ABSTRACT
The riboflavin analogs roseoflavin (RoF) and 8-demethyl-8-aminoriboflavin (AF) are produced by the bacteria Streptomyces davawensis and Streptomyces cinnabarinus. Riboflavin analogs have the potential to be used as broad-spectrum antibiotics, and we therefore studied the metabolism of riboflavin (vitamin B2), RoF, and AF in the human pathogen Listeria monocytogenes, a bacterium which is a riboflavin auxotroph. We show that the L. monocytogenes protein Lmo1945 is responsible for the uptake of riboflavin, RoF, and AF. Following import, these flavins are phosphorylated/adenylylated by the bifunctional flavokinase/flavin adenine dinucleotide (FAD) synthetase Lmo1329 and adenylylated by the unique FAD synthetase Lmo0728, the first monofunctional FAD synthetase to be described in bacteria. Lmo1329 generates the cofactors flavin mononucleotide (FMN) and FAD, whereas Lmo0728 produces FAD only. The combined activities of Lmo1329 and Lmo0728 are responsible for the intracellular formation of the toxic cofactor analogs roseoflavin mononucleotide (RoFMN), roseoflavin adenine dinucleotide (RoFAD), 8-demethyl-8-aminoriboflavin mononucleotide (AFMN), and 8-demethyl-8-aminoriboflavin adenine dinucleotide (AFAD). In vivo reporter gene assays and in vitro transcription/translation experiments show that the L. monocytogenes FMN riboswitch Rli96, which controls expression of the riboflavin transport gene lmo1945, is negatively affected by riboflavin/FMN and RoF/RoFMN but not by AF/AFMN. Treatment of L. monocytogenes with RoF or AF leads to drastically reduced FMN/FAD levels. We suggest that the reduced flavin cofactor levels in combination with concomitant synthesis of inactive cofactor analogs (RoFMN, RoFAD, AFMN, and AFAD) explain why RoF and AF contribute to antibiotic activity in L. monocytogenes.
IMPORTANCE The riboflavin analogs roseoflavin (RoF) and 8-demethyl-8-aminoriboflavin (AF) are small molecules which are produced by Streptomyces davawensis and Streptomyces cinnabarinus. RoF and AF were reported to have antibacterial activity, and we studied how these compounds are metabolized by the human bacterial pathogen Listeria monocytogenes. We found that the L. monocytogenes protein Lmo1945 mediates uptake of AF and RoF and that the combined activities of the enzymes Lmo1329 and Lmo0728 are responsible for the conversion of AF and RoF to toxic cofactor analogs. Comparative studies with RoF and AF (a weaker antibiotic) suggest that the reduction in FMN/FAD levels and the formation of inactive FMN/FAD analogs explain to a large extent the antibiotic activity of AF and RoF.
INTRODUCTION
Riboflavin (vitamin B2) is not synthesized by mammals but is synthesized by many microorganisms and by all plants (1). The genomes of the human bacterial pathogens Listeria monocytogenes, Streptococcus pyogenes, and Enterococcus faecalis do not contain genes encoding riboflavin biosynthetic enzymes (2), and thus these microorganisms are riboflavin auxotrophs. L. monocytogenes, Streptococcus pyogenes, and Enterococcus faecalis produce energy-coupling factor (ECF) transporters which combine with a riboflavin-specific binding subunit (subunit EcfS or RibU) (3), and ECF-RibU-mediated uptake is the sole source of riboflavin (4).
Riboflavin cannot be detected in cell extracts of bacteria (5–7), indicating that it is completely metabolized by flavokinases (RibF) (EC 2.7.1.26) and FAD synthetases (RibC) (EC 2.7.7.2). These enzymes catalyze the formation of FMN (from riboflavin and ATP) and FAD (from FMN and ATP) (see Fig. S1 in the supplemental material) and play an important role in metabolic trapping of riboflavin (8). In bacteria, bifunctional flavokinases/FAD synthetases have been found, whereas in eukarya and archaea, monofunctional enzymes seem to be the rule (9). FMN and FAD are cofactors of flavoproteins/flavoenzymes, which have a wide variety of different biological functions (10). The number of flavin-dependent proteins varies greatly in different organisms (and among pathogens) and covers a range from approximately 0.1% to 3.5% of the proteome (11). In Escherichia coli, FMN and FAD are present at levels which are about 30 times lower than those of the highly abundant amino acids or nucleotides (12), whereby FAD (170 μM) clearly is present at higher levels than FMN (54 μM) (12). This is in line with the fact that about 90% of all flavoproteins use FAD as a cofactor and only 10% use FMN (11).
The Gram-positive soil bacteria Streptomyces davawensis and Streptomyces cinnabarinus are the only organisms known to produce the antibiotic roseoflavin (RoF), a structural riboflavin analog (see Fig. S1 in the supplemental material) (13). RoF biosynthesis in S. davawensis is carried out by three enzymes. The 8-demethyl-8-aminoriboflavin-5′-phosphate synthase RosB generates 8-demethyl-8-aminoriboflavin mononucleotide (AFMN) (the key intermediate of RoF biosynthesis) from FMN (14). An as yet unknown phosphatase produces AF from AFMN. Finally, the N,N-8-demethyl-8-aminoriboflavin dimethyltransferase RosA (15, 16) catalyzes the formation of RoF from AF and S-adenosylmethionine. Inactivation of rosA generated a recombinant S. davawensis strain that produces AF (see Fig. S1) (15), which can be employed to synthesize this compound.
Growth of L. monocytogenes was found to be profoundly inhibited by RoF (17), and this riboflavin auxotroph appeared to be an ideal organism to study the antibiotic effect of flavin analogs. It was shown by Northern blotting experiments that addition of RoF reduced expression of the riboflavin transporter gene lmo1945 (ribU), and it was speculated that the resulting riboflavin deficiency was responsible for the growth-inhibiting effect of RoF (17). The same work showed that RoF stimulated L. monocytogenes virulence gene expression and infection abilities in a mechanism independent of the FMN riboswitch (17). These interesting results prompted us to study riboflavin metabolism and the mechanism of action of RoF and AF in L. monocytogenes in more detail.
MATERIALS AND METHODS
Chemicals and materials.
RoF was obtained from MP Biomedicals (Heidelberg, Germany) and was dissolved in dimethyl sulfoxide (DMSO). AF was a gift from Peter Macheroux (Technical University of Graz, Austria) and was dissolved in DMSO. Roseoflavin mononucleotide (RoFMN), AFMN, roseoflavin adenine dinucleotide (RoFAD), and 8-demethyl-8-aminoriboflavin adenine dinucleotide (AFAD) were prepared enzymatically using purified recombinant human flavokinase and human FAD synthetase as described previously (18). All other chemicals were from Sigma-Aldrich (Munich, Germany).
Bacterial strains and plasmids.
