish. Application of the sampling-based methods instead of exhaustive enumeration of causal configurations can further reduce the computing time (Benner et al. 2016).
Lipid data fine mapping
The lipid meta-analysis summary statistics have been used in previous studies for either improving the power of GWAS discovery (Pickrell 2014) or fine mapping (Kichaev et al. 2014). Therefore it is natural to test our proposed method on this data set and compare it with other methods (see Methods for more details). After some initial investigation of the marginal Z-test statistics from meta-analysis and the correlation matrix from EUR population in the 1000 Genomes Project, we found that there are inconsistencies, see more in Discussion. To alleviate spurious findings due to mismatched Z statistics and the LD matrix, we set the maximal number of causal variants to 1, therefore not influenced by the potential mismatches. The sample sizes for each variant are relatively stable, ranging from 80,000 to ∼100,000. Therefore we simply set the sample size parameter to 80,000 for CAVIARBF; setting to 100,000 showed similar results. Table 3 shows the top 10 ranked annotations by different methods for HDL trait and ranked by the absolute value of the standardized coefficients from CAVIARBF_ENET_CV. The top-ranked annotations are generally different among different methods. For example, Repressed (k562), a repressed region in the k562 cell line, is ranked as the second top annotation by CAVIARBF_ENET_CV and included by CAVIARBF_top10t, but not by other methods. DHSs in the fMuscle (arm) cell line is top ranked by all methods considered. Among the top 10 ranked annotations by CAVIARBF_ENET_CV, Repressed (k562), Repressed (hepg2), Transcribed (k562), Repressed (hela), DHSs (fetal large intestine), and DHSs (fetal muscle), are also identified as top annotations in Pickrell’s results (Pickrell 2014).The effect sizes from CAVIARBF_ENET_CV are much smaller than that from fitting each annotation individually. This is because the effect size from CAVIARBF_ENET_CV is the additional effect after accounting for other correlated annotations. The penalization used in the model also shrinks the effect size. Due to the correlation among annotations, top annotations from the top10t strategy are in general not the same as the top 10 annotations by individual P-values. For CAVIARBF and PAINTOR, top annotations and annotations from the top10t strategy are similar, showing three common annotations with similar effect size. We note that the top annotations in Table 3 are not all the same as those from Figure 1 in Pickrell (2014) or as those in Table 6 from Kichaev et al. (2014). This may be due to several differences among the analyses: First, the methods used are different, even though they share some similarities. Second, the final data sets used for fine mapping are different. In Pickrell (2014), all regions were used to fit the model no matter whether there were significant signals. Even though both our and the analyses of Kichaev et al. (2014) were restricted to regions showing significant signals, the regions selected were different; 41 regions in our analysis vs. 37 in Kichaev et al. (2014), and the maximal number of causal variants assumed may also differ. The individual annotation results for LDL, TC, and TG can be found in File S1.
Table 3. Top-ranked annotations for HDL trait.
| Cell line | Type | CAVIARBF_ENET_CV | CAVIARBF_Individual_Annotation | CAVIARBF_top10t | PAINTOR_Individual_Annotation | PAINTOR_top10t | |||
|---|---|---|---|---|---|---|---|---|---|
| γ | standardized γ | P-value | γ | γ | P-value | γ | γ | ||
| — | distance to TSS | −0.053 | −0.053 | 0.290 | −0.210 | 0.000 | 0.186 | 0.313 | 0.000 |
| K562 | repressed | −0.060 | −0.030 | 0.041 | −0.755 | −0.369 | 0.096 | 0.650 | 0.000 |
| hepg2 | repressed | −0.045 | −0.023 | 0.062 | −0.715 | 0.000 | 0.121 | 0.641 | 0.000 |
| LCL | repressed | −0.038 | −0.019 | 0.138 | −0.512 | 0.000 | 0.243 | 0.455 | 0.000 |
| k562 | transcribed | 0.037 | 0.016 | 0.116 | 0.634 | 0.000 | 0.140 | −0.672 | 0.000 |
| hela | repressed | −0.029 | −0.014 | 0.220 | −0.477 | 0.000 | 0.316 | 0.444 | 0.000 |
| H1hesc | repressed | −0.029 | −0.014 | 0.185 | −0.507 | 0.000 | 0.315 | 0.424 | 0.000 |
| fIntestine (Lg) | DHS | 0.051 | 0.013 | 0.005 | 1.721 | 0.000 | 0.012 | −1.760 | 0.000 |
| fMuscle (trunk) | DHS | 0.038 | 0.010 | 0.014 | 1.641 | 0.000 | 0.032 | −1.743 | 0.000 |
| fMuscle (arm) | DHS | 0.039 | 0.010 | 0.010 | 1.738 | 1.401 | 0.019 | −1.877 | −1.064 |
| Conjunctival fibroblasts | DHS | 0.043 | 0.008 | 0.005 | 1.945 | 0.000 | 0.019 | −1.940 | 0.000 |
| fThymus | DHS | 0.034 | 0.007 | 0.016 | 1.822 | 0.000 | 0.042 | −1.886 | 0.000 |
| Villous mesenchymal fibroblast | DHS | 0.031 | 0.007 | 0.031 | 1.468 | 0.000 | 0.035 | −1.601 | 0.000 |
| hepg2 | TSS | 0.033 | 0.005 | 0.004 | 2.145 | 1.041 | 0.005 | −2.249 | −1.341 |
| — | coding exons | 0.028 | 0.004 | 0.008 | 2.190 | 1.552 | 0.017 | −2.238 | 0.000 |
| — | 3′ UTR exons | 0.025 | 0.003 | 0.022 | 2.211 | 2.007 | 0.010 | −2.482 | −2.316 |
| — | nonsynonymous | 0.024 | 0.002 | 0.009 | 1.923 | 0.000 | 0.003 | −3.038 | −2.394 |
Rows in boldface font indicate the top 10 annotations by different methods. To be consistent with the annotation parameterization as in CAVIARBF, the sign of effect sizes from PAINTOR need to be flipped.
