ABSTRACT
Horizontal acquisition of novel chromosomal genes is considered to be a key process in the evolution of bacterial pathogens. However, the identification of gene presence or absence could be hindered by the inconsistencies in bacterial genome annotations. Here, we performed a cross-annotation of omnipresent core and mosaic accessory genes in the chromosome of Salmonella enterica serovar Typhimurium across a total of 20 fully assembled genomes deposited into GenBank. Cross-annotation resulted in a 32% increase in the number of core genes and a 3-fold decrease in the number of genes identified as mosaic genes (i.e., genes present in some strains only) by the original annotation. Of the remaining noncore genes, the vast majority were prophage genes, and 255 of the nonphage genes were actually of core origin but lost in some strains upon the emergence of the S. Typhimurium serovar, suggesting that the chromosomal portion of the S. Typhimurium genome acquired a very limited number of novel genes other than prophages. Only horizontally acquired nonphage genes related to bacterial fitness or virulence were found in four recently sequenced isolates, all located on three different genomic islands that harbor multidrug resistance determinants. Thus, the extensive use of antimicrobials could be the main selection force behind the new fitness gene acquisition and the emergence of novel Salmonella pathotypes.
IMPORTANCE Significant discrepancies in the annotations of bacterial genomes could mislead the conclusions about evolutionary origin of chromosomal genes, as we demonstrate here via a cross-annotation-based analysis of Salmonella Typhimurium genomes from GenBank. We conclude that despite being able to infect a broad range of vertebrate hosts, the genomic diversity of S. Typhimurium strains is almost exclusively limited to gene loss and the transfer of prophage DNA. Only nonphage chromosomal genes acquired after the emergence of the serovar are linked to the genomic islands harboring multidrug resistance factors. Since the fitness factors could lead to increased virulence, this poses an important research question: could overuse or misuse of antimicrobials act as selection forces for the emergence of more pathogenic strains of Salmonella?
INTRODUCTION
Horizontal gene transfer in bacteria is a major force in adaptation to novel environments, in genome diversification, and, in particular, in the evolution of bacterial virulence (1–4). Horizontally transferred genes create a mosaic structure of the species pangenome, where the so-called accessory genes are present only in some strains, in contrast to the core genes that are typically present in all strains (5–7). The chromosomal accessory genes can be subdivided into genes of phage origin and nonphage genes. The latter usually are acquired in the form of genomic islands, episomes, or transposons that are often related to bacterial physiology, antigenic diversification, and thus fitness (4, 7, 8). In contrast, the phage genes are directly related to phage physiology, although many prophages are also known to contribute virulence and fitness factors to bacteria (9, 10). Distinguishing the accessory genes from the core genes and defining their nature is of critical importance for understanding the evolutionary dynamics and adaptive mechanisms of bacterial species.
Salmonella enterica subsp. enterica represents one of the most important and widely distributed bacterial pathogens to both humans and domesticated animals (11–14). Salmonella serovar Typhimurium represents a broad-host-range spectrum and is one of the most commonly isolated serovars from human, retail meats of diverse origins, and the environment. Although S. Typhimurium is primarily known to cause self-limiting gastroenteritis, it can also be systemically invasive and exhibit a multidrug resistance (MDR) phenotype (15–17). Because Typhimurium serves as a complex serovar involving multiple eco-/pathovars, the horizontal acquisition of genetic material might play a pivotal role in the emergence and evolution of different S. Typhimurium strains. However, systematic analysis of the extent and nature of accessory genes in the S. Typhimurium pangenome has not yet been performed despite the availability of many fully assembled high-quality genomes representing this serovar.
A major hindrance for gene content analysis is inconsistency in existing annotations of even fully assembled, high-quality sequenced genomes, with genes that are annotated in some strains but unannotated or misannotated in other strains. Such inconsistent and incomplete gene annotations can severely limit genome-wide comparison and profiling of core and accessory fractions based on annotated genes, proteins, or functional clusters. Therefore, a lack of completeness and uniformity in annotations considerably restrains the usefulness of the availability of genome sequences in understanding the mechanisms of bacterial evolution, especially the steps related to pathogenesis.
Here, we compare gene content across chromosomal portions of the genomes of eight S. Typhimurium strains that are fully assembled and currently available in GenBank. We detect that there are significant discrepancies in the annotation of, on average, hundreds of genes per strain, leading to highly misleading conclusions about the extent of S. Typhimurium genome openness to the gene transfer. Based on cross-annotation of the existing annotated genomes, we determined here that the diversification of genomes in the S. Typhimurium strains is driven almost exclusively by phages and via core gene loss. It appears that, since the emergence of the serovar, with the exception of one strain, none of the S. Typhimurium strains has acquired any novel genes that are not directly related to the phage biology. This study indicates that the acquisition of nonphage genes plays only a small role in the fitness diversification of S. Typhimurium strains and highlights a critical need for the cross-annotation of existing bacterial genomes.
MATERIALS AND METHODS
Bacterial genomes and phylogeny.
Eight fully assembled genomes of Salmonella enterica serovar Typhimurium (strains D23580, 798, ST4/74, T000240, UK-1, SL1344, LT2, and 14028S) were downloaded from GenBank (National Center for Biotechnology Information). For comparison, complete sequences of fully assembled genomes from 12 other S. Typhimurium strains and 15 non-S. Typhimurium strains were selected and downloaded. Among the non-S. Typhimurium strains, except for two strains each from the Paratyphi A and Typhi serovars, the strains were clonally distinct according to multilocus sequence typing (MLST) analysis (http://mlst.ucc.ie) (see Fig. S1 in the supplemental material).
Pangenomic analysis and reannotation of genomes.
We used our recently developed tool PanCoreGen (18) that, apart from several other features, generates pangenomic profiles of chromosomal genes via reannotations of each annotated genome using rest of the annotated genomes as references. Based on user-defined threshold values of nucleotide sequence identity and length coverage, this tool distinguishes each gene in the analyzed set of genomes as “core” (i.e., present in all genomes of the analyzed data set), “mosaic” (i.e., present in multiple but not all genomes), or “strain specific” (i.e., present only in one of the annotated genomes). We applied the PanCoreGen tool to eight annotated chromosomes of S. enterica serovar Typhimurium to create the pangenomic profile of serovar Typhimurium. For a BLAST (blastn) search of orthologs, we used 95% nucleotide sequence identity and gene length coverage as the lower limit. All of the analyses were restricted to the chromosomal genes, not considering the plasmids. We found a pangenome size of 5,982 genes, 5,345 of which were core genes. The gene distribution for each genome resulting from the pangenomic profile was used for reannotation. We reannotated each genome based on the following four rigorous steps.
(i) Each gene detected by PanCoreGen for a genome was checked to determine whether it was already annotated or not in the existing gene annotations for that genome. We used a BLAST analysis that yielded 100% sequence identity and at least 50% length coverage for a gene to be considered a newly annotated gene. A newly annotated gene might be either completely unannotated previously or partially annotated, where the gene length was less than half the length observed in a new annotation.
