Abstract
The 2-oxoglutarate-dependent iron enzyme ALKBH3 is an antitumor target and a potential diagnostic marker for several tumor types, including prostate cancer. However, there is at present no simple way to measure this enzyme’s activity. Here we describe a fluorogenic probe design (MAQ) that is directly responsive to ALKBH3 repair activity. It makes use of the fluorescence-quenching properties of 1-methyladenine; removal of the alkyl group results in a >10-fold light-up signal. The probe is specific for ALKBH3 over its related homologue ALKBH2 and can be used to identify and measure the effectiveness of enzyme inhibitors. Measurements of the enzyme substrate parameters show that MAQ displays Km and kcat values essentially the same as those of the native substrate. Finally, we show that the probe functions efficiently in cells, allowing imaging and quantitation of ALKBH3 activity by microscopy and flow cytometry. We expect that MAQ probes will be broadly useful in the study of the basic biology of ALKBH3 and in clinical cancer applications as well.
The AlkB family of oxidative enzymes is broadly important in biology and medicine. While bacteria such as E. coli express only one such enzyme (AlkB), humans express at least nine homologues (ALKBH1–8 and FTO).1–3 Of these, five are documented to act on nucleic acid substrates. ALKBH5 and FTO have been shown to remove the methyl groups from N6-methyladenosine in RNA.4,5 ALKBH2 and -3 remove methyl groups from 1-methyladenosine and 3-methylcytosine in DNA and/or RNA.6 The alkyl substitution in these last two examples can arise from exposure to extraneous and intrinsic methylating agents, and results in cytotoxicity, so mechanisms for repair of this damage are essential to normal cellular growth.1,7
ALKBH3, also referred to as prostate cancer antigen-1 (PCA-1), is a 37.9 kDa enzyme, adopting a β-strand jellyroll fold,8 and requiring 2-oxoglutarate (2OG) and Fe(II) for activity. It is reported to be localized both in the cell nucleus and in the cytosol.6 The enzyme removes alkyl groups from nucleic acid bases via an oxidative mechanism involving a putative hydroxymethyl intermediate, which rapidly eliminates to yield formaldehyde and the repaired nucleobase.9 ALKBH3 exhibits a strong preference for single-stranded DNA or RNA, which distinguishes it from its close homologue ALKBH2, which removes similar alkyl lesions preferentially in double-stranded DNA.6,10 The preferred substrates for ALKBH3 have been documented: 1-methyladenine (m1A, Figure 1) and 3-methylcytosine are good substrates, while low levels of activity have also been observed for 3-methylthymine and ethenoadenine.11
Figure 1.
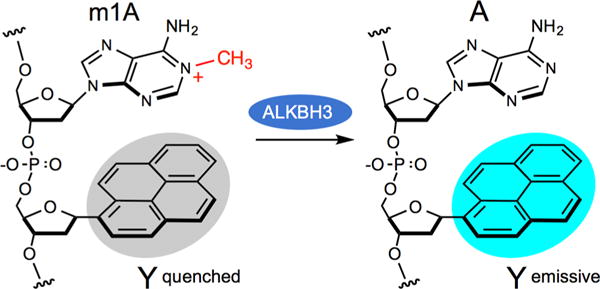
Demethylation of 1-methyladenine (m1A) by ALKBH3 in a MAQ probe that employs an adjacent fluorescent deoxynucleoside (Y) that is selectively quenched by the cationic alkylated base.