Table 6. Top-ranked variants with PIPs of different methods for the eQTL study.
| Chr | Target gene | rsid | Position | Dosage allelea | z | CAVIARBF_ENET_CV | CAVIARBF_non | CAVIARBF_top10t |
|---|---|---|---|---|---|---|---|---|
| 5 | IRX4 | rs12655062 | 1890877 | A | −26.758 | 1.0000 | 1.0000 | 1.0000 |
| 10 | SFXN2 | rs2902548 | 104487382 | T | −16.385 | 1.0000 | 1.0000 | 1.0000 |
| 10 | NCOA4 | rs10993994 | 51549496 | T | 22.665 | 1.0000 | 1.0000 | 1.0000 |
| 17 | HNF1B | rs3760511 | 36106313 | G | 6.623 | 1.0000 | 0.0343 | 1.0000 |
| 22 | TBX1 | rs1978060 | 19749525 | A | 21.090 | 1.0000 | 1.0000 | 1.0000 |
| 2 | SEPT2 | rs115271170 | 242347614 | T | 29.950 | 1.0000 | 1.0000 | 1.0000 |
| 2 | RAB17 | rs13404216 | 238491258 | A | −11.072 | 1.0000 | 0.9999 | 1.0000 |
| X | GJB1 | rs747181 | 70432708 | A | −12.501 | 0.9999 | 0.9999 | 0.9999 |
| 5 | IRX4 | rs6890484 | 1890577 | G | 6.966 | 0.9989 | 0.9960 | 0.9960 |
| 6 | HLA-L | rs114620415 | 31239821 | G | −20.153 | 0.9986 | 0.9794 | 0.9971 |
| 6 | HLA-L | rs116373865 | 31239296 | G | −23.825 | 0.9983 | 0.9782 | 0.9967 |
| 6 | HLA-L | rs114172808 | 31303275 | G | 12.760 | 0.9980 | 0.9968 | 0.9972 |
| 7 | HOTTIP | rs10486567 | 27976563 | A | 6.358 | 0.9967 | 0.1844 | 0.9975 |
| 2 | C2orf43 | rs72784331 | 20877436 | A | 7.622 | 0.9951 | 0.8203 | 0.9963 |
| 10 | SFXN2 | rs3818708 | 104503584 | T | −14.792 | 0.9899 | 0.1126 | 0.9985 |
| 17 | HNF1B | rs4430796 | 36098040 | G | −7.334 | 0.9881 | 0.0835 | 1.0000 |
| 21 | TMPRSS2 | rs8134378 | 42893758 | A | −14.832 | 0.9858 | 0.3305 | 0.9578 |
| 21 | TMPRSS2 | rs56095453 | 42893808 | A | 7.872 | 0.9841 | 0.3941 | 0.9756 |
| 17 | FAM57A | rs2644715 | 632686 | A | 11.924 | 0.9800 | 0.9542 | 0.9552 |
| 17 | FAM57A | rs2474694 | 618039 | A | −19.853 | 0.9643 | 0.7060 | 0.7953 |
| 4 | BMPR1B | rs6853490 | 95544718 | G | −10.289 | 0.9602 | 0.0319 | 0.9862 |
| 5 | COMMD10 | rs804152 | 115670932 | C | 10.097 | 0.9577 | 0.9558 | 0.9586 |
| 1 | LOC284581 | rs9438393 | 205782718 | G | 52.062 | 0.9500 | 0.6991 | 0.9143 |
| 2 | NOL10 | rs7567304 | 10790029 | T | 12.020 | 0.9331 | 0.9001 | 0.9329 |
| 14 | SYNJ2BP | rs1044527 | 70833819 | G | 13.428 | 0.9044 | 0.5466 | 0.6407 |
| 10 | SFXN2 | rs2297451 | 104503459 | A | 13.710 | 0.8727 | 0.0393 | 0.9808 |
| No. of SNPs (PIP >0.9) | 25 | 14 | 24 |
Variants have at least one PIP >0.9 from different methods. Rows in boldface font indicate at least one method outputs PIP <0.9. The variants are sorted by PIPs from CAVIARBF_ENET_CV in descending order. Chr, chromosome.
Dosage value is the count of this allele.