(ii) All newly annotated genes were included only if no premature stop codons were present. Otherwise, the genes were discarded to avoid the inclusion of pseudogenes.
(iii) We checked all of the newly annotated genes by using BLAST (blastn) against all annotated pseudogenes in eight S. Typhimurium strains with the constraints of 95% sequence identity and 20% length coverage. This approach would avoid the inclusion of smaller fractions of any pseudogene annotated as an open reading frame (ORF) in some genomes, along with pseudogenes that did not accumulate any premature stop codons (e.g., disabled genes or unitary pseudogenes).
(iv) All partially annotated genes were detected with a further orthology search (100% sequence identity and 10% length coverage) among previously annotated genes.
Pangenomic profiling.
We created the pangenomic profile of eight S. Typhimurium strains via serial inclusion of n genomes (where n goes from 1 through 7) using eight random combinations for n = 2, 3, … 7. This profile was generated for three sets: genomes with existing GenBank annotations, genomes after reannotation, and reannotated genomes without prophage regions. Using Prism software, we performed least-squares curve fitting based on the power law “n = κNγ” to median values. The exponent “γ ≈ 0” indicates a closed pangenome (19).
Phage region identification.
In each of eight S. Typhimurium strains, we identified prophage sequences using the PHAST web server (a phage search tool [http://phast.wishartlab.com/]) (20) by uploading the reannotated GenBank formatted files. We considered all the regions identified as either “intact,” “incomplete,” or “questionable” by PHAST to be the probable prophage regions for all the strains under study. We also considered the phage genes annotated in each of the existing GenBank files. The orthologous sequences of these phage genes were extracted across all of the S. Typhimurium strains under study.
Functional enrichment analysis.
We used DAVID software (21) for clustering based on the protein functions of the DT12 genomic island genes of strain T000240. The classification stringency was set to “medium” for the analysis.
RESULTS
Large fraction of unannotated genes in genomes of S. Typhimurium strains.
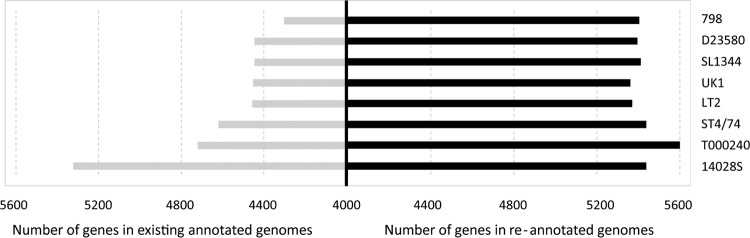
We analyzed gene presence or absence in the chromosomal portions of fully assembled genomes of eight archetypal S. Typhimurium strains: 14028S, 798, D23580, LT2, SL1344, ST474, T000240, and UK-1. Based on seven housekeeping loci that are used for MLST, these strains formed a single tight clade within Salmonella enterica subspecies I (see Fig. S1 in the supplemental material). The genome size variability of the S. Typhimurium strains ranged from 4.82 Mb (UK-1) to 4.95 Mb (T000240), with only a 0.81% ± 0.14% average pairwise difference between the strains. In contrast, based on the GenBank annotation of protein-coding genes, the number of genes in the S. Typhimurium genomes varied between 4,326 (strain 798) and 5,323 genes (strain 14028S), with a 7.14% ± 1.36% average pairwise difference between the strains (Fig. 1, gray bars). The number of annotated genes in individual genomes did not correlate (R2 = 0.03) with the size of the genomes (see Fig. S2 in the supplemental material).
FIG 1.
Comparison of the numbers of genes present in eight S. Typhimurium strains. The left panel represents the numbers of protein-coding genes present in existing annotation, and the right panel shows the numbers of genes after reannotation.
We reannotated protein-coding genes in each S. Typhimurium strain by cross-annotating their genomes using recently developed PanCoreGen software (18). After the cross-annotation, the pairwise difference in the gene content between the strains decreased 5-fold, i.e., to 1.43% ± 0.26% (P < 0.0001). The average gene content per genome increased from 4,600 ± 112 in GenBank annotated genomes to 5,430 ± 26 genes after the cross-annotation, which is higher than the number of originally annotated genes in the genome of strain 14028S, with the highest number of genes according to the GenBank (Fig. 1, black bars). The median lengths of ORFs missed by the original annotations was relatively small and ranged from 132 to 147 bp (see Tables S1 and S2 in the supplemental material). However, each reannotated genome had, on average, 34 newly annotated genes that were ≥300 bp long. The longest such gene was ftsK (4,086 bp) encoding DNA translocase that was missed by the original annotation in strain UK-1. Importantly, after the cross-annotation, the number of genes per genome was well correlated (R2 = 0.86) with the overall sizes of corresponding genomes (see Fig. S2 in the supplemental material).
Thus, there were substantial discrepancies in the GenBank annotations of fully assembled genomes of eight archetypal S. Typhimurium strains. Although the discrepancies mostly involved small ORFs, this resulted in underestimation of the gene number in every genome, as well as overestimation of the differences in gene content between the strains.
Significant reduction in the number of noncore genes after cross-annotation.
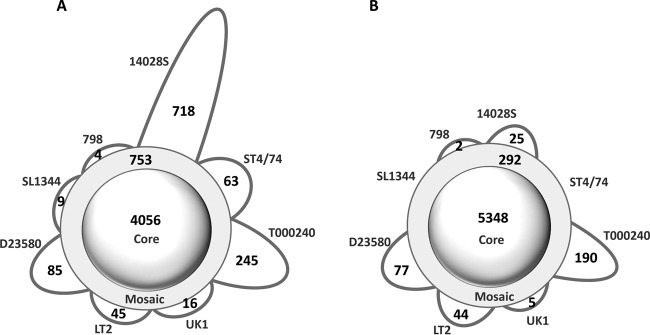
We next assessed the number of the “core genes” present in all eight strains, the “mosaic genes” present in multiple but not all strains, and the “strain-specific genes” present in only one of the strains. Based on the GenBank annotations, there were 4,056 core, 753 mosaic, and 1,185 strain-specific genes (Fig. 2A), with the chicken-derived strain 14028S showing the highest number of strain-specific genes (718 genes). Thus, the core genes represented only 67.7% of all chromosomal genes (pangenome) and, on average, comprised 88.5% of the individual genomes. Furthermore, according to the original annotations, the Typhimurium serovar appeared to have an “open genome,” where the pangenome size was notably increasing with the increase in the number of genomes analyzed (Fig. 3A). The curve-fitting analysis yielded an exponent γ value of 0.16 ± 0.02 that was significantly above zero (γ = 0 is indicative of a completely “closed genome” [see Materials and Methods for details]).
FIG 2.
Schematic representation of the pangenomic profile for eight S. Typhimurium strains. The numbers of core-, mosaic-, and strain-specific genes are shown based on existing annotation (A) and reannotation (B) using the 95% nucleotide sequence identity and gene length coverage thresholds for orthologous gene identification.
FIG 3.