Recent studies have associated the expression of ALKBH3 with tumor growth, thus heightening the biological and clinical interest in this enzyme. Elevated levels of the enzyme were found in prostate tumor sections but not in normal epithelial tissue (as measured by immunohistochemical (IHC) staining), and expression of the protein was correlated with prostate cancer cell survival and increased invasiveness.12,13 siRNA-mediated knockdown of ALKBH3 expression induced apoptosis in the DU145 prostate cell line, and administration of ALKBH3-targeted siRNA in a murine model with prostate tumor xenografts resulted in suppression of tumor growth.14 Similar results have also been documented in pancreatic cancer, where IHC analysis of tumors from pancreatic cancer patients revealed a correlation of protein levels with advanced tumor status.15,16 Mouse models of pancreatic cancer, nonsmall-cell lung cancer, and urothelial carcinoma have all shown reduced tumor growth and/or invasiveness as a result of siRNA knockdown of ALKBH3.13–16 Such studies suggest the possibility of inhibiting ALKBH3 as a therapeutic approach for solid tumors, and a search for selective small-molecule inhibitors has recently begun.17,18
Despite the growing clinical significance of this enzyme, methods to measure ALKBH3 activity are laborious and indirect. For example, immunohistochemistry requires multiple steps involving tissue preparation and staining and washing steps, and measures protein quantities rather than enzyme activity.14 Assays of enzyme activity in vitro or in cell lysates have typically involved the use of radiolabeled substrate DNA or RNA, followed by quantitation of radioactive products.6 A recent screen of potential ALKBH3 inhibitors utilized an assay requiring a separate quantitative PCR analysis of alkylated DNA for every time point or dosage.17 To date, no fluorescence assay of this enzyme exists, nor is there a way to quantify or image its repair activity in living cells.
We describe here the design, synthesis, and applications of novel 1-methyadenine quenched (MAQ) probes that directly measure ALKBH3 activity in minutes. We show that a MAQ probe can be used conveniently to quantify protein activity and to evaluate the efficiency of inhibitors. Moreover, a nuclease-protected version of the probe functions efficiently and specifically in live tumor cell lines, imaging ALKBH3 activity in vivo.
Our design of fluorogenic probes for ALKBH3 relies on the unusual structure of 1-methyladenine. At neutral pH the alkylated adenine is an electron-deficient cationic heterocycle (Figure 1).19 Electron-poor aromatic groups are known to act as fluorescence quenching groups via photoinduced charge transfer in which the excited-state fluorophore donates an electron to the quencher.20 Thus, we considered the possibility that, when placed near an appropriate fluorescent DNA base, this damaged DNA base might serve to quench emission. Enzymatic removal of the methyl group would restore native adenine, which is generally not a quencher of fluorescence.21 As a result, fluorescence would increase, signaling enzymatic activity in real time.
To identify a fluorescent label that is strongly quenched by m1A, we synthesized several dinucleotides containing the damaged base coupled to a fluorescent DNA base. Fluorescence measurements revealed at least two fluorescent nucleosides that are quenched by at least a factor of 4 when incorporated adjacent to m1A relative to their emission alone (Table S1 in Supporting Information). Because of its strong response and tunable wavelength via monomer/excimer emission,22 we chose to proceed with pyrene nucleoside Y, and we investigated the effects of structure on activity by synthesizing 19 oligonucleotides containing m1A and Y at adjacent positions, varying length and position by incorporation of multiple deoxyadenosines (see Tables S2 and S3 for sequences and characterization data).
These candidate probes were tested as substrates for ALKBH3 since little data yet exists on length and sequence preferences of the enzyme. First we confirmed the activity of the enzyme by MALDI-MS analysis of an oligomer containing a single m1A, which showed that the enzyme restores the mass of the unmodified DNA (Figure S1). We then proceeded to test the enzyme with the candidate fluorogenic probes. The results showed that fluorescence responses were faster (signaling more rapid enzymatic reaction) when m1A is located to the 5′ side of Y and light-up responses were greater in oligomers 6 nt or longer. Enzymatic rates were slower on the shortest oligomers (Figure S2) and increased at 10–12 nt lengths, a characteristic consistent with previous findings for AlkB and ALKBH3.23 Fluorescence enhancements were as large as 10.5-fold (for the 10mer). Given this preferred length, we then replaced an adenine with a second pyrene or perylene monomer to engender long-wavelength excimer/exciplex emission22 for more convenient detection (Table S3). Based on these results, we chose the optimized 10mer oligomer MAQ (5′-dAAAA1MeAYYAAA) for further studies; it contains two Y residues for cyan emission adjacent to the quenching m1A.