Next we compared the top ranked variants by different methods (Table 4). CAVIARBF with and without annotations (CAVIARBF_ENET_CV vs. CAVIARBF_non) do not show much difference, even though for rs2923084, the PIP is ∼0.95 from CAVIARBF_ENET_CV and 0.86 from CAVIARBF_non. CAVIARBF_ENET_CV has similar PIPs as CAVIARBF_top10t for most SNPs; but for rs1800961, the PIP is ∼0.06 from CAVIARBF_ENET_CV but ∼0.58 from CAVIARBF_top10t. This is likely due to overoptimism of the top10t strategy. To illustrate this, we analyzed the simulated data sets with moderate annotation effects and 40 initial loci, setting the maximal number of causal variants to 1. We calculated the percentage of causal variants where both PIPs from CAVIARBF_non and CAVIARBF_ENET_CV fell within 0–0.1 and PIPs from CAVIARBF_top10t fell within 0.5–0.7. The percentage is 0.23 (17 out of 74). When adding the condition that PAINTOR_top10t fell within 0.8–1 and PAINTOR_non fell within 0–0.2, the percentage is 0.15 (7 out of 47). This illustrates the overoptimism pertaining to the top10t strategy. On the other hand, for SNPs where both PIPs from CAVIARBF_non and CAVIARBF_top10t fell within 0–0.1 and PIPs from CAVIARBF_ENET_CV fell within 0.5–0.7, the percentage of causal variants is 0.57 (29 out of 51), calibrated well with the PIP range. Comparing CAVIARBF and PAINTOR, for some SNPs there are large PIP differences even though in general PIPs are similar. For example, for rs2923084, the PIP from CAVIARBF_non is ∼0.86, but it is ∼0.20 from PAINTOR_non. Since the causal state needs confirmation from laboratory experiments, we cannot conclude which method is more accurate on this SNP, but being aware of the differences may be helpful for future application and comparison. Results for LDL, TC, and TG can be found in Table S3.
Table 4. Top-ranked variants with PIPs of different methods for HDL trait.
| Chr | rsid | Position | Dosage allelea | Imputation qualityb | z | CAVIARBF_ENET_CV | CAVIARBF_non | CAVIARBF_top10t | PAINTOR_top10t | PAINTOR_non |
|---|---|---|---|---|---|---|---|---|---|---|
| 11 | rs964184 | 116648917 | C | 0.986 | −13.858 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 16 | rs2925979 | 81534790 | C | 1 | −6.700 | 0.9979 | 0.9838 | 0.9959 | 0.9018 | 0.7769 |
| 19 | rs4420638 | 45422946 | G | 1 | 9.423 | 0.9955 | 0.9952 | 0.9895 | 0.9237 | 0.9710 |
| 11 | rs2923084 | 10388782 | G | 1 | 5.465 | 0.9458 | 0.8566 | 0.9673 | 0.3826 | 0.2019 |
| 4 | rs13107325 | 103188709 | T | 1 | 6.516 | 0.9196 | 0.9077 | 0.9853 | 0.9377 | 0.5851 |
| 8 | rs9987289 | 9183358 | G | 1 | −10.309 | 0.9022 | 0.9013 | 0.9013 | 0.5744 | 0.5744 |
| 20 | rs1800961 | 43042364 | T | 0.670 | 3.246 | 0.0661 | 0.0700 | 0.5837 | 0.9080 | 0.1207 |
Variants have at least one PIP >0.9 from different methods. Rows in boldface font indicate at least one method outputs PIP <0.9. The variants are sorted by PIPs from CAVIARBF_ENET_CV in descending order. Chr, chromosome.
Dosage value is the count of this allele.
This is the r2pred output from ImpG indicating the quality of imputed z statistics, 1 indicates the original z statistics from meta-analysis.
cis-eQTL fine mapping
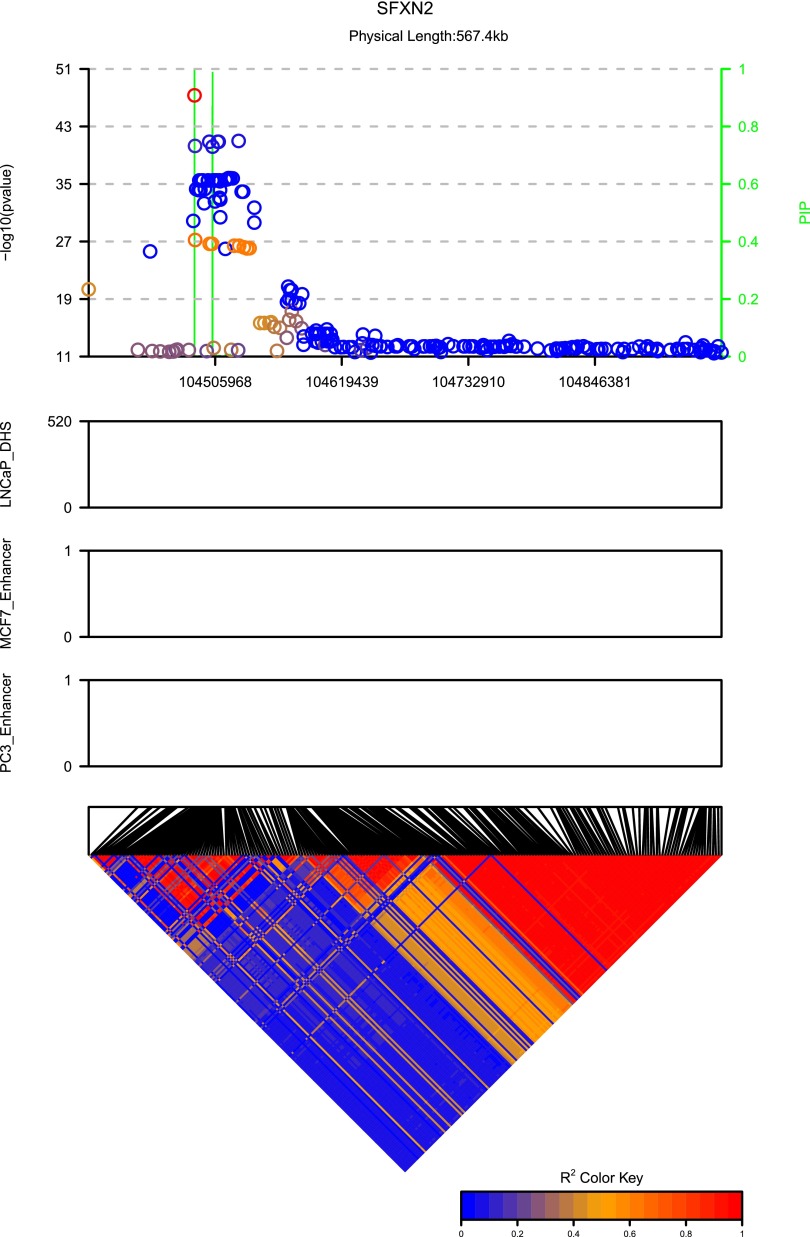
The objective of this study was to identify cis-eQTLs for genes related to PrCa risk SNPs (Thibodeau et al. 2015) using genotypes and RNA-seq data from normal prostate tissue samples from 471 individuals. Figure 10 shows the advantage of using PIPs instead of P-values in identifying cis-eQTLs on gene SFXN2. It is difficult to identify the potential causal variants from the P-values due to the large number of SNPs in LD. The green lines show the PIPs by applying our proposed method by taking both the LD information and the annotation information into account. It is clear that there are at least two highly likely causal variants influencing the gene expression. Actually there are three potential causal variants, two of which are very close in position.
Figure 10.
P-values, PIPs, annotations, and the LD for cis-eQTL analysis of SFXN2. The green lines are PIPs. The LD between the peak SNP and the remaining SNPs are color coded in the top panel. The three middle panels illustrated the annotations for individual SNPs. The bottom diagonal matrix shows the LD pattern among all SNPs in this locus.