A Bacillus subtilis strain overexpressing L. monocytogenes lmo1945 was generated using pHT01 (Mobitech, Göttingen, Germany), PCR, and the modifying oligonucleotides 5′-ATA TAT GGA TCC ATG AAG AAT TAT TCA ATG AA-3′ and 5′-ATT GAC GTC TTA ATG GCT TAT TTC TTG TTG TCT TTT-3′. The corresponding plasmid was named pHT01-lmo1945. Construction of B. subtilis ΔribB::Ermr ΔribU::Kanr and B. subtilis ΔribU::Kanr was described earlier (19). An E. coli BL21(DE3) strain overexpressing L. monocytogenes lmo1329 was generated using pET-24a(+) (Thermo Fisher, Darmstadt, Germany), PCR, and the modifying oligonucleotides 5′-AAT AAT GCT AGC ATG AAG ACG ATA TAC TTA CA-3′ and 5′-AAT ATA GCG GCC GCA TCT TCT AAT TTA GCA A-3′. The resulting strain containing pET-24a(+)lmo1329-His produced a C terminally His6-tagged version of Lmo1329 (Lmo1329-His6). An E. coli BL21(DE3) strain overexpressing L. monocytogenes lmo0728 was constructed as follows. The gene lmo0728 was amplified by PCR using the modifying oligonucleotides 5′-ATA GCC ATG GAA GTA TCG CAT GTA AC-3′ and 5′-TAT CGG ATC CTT ACT CGG AAA GTT CGT TT−3′. The resulting PCR product was digested and ligated to pET-MBP-1a (20), and the novel plasmid was named pET-MBP-lmo0728. A linker (5′-CAT GGT GGC GG TGG CGG TGG C-3′; 5′-CAT GGC ACC GCC ACC GCC ACC-3′) representing the tobacco etch virus (TEV) protease recognition site was introduced into the NcoI site of pET-MBP-lmo0728, resulting in pET-MBP-lmo0728TEV, which in turn was used to transform E. coli BL21(DE3). The corresponding strain produced an N-terminal His6-tagged version of a maltose binding protein (MBP)-Lmo0728 fusion (His6-MBP-Lmo0728) following treatment with TEV. For generation of PribLm-Rli96-lacZ fusions, pDG268 was employed. This plasmid contains a promoterless lacZ gene and a chloramphenicol resistance cassette flanked by 5′ and 3′ parts of the B. subtilis amyE gene, allowing integration into the amyE site of the B. subtilis chromosome. The rib promoter region, including the FMN riboswitch Rli96 from L. monocytogenes was amplified by PCR using the modifying oligonucleotides 5′-CGC GAA TTC ATA AAT AAA ACC AGC TAA TT-3′ and 5′-CGC GGA TCCGAT GTT CAC CAA GAA GCG AG-3′. The resulting PCR product was treated with EcoRI and BamHI and ligated to EcoRI- and BamHI-digested pDG268. The resulting plasmid pPribLm-Rli96-lacZ (1 μg) was linearized using XhoI and used to transform B. subtilis 168. The plasmid pPrib-RFN-lacZ contains the B. subtilis ribDG promoter region, including the B. subtilis ribDG-FMN riboswitch fused to lacZ (PribDG-ribDG-FMN riboswitch-lacZ) and was constructed earlier (21). For in vitro transcription/translation assays, the plasmid pT7-Rli96-luc was constructed as follows. A DNA fragment coding for Rli96 was produced by PCR using the modifying oligonucleotides 5′-CGC AAA GCT TAT AAA TAA AAC CAG CTA ATT-3′ and 5′-ATA TAT GCG GCC GCT CAT TGA ATA ATT CTT CAT TG-3′. The PCR product was digested and ligated to linearized pT7-luc (7). Construction of the recombinant strain L. monocytogenes M1 (a L. monocytogenes EGDe derivative) was described earlier (17). The strain M1 contains two nucleotide exchanges in Rli96 (see Fig. S2 in the supplemental material), which were reported to lead to expression of lmo1945 even in the presence of high flavin levels (riboflavin or roseoflavin). The strain is deregulated with regard to flavin uptake and was found to grow better in the presence of 100 μM RoF (17). The B. subtilis strain expressing ribU from pDG148 was constructed in an earlier work (21).
Growth conditions.
E. coli was aerobically grown at 37°C in LB medium. B. subtilis was aerobically cultivated at 37°C in LB or Spizizen minimal medium supplemented with glucose (0.4%), Casamino Acids (0.02%), and tryptophan (0.05 mg/ml). Antibiotics were added to the growth medium when required. Expression of LacI-controlled genes was stimulated by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. L. monocytogenes was grown in either brain heart infusion (BHI) broth (Becton Dickinson, Heidelberg, Germany) or in a minimal medium (22). For growth assays in the presence/absence of flavin analogs, overnight cultures of L. monocytogenes (BHI broth) were diluted 100-fold in either BHI broth or minimal medium. The bacteria were grown for 10 h at 37°C with aeration.
Preparation of cell extracts of riboflavin-, RoF-, and AF-treated L. monocytogenes cells.
For the determination of the intracellular flavin concentration in L. monocytogenes, 10 ml of a liquid culture was harvested by centrifugation (8,000 × g, 4°C, 10 min). The cells were washed three times with 1 ml of washing buffer (50 mM NaCl, 50 mM Tris-HCl [pH 7.5]) to remove all residual flavins. The cells were suspended in 50 μl of washing buffer, and the optical density at 600 nm (OD600) was measured and correlated to the total cell count. Then 100 μl of GES (5 M guanidinium thiocyanate, 0.1 M EDTA, 0.5% [mass/vol] sodium lauroyl sarcosinate [pH 8]) was incubated at 70°C. To the heated GES mixture, 40 μl of the sample was added and mixed vigorously for 5 s. The mixture was incubated for 1 min at 70°C to release all flavins from flavoproteins. The reaction was stopped by addition of 860 μl of ice-cold water. The mixture was again centrifuged, and the supernatant was subjected to liquid chromatography (LC) analysis. The flavins detected represent the total flavin content of the cells.
LC analysis of flavins.
Flavins were analyzed by LC using a ReproSil-Pur C18-AQ column (5-μm particle size, 250 mm by 4.6 mm; Dr. Maisch, Ammerbuch-Entringen, Germany). The following mixture was used at a flow rate of 1 ml/min to equilibrate and wash the column: 15% (vol/vol) methanol-20 mM formic acid-20 mM ammonium formate (pH 3.7). After sample injection, the methanol concentration was increased in a linear gradient over 8 min to 42% methanol (vol/vol). This concentration was maintained for 2 min. Detection of flavins was carried out using a photometer (riboflavin, FMN, and FAD, λ = 445 nm; AF, AFMN, and AFAD, λ = 480 nm; RoF, RoFMN, and RoFAD, λ = 503 nm).
Analysis of chromosomal FMN riboswitch-lacZ fusions.