We proceeded to characterize the responses of the MAQ probe in solution. In the presence of the purified ALKBH3 enzyme, the probe gave rapid and robust responses, yielding full signal in ~120 min (Figure 2A) with an 8-fold increase in fluorescence (Figure S3). Spectral studies of the fully repaired probe showed that it has a robust quantum yield of 0.35 in PBS buffer, with an absorption maximum at 345 nm (ε = 86,000 M−1 cm−1) and an emission maximum at 480 nm (Figure S3). Next we varied probe concentrations and measured reaction rates to quantify enzymatic parameters (Figure S4). The data show that the probe functions with efficiency nearly the same as that of the native substrate,24 exhibiting a KM of 400 ± 50.0 nM and a kcat of 2.4 min−1.
Figure 2.
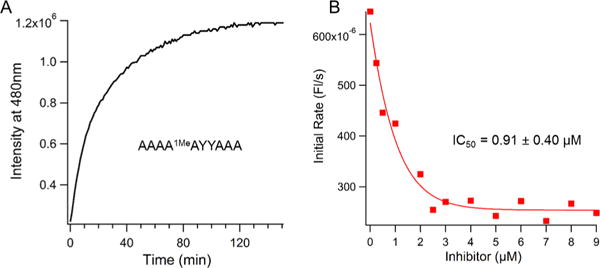
Fluorescence responses of MAQ probe to ALKBH3 and to an inhibitor. (A) Response of MAQ probe (1 μM) to ALKBH3. (B) Plot showing inhibition curve generated by MAQ probe for a recently reported inhibitor of ALKBH3.17,18 Y = α-pyrene deoxynucleoside.
Fluorescent enzyme probes are commonly useful in screening and characterizing enzyme inhibitors. In this light, we tested the probe MAQ for its ability to respond to a recently identified ALKBH3 inhibitor (HUHS015).17,18 We found that the probe did yield decreasing ALKBH3 signal with increasing inhibitor concentration; we used initial rates of reaction to independently determine the inhibition constant (Figures 2B and S5). The Ki value was found to be 0.91 ± 0.40 μM, similar to the reported value of 0.67 μM, determined previously by a relatively indirect PCR assay.17,18
ALKBH3 is known to vary widely in expression in different cell types25 and is reported to be overexpressed in certain solid tumors.13–16 To test whether the MAQ probe design can function to measure enzyme activity in cells, we designed a nuclease-protected version of MAQ, designated PMAQ, using 2′-O-methyl groups to block nuclease activity (Figure 3). In vitro, this modification has little or no effect on probe response except that its light-up response was somewhat greater (12-fold, Figure S6). Incubation of this probe in lysates of MCF7 and TK6 cells revealed signals reaching a maximum in ~30 min (Figure S7). Incubation with an inhibitor in the second cell line ablated the majority of the signal, confirming that the signals are in fact generated from ALKBH3 activity.
Figure 3.
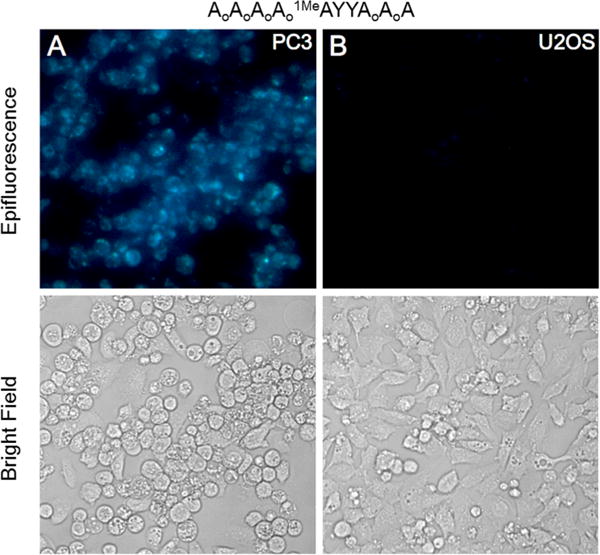
Fluorescence responses of nuclease-protected probe PMAQ (2 μM, shown above) to variable levels of ALKBH3 expression in prostate cancer cell lines PC3 (A, high expression) and U2OS (B, low expression).