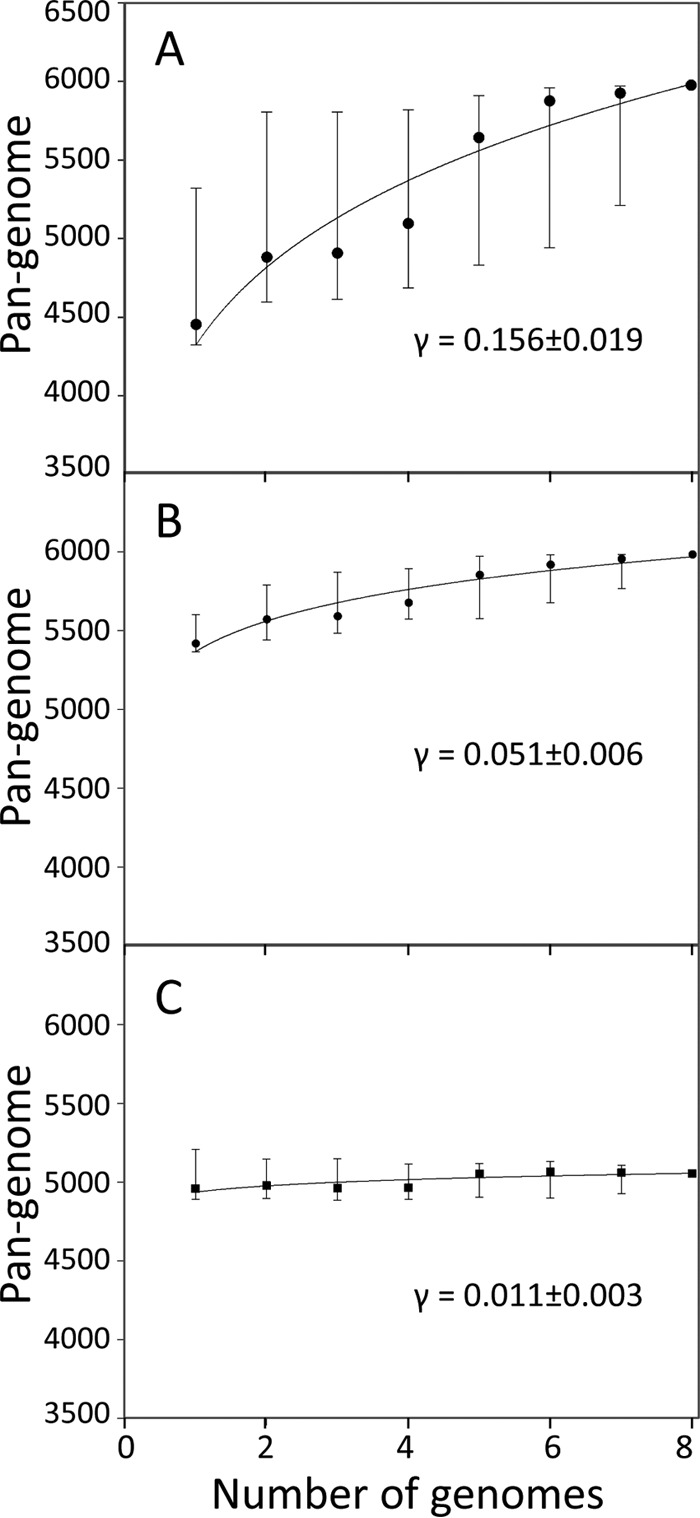
Pangenome size distribution with increasing numbers of genomes in a set of eight S. Typhimurium strains. The pangenome size is depicted by the number of protein-coding genes based on existing annotation (A), reannotation (B), and reannotation excluding the prophage regions (C). The power law fit (n = κNγ) was performed using median values (black dots).
After the cross-annotation by PanCoreGen, the number of the S. Typhimurium core genes increased 1.3-fold, i.e., to 5,348 genes, whereas the number of mosaic and strain-specific genes decreased ∼3-fold, i.e., to 292 and 343 genes, respectively (Fig. 2B). The relative distribution of strain-specific genes also changed significantly. Only 25 genes were unique in the strain 14028S, whereas the highest number of strain-specific genes was in the multidrug-resistant strain T000240 (190 genes), followed by the systemically invasive strain D23580 (77 genes). On the other end, the calf salmonellosis strain ST474 and the model gastroenteritis strain SL1344 (which is a ST474 auxotroph derivative) did not have any genes uniquely present in them. In contrast to the original annotation, 98.5% of the genome contents, on average, were determined to be of core origin based on our cross-annotation analyses. Overall, the core genes comprised a significantly larger portion of the pangenome (89.4%) than before the cross-annotation. As a result, the “openness” of the Typhimurium serovar's pangenome became significantly less obvious (Fig. 3B), with the γ value decreasing by >3-fold (γ = 0.05 ± 0.01, P < 0.0001).
Thus, cross-annotation of the S. Typhimurium strain genomes significantly increased the proportion of core genes in both the serovar pangenome and individual genomes. Although after the cross-annotation the openness of the S. Typhimurium genome became much less obvious, it was not completely closed, indicating a certain level of horizontal gene movement within the serovar. The remainder of the study was performed using the reannotated S. Typhimurium genomes.
Noncore predominance and active transfer of prophages in the serovar.
We determined the genes of prophage origin based on existing annotations of phage genes and also by using the PHAST program (20) to predict the prophage clusters. Among the genes that were newly annotated by PanCoreGen at least in one genome, about 10% were of prophage origin (not shown). Of the 922 prophage genes identified in the cross-annotated pangenome, 456 were core genes, 238 were mosaic genes, and 228 unique genes (Fig. 4A). Thus, the prophage genes comprised only 8.5% of the core genes, but 81.5% of the mosaic genes and 66.5% of the strain-specific genes. Also, without the prophage genes, the core genes comprised 96.5% of the pangenome and 99% of individual genomes on average. After exclusion of the prophage genes from the analysis of the genome openness, the γ value dropped further by >2-fold (γ = 0.011 ± 0.003, P < 0.0001) (Fig. 3C) and was only marginally above zero.
FIG 4.
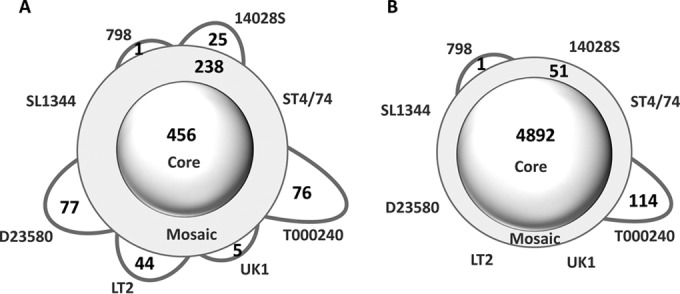
Schematic representation of the pangenomic profile for different genomic fractions of S. Typhimurium strains after reannotation. The numbers of core-, mosaic-, and strain-specific genes are shown for only the prophage regions (A), and genomes excluding the prophage regions (B), using 95% nucleotide sequence identity and gene length coverage thresholds for orthologous gene identification.