We set the maximal number of causal variants to 3 because many loci show multiple eQTL signals. This can be illustrated using the sum of PIPs for each loci, an estimation of expected causal variants. Figure S9 shows the histogram of added final PIPs (using CAVIARBF_ENET_CV) for each target gene, which supports the multiple eQTL signal assumption for many loci.
We first compare estimated PIPs using approximate and exact calculations on data sets showing different proportions of variance explained in gene expression traits. We selected two target genes, one with a relatively small PVE by individual variants and the other with a large PVE. The maximal number of causal variants was set to 3. We then ran different fine-mapping options/methods with results displayed in Figure S10. When variants explain a small proportion of the variance (left panel), methods using the approximation, such as PAINTOR and CAVIARBF using the approximate calculation, have similar PIPs as CAVIARBF using the exact calculation. In this scenario, PIPs are also not sensitive to the parameter of . However, when variants explain a large proportion of the variance (right panel), PAINTOR and CAVIARBF using the approximation have similar PIPs, but their results are different from CAVIARBF using the exact calculation. PIPs are also sensitive to the parameter of . Therefore for all eQTL fine mapping, we did not run PAINTOR and used CAVIARBF with the exact calculation and chose a set of values for (0.1, 0.2, 0.4, 0.8, 1.6) to calculate the average Bayes factors, similar to those used in Wen et al. (2015).
The distance to TSS has been shown to associate with the enrichment of cis-eQTLs (Gaffney et al. 2012; Wen et al. 2015). As a validation, we tested the significance of this annotation in our eQTL data. Surprisingly, the P-value for the distance to the TSS is 0.3215, which appears inconsistent with previous findings. Plotting the final fitted PIPs vs. the distance to the TSS (Figure S11) and further examination provide an explanation. There is an obvious decreasing pattern for PIPs when the distance to the TSS is <200 kb, however, PIPs are almost flat further away. In other words, the size of the region selected around the TSS can influence the significance test. We verified this by using the same fitted PIPs but picking SNPs within different distances from the TSS. The P-values from linear regression testing are 0.311 for the threshold of 1.1 Mb, 0.0624 for 200 kb, 0.000304 for 100 kb and 0.0091 for 50 kb. This is consistent with previous significant results using 100 kb to the TSS as the boundary of selected regions (Gaffney et al. 2012; Wen et al. 2015).
Table 5 shows the results of top-ranked annotations by different methods and sorted by the magnitude of the estimated effect size by CAVIARBF_ENET_CV. Androgen receptor (AR) binding sites and DHSs in LNCaP cells are the top two ranked annotations by CAVIARBF_ENET_CV; where DHSs in LNCaP cells, the second top annotation, has a P-value of 3.43 × 10−09, the smallest P-value among all annotations. These two annotations are also selected by CAVIARBF_top10t. Most annotations selected by CAVIARBF_top10t have a large standardized effect size γ in CAVIARBF_ENET_CV, even though in general the annotations selected by these two methods are not the same. Other top-ranked annotations from CAVIARBF_ENET_CV include the enhancer region, transcription factor binding sites, the heterochromatin state, the repressed and promoter region, and histone modification annotations. Similarly as in the lipid data, due to the correlation among annotations, annotations from the top10t strategy are in general not the same as the top 10 annotations by individual P-values. The effect sizes from CAVIARBF_ENET_CV are also smaller than that from fitting each annotation individually. In general, the sign of the effect sizes matched well with the function of annotations for both CAVIARBF_ENET_CV or CAVIARBF using individual annotations. For example, the heterochromatin state annotation has a negative value, therefore decreasing the probability of being eQTLs. This is intuitive because under the heterochromatin state, DNA sequences are tightly packed and hard to access. On the other hand, DHSs, except one from the PrEC cell line which is very small in magnitude (−0.0073), and H3K4me3 all have positive values, consistent with their roles of DHSs and promoting gene regulation.