For lacZ reporter assays, B. subtilis was cultivated to an OD600 of 0.4 in Spizizen minimal medium. Flavins were added, and the cultures were grown for another 4.5 h. The cells were collected by centrifugation, washed in PM (10 mM NaH2PO4, 90 mM Na2HPO4, 1 mM MgSO4 [pH 7.8]), suspended in 0.05 volume of PM, and disrupted using a vibratory tube mill at maximum speed in the presence of glass beads (0.3 mm in diameter). The specific LacZ activity was determined with 2-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate and is expressed as nanomoles of ONPG hydrolyzed per minute per microgram of total protein (milliunits per microgram). Protein was determined according to Bradford using bovine serum albumin as a standard.
In vitro transcription/translation assays.
The coupled transcription/translation assays were performed using the E. coli T7 S30 extract system for circular DNA kit (Promega, Mannheim, Germany) according to the recommendations of the manufacturer. Luciferase activity was determined by employing the luciferase assay reagent (luciferase assay system; Promega) in a microtiter plate reader (Tecan Genios Pro microplate reader; Tecan, Mainz, Germany) as described previously (23). The template plasmids were isolated from the corresponding recombinant E. coli strains using the GeneJET plasmid miniprep kit (Fermentas, Heidelberg, Germany). The isolated plasmid was again purified using the same kit and eluted from the anion exchange columns with nuclease-free water.
Production and purification of recombinant Lmo1329 and Lmo0728.
Lmo1329-His6 was produced in E. coli BL21(DE3) transformed with pET-24a(+)lmo1329-His. E. coli BL21(DE3) harboring pET-24a(+)lmo1329-His was grown to an OD600 of 0.6. Synthesis of recombinant Lmo1329-His6 was stimulated by adding IPTG, and the cultures were grown for another 3 h. Cells were harvested by centrifugation (3,500 × g) and stored at −20°C. Frozen cell paste was suspended in 30 ml of binding buffer (50 mM Na2HPO4, 500 mM NaCl, 20 mM imidazole [pH 7.4]) to which 1 tablet of cOmplete (EDTA-free protease inhibitor cocktail) was added (Roche, Mannheim, Germany). Cells were passed twice through a French press at 2,000 × 105 Pa. Centrifugation (10,000 × g, 4°C, 20 min) removed cell debris and unbroken cells. The lysate was cleared by ultracentrifugation (106,000 × g, 4°C, 30 min) and applied to a 5-ml HisTrap column after equilibration with binding buffer. Chromatographic steps were performed using the ÄKTApurifier system (GE Healthcare). When the UV signal returned to baseline elution of the His6-tagged protein was stimulated by continuously increasing the concentration of the elution buffer (50 mM Na2HPO4, 500 mM NaCl, 200 mM imidazole [pH 7.4]) (linear gradient over 10 column volumes). Aliquots of the fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and staining with Coomassie brilliant blue G-250. His6-MBP-Lmo0728 was produced in E. coli BL21(DE3) transformed with pET-MBP-lmo0728TEV. Gene expression was carried out for 16 h after stimulation with 1 mM IPTG (final concentration) at an OD600 of 0.8, and the incubation temperature for the protein production phase was reduced to 25°C (from 37°C). The cell extract (see above) was applied to a HisTrap-column and a linear imidazole gradient was used for elution (see above). The eluted fusion protein was subjected to a proteolytic digest (4°C, 16 h) with a His6-tagged version of TEV protease (produced by employing pTH24 [24]), which was removed by affinity chromatography together with the His6-MBP part of the fusion protein. The flowthrough contained Lmo0728.
Flavokinase/FAD synthetase assay.
Flavokinase activity was measured in a final volume of 2 ml of 50 mM potassium phosphate (pH 7.5) containing 50 μM flavin, 1 mM ATP, 12 mM NaF, 6 mM MgCl2, and 24 mM Na2SO3. The mixture was incubated at 37°C for 5 min, and the reaction was started by addition of the enzyme. After appropriate time intervals, an aliquot was removed and analyzed by LC. Flavokinase activity is expressed as nanomoles of phosphorylated flavin formed from flavin and ATP. The reaction velocity v was determined separately for each substrate concentration by linear regression using multiple data points. The substrate concentrations were tested in triplicate. The kinetic constants Km and Vmax were evaluated with the Michaelis-Menten equation using SigmaPlot (Erkrath, Germany). The turnover numbers, kcat, were calculated with the subunit molecular mass of 35.5 kDa for Lmo1329. The FAD synthetase Lmo0728 was measured accordingly using phosphorylated flavins and ATP as substrates. kcat was calculated with the subunit molecular mass of 27.7 kDa for Lmo0728.
RESULTS
The riboflavin analogs RoF and AF inhibit growth of L. monocytogenes.
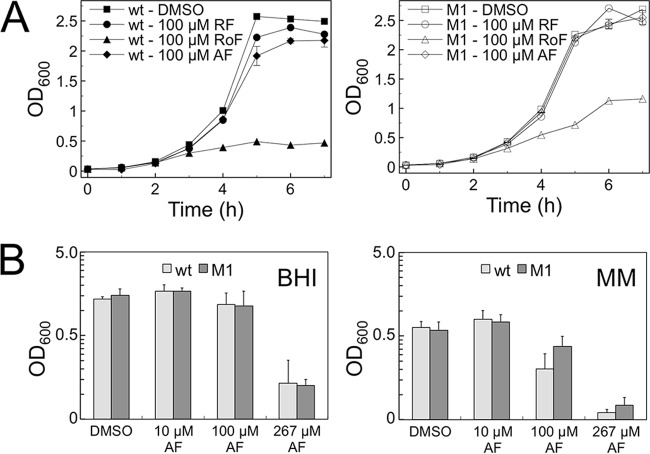
The objective of the following experiment was to validate the previous finding that growth of L. monocytogenes is reduced in the presence of the antibiotic RoF (17) and to investigate the putative antibacterial activity of AF. Two different L. monocytogenes strains (17), were employed in the growth experiments, L. monocytogenes wild type and L. monocytogenes M1. The latter strain contains mutations in the FMN riboswitch Rli96 (see Fig. S2 in the supplemental material), which were reported to reduce RoF sensitivity by deregulating expression of the putative riboflavin transporter gene lmo1945 (17). Growth of L. monocytogenes wild type was strongly reduced in the presence of 100 μM RoF but not in the presence of 100 μM AF (Fig. 1A). At higher AF concentrations (267 μM), a growth-inhibiting effect of AF was observed in a rich medium (BHI broth) as well as in a minimal medium (Fig. 1B). Growth of L. monocytogenes M1 was also reduced in the presence of 100 μM RoF, although approximately a doubling of the final OD600 was reached compared to that for the wild-type strain (Fig. 1A). AF reduced growth of M1 at 267 μM but not at 100 μM (Fig. 1). No difference with regard to growth inhibition by AF was found in the two strains, indicating that AF does not affect the FMN riboswitch Rli96.
FIG 1.