Next we tested the PMAQ probe in intact cells, employing three different cell lines and using a commercial cationic lipid for intracellular delivery. The experiments revealed fluorescence readily observable by epifluorescence microscope (Figures 3 and S8). Signals varied in correlation with the reported variations in ALKBH3 levels. For example, PC3 prostate cells are reported to have high ALKBH3 expression as measured by Western blot, while U2OS cells have very low levels.25 This difference is clearly observable under the microscope (Figure 3). Mouse embryonic fibroblasts (MEFs) engineered to lack ALKBH3 expression26 showed little fluorescence, whereas MEFs with the same genetic background but expressing the enzyme exhibited a clear positive signal (Figure S9). Examination of the cells at higher magnification revealed signals appearing primarily in the cytosol and only weakly in the nucleus (Figure S10); previous studies have observed localization in both cellular compartments,6 although they measured protein location rather than enzyme activity. It is possible that the PMAQ probe is more efficiently delivered to the cytosol than the nucleus by the cationic lipid since introduction of the PMAQ product strand also localizes mainly in the cytosol (Figure S10). Although nuclear localization is needed for activity on chromosomal DNA, Krokan showed previously that the enzyme also has activity on m1A demethylation in RNA.6 In addition, very recent studies have identified m1A at conserved positions in mammalian mRNAs27,28 and have shown that ALKBH3 can remove these methyl groups, thus supporting a function of cytosolic enzyme. In addition to observation by microscopy, we found that PMAQ fluorescence in MEFs could also be quantified by flow cytometry (Figure 4) via 355 nm laser excitation; the data showed differential fluorescence for wild-type cells versus those with the ALKBH3 gene deleted.
Figure 4.
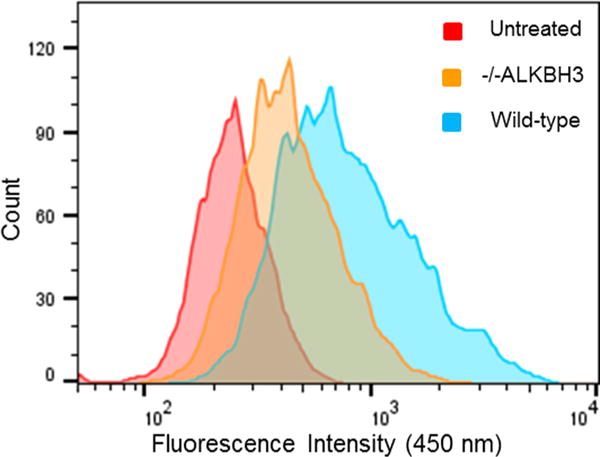
Quantitation of ALKBH3 signals by flow cytometry. Data are obtained with 2 μM probe in wild-type mouse embryonic fibroblasts and in cells with ALKBH3 knocked out. See SI for methods.
Because ALKBH3 and ALKBH2 are closely related in sequence and both repair the same damaged bases,6 we tested whether the MAQ probes are in fact selective for the former enzyme. We tested the PMAQ probe in vitro with the two enzymes and another homologue FTO; data are shown in Figure S6. The results show essentially complete selectivity (>900-fold preference) of the probe for ALKBH3 over ALKBH2 and FTO. This confirms that the reported preference11 of ALKBH2 for double-stranded substrates is quite strong. We proceeded to test the same question in live cells, employing MEFs expressing either enzyme separately or both enzymes.26 The data show that cells expressing only ALKBH2 and not ALKBH3 give little signal with the PMAQ probe (see SI Figure S9), thus establishing that the probe responds selectively for ALKBH3 not only in vitro but also in mammalian cells.
Taken together, our data show that MAQ probes can function conveniently and specifically to measure ALKBH3 activity both in solution and in intact cells. The robust signal increase observed here enables the application of the probes by simple addition to the sample, without need for washing steps. The cyan emission of the probes occurs with a large (135 nm) Stokes shift, thus avoiding issues of overlap of excitation light with emission. The MAQ probes enable the experimenter to measure enzyme activity directly, rather than simply protein quantity as is done by classical IHC and Western blot methods. Our fluorescence probing method also avoids the need for radiolabeling or for indirect and nonlinear PCR-based assays. We are unaware of any prior example of a fluorescence reporter for the AlkB class of oxidative repair enzymes, although probes of other classes of repair enzymes, including UDG and OGG1, have been reported.29–32 Since several distinct enzymes in the AlkB family are known,1–3 it will be of interest to see whether similar design principles might be extended to others in this broad class.