Table 5. Top-ranked annotations for the eQTL study.
| Data ID | Cell line | Type | CAVIARBF _ENET_CV | CAVIARBF _Individual_Annotation | CAVIARBF_top10t | ||
|---|---|---|---|---|---|---|---|
| γ | Standardized γ | P-value | γ | γ | |||
| GSM698597 | LNCaP | AR | 0.0760 | 0.5885 | 2.16 × 10−03 | 0.4260 | 0.2595 |
| GSM736565 | LNCaP | DHS | 0.1193 | 0.1529 | 2.30 × 10−09 | 0.2565 | 0.2272 |
| GSM686926 | LNCaP | FoxA1 | −0.0667 | −0.1112 | 5.76 × 10−01 | −0.1945 | 0.0000 |
| GSE57498 | MCF7 | ChromHMM.Enhancer | −0.0847 | −0.1085 | 5.47 × 10−02 | −0.5081 | 0.0000 |
| GSE57498 | PC3 | ChromHMM.Enhancer | 0.0836 | 0.1005 | 4.33 × 10−02 | 0.1996 | 0.0000 |
| GSM989640 | LNCaP | NKX3-1 | 0.1077 | 0.0963 | 2.97 × 10−05 | 0.2423 | 0.0000 |
| GSM1410771 | VCaP | RUNX1 | 0.0808 | 0.0955 | 3.37 × 10−04 | 0.2369 | 0.0000 |
| GSE57498 | PC3 | ChromHMM.Heterochromatin | −0.0820 | −0.0941 | 3.45 × 10−03 | −0.3140 | −0.2469 |
| GSM816637 | LNCaP | DHS | 0.0688 | 0.0930 | 1.26 × 10−03 | 0.1303 | 0.0000 |
| GSM736603 | LNCaP | DHS | 0.0691 | 0.0849 | 2.99 × 10−08 | 0.2658 | 0.0000 |
| GSE57498 | HMEC | ChromHMM.Repressed | 0.0591 | 0.0657 | 3.65 × 10−02 | 0.1652 | 0.1612 |
| GSM686935 | LNCaP | H3K4me3 | 0.0620 | 0.0641 | 1.28 × 10−03 | 0.2240 | 0.1453 |
| GSE57498 | MCF7 | ChromHMM.Transcribed | 0.0532 | 0.0635 | 2.51 × 10−02 | 0.2184 | 0.1576 |
| GSE57498 | MCF7 | ChromHMM.CTCF | 0.0364 | 0.0363 | 2.74 × 10−02 | 0.2229 | 0.1850 |
| GSM816634 | LNCaP | DHS | 0.0221 | 0.0294 | 1.57 × 10−04 | 0.1662 | 0.0000 |
| GSM699633 | LNCaP | NKX3-1 | 0.0204 | 0.0236 | 7.31 × 10−04 | 0.1252 | 0.0000 |
| GSM1024742 | PrEC | DHS | −0.0073 | −0.0069 | 1.12 × 10−03 | 0.2143 | 0.0000 |
| GSE57498 | PrEC | ChromHMM.Promoter | −0.0008 | −0.0009 | 1.76 × 10−03 | 0.2307 | 0.0000 |
Rows in boldface font indicate the top 10 annotations ranked by different methods. CAVIARBF_ENET_CV ranks annotations based on the absolute value of coefficients, CAVIARBF_Individual_Annotation ranks annotations based on individual P-values. LNCaP, a prostate adenocarcinoma cell line; PC3, a prostate cancer cell line; HMEC, a normal mammary epithelial cell line; MCF7, a breast cancer cell line; PrEC, a normal prostate epithelial cell line.
Finally, we compare the top ranked SNPs and PIPs using different methods in Table 6. Clearly, incorporating annotations increased the number of potential eQTLs with high PIPs. There are 25 SNPs with PIP >0.9 using CAVIARBF_ENET_CV, compared to 14 SNPs with PIP >0.9 using CAVIARBF without annotations (CAVIARBF_non). For most SNPs listed in Table 6, CAVIARBF_ENET_CV and CAVIARBF_top10t have similar PIPs. However, there is still a relatively large PIP difference for SNP rs1044527, with 0.9044 from CAVIARBF_ENET_CV and 0.6407 from CAVIARBF_top10t. Given the better performance of CAVIARBF_ENET_CV over the top10t strategy based on simulation studies, the results from CAVIARBF_ENET_CV may be more reliable. We further examined several SNPs with large increase of PIPs when incorporating functional annotations. The major annotation components are presented in Table 7. Several SNPs only rely on one major annotation component to increase the PIPs, e.g., rs3760511 within an androgen response element (Clinckemalie et al. 2013). Other SNPs combine multiple annotations together to improve the PIPs. Among these SNPs, rs10486567 is reported as a risk enhancer affecting both NKX3-1 and FOXA-AR motifs (Hazelett et al. 2014), SNP rs8134378 was reported within an androgen response element and reduces binding and transactivation by the androgen receptor (Clinckemalie et al. 2013).