The riboflavin (RF) analogs roseoflavin (RoF) and 8-demethyl-8-aminoriboflavin (AF) negatively interfere with growth of different Listeria monocytogenes strains. (A) Growth of L. monocytogenes wild type (wt) (left panel) and L. monocytogenes M1 (M1) (right panel) in brain heart infusion (BMI) broth in the presence of DMSO (control), RF (100 μM), RoF (100 μM), or AF (100 μM) was recorded at λ = 600 nm. L. monocytogenes M1 contains chromosomal mutations in the FMN riboswitch Rli96 which were reported to confer partial RoF resistance (17). (B) Both strains were grown in BHI broth or a minimal medium (MM) to the stationary phase in the presence or absence (DMSO control) of indicated amounts of AF and the final OD600 was determined. At 267 μM, AF growth was reduced in both strains, and the effect of AF was most pronounced in a minimal medium. Mean values of three independent experiments are given.
The gene product of lmo1945 is responsible for flavin uptake.
In a recent paper, the structure of the riboflavin-binding subunit Lmo1945 (EcfS or RibU) of the riboflavin uptake system was reported (25). Physiological experiments showing that Lmo1945 is indeed involved in riboflavin transport were not performed (25), and we set out to validate this anticipated function. As a test strain, a B. subtilis riboflavin auxotrophic double mutant (ΔribB::Ermr ΔribU::Kanr) was employed. This test strain carries deletions in ribB (encoding essential riboflavin synthase, EC 2.5.1.9) and in ribU (the lmo1945 homolog in B. subtilis). B. subtilis ΔribB::Ermr ΔribU::Kanr is able to grow only when riboflavin (100 μM) is present. At a concentration of 100 μM, the vitamin is able to permeate in sufficient amounts over the cytoplasmic membrane in the absence of a dedicated riboflavin transport system (Fig. 2D). The gene lmo1945 was overexpressed in B. subtilis ΔribB::Ermr ΔribU::Kanr using the plasmid pHT01-lmo1945. The corresponding recombinant strain grew on LB medium (which contains about 4 μM riboflavin) (Fig. 2A). Since a control strain containing empty pHT01 was not able to grow under these conditions, the results of these experiments suggested that Lmo1945 facilitates riboflavin transport (Fig. 2). Similar experiments were performed in liquid minimal medium containing glucose and different amounts of riboflavin (see Fig. S3 in the supplemental material). Again, we observed that the presence of pHT01-lmo1945 allowed growth of B. subtilis ΔribB::Ermr ΔribU::Kanr even at low riboflavin levels (10 μM), which validated the above results.
FIG 2.
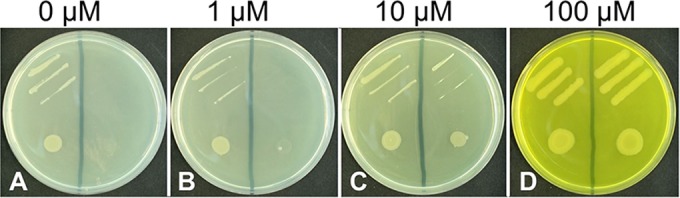
Lmo1945 facilitates riboflavin (RF) uptake. Streaks (top sections of the plates) and drops (bottom sections, about 50,000 cells) of RF auxotrophic Bacillus subtilis ΔribB::Ermr ΔribU::Kanr cells expressing lmo1945 from plasmid pHT01-lmo1945 (left sections) were applied to LB plates (about 4 μM RF) containing the indicated (additional) amount of RF and IPTG (to stimulate expression of lmo1945). Growth was recorded after incubation for 36 h at 37°C. As a control, strains were transformed with the empty expression vector pHT01 (right sections of the plates). (A and B) Only the cells transformed with pHT01-lmo1945 grew in the presence of small amounts of RF. (C) At 10 μM RF, the control strain showed reduced growth compared to that of the pHT01-lmo1945-containing strain. (D) At 100 μM RF, no difference in growth was observed.
The presence of Lmo1945 increases RoF sensitivity of a recombinant B. subtilis strain.
The results of the following experiment suggest that Lmo1945 not only facilitates riboflavin transport but also is responsible for uptake of toxic riboflavin analogs. A B. subtilis strain deficient in flavin uptake (ΔribU::Kanr) was transformed with pHT01-lmo1945 (see previous section) and tested with regard to RoF sensitivity on LB plates containing different amounts of RoF (Fig. 3). Growth of the control strain harboring empty pHT01 was slightly reduced at 100 μM but not at 50 μM RoF (Fig. 3B and C). In contrast, growth of the pHT01-lmo1945-containing strain was strongly reduced at 100 μM and slightly reduced at 50 μM RoF (Fig. 3B and C). Growth experiments using liquid minimal medium containing different amounts of roseoflavin produced similar results and revealed that at 100 μM, RoF growth of the pHT01-lmo1945 containing B. subtilis strain ΔribU::Kanr was strongly reduced (in contrast to the control) (see Fig. S4 in the supplemental material).
FIG 3.
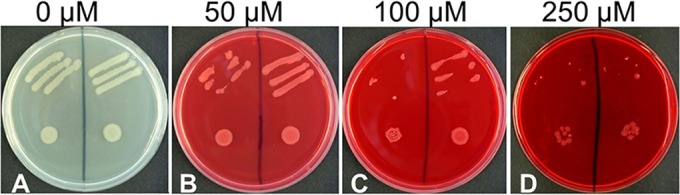
Lmo1945 facilitates roseoflavin (RoF) transport. Streaks (top sections of the plates) and drops (bottom sections, about 50,000 cells) of riboflavin prototrophic Bacillus subtilis ΔribU::Kanr cells (but deficient in the endogenous flavin transporter component RibU) expressing lmo1945 from plasmid pHT01-lmo1945 (left sections) were applied to LB plates containing the indicated amount of roseoflavin and IPTG (to stimulate expression of lmo1945). Growth was recorded after incubation for 36 h at 37°C. As a control, strains were transformed with the empty expression vector pHT01 (right sections of the plates). The strain transformed with pHT01-lmo1945 showed reduced growth compared to that of the control at 50 μM RoF and 100 μM RoF, indicating that Lmo1945 promotes RoF uptake. At 250 μM RoF, both strains were sensitive to RoF, indicating that at these RoF levels the antibiotic is able to cross the cytoplasmic membrane in the absence of a flavin transporter.
Flavin cofactors and flavin cofactor analogs are present in cell lysates of L. monocytogenes.