Given the strong current trend toward targeted cancer therapies, it seems possible that measurement of ALKBH3 activity in tissue samples may become a clinically useful way to select patients who may be more susceptible to therapy and for measuring patient responses to treatment. Such approaches are now common for a number of cancers; for example, treatments of melanoma and acute myeloid leukemia currently involve targeted patient selection by measurements of DNA repair activity.33,34 If ALKBH3-targeted therapies17,18 proceed to clinical stages, MAQ probes may make clinical assessments of such factors in ALKBH3-dependent tumors more direct, quantitative, rapid, and practical. Future work will be required to test this possibility.
In addition to their use in measuring this enzyme’s activity for biological and clinical studies, MAQ probes are expected to have considerable utility in the ongoing search for inhibitors of ALKBH3. Both in vitro and whole cell assays should be possible since we have demonstrated both a functional MAQ probe design for in vitro use and PMAQ probes for cellular application. Given the relatively modest binding of known inhibitors, it seems likely that compounds with greater potency will be sought.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (GM110050 and GM067201 to E.T.K. and CA176622, CA107399 (Project 2). and P30CA33572 to T.R.O.) and the Army Research Office (W911NF-13-1-0181) for support. A.A.B. is supported by a fellowship from the Human Frontiers Science Program.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b00986.
Experimental methods and materials, additional data, and characterization of synthetic probes (PDF)
Notes
The authors declare no competing financial interest.
References
- 1.Drabløs F, Feyzi E, Aas PA, Vaagbø CB, Kavli B, Bratlie MS, Peña-Diaz J, Otterlei M, Slupphaug G, Krokan HE. DNA Repair. 2004;3:1389. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. DNA Repair. 2007;6:429. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. J Biol Chem. 2015;290:20734. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang Y-G, He C. Nat Chem Biol. 2011;7:885. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, Vågbø CB, Shi Y, Wang W-L, Song S-H, Lu Z, Bosmans RPG, Dai Q, Hao Y-J, Yang X, Zhao W-M, Tong W-M, Wang X-J, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang Y-G, He C. Mol Cell. 2013;49:18. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, Krokan HE. Nature. 2003;421:859. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 7.Delaney JC, Essigmann JM. Proc Natl Acad Sci U S A. 2004;101:14051. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundheim O, Vågbø CB, Bjørås M, Sousa MML, Talstad V, Aas PA, Drabløs F, Krokan HE, Tainer JA, Slupphaug G. EMBO J. 2006;25:3389. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Begley TJ, Samson LD. Trends Biochem Sci. 2003;28:2. doi: 10.1016/s0968-0004(02)00010-5. [DOI] [PubMed] [Google Scholar]
- 10.Chen B, Liu H, Sun X, Yang C-G. Mol BioSyst. 2010;6:2143. doi: 10.1039/c005148a. [DOI] [PubMed] [Google Scholar]
- 11.Mishina Y, Yang C-G, He C. J Am Chem Soc. 2005;127:14594. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimada K, Nakamura M, Ishida E, Higuchi T, Yamamoto H, Tsujikawa K, Konishi N. Cancer Sci. 2008;99:39. doi: 10.1111/j.1349-7006.2007.00655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamato I, Sho M, Shimada K, Hotta K, Ueda Y, Yasuda S, Shigi N, Konishi N, Tsujikawa K, Nakajima Y. Cancer Res. 2012;72:4829. doi: 10.1158/0008-5472.CAN-12-0328. [DOI] [PubMed] [Google Scholar]