Table 7. Major annotation components that largely improve the PIPs of SNPs in the eQTL study.
| rsid | Annotations |
|---|---|
| rs3760511 | AR binding in LNCaP |
| rs10486567 | DHS, NKX3-1 binding in LNCaP |
| rs3818708 | DHS in LNCaP |
| rs4430796 | AR binding peak in LNCaP |
| rs8134378 | DHS, NKX3-1, AR binding in LNCaP, RUNX1 binding in VCaP |
| rs56095453 | DHS, NKX3-1in LNCaP, RUNX1 binding in VCaP |
| rs6853490 | DHS, NKX3-1in LNCaP |
| rs1044527 | RUNX1 binding in VCaP, NKX3-1, AR binding in LNCaP |
| rs2297451 | DHS in LNCaP |
| rs2474694 | DHS, H3K4me3in LNCaP, RUNX1 binding in VCaP |
| rs9438393 | H3K4me3, DHS in LNCaP, ChromHMM.Heterochromatin in PC3 and HMEC, ChromHMM.Promoter in PC3, RUNX1 binding in VCaP |
Discussion
In this study, we proposed a framework to systematically incorporate a large number of annotations in association studies. By using CAVIARBF to calculate the PIPs, the framework only requires summary statistics. For quantitative traits, when the explained variance of the traits is high, e.g., for some eQTLs, calculating Bayes factors from the summary statistics using approximation may produce misleading results. Instead we derived an exact calculation. Further investigation for binary traits with high explained variances may be useful.
Our method shares similarities with some existing methods, but also differs significantly from them. The most significant difference is that our method is a general framework to systematically incorporate functional annotations, and it uses the ENET in the penalized model with CV for parameter selection. In principle, any method that can output PIPs which take into account the prior probability of each variant being causal can be used within this framework. Compared to fgwas (Pickrell 2014), our method results in a more robust way to automatically select sparse annotations, it also allows multiple causal variants in each locus where fgwas assumes no more than one. The performance of fgwas is similar to that of CAVIARBF_L1_CV, except that CAVIARBF_L1_CV does not show decreased performance under the null annotations. There are also other differences: fgwas only supports binary annotations or a distance model using integers, while CAVIARBF can use any numeric annotations (binary or continuous). fgwas uses a penalized likelihood in CV for the testing fold while CAVIARBF uses the likelihood directly. Compared to the top5t or top10t strategy recommended by PAINTOR (Kichaev et al. 2014), the proposed method does not have the dilemma of choosing the significance threshold: if the P-value threshold is too stringent, we may lose informative annotations; if it is too liberal, we may include too many noninformative annotations. Fitting the model with top-ranked annotations without penalization may also result in overfitting (Pickrell 2014). FM-QTL (Wen et al. 2015) uses the full genotype data, assumes a small number of annotations and uses the same maximum likelihood-based method to incorporate annotations as in PAINTOR. iBRI (Quintana and Conti 2013) uses an L2 penalty with a fixed parameter, which may not be optimal. For PIP calculation, both FM-QTL and iBRI use an MCMC-based approach, which may be faster when there are a relatively larger number of causal variants in a large region. Therefore, an alternative is to use MCMC to perform genome-wide association analysis and fine mapping simultaneously, such as in Zhu and Stephens (2016). There are also other sampling-based methods showing highly reduced computing cost (Benner et al. 2016).
The proposed model assumes independence among different loci. Some eQTLs may regulate more than one gene, thus are shared for more than one locus. This may create dependencies among loci but the extent and how much it will influence the final results are not clear. More investigation is needed to accommodate the overlapping scenario.
When the number of loci is small, e.g., ∼20 loci, the estimated PIP may be overly optimistic. One reason may be that we use a point estimation (MAP) from glmnet instead of a full Bayesian method. A full Bayesian method for the proposed graphical model may show better calibration. Another possible cause is that CV among loci may not be very effective if the number of loci is small. This may result in overfitting with the grid search to tune parameters. If full genotype data are available, CV by partitioning individuals instead of partitioning loci would be a better solution, and the assumption of independence between individuals is much easier to satisfy than the assumption of independence between loci. Otherwise, better methods may be needed to tune the parameters. In the extreme case, when the number of loci is very small, e.g., <5, the benefit from annotations is not likely to be obvious. In this case we can perform fine mapping without using annotations.
When ranking annotations based on the MAP point estimate of γ, there is no direct quantification of the uncertainty of this estimate. To measure the estimation variance, we can use the bootstrap method by resampling the loci as previously proposed (Kichaev et al. 2014). Because we use CV to choose the best penalty parameters, different partitioning of the loci may select different penalty parameters resulting in different MAP estimates. In this case, we may need to run the CV several times and pick the penalty parameters with the highest frequency, i.e., the mode of all selected penalty parameters. We ran CV five times for real data application in this paper, except the HDL data, where we ran CV 20 times to pick the mode because several parameters show similar frequencies and the mode is not very sharp.
The proposed framework can also be used to incorporate pathway information. One simple method is to code each pathway as an annotation, where 1 indicates the variant is in the pathway and 0 otherwise. How much the pathway information can improve the performance needs further study.