The results of the following experiments show that riboflavin, RoF, and AF are metabolized by flavokinases/FAD synthetases present in L. monocytogenes. L. monocytogenes wild type and the deregulated strain L. monocytogenes M1 were grown to the stationary phase in BHI broth (containing 4 μM riboflavin) and also in the presence of (additional) riboflavin (100 μM), RoF (100 μM), or AF (100 μM). The cells were harvested, washed, and lysed. The proteins within the resulting cell extracts were fully denatured, and the flavins released (nonphosphorylated/adenylylated flavins, FMN analogs, and FAD analogs) were analyzed by liquid chromatography (Table 1). In the untreated L. monocytogenes wild-type strain (no additional riboflavin was added to the BHI cultures), riboflavin was not found indicating that flavokinases/FAD synthetases completely converted cytoplasmic riboflavin to FMN (7 μM) and FAD (60 μM), whereby the higher FAD levels reflect the higher cellular demand of FAD. The addition of riboflavin (100 μM) only slightly increased the amount of FMN and FAD; however, the FMN/FAD ratio changed in favor of FMN (which can be explained by a comparably low kcat of the L. monocytogenes flavokinase) (Table 2). Nonphosphorylated/adenylylated riboflavin was not found. Upon addition of RoF and AF, the cofactor analogs RoFMN/RoFAD and AFMN/AFAD were found, showing that flavokinases and FAD synthetases within L. monocytogenes accept these flavins as substrates (see the reaction scheme in Fig. S1 in the supplemental material). Nonmetabolized RoF and AF were detected, indicating that these flavins are not as efficiently phosphorylated/adenylylated as riboflavin. The presence of AFMN/AFAD and RoFMN/RoFAD (indirectly) confirmed that the flavin analogs AF/RoF are taken up by L. monocytogenes from the culture medium (most likely by Lmo1945), whereby the higher AF/AFMN/AFAD levels (compared to the RoF/RoFMN/RoFAD levels) suggest that AF is a better substrate for uptake (compared to RoF). Notably, no FMN and drastically reduced FAD levels were detected in AF- and RoF-treated cells.
TABLE 1.
Flavin levels for an assumed cellular volume of 1 μm3 in Listeria monocytogenes strains grown on BHI broth
| Strain and treatmenta | Level (μM) of: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| RF | FAD | FMN | RoF | RoFAD | RoFMN | AF | AFAD | AFMN | |
| wt | |||||||||
| DMSO | 0 | 60 | 7 | 0 | 0 | 0 | 0 | 0 | 0 |
| RF | 0 | 84 | 17 | 0 | 0 | 0 | 0 | 0 | 0 |
| RoF | 0 | 11 | 0 | 4 | 63 | 7 | 0 | 0 | 0 |
| AF | 0 | 18 | 0 | 0 | 0 | 0 | 35 | 64 | 26 |
| M1 | |||||||||
| DMSO | 0 | 74 | 11 | 0 | 0 | 0 | 0 | 0 | 0 |
| RF | 14 | 140 | 24 | 0 | 0 | 0 | 0 | 0 | 0 |
| RoF | 0 | 24 | 0 | 20 | 113 | 16 | 0 | 0 | 0 |
| AF | 0 | 29 | 0 | 0 | 0 | 0 | 65 | 88 | 28 |
Strains (wt, wild type; M1, deregulated Rli96-defective strain) were treated with 100 μM riboflavin (RF), 100 μM roseoflavin (RoF), or 100 μM 8-demethyl-8-aminoriboflavin (AF).
TABLE 2.
Kinetic constants for the flavokinase/FAD synthetase reactions of Lmo0728 (27.7 kDa) and Lmo1329 (35.5 kDa) from Listeria monocytogenes
| Enzymea | Substrate | Km (μM) | Vmax (U mg−1)b | kcat (s−1) | kcat/Km (μM−1 s−1) |
|---|---|---|---|---|---|
| Lmo1329 (FK) | Riboflavin | 6.9 | 668 | 0.40 | 0.057 |
| Lmo1329 (FS) | FMN | 29.2 | 3,706 | 4.39 | 0.150 |
| Lmo0728 (FS) | FMN | 12.9 | 1,093 | 0.51 | 0.040 |
FK, flavokinase; FS, FAD synthase.
Specific activities (U mg−1) are in nmol min−1 mg−1 protein.
Addition of riboflavin to strain M1 (which contains a mutation in the FMN riboswitch Rli96) (see Fig. S2 in the supplemental material) resulted in higher flavin levels than in the wild-type strain. This is in line with the idea that the mutation within Rli96 generates an FMN riboswitch which does not terminate transcription and thus does not reduce expression of the flavin transporter gene lmo1945 even though the Rli96-regulating ligand FMN is present in the cytoplasm (17). In contrast to the wild-type strain, nonmetabolized riboflavin was found, suggesting that flavokinases/FAD synthetases within L. monocytogenes were overloaded. The FAD levels in RoF- and AF-treated M1 cells again were strongly reduced and FMN was not detected.
The following in vitro experiments were initiated to characterize the flavokinases/FAD synthetases of L. monocytogenes in the presence of different flavin substrates to get a more complete picture with regard to the metabolism of RoF and AF.
The gene product of lmo1329 is a bifunctional flavokinase/FAD synthetase.
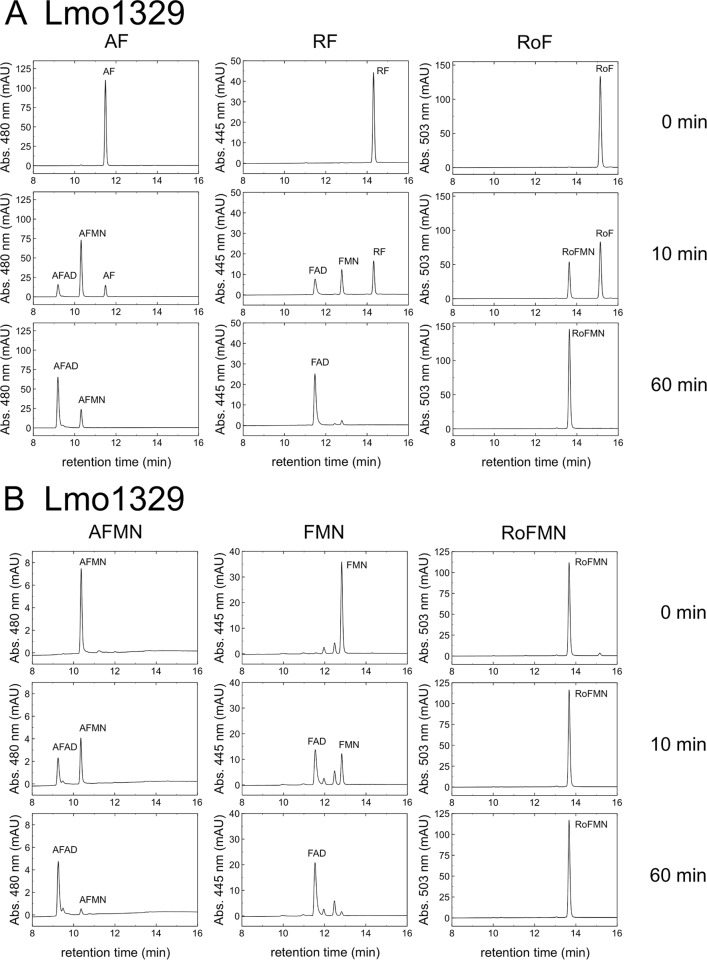
The well-characterized bifunctional flavokinase/FAD synthetase from B. subtilis (26), a close relative of L. monocytogenes, was compared to all known and putative L. monocytogenes proteins using BLASTp. Two putative flavokinases/FAD synthetases (encoded by lmo1329 and lmo0728) were identified. A recombinant version of Lmo1329, Lmo1329-His6, was overproduced in E. coli and purified to apparent homogeneity. Lmo1329-His6 was tested for flavokinase and FAD synthetase activity using different flavin substrates. The enzyme was found to synthesize FMN and FAD (from riboflavin and ATP) and thus is a bifunctional flavokinase/FAD synthetase (Fig. 4A and B). When the substrate riboflavin was replaced by AF, AFMN and AFAD were found. When RoF was used as a substrate, only the formation of RoFMN (but not of RoFAD) was observed (Fig. 4A). The FAD synthetase activity of Lmo1329 was also tested with FMN, AFMN, and RoFMN as substrates (Fig. 4B). As expected, FAD and AFAD were synthesized from phosphorylated flavins and ATP but not RoFAD. In Table 2, the kinetic parameters for the substrates riboflavin and FMN are summarized. The sensitivity of our noncontinuous flavokinase/FAD synthetase assay did not allow the determination of kinetic parameters for the flavin substrates AF and RoF. From the existing data, we were able to estimate that the apparent Km values for both substrates were less than 1 μM. The data shown in Fig. 4 indicate that RoF/AF/AFMN were metabolized by Lmo1329 much slower than riboflavin/FMN, which explains why nonphosphorylated/adenylylated RoF/AF were present in comparably large amounts.