- 14.Koike K, Ueda Y, Hase H, Kitae K, Fusamae Y, Masai S, Inagaki T, Saigo Y, Hirasawa S, Nakajima K, Ohshio I, Makino Y, Konishi N, Y H, Tsujikawa K. K Curr Cancer Drug Targets. 2012;12:847–856. doi: 10.2174/156800912802429283. [DOI] [PubMed] [Google Scholar]
- 15.Tasaki M, Shimada K, Kimura H, Tsujikawa K, Konishi N. Br J Cancer. 2011;104:700. doi: 10.1038/sj.bjc.6606012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimada K, Fujii T, Tsujikawa K, Anai S, Fujimoto K, Konishi N. Clin Cancer Res. 2012;18:5247. doi: 10.1158/1078-0432.CCR-12-0955. [DOI] [PubMed] [Google Scholar]
- 17.Nakao S, Mabuchi M, Shimizu T, Itoh Y, Takeuchi Y, Ueda M, Mizuno H, Shigi N, Ohshio I, Jinguji K, Ueda Y, Yamamoto M, Furukawa T, Aoki S, Tsujikawa K, Tanaka A. Bioorg Med Chem Lett. 2014;24:1071. doi: 10.1016/j.bmcl.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Mabuchi M, Shimizu T, Ueda M, Sasakawa Y, Nakao S, Ueda Y, Kawamura A, Tsujikawa K, Tanaka A. In Vivo (Brooklyn) 2015;29:39. [PubMed] [Google Scholar]
- 19.Mikhailov SN, Rozenski J, Efimtseva EV, Busson R, Van Aerschot A, Herdewijn P. Nucleic Acids Res. 2002;30:1124. doi: 10.1093/nar/30.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manoharan M, Tivel KL, Zhao M, Nafisi K, Netzel TL. J Phys Chem. 1995;99:17461. [Google Scholar]
- 21.Wilson JN, Cho Y, Tan S, Cuppoletti A, Kool ET. ChemBioChem. 2008;9:279. doi: 10.1002/cbic.200700381. [DOI] [PubMed] [Google Scholar]
- 22.Wilson JN, Gao J, Kool ET. Tetrahedron. 2007;63:3427. doi: 10.1016/j.tet.2006.07.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koivisto P, Duncan T, Lindahl T, Sedgwick B. J Biol Chem. 2003;278:44348. doi: 10.1074/jbc.M307361200. [DOI] [PubMed] [Google Scholar]
- 24.Lee D-H, Jin S-G, Cai S, Chen Y, Pfeifer GP, O’Connor TR. J Biol Chem. 2005;280:39448. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 25.Dango S, Mosammaparast N, Sowa ME, Xiong L-J, Wu F, Park K, Rubin M, Gygi S, Harper JW, Shi Y. Mol Cell. 2011;44:373. doi: 10.1016/j.molcel.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nay SL, Lee D-H, Bates SE, O’Connor TR. DNA Repair. 2012;11:502. doi: 10.1016/j.dnarep.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. Nat Chem Biol. 2016 doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 28.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, Zheng G, Pan T, Solomon O, Eyal E, Hershkovitz V, Han D, Doré LC, Amariglio N, Rechavi G, He C. Nature. 2016;530:441. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang YL, Kwon K, Stivers JT. J Biol Chem. 2001;276:42347. doi: 10.1074/jbc.M106594200. [DOI] [PubMed] [Google Scholar]
- 30.Ono T, Wang S, Koo C-K, Engstrom L, David SS, Kool ET. Angew Chem, Int Ed. 2012;51:1689. doi: 10.1002/anie.201108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono T, Edwards SK, Wang S, Jiang W, Kool ET. Nucleic Acids Res. 2013;41:e127. doi: 10.1093/nar/gkt309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards SK, Ono T, Wang S, Jiang W, Franzini RM, Jung JW, Chan KM, Kool ET. ChemBioChem. 2015;16:1637. doi: 10.1002/cbic.201500184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tawbi HA, Buch SC. Clin Adv Hematol Oncol. 2010;8:259. [PubMed] [Google Scholar]
- 34.Brandwein JM, Kassis J, Leber B, Hogge D, Howson-Jan K, Minden MD, Galarneau A, Pouliot J-F. Br J Haematol. 2014;167:664. doi: 10.1111/bjh.13094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