In our lipid data analysis, we found inconsistencies between the available Z statistics and the LD from the EUR population in 1000 Genomes Project. For example, rs2144300 and rs4846914 are in perfect LD, correlation coefficient is 1, in EUR. However, the meta-analysis z-scores are 9.16 and 9.442. Without using annotation and assuming one maximal causal variant, this difference results in rs4846914 with PIP 0.61, much higher than rs2144300 with PIP 0.05, even though it makes more sense that these two should have the same or at least very similar PIPs. If the difference of z-scores is not due to computing error, there are two potential sources which may also be common for other meta-analysis results. First, there may be other populations that show different LD patterns from the EUR population. To address this problem, previous studies (Kichaev and Pasaniuc 2015; Wen et al. 2015) have developed different ways to handle multiple populations. Our framework can also be extended similarly, which is an interesting future direction. Second, and more likely to be the source, imputed genotypes or meta-analyzed z-scores were not computed for the same set of individuals, given that usually an r2 threshold is used for imputed genotypes, e.g., 0.3 for the meta-analysis results (Teslovich et al. 2010). This is discussed in detail in Zhu and Stephens (2016). Therefore, a better way is to report summary statistics using all imputed genotypes without applying a threshold. In our method, we assume the sample sizes are the same or at least similar. Strictly speaking, it is not fair to compare two SNPs if their sample size is different because the Bayes factors depend on the sample size. However, if the sample size difference is small, it might not influence the comparison much.
Acknowledgments
This research was supported by the United States Public Health Service; National Institutes of Health, contract grant number GM-065450 and CA-151254; and by the Department of Defense (W81XWH-11-1-0261). Informed consent was obtained from all subjects and the study was approved by the Mayo Clinic Institutional Review Board.
Appendix
CAVIARBF Reduces to the ABF When There Is at Most One Causal Variant
We derive the equivalence using the ABF Equation 3 in Chen et al. (2015). When , Equation 3 reduces to
Therefore,
The corresponding ABF−1 (H1 vs. H0, from Equation 2 in Wakefield 2009) is
Therefore, CAVIARBF in the case and ABF have the same form. If , then CAVIARBF and ABF are exactly the same. This is the case in the application of ABF in fgwas (Pickrell 2014) when V is not available from data. For quantitative traits, it is estimated as , where f is the allele frequency (this is half of the value in Equation 14 from Pickrell 2014, and is used in fgwas code). We have . The Bayes factor in CAVIARBF and fgwas are exactly the same if . This can be done in CAVIARBF by setting and setting the weights to the variance of the SNP using . For binary traits, , where n is the total sample size, is the number of cases, and is the number of controls. Therefore . The Bayes factor in CAVIARBF and fgwas are exactly the same if . This can be done in CAVIARBF by setting to and setting the weights to the variance of the SNP using . From the above derivation, we can also see that fgwas assumes the prior normal distribution of the variant effect size on the original scale as in BIMBAM instead of the standardized scale (Chen and Schaid 2014). The only difference between CAVIARBF and ABF is that in our previous derivation we assumed z was a score statistic but Wakefield assumed a Wald statistic, however, these two statistics are asymptotically equivalent under the null or small deviation from the null, thus very similar due to the large sample size and small effects in GWAS.
Exact Bayes Factors Using Marginal Statistics for Quantitative Traits
In this section we derive the exact Bayes factors using a D2 prior (Servin and Stephens 2007), which only depends on the marginal statistics and the correlation matrix. Suppose is an n×m matrix with coded genotypes, where n is the number of individuals and m is the number of coded columns from putative causal variants. Without loss of generality, we assume is centered, i.e., , where is the element of row i and column j in . Let be the sample variance of the jth column, i.e., . The quantitative phenotype y is an n×1 vector. We assume a linear model as follows:
| (1) |
where β is the effect size and . Here denotes the n×n identity matrix. The D2 prior assumes that β has a prior normal distribution N(0, , where is a diagonal matrix and β and are independent. Let , where empty blocks are simply 0s. Then the Bayes factor using a D2 prior (Servin and Stephens 2007) can be written as:
| (2) |
where , i.e., the augmented matrix with all 1s in the first column, is the mean value of y. The superscript T means the transpose of the corresponding matrix or vector. Equation 2 can be further simplified to
Let S be the diagonal matrix with diagonal entries . Then the correlation matrix of is , therefore . Then we have
and
Let be the marginal test statistics corresponding to the jth column of , denoted by , then , where . Because , we have
Then can be written as
Therefore we have
where we have used the fact that has the same sign as . Let be the column vector where the jth component is , the correlation coefficient between and , then we have:
Therefore the Bayes factor can be written using only the marginal test statistics and correlation matrix as follows:
Footnotes
Communicating editor: N. Yi
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.188953/-/DC1.