FIG 4.
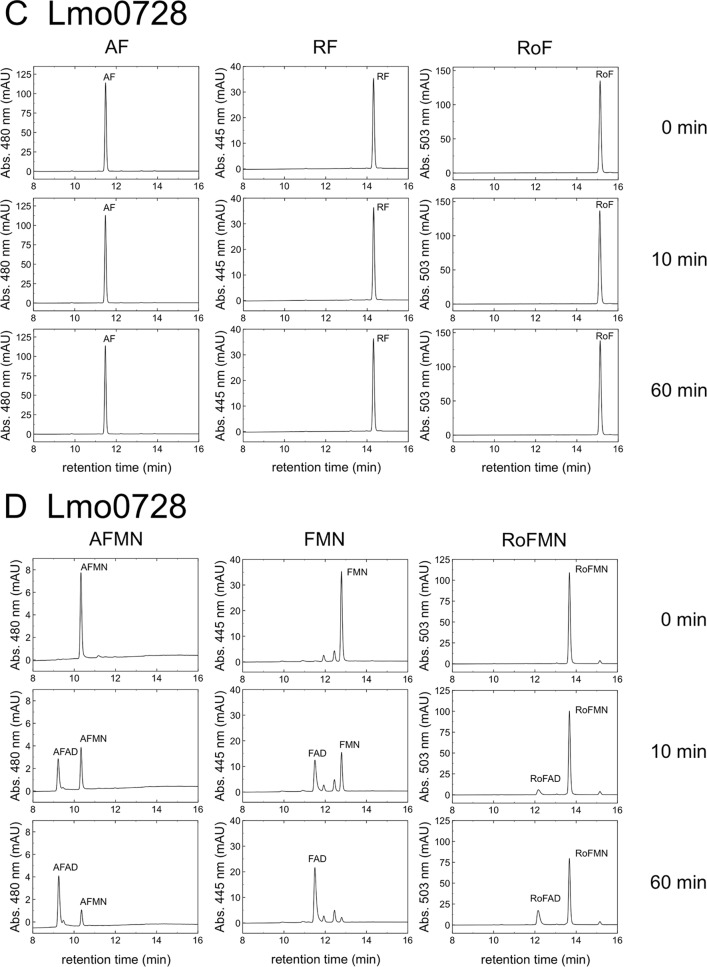
Lmo1329-catalyzed (A and B) and Lmo0728-catalyzed (C and D) reactions. Different flavin substrates (8-demethyl-8-aminoriboflavin [AF], riboflavin [RF], roseoflavin [RoF]) were used for testing the two different Listeria monocytogenes enzymes, Lmo1329 and Lmo0728, with regard to flavokinase/FAD synthetase activity. Upon reaction of these flavins with ATP, the products 8-demethyl-8-aminoriboflavin mononucleotide (AFMN), flavin mononucleotide (FMN), roseoflavin mononucleotide (RoFMN), 8-demethyl-8-aminoriboflavin adenine dinucleotide (AFAD), flavin adenine dinucleotide (FAD), and roseoflavin adenine dinucleotide (RoFAD) were formed. Assay mixtures containing 50 μM flavins, 1 mM ATP, 12 mM NaF, 6 mM MgCl2, and 24 mM Na2SO3 were incubated at 37°C for 5 min. Purified recombinant Lmo1329 or Lmo0728 (1 μg/ml) was added, and the mixtures were incubated for 0 min, 10 min, or 60 min. An aliquot was removed from the assay mixtures, and flavins were analyzed by high-performance liquid chromatography (HPLC). Flavins was detected photometrically at 445 nm (RF and FMN), 480 nm (AF and AFMN), or 503 nm (RoF and RoFMN). Peak intensity is given in arbitrary absorbance units (mAU).
The turnover number of Lmo1329 for FMN is about 13 times higher than that of the bifunctional flavokinase/FAD synthetase from Corynebacterium ammoniagenes (27), which in turn has the highest turnover number of all FAD synthetases analyzed so far (28). The turnover number for riboflavin of Lmo1329 is comparably low and explains why nonphosphorylated/adenylylated RF/RoF/AF were present in cells treated with these flavins.
The gene product of lmo0728 is a monofunctional FAD synthetase.
Lmo1329 was found to not synthesize RoFAD. Since RoFAD was present in RoF-treated cells, another enzyme had to be responsible for the formation of RoFAD, which prompted us to investigate Lmo0728 as the second putative flavokinase/FAD synthetase of L. monocytogenes (see preceding section). Lmo0728 was overproduced in E. coli as a His6-MBP-tagged protein and purified to apparent homogeneity. The tags were removed, and Lmo0728 was tested for flavokinase and FAD synthetase activity using different flavin substrates and ATP (Fig. 4C). The enzyme was found to not generate FMN, AFMN, or RoFMN from RF, AF, or RoF and ATP, showing that Lmo0728 is not a flavokinase. Lmo0728 was tested with FMN and ATP as substrates (Fig. 4D), and the formation of FAD showed that Lmo0728 is an FAD synthetase. In Table 2, the kinetic parameters for the FAD synthetase activity of Lmo0728 are compared to those for the FAD synthetase activity of Lmo1329. The apparent Km for FMN is lower in Lmo0728, indicating that this enzyme is able to generate FAD at lower concentrations of FMN. Lmo0728 is less active on the substrates AFMN and RoFMN (Fig. 4D), and the resulting “occupied” active site of Lmo0728 may contribute to the reduced FMN/FAD levels in AF/RoF-treated L. monocytogenes cells (Table 1).
The FMN riboswitch Rli96 is a target for FMN and RoFMN but not for AFMN in vivo.