Ninety-eight percent of the prophage genes were located on the chromosome in 19 clusters, each incorporating at least two genes, with a median size of 28 genes (see Table S3 in the supplemental material). Ten of the clusters were designated by PHAST as intact prophages and contained 68% of all phage genes. Another six clusters, with 157 genes, were designated incomplete prophage regions. The remaining three clusters, with 118 genes, were designated by PHAST as questionable.
In order to understand the evolutionary origin and dynamics of phage genes, we analyzed in detail their nature, their strain distribution, and in particular their presence in Salmonella enterica serovar Heidelberg strain SL476. This S. Heidelberg strain was chosen as the closest relative based on evolutionary distance of the MLST sequences across serovars (see Fig. S1 in the supplemental material).
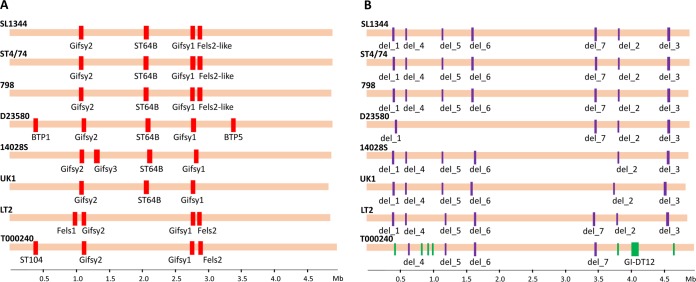
Of 10 intact prophages, only ϕGifsy1 and ϕGifsy2 were found in all S. Typhimurium strains (Fig. 5A), suggesting their ancestral nature in the serovar. Interestingly, the intact core prophages were either absent (ϕGifsy1) or only partially present (ϕGifsy2) in the S. Heidelberg strain. In contrast, five intact prophages—ϕGifsy3 (strain 14028S), ϕFELS1 (LT2), ϕST104 (T000240), ϕBTP1, and ϕBTP5 (D23580)—were found only in one strain. A detailed examination of the chromosomal regions corresponding to the insertion sites of the strain-specific prophages showed no remnant scars in S. Typhimurium strains that lack these phages or in S. Heidelberg, strongly suggesting that these phages were acquired after the serovar had emerged.
FIG 5.
Map of accessory nonphage and “intact” prophage clusters in eight S. Typhimurium genomes. “Intact” prophage regions (as identified by PHAST) (A) and deleted/inserted regions (B) are marked in a multiple alignment. Red bars show the intact prophage regions present in the genomes; purple and green bars designate the regions deleted from and inserted into the genomes, respectively.
Three remaining intact prophages—ϕST64B, ϕFELS2, and ϕFELS2-like—had the mosaic distribution, i.e., they were found in multiple strains but not in all strains. None of these phages or their remnants was present in the S. Heidelberg strain. ϕST64B was present in all but the LT2 and T000240 strains. However, close examination detected remnant sequences of ϕST64B in the corresponding position in the latter strains, suggesting that ϕST64B was originally present in LT2 and T000240, too, but was lost. Another intact mosaic prophage, ϕFELS-2, was present only in LT2 and T000240. However, again, in 14028S, UK-1, and D23580 the remnant scars were present, suggesting ϕFELS-2 loss from the strains. Interestingly, in strains SL1344, ST474, and 798 there was another prophage in the position of the ϕFELS-2 prophage. This phage had 44% genes homologous to ϕFELS-2 (based on 95% sequence identity and length coverage between SL1344 and LT2) and thus was designated previously a “ϕFELS-2-like” prophage. Therefore, both the ϕST64B and the ϕFELS-2 phages appeared to be originally core phages but underwent partial loss or replacement, whereas the ϕFELS2-like prophage was likely the result of an insertion event that occurred later during the evolution of the serovar.
Analysis of phage clusters designated incomplete indicated that four of them were clearly of a core nature (data not shown). Interestingly, this determination could be made only after cross-annotation of the genomes since the GenBank annotation missed them in multiple strains, again causing some misleading conclusions. The other 18 phage genes were not clustered, and all of them were detected as core genes, being present in the same locations across the genomes.
Thus, prophage genes comprise the largest portion of noncore genes, and without them the S. Typhimurium pangenome appears to be almost completely closed. Altogether, the comparative map of clusters of phage origins suggests a continuous acquisition and a loss of phage materials across the S. Typhimurium strains.
Evolutionary origin of the nonphage accessory genes.
Upon removal of the prophage genes and the genes from the repeat regions, 51 genes remained in the mosaic category and 115 genes remained in the strain-specific category, with all but 1 of the latter found in strain T000240 (Fig. 4B). To analyze the origin of nonphage genes, we analyzed their nature in detail, again using S. Heidelberg strain SL476 as the closest relative of the S. Typhimurium clade in the MLST phylogeny (see Fig. S1 in the supplemental material).
Among the 51 mosaic genes of nonphage origin, 34 genes were located in seven clusters of 2 or more genes (see Table S4 in the supplemental material). All of these clusters, however, were designated as being mosaic by being absent only in one or two strains (Fig. 5B): del_1, del_2, and del_3 were absent only in T000240; del_4, del_5, and del_6 were absent only in D23580; and del_7 was absent in UK-1 and 14028S. Moreover, in the S. Heidelberg strain, all of the mosaic gene clusters were present in full and at 99% nucleotide sequence identity, except for del_7, of which only 59% of the cluster was identical in the S. Heidelberg strain at an identity level of ≥98%. Close examination of the remaining 17 mosaic genes that were not in clusters has shown that they were actually present in all strains (i.e., had a core nature), but in certain strains their copies were either truncated by >5% due to the insertional disruption or to partial deletion (see Table S4 in the supplemental material). This led PanCoreGen to miss their actual presence in some strains during the reannotation using 95% cutoffs for both nucleotide sequence identity and gene length coverage. Also, 16 of these genes were present at full length in the S. Heidelberg strain as well, suggesting that their presence was ancestral to S. Typhimurium. Thus, it appears that nonphage genes that were designated mosaic in nature were deemed mosaic due either to their loss in clusters or to partial truncation in some S. Typhimurium strains rather than due to a de novo acquisition.
As mentioned above, only two strains were found to have strain-specific genes of nonphage origin: there were 114 such genes in strain T000240 and only 1 gene in strain 798. The strain 798-specific gene was the 2,097-bp rnfC gene encoding an electron transport complex protein. However, further analysis showed that rnfC was essentially a core gene present in all S. Typhimurium strains, with the rest of the strains carrying a longer 2,208-bp gene version. Due to the length difference, PanCoreGen mistakenly designated the shorter version as specific to strain 798 and the longer one as mosaic in the other strains. Interestingly, however, the S. Heidelberg strain had the longer gene, whereas the shorter version was found in several other serovars, such as Enteritidis, Gallinarum, and Pullorum (data not shown).
A detailed examination of the 114 strain-specific genes in T000240 showed that 100 genes were clustered in a single 82-kb chromosomal island GI-DT12 identified previously (17). The insertion site of GI-DT12 was in the middle of the putative regulatory gene STM14_4564 (as annotated in strain 14028S). This gene remained intact, without any “scars,” in the rest of the S. Typhimurium strains, as well as in the S. Heidelberg strain SL476. This observation strongly suggested that the island was acquired by strain T000240 rather than lost by other strains.