Literature Cited
- Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., et al. , 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner C., Spencer C. C., Havulinna A. S., Salomaa V., Ripatti S., et al. , 2016. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32: 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert S. A., Robinson M. D., Strbenac D., Statham A. L., Song J. Z., et al. , 2013. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell 23: 9–22. [DOI] [PubMed] [Google Scholar]
- Bishop C. M., 2006. Pattern Recognition and Machine Learning. Springer, New York. [Google Scholar]
- Carbonetto P., Stephens M., 2013. Integrated enrichment analysis of variants and pathways in genome-wide association studies indicates central role for IL-2 signaling genes in type 1 diabetes, and cytokine signaling genes in Crohn’s disease. PLoS Genet. 9: e1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Schaid D. J., 2014. PedBLIMP: extending linear predictors to impute genotypes in pedigrees. Genet. Epidemiol. 38: 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Larrabee B. R., Ovsyannikova I. G., Kennedy R. B., Haralambieva I. H., et al. , 2015. Fine mapping causal variants with an approximate bayesian method using marginal test statistics. Genetics 200: 719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinckemalie L., Spans L., Dubois V., Laurent M., Helsen C., et al. , 2013. Androgen regulation of the TMPRSS2 gene and the effect of a SNP in an androgen response element. Mol. Endocrinol. 27: 2028–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kellis M., 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kellis M., 2015. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol. 33: 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J., Hastie T., Tibshirani R., 2010. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33: 1–22. [PMC free article] [PubMed] [Google Scholar]
- Gaffney D. J., Veyrieras J. B., Degner J. F., Pique-Regi R., Pai A. A., et al. , 2012. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 13: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Stephens M., 2008. Practical issues in imputation-based association mapping. PLoS Genet. 4: e1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y. T., Stephens M., 2011. Bayesian variable selection regression for genome-wide association studies and other large-scale problems. Ann. Appl. Stat. 5: 1780–1815. [Google Scholar]
- Gusev A., Lee S. H., Trynka G., Finucane H., Vilhjalmsson B. J., et al. , 2014. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 95: 535–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelett D. J., Rhie S. K., Gaddis M., Yan C., Lakeland D. L., et al. , 2014. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 10: e1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H. H., Meyer C. A., Shin H., Bailey S. T., Wei G., et al. , 2010. Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 42: 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M. M., Ernst J., Wilder S. P., Kundaje A., Harris R. S., et al. , 2013. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 41: 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F., Kostem E., Kang E. Y., Pasaniuc B., Eskin E., 2014. Identifying causal variants at loci with multiple signals of association. Genetics 198: 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G., Pasaniuc B., 2015. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 97: 260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G., Yang W. Y., Lindstrom S., Hormozdiari F., Eskin E., et al. , 2014. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 10: e1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyung M., Gill J., Ghosh M., Casella G., 2010. Penalized regression, standard errors, and Bayesian lassos. Bayesian Anal. 5: 369–411. [Google Scholar]
- Maurano M. T., Humbert R., Rynes E., Thurman R. E., Haugen E., et al. , 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. Y., Hastie T., 2008. Penalized logistic regression for detecting gene interactions. Biostatistics 9: 30–50. [DOI] [PubMed] [Google Scholar]
- Pasaniuc B., Zaitlen N., Shi H., Bhatia G., Gusev A., et al. , 2014. Fast and accurate imputation of summary statistics enhances evidence of functional enrichment. Bioinformatics 30: 2906–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., 2014. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 94: 559–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana M. A., Conti D. V., 2013. Integrative variable selection via Bayesian model uncertainty. Stat. Med. 32: 4938–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin B., Stephens M., 2007. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 3: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N. L., Massie C. E., Ramos-Montoya A., Zecchini V., Scott H. E., et al. , 2013. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell 23: 35–47. [DOI] [PubMed] [Google Scholar]
- Su Z., Marchini J., Donnelly P., 2011. HAPGEN2: simulation of multiple disease SNPs. Bioinformatics 27: 2304–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay P. C., Statham A. L., Kelly T. K., Clark S. J., Jones P. A., 2014. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res. 24: 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K., Suzuki T., Fujimura T., Urano T., Takahashi S., et al. , 2014. CtBP2 modulates the androgen receptor to promote prostate cancer progression. Cancer Res. 74: 6542–6553. [DOI] [PubMed] [Google Scholar]
- Tan P. Y., Chang C. W., Chng K. R., Wansa K. D., Sung W. K., et al. , 2012. Integration of regulatory networks by NKX3–1 promotes androgen-dependent prostate cancer survival. Mol. Cell. Biol. 32: 399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., et al. , 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau S. N., French A. J., McDonnell S. K., Cheville J., Middha S., et al. , 2015. Identification of candidate genes for prostate cancer-risk SNPs utilizing a normal prostate tissue eQTL data set. Nat. Commun. 6: 8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R., 1996. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. B 58: 267–288. [Google Scholar]
- Tibshirani R. J., Taylor J., 2012. Degrees of freedom in lasso problems. Ann. Stat. 40: 1198–1232. [Google Scholar]
- Wakefield J., 2009. Bayes factors for genome-wide association studies: comparison with P-values. Genet. Epidemiol. 33: 79–86. [DOI] [PubMed] [Google Scholar]
- Wang D., Garcia-Bassets I., Benner C., Li W., Su X., et al. , 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Stephens M., 2010. Using linear predictors to impute allele frequencies from summary or pooled genotype data. Ann. Appl. Stat. 4: 1158–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Luca F., Pique-Regi R., 2015. Cross-population joint analysis of eQTLs: fine mapping and functional annotation. PLoS Genet. 11: e1005176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Carbonetto P., Stephens M., 2013. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 9: e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Stephens M., 2016. Bayesian large-scale multiple regression with summary statistics from genome-wide association studies. bioRxiv, DOI: http://dx.doi.org/10.1101/042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Hastie T., 2005. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol. 67: 301–320. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The detailed annotation list and Gene Expression Omnibus track numbers are shown in File S2. The summary statistics and annotation matrix used of the eQTL data are available upon request. Software is available at https://bitbucket.org/Wenan/caviarbf.