Expression of the riboflavin transporter gene lmo1945 is directed by the promoter PribLm (29). The 5′ untranslated part of the monocistronic lmo1945-mRNA contains the FMN riboswitch Rli96, which controls expression of lmo1945 (17) from PribLm. Rli96 consists of an FMN-responsive aptamer and an overlapping expression platform (see Fig. S2 in the supplemental material). Its predicted secondary structure in the presence of FMN suggests that FMN binding to the aptamer portion leads to the formation of an intrinsic terminator and thus abolishes transcription of lmo1945 (transcriptional control). In contrast, when FMN levels are low, an alternative structure forms, which results in formation of a full-length transcript of lmo1945. To test the predicted function of Rli96 in vivo, a chromosomal PribLm-Rli96-lacZ transcriptional reporter gene fusion was constructed and inserted into the amyE locus of a specialized B. subtilis test strain (“Rli96”). To ensure that flavins added to the culture medium were completely taken up, this specialized test strain also overexpressed B. subtilis ribU from the plasmid pDG148. This is necessary since chromosomal ribU is under the control of the ribU FMN riboswitch (30), which shuts down flavin transport in the presence of high flavin levels. The specialized test strain Rli96 was challenged with riboflavin (50 μM and 100 μM), and PribLm-Rli96-driven β-galactosidase (LacZ) synthesis was monitored in cell extracts by measuring LacZ activity (Fig. 5). A decrease in specific LacZ activity by 25% upon addition of riboflavin (50 μM and 100 μM) indicated that physiological amounts of riboflavin reduced Rli96-controlled transcription. Rli96 apparently was less responsive to riboflavin treatment than the ribDG-FMN riboswitch from B. subtilis (31), which in a similar experimental setting (employing the B. subtilis promoter PribDG and a chromosomal PribDG-ribDG-FMN riboswitch-lacZ construct) showed a reduction by 66% upon treatment of the corresponding test strain RFNbs with riboflavin (Fig. 5). The strain containing the PribLm-Rli96-lacZ transcriptional fusion was, in addition, challenged with AF and RoF, and the data strongly suggest that AF, in contrast to RoF and riboflavin, does not reduce Rli96-controlled expression (Fig. 6). In contrast, AF positively affects Rli96, which might explain why more AFMN/AFAD is present in AF-treated cells (than in RoFMN/RoFAD in RoF-treated cells) (Table 1). Notably, the ribB FMN riboswitch of S. davawensis is also positively affected by RoF (7).
FIG 5.
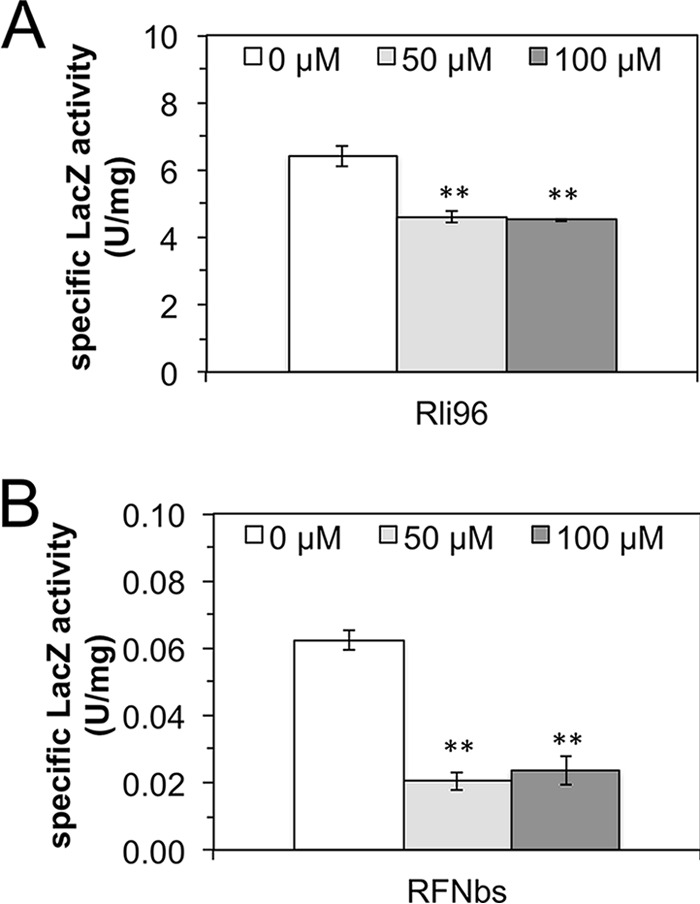
The FMN riboswitch Rli96 of Listeria monocytogenes in vivo is less responsive to FMN than the corresponding ribDG FMN riboswitch from Bacillus subtilis. Specific β-galactosidase (LacZ) activities in cell extracts of different B. subtilis test strains (Rli96 [A] and RFNbs[B]) treated with the indicated concentrations of riboflavin were determined. The strain Rli96 harbored a chromosomal PribLm-Rli96-lacZ transcriptional fusion. The L. monocytogenes promoter PribLm drives expression of the riboflavin transporter component lmo1945 and was found to also be active in B. subtilis. The strain RFNbs contained a chromosomal PribBs-ribDG-FMN transcriptional fusion. The B. subtilis promoter PribBs drives expression of the riboflavin biosynthetic genes ribDGEABHT (43), and the corresponding ribDG FMN riboswitch regulates this gene cluster by prematurely terminating transcription in the presence of high levels of FMN. Both strains expressed the B. subtilis riboflavin transporter gene ribU from the plasmid pDG148 to ensure constitutive riboflavin uptake. Upon addition of riboflavin to the cultures, LacZ activities decreased due to accumulating FMN (which was produced from riboflavin by the flavokinase RibC [26]). The decrease was more prominent in the case of RFNbs, indicating that the ribDG FMN riboswitch is more effective than Rli96. The white bar shows the LacZ activity of untreated controls. The data represent mean values from three independent experiments with the indicated standard deviations (**, P ≤ 0.01).
FIG 6.
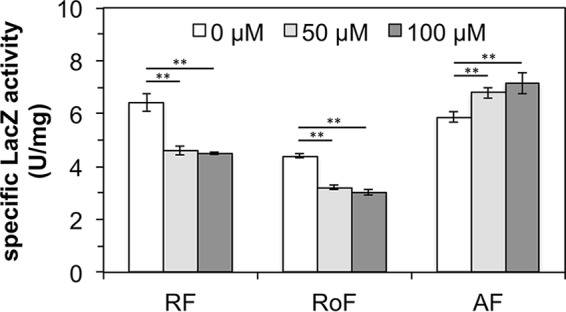
The FMN riboswitch Rli96 of Listeria monocytogenes is affected by riboflavin (RF) and roseoflavin (RoF) but not by 8-demethyl-8-aminoriboflavin (AF) in vivo. The specific β-galactosidase (LacZ) activities in cell extracts of the B. subtilis test strain Rli96 (Fig. 5) are shown upon treatment of the cells with RF, RoF, and AF. Upon addition of RF to the cultures, LacZ activities decreased. The addition of RoF to the cultures had a similar effect. The addition of AF enhanced Rli96-controlled gene expression. The data represent mean values from three independent experiments with the indicated standard deviations (**, P ≤ 0.01).