Of the remaining 14 T000240-specific genes, 12 were actually represented by six identical copies of two overlapping genes (504 and 276 bp) encoding IS1 transposases that propagated across the genome, disrupting different gene regions and thus resulting in some genes being mosaic in nature. Two other T000240-specific genes, the 309-bp STMDT12_C29860 and the 387-bp STMDT12_C26860, encoded a hypothetical protein. The former one replaced a 117-bp hypothetical gene present in the rest of the strains except LT2, while the latter was detected immediately upstream of T000240-specific “questionable” phage cluster Q3 (see Table S3 in the supplemental material). Thus, together with the island genes, 14 other T000240-specific genes also appear to be acquired horizontally by the T000240 strain.
Altogether, it is evident that only one of the S. Typhimurium strains analyzed, T000240, had undergone genomic acquisition events that did not involve prophages, primarily the 82-kb genomic island. The rest of the nonphage genes that were originally defined here as mosaic or unique in the S. Typhimurium strains were actually core genes either completely or partially lost in some strains after the serovar had emerged.
Identification of the complete genomic core of the Typhimurium serovar.
Based on the analysis of eight cross-annotated genomes presented above, we could show that, upon exclusion of prophage genes, the ancestral (“total”) chromosomal core of the Typhimurium serovar is comprised of 4,944 genes, including 4,892 omnipresent genes (“stable core”) and 52 partially lost genes (“unstable core”).
To assess the completeness of the genomic core identified, we added to the analysis 12 more completely sequenced S. Typhimurium genomes that became available in GenBank during the course of this study (see Table S5 in the supplemental material). Upon cross-annotation of all 20 genomes together, 4,694 genes remained omnipresent, whereas 198 genes of the original stable core were completely or partially lost from some of 12 genomes. A total of 84% of all of these genes belonged to eight clusters (of two or more consecutive genes), with the largest deletion event occurring in strain VNP20009, a modified derivative of 14028S (22), involving 102 genes across a 108-kb region. In this new set of genomes, we also found the presence of two of seven deletion events noted previously for the set of 52 mosaic genes. The event del_7 appeared to occur in the common ancestor of UK-1, 14028S, and VNP20009. Interestingly, del_4, which was earlier detected in the MDR and invasive strain D23580, was also present in two other MDR strains, DT104 and 138736.
Upon the cross-annotation of 20 genomes of S. Typhimurium, 29 new genes were added to the total core. All of them were missed in the original set of eight genomes because of their absence in the annotations rather than genes per se. Of these newly annotated genes, 24 were omnipresent (stable core), and the remaining 5 showed a deletion in some of the genomes.
Thus, the originally defined set of core genes proved to be highly representative for the Typhimurium serovar. Upon the cross-annotation of 20 fully assembled genomes deposited in GenBank, the nonphage total core of S. Typhimurium stands at 4,973 genes, with the omnipresent stable core comprised of 4,718 genes (95%) and partially lost unstable core of 255 genes (5%), with the latter represented by 15 to 19 strains.
Insertion of genomic islands: limited, but all with MDR and fitness genes.
As described above, in the originally analyzed set of eight genomes, a horizontally acquired set of genes was found only in one strain: the MDR strain T000240 with the 100-gene genomic island DT12. Functional enrichment analysis (see Table S6 in the supplemental material) of the DT12 island revealed genes that code for resistance to chloramphenicol, sulfonamides, tetracycline, etc. [i.e., bla(oxa-30), aadA1, qacEΔ1, and sul1, cat, and tetA], as well as fitness- and (potentially) virulence-associated genes such as aerobactin iron-acquisition siderophore system (lutA and lucABC) and iron transporter (sitABCD) genes.
Analysis of the 12 additional genomes of S. Typhimurium revealed two additional genomic islands in three strains; both of these islands were reported previously (23, 24). One of them was GI-VII-6, a 125-kb island that was incorporated in an MDR strain L-3553 isolated from cattle in Japan and was found to code for several antibiotic resistance genes (aadA, strA, strB, sul1, sul2, tetA, floR, and dfrA12), along with the blaCMY-2 gene (for extended-spectrum cephalosporin resistance). In addition, this island gene coded a number of transcriptional regulators (STL3553_RS05185, STL3553_RS05225, STL3553_RS05270, and STL3553_RS05280), as well as siderophore transporter proteins (STL3553_RS05320), that likely affect the overall fitness and possibly also the virulence of the strains. Identical copies of another genomic island, SGI-1 (43 kb), were found in two closely related strains: DT104 and 138736. This island again coded genes responsible for the MDR phenotype (i.e., resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, tetracycline, fluoroquinolones, etc.) for both of these strains that was attributed to a 13-kb region of the island carrying MDR genes (e.g., aadA2, floR, tetG, pse-1, sul1, etc.). We also found potential fitness-related genes encoding transcriptional regulators (e.g., tetR, orf1, etc.) and recombination proteins (e.g., tnpA, tnpR, int1, and orf2).
The only other set of noncore nonphage genes identified in the newly analyzed genomes were a 24.6-kb genomic insertion in position between genes CFSAN001921_RS03395 and CFSAN001921_RS03550 of another MDR strain, CFSAN001921 (25). In the insertion, 13 of 27 genes were of hypothetical origin, with none of the remaining genes being related to known resistance or fitness factors but primarily related to the genomic stability cassettes. Two of the genes were annotated to encode phage functions, and the insertion itself was located in the insertion site of Fels-2 prophage of LT2. Thus, the phage origin of the 24.6-kb genomic insertion could not be excluded. Altogether, the cross-annotation analysis of 20 genomes of S. Typhimurium strains revealed only three genomic regions (in four strains) that were clearly of nonphage origin being horizontally transferred, and all of them carried multiple antibiotic resistance- and fitness-related genes.
DISCUSSION
In this study, we eliminated discrepancies in the annotation of genes in 20 fully assembled genomes of Salmonella Typhimurium strains deposited in GenBank. We performed cross-annotation using PanCoreGen software package to add, on average, hundreds of genes to each genome that were missed by the original annotation and to identify genes present in all, some, or only one of the S. Typhimurium strains. We determined that chromosomal portion of the S. Typhimurium genome is highly conserved, with limited influx of novel fitness-related genes via horizontal transfer, except for the horizontal movements of phage clusters. Since the serovar has emerged, only a few nonphage horizontal transfer events could be recorded, all of them related to the MDR phenotype. However, the acquisition of antibiotic resistance brings genes that might change the fitness and, potentially, the virulence of the strains. Moreover, the fitness evolution in S. Typhimurium could also be driven in certain strains by complete or partial loss of some of the original core genes.
High gene content diversity in Salmonella enterica subsp. enterica genomes reflects the considerable extent of horizontal gene transfer events. Previous works have demonstrated the importance of gene acquisition in virulence, antibiotic resistance, novel metabolic pathways, and other adaptive traits. Gene acquisition in the form of islands or as parts of phages has been suggested to play key role in the emergence of different Salmonella serovars. For example, genes harboring an array of genomic islands and prophages (SPI-6, Gifsy-1, Gifsy-2, etc.) are important for the intracellular replication of S. Typhimurium strains (26). SPI-12, encoding a remnant phage in S. Typhimurium, includes at least four genes for transcriptional activation/regulation and fitness, thereby facilitating bacterial survival in the host (27). Both SPI-1 and SPI-2 encode type III secretion systems (T3SS) involved in the translocation of virulence proteins into host cells, whereas multiple T3SS effectors are part of SPI-5. In addition, SPI-3 and SPI-4 promote the intestinal colonization via MisL (a member of the autotransporter protein family) and SiiE (a nonfimbrial adhesin as part of a type I secretion system), respectively. All of these insertion events are known to be ancestral to the Typhimurium serovar. However, whether or not gene transfer plays a significant role in the genome and fitness diversification after the serovars have emerged has not been studied in detail.
Here, we investigated the intraserovar evolution of Salmonella Typhimurium that arguably exhibits the broadest host and pathogenicity range among all serovars of S. enterica subsp. enterica. This serovar is able to infect both livestock and human hosts (e.g., pathovar DT104) and exhibits a restricted host range (e.g., pathovars DT2 and DT99) (28). The importance and diversity of the Typhimurium serovar is reflected in the number of strains with sequenced genomes of a high-quality assembly that were deposited in GenBank. This includes one of the first Salmonella strains sequenced 15 years ago—that of the model strain LT2 (12)—and strains from invasive human infections (D23580, 33676, and YU39) (15, 29, 30), or harboring MDR phenotype and isolated from various animals (CFSAN001921, L-3553, etc.) (25, 31).
One could expect that the large time period over which the S. Typhimurium genomes had been obtained and the diversity of research groups who contributed them to the public domain would certainly result in differences in the genome annotations. First, the algorithms for the recognition of ORFs have been evolving constantly, and different research groups could use different stringency criteria (such as a minimal sequence length) for defining an ORF as a functional gene. Second, in some genomes certain genes could be absent due to either gene deletion or horizontal acquisition. Third, nonsense or frameshift mutations, small deletions, and insertional disruptions could cause certain genes to be missed by the annotation. Finally, the use of different reference genomes and/or annotation databases could result in giving the same gene a different name or hypothetical function status. One way or another, all of this could lead to significant differences in the annotation of different genomes that could be mere artifacts, with many genes missed or given various names. This problem is not limited to Salmonella Typhimurium and is likely to be a general problem in bacterial genomics.
A couple of recent pangenomic profiles of Salmonella enterica were based on the construction of orthologous groups (32) or on the de novo annotation of genes (14). Over the last decade, a number of powerful analysis tools have been developed for pangenomic profiling that can successfully identify homologous genes and their core, mosaic, or strain-specific nature in a given data set, along with plotting and visualization of the profiles, both sequence and function-based annotation and curation of genomes, reconstructing phylogenetic relationships of orthologous genes or families, etc. (33, 34). However, an important factor yet to be integrated into existing pangenomic analysis approaches appears to be gene gain and gene loss that occurs during specific lineage evolution, i.e., to differentiate the accessory genes in the pangenome derived via horizontal gene acquisition from the “ancestrally core” genes that have been lost from specific strains or lineages over the course of evolution and thus are part of the accessory fraction (which we designate here as unstable core genes). Such information in the assortment of genes would provide insights on possible functional adaptation via pangenomic evolution. Here, we consider each annotated S. Typhimurium genome as a reference relying on existing annotations to perform cross-annotation among annotated genomes to identify unannotated genes, and finally we reconstruct the pangenome based on uniformly reannotated individual genomes. We identify a total of 4,718 genes found in all strains (stable core) and 255 genes found in most isolates (unstable core) in the analyzed S. Typhimurium strains. The determination of the total core gene set that was originally present when the serovar had emerged offers the research field with an evolutionary “Eve” of S. Typhimurium strains that could be used for the annotation of newly sequenced genomes to achieve better understanding of their evolutionary dynamics.
Upon cross-annotation of the originally analyzed 8 and then 12 additional genomes, it has become clear that the accessory fraction of the S. Typhimurium genomes is predominantly contributed by the continuous inflow and outflow of phage elements. In the absence of the horizontal transfer of phage clusters, the Typhimurium serovar would have been considered to have almost a closed pangenome. Although we did not analyze here the plasmid diversity of the S. Typhimurium strains, whether or not plasmids play a significant role in the adaptive evolution of Salmonella Typhimurium, it is astounding that the serovar's genome appears to be so restricted to the acquisition of nonphage genomic islands by horizontal transfer. Also, there is always a possibility that a genomic island, instead of being a result of true horizontal transfer event, is a shift from an unanalyzed plasmid, thereby leading to an even more closed structure of the S. Typhimurium pangenome. Keeping in mind the broadest host and pathogenicity ranges of S. Typhimurium among all S. enterica serovars, one can suggest a possibility of similar scenarios for other S. enterica serovars as well. This presents a great contrast, for example, to the continuous movement of genomic islands in Escherichia coli strains that belong to the same serotypes and/or multilocus sequence types (STs). These closely related clonal groups might be considered equivalent to specific serovars of Salmonella from the perspectives of population and evolutionary genetics. For example, a single avian-pathogenic E. coli strain APECO1 from ST95 has been shown to harbor 43 genomic islands that differ greatly in content from strains representing the same serotype and/or ST (35). Also, E. coli strains of the MDR clonal group ST131 vary considerably in the acquisition of several genomic islands that carry various pathogenicity-relevant genes (with no antibiotic resistance) (36). Another example, ST73, combines strains with highly diverse genomic content and includes the model uropathogenic strain CFT073, the asymptomatic bacteriuria strain 83972, and the probiotic strain Nissle 1917 (37, 38). This suggests strong physiological barriers in Salmonella compared to E. coli. Alternatively, limited genomic diversity in S. Typhimurium strains could also be the result of the relatively recent emergence of this serovar, which did not allow enough time for the frequency of horizontal transfer events to be comparable to some of E. coli serotypes or STs that were of much older origin. Therefore, additional studies on accurate molecular clock estimation of the clonal group ages are warranted in order to understand the basis of the difference in the gene transfer rates in different species.
Majority of the prophage gene clusters seemed to be comprised of genes related to the phage movement, biogenesis and structural components. However, this does not mean that phage acquisition or loss could not have an adaptive effect on the S. Typhimurium strains, including their virulence. On one hand, disruption of the genomic region of the prophage insertion could affect the function of nearby genes. Interestingly, two different phage clusters, ϕST104 and ϕBTP1, are inserted into the same spot in the MDR strain T000240 and the systemically invasive strain D23580, respectively. In both strains, the insertion has happened downstream of the proA gene (encoding gamma-glutamyl phosphate reductase) and upstream of the IS3 transposase that is itself positioned immediately upstream of the stb fimbrial gene cluster. If the function of the surrounding genes is affected one way or another by the phage insertion, this could have significant effects on the bacterial physiology. On the other hand, the presence of prophages in the bacterial chromosome could have a pleiotropic effect on the expression of chromosomal genes by, for example, transcriptional cross-regulation. Thus, these phages might have a similar effect on corresponding host strains not only by being inserted into the same genomic site but also by having a similar in trans effect. It is noteworthy that we detected the presence of a phage cluster at exactly the same location as that of ϕST104 and ϕBTP1 (in strains T000240 and D23580, respectively) exclusively in all of the MDR and/or invasive strains among the additional 12 S. Typhimurium strains analyzed (i.e., U288, CFSAN001921, DT104, 138736, L-3553, 33676, and YU39). In the remaining 11 non-MDR and/or noninvasive strains, this genomic location remained uninterrupted where proA was immediately followed by the IS3 transposase. The insertion of different phage clusters suggested a hot spot nature of this genomic region, allowing independent acquisition of the phage genes by MDR and/or invasive strains of this serovar. However, detailed experimental and population studies are warranted to determine whether there is any direct or indirect role of prophage genes in S. Typhimurium physiology, virulence, and/or drug resistance phenotypes.
A detailed analysis for gene presence or absence in the cross-annotated S. Typhimurium strains showed that about 5% of the original core genes were completely or partially lost in some strains after the serovar emerged. Gene loss by deletion or the formation of pseudogenes is thought to be driven by two contrasting evolutionary processes. On one hand, genes can be lost as part of “use-or-lose” evolutionary dynamics, i.e., if the function of these genes are no longer needed for a particular lifestyle of the bacterial strain. Such a loss is accumulated as a result of genetic drift, i.e., via random events not driven by positive selection. On the other hand, gene loss could be driven by a “die-or-lose” evolutionary mechanism that removes genes whose functions reduce fitness in certain environments. In the course of clinical infection, for example, some genes might interfere with the expression or function of virulence-promoting factors or increase the liability of the pathogen by expressing traits recognizable by the host defenses.
The complete or partial loss of core genes was noted primarily in invasive strain D23580, MDR strain T000240, and in two closely related strains, UK-1 (called the “universal killer” for its pronounced invasion and virulence properties) and 14028S (15, 17, 39, 40). This finding highlights a possible central role for gene inactivation in the evolution of fitness diversity in Salmonella Typhimurium strains. Interestingly, previous work has suggested an adaptive convergence of the loss of the del_6 cluster from systemically invasive S. Typhimurium D23580 with the same event in the systemically invasive serovar Typhi (15). However, we detected here that all non-Typhimurium serovars (invasive and noninvasive) showed either a partial or a complete loss or a truncation of genes in this cluster (see Fig. S3 in the supplemental material). Also, earlier work demonstrated that disruption of stbC gene in the stb fimbrial operon could lead strain LT2 to a flagellated strain exhibiting constitutively mannose-sensitive agglutinating and MDR phenotype (41). Interestingly, we detected that, although stbC gene has remained intact, other genes of the stb fimbrial operon, such as stbB, stbA, etc., are lost in the MDR strain T000240 due to the absence of the del_1 cluster. It would be worth finding out whether the loss of these genes might have somehow contributed to the resistance phenotype of this strain. Our analysis of an additional 12 strains of Salmonella serovar Typhimurium revealed the loss of a few other ancestrally core gene clusters in some of them (see Table S7 in the supplemental material). However, understanding the functional and adaptive significance of the gene loss and inactivation in S. Typhimurium strains is beyond the scope of this study and will require an expanded analysis of S. Typhimurium and non-S. Typhimurium strains, as well as experimental studies.
In the original set of eight genomes, the only true transfer of gene cluster of nonphage origin was the T000240-specific genomic island GI-DT12 that possesses determinants of antibiotic resistance, mercury resistance, iron acquisition, heavy metal tolerance, etc. Previous work demonstrated that this island has most likely helped the isolate to adapt to adverse environmental conditions such as extremely polluted sewage (17). In the additional 12 genomes, only two more genomic islands (in strains DT104, 138736, and L-3553) were identified that also carried both MDR and fitness genes. Although we identified one more potential island (in another MDR strain, CFSAN001921) of obscure origin and function, our observations indicate strongly that, in the course of the evolution of the Typhimurium serovar, antibiotic resistance might be the major selection factor behind the acquisition of genomic islands that also brings some fitness-associated genes into the recipient genomes.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by National Institutes of Health grant R01 AI106007.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00545-16.
REFERENCES
- 1.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV, Makarova KS, Aravind L. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol 55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutta C, Pan A. 2002. Horizontal gene transfer and bacterial diversity. J Biosci 27:27–33. doi: 10.1007/BF02703681. [DOI] [PubMed] [Google Scholar]
- 4.Wiedenbeck J, Cohan FM. 2011. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev 35:957–976. doi: 10.1111/j.1574-6976.2011.00292.x. [DOI] [PubMed] [Google Scholar]
- 5.Mira A, Martin-Cuadrado AB, D'Auria G, Rodriguez-Valera F. 2010. The bacterial pan-genome: a new paradigm in microbiology. Int Microbiol 13:45–57. [DOI] [PubMed] [Google Scholar]
- 6.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome. ” Proc Natl Acad Sci U S A 102:13950–13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: building the web of life. Nat Rev Genet 16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 8.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33:376–393. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brussow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Valera F, Martin-Cuadrado AB, Rodriguez-Brito B, Pasic L, Thingstad TF, Rohwer F, Mira A. 2009. Explaining microbial population genomics through phage predation. Nat Rev Microbiol 7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2013. National Salmonella surveillance annual report, 2011. US Department of Health and Human Services/Centers for Disease Control and Prevention, Atlanta, GA. [Google Scholar]
- 12.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 13.Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, Quail MA, Stevens M, Jones MA, Watson M, Barron A, Layton A, Pickard D, Kingsley RA, Bignell A, Clark L, Harris B, Ormond D, Abdellah Z, Brooks K, Cherevach I, Chillingworth T, Woodward J, Norberczak H, Lord A, Arrowsmith C, Jagels K, Moule S, Mungall K, Sanders M, Whitehead S, Chabalgoity JA, Maskell D, Humphrey T, Roberts M, Barrow PA, Dougan G, Parkhill J. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res 18:1624–1637. doi: 10.1101/gr.077404.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobsen A, Hendriksen RS, Aaresturp FM, Ussery DW, Friis C. 2011. The Salmonella enterica pan-genome. Microb Ecol 62:487–504. doi: 10.1007/s00248-011-9880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, Harris D, Clarke L, Whitehead S, Sangal V, Marsh K, Achtman M, Molyneux ME, Cormican M, Parkhill J, MacLennan CA, Heyderman RS, Dougan G. 2009. Epidemic multiple drug-resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res 19:2279–2287. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, Nunes J, Tsolis RM, Adams LG, Baumler AJ. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect Immun 71:1–12. doi: 10.1128/IAI.71.1.1-12.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izumiya H, Sekizuka T, Nakaya H, Taguchi M, Oguchi A, Ichikawa N, Nishiko R, Yamazaki S, Fujita N, Watanabe H, Ohnishi M, Kuroda M. 2011. Whole-genome analysis of Salmonella enterica serovar Typhimurium T000240 reveals the acquisition of a genomic island involved in multidrug resistance via IS1 derivatives on the chromosome. Antimicrob Agents Chemother 55:623–630. doi: 10.1128/AAC.01215-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul S, Bhardwaj A, Bag SK, Sokurenko EV, Chattopadhyay S. 2015. PanCoreGen: profiling, detecting, annotating protein-coding genes in microbial genomes. Genomics 106:367–372. doi: 10.1016/j.ygeno.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Broadway KM, Modise T, Jensen RV, Scharf BE. 2014. Complete genome sequence of Salmonella enterica serovar Typhimurium VNP20009, a strain engineered for tumor targeting. J Biotechnol 192:177–178. doi: 10.1016/j.jbiotec.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Boyd D, Peters GA, Cloeckaert A, Boumedine KS, Chaslus-Dancla E, Imberechts H, Mulvey MR. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol 183:5725–5732. doi: 10.1128/JB.183.19.5725-5732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee K, Kusumoto M, Sekizuka T, Kuroda M, Uchida I, Iwata T, Okamoto S, Yabe K, Inaoka T, Akiba M. 2015. Extensive amplification of GI-VII-6, a multidrug resistance genomic island of Salmonella enterica serovar Typhimurium, increases resistance to extended-spectrum cephalosporins. Front Microbiol 6:78. doi: 10.3389/fmicb.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann M, Muruvanda T, Allard MW, Korlach J, Roberts RJ, Timme R, Payne J, McDermott PF, Evans P, Meng J, Brown EW, Zhao S. 2013. Complete genome sequence of a multidrug-resistant Salmonella enterica serovar Typhimurium var. 5 strain isolated from chicken breast. Genome Announc 1:e01068-13. doi: 10.1128/genomeA.00294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klumpp J, Fuchs TM. 2007. Identification of novel genes in genomic islands that contribute to Salmonella Typhimurium replication in macrophages. Microbiology 153:1207–1220. doi: 10.1099/mic.0.2006/004747-0. [DOI] [PubMed] [Google Scholar]
- 27.Tomljenovic-Berube AM, Henriksbo B, Porwollik S, Cooper CA, Tuinema BR, McClelland M, Coombes BK. 2013. Mapping and regulation of genes within Salmonella pathogenicity island 12 that contribute to in vivo fitness of Salmonella enterica serovar Typhimurium. Infect Immun 81:2394–2404. doi: 10.1128/IAI.00067-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabsch W, Andrews HL, Kingsley RA, Prager R, Tschape H, Adams LG, Baumler AJ. 2002. Salmonella enterica serotype Typhimurium and its host-adapted variants. Infect Immun 70:2249–2255. doi: 10.1128/IAI.70.5.2249-2255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calva E, Silva C, Zaidi MB, Sanchez-Flores A, Estrada K, Silva GG, Soto-Jimenez LM, Wiesner M, Fernandez-Mora M, Edwards RA, Vinuesa P. 2015. Complete genome sequencing of a multidrug-resistant and human-invasive Salmonella enterica serovar Typhimurium strain of the emerging sequence type 213 genotype. Genome Announc 3:e00663-15. doi: 10.1128/genomeA.00663-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva C, Calva E, Calva JJ, Wiesner M, Fernandez-Mora M, Puente JL, Vinuesa P. 2015. Complete genome sequence of a human-invasive Salmonella enterica serovar Typhimurium strain of the emerging sequence type 213 harboring a multidrug resistance IncA/C plasmid and a blaCMY-2-carrying IncF plasmid. Genome Announc 3:e01323-15. doi: 10.1128/genomeA.01323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shahada F, Sekizuka T, Kuroda M, Kusumoto M, Ohishi D, Matsumoto A, Okazaki H, Tanaka K, Uchida I, Izumiya H, Watanabe H, Tamamura Y, Iwata T, Akiba M. 2011. Characterization of Salmonella enterica serovar Typhimurium isolates harboring a chromosomally encoded CMY-2 beta-lactamase gene located on a multidrug resistance genomic island. Antimicrob Agents Chemother 55:4114–4121. doi: 10.1128/AAC.00560-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordienko EN, Kazanov MD, Gelfand MS. 2013. Evolution of pan-genomes of Escherichia coli, Shigella spp., and Salmonella enterica J Bacteriol 195:2786–2792. doi: 10.1128/JB.02285-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vernikos G, Medini D, Riley DR, Tettelin H. 2015. Ten years of pan-genome analyses. Curr Opin Microbiol 23:148–154. doi: 10.1016/j.mib.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 34.Xiao J, Zhang Z, Wu J, Yu J. 2015. A brief review of software tools for pangenomics. Genomics Proteomics Bioinformatics 13:73–76. doi: 10.1016/j.gpb.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson TJ, Wannemuehler Y, Kariyawasam S, Johnson JR, Logue CM, Nolan LK. 2012. Prevalence of avian-pathogenic Escherichia coli strain O1 genomic islands among extraintestinal and commensal E. coli isolates. J Bacteriol 194:2846–2853. doi: 10.1128/JB.06375-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul S, Linardopoulou EV, Billig M, Tchesnokova V, Price LB, Johnson JR, Chattopadhyay S, Sokurenko EV. 2013. Role of homologous recombination in adaptive diversification of extraintestinal Escherichia coli. J Bacteriol 195:231–242. doi: 10.1128/JB.01524-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hancock V, Vejborg RM, Klemm P. 2010. Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073: comparison of transcriptomes, growth, and biofilm formation. Mol Genet Genomics 284:437–454. doi: 10.1007/s00438-010-0578-8. [DOI] [PubMed] [Google Scholar]
- 38.Vejborg RM, Friis C, Hancock V, Schembri MA, Klemm P. 2010. A virulent parent with probiotic progeny: comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol Genet Genomics 283:469–484. doi: 10.1007/s00438-010-0532-9. [DOI] [PubMed] [Google Scholar]
- 39.Jarvik T, Smillie C, Groisman EA, Ochman H. 2009. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J Bacteriol 192:560–567. doi: 10.1128/JB.01233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y, Kong Q, Yang J, Mitra A, Golden G, Wanda SY, Roland KL, Jensen RV, Ernst PB, Curtiss R III. 2012. Comparative genome analysis of the high pathogenicity Salmonella Typhimurium strain UK-1. PLoS One 7:e40645. doi: 10.1371/journal.pone.0040645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu KH, Wang KC, Lee LW, Huang YN, Yeh KS. 2012. A constitutively mannose-sensitive agglutinating Salmonella enterica subsp. enterica serovar Typhimurium strain, carrying a transposon in the fimbrial usher gene stbC, exhibits multidrug resistance and flagellated phenotypes. Sci World J 2012:280264. doi: 10.1100/2012/280264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.