L. monocytogenes Rli96 is affected by FMN but not by AFMN in vitro.
To validate the in vivo gene expression results, in vitro transcription/translation assays employing the plasmid pT7luc (7) were performed. Rli96 was placed between the bacteriophage T7 promoter and a luciferase reporter gene (luc) to generate a transcriptional fusion (31). The plasmid was used as a template in T7 RNA polymerase-based in vitro transcription/translation assays, which were run in the presence of FMN, RoFMN, or AFMN. These cofactor analogs have been shown in this work to be produced within the cell upon treatment with riboflavin, RoF, or AF. FMN caused reductions in luciferase activity of 27% and 46%, depending on the amount of FMN (Fig. 7). RoFMN caused reductions in luciferase activity of 48% and 56%. AFMN had no effect on the activity of Rli96.
FIG 7.
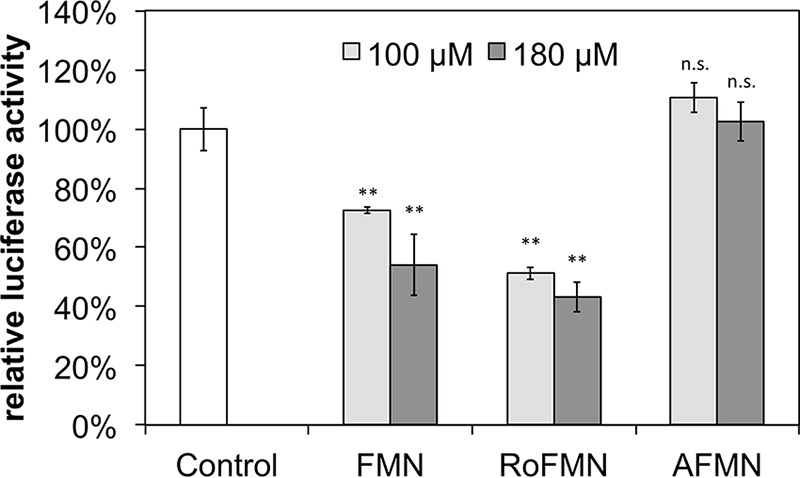
The FMN riboswitch Rli96 of Listeria monocytogenes is affected by RoFMN but not by AFMN in vitro. A reporter plasmid (pT7-Rli96-luc) producing a transcriptional fusion of Rli96 and the reporter gene luc (coding for firefly luciferase) were used as a DNA template for an in vitro transcription/translation assay driven by RNA polymerase of bacteriophage T7. The specific luciferase activities were determined, and the data were normalized to 100%. Mean values from three independent experiments are shown. The addition of FMN and RoFMN resulted in a reduction of Luc activity. The addition of AFMN, however, did not reduce luciferase activity. The data represent mean values from three independent experiments with the indicated standard deviations (**, P ≤ 0.01; n.s., not different from the control).
DISCUSSION
Treatment of L. monocytogenes infections is difficult since these bacteria thrive within the host cell (32). Toxic riboflavin analogs such as RoF and AF are readily taken up by all cells (also by human cells [33]) and therefore meet an important requirement for being antimicrobials effective against L. monocytogenes. Our results employing specialized B. subtilis test strains show that L. monocytogenes Lmo1945 mediates uptake of flavins and flavin analogs. Lmo1945 represents the membrane-embedded substrate-binding subunit (EcfS or RibU) of the ECF riboflavin uptake complex, which also contains the transmembrane coupling subunit EcfT and two ATP-binding cassettes EcfA and EcfA′. In our experiments Lmo1945 either facilitated riboflavin uptake alone or utilized the compatible ECF-components EcfT/EcfA/EcfA′ (34) of the B. subtilis test strain. Results of a L. monocytogenes transcriptome study (in BHI broth) revealed that full-length transcripts of lmo1945 are present in the exponential growth phase and also during stationary growth (29). Since no other putative riboflavin transporter was detected (2) and since L. monocytogenes is not able to synthesize riboflavin, we suggest that Lmo1945 represents an indispensable protein.
Lmo1329 and Lmo0728 are the only enzymes responsible for the metabolism of flavins in L. monocytogenes, and Lmo1329 is the only enzyme which synthesizes essential FMN. This is in line with the transcriptome data showing that lmo1329 is highly expressed under all conditions tested (29). According to a new nomenclature (35), we propose the name RibCF for the bifunctional flavokinase/FAD synthetase Lmo1329 whereby RibC represents the N-terminal FAD synthetase domain and RibF represents the C-terminal flavokinase domain. Lmo1329 (RibCF) produces AFMN, RoFMN, and AFAD but not RoFAD and thus is different from all other flavokinases/FAD synthetases analyzed so far (5, 18, 36). Lmo0728 (RibC) is an FAD synthetase and is responsible for the formation of toxic RoFAD in L. monocytogenes. Inspection of transcriptome data revealed that lmo0728 is expressed under all conditions tested (29), however, at a 5 times lower level than lmo1329. We were not able to generate lmo0728 deletion strains and therefore tentatively suggest that this gene is essential. FAD is the predominant flavin-derived cofactor in all cells (11), and we speculate that the physiological role of Lmo0728 (RibC) is to support Lmo1329 (RibCF) with regard to the synthesis of FAD. To our knowledge, this is the first experimental proof of a monofunctional FAD synthetase in bacteria, although the presence of such an enzyme has been proposed (37). The only other known bacterial monofunctional flavin-metabolizing enzyme is the regulator RibR from B. subtilis, which, in addition to being a regulator, is a flavokinase. In contrast to Lmo0728 (RibC), B. subtilis RibR has a very low turnover number and does not contribute to flavin cofactor synthesis (38). Lmo0728 (RibC) misses two highly conserved domains present in all flavokinases (39), which explains why Lmo0728 (RibC) is not able to synthesize FMN from riboflavin and ATP (see Fig. S5 in the supplemental material). Interestingly, Lmo0728 appears to be more closely related to the E. coli bifunctional flavokinase/FAD synthetase RibCF than to Lmo1329 (RibCF).
The very limited response of the L. monocytogenes FMN riboswitch Rli96 to riboflavin (in vivo) or FMN (in vitro) compared to that of the B. subtilis ribDG-FMN riboswitch indicates that L. monocytogenes has adapted to the relatively high and constant levels of riboflavin present in its environment and only reduces riboflavin uptake when there is a danger of accumulating too high (and possibly toxic) levels of riboflavin (40). Notably, the corresponding data were generated using B. subtilis (a close relative of L. monocytogenes), and we cannot rule out the possibility that Rli96 behaves differently in L. monocytogenes.
In light of our present and previous results, we suggest the following mechanism of action of RoF in L. monocytogenes. Lmo1945 is responsible for uptake of RoF, and the combined activities of Lmo0728 and Lmo1329 generate the cofactor analogs RoFMN and RoFAD. RoFMN reduces Rli96-controlled expression of lmo1945 (Fig. 7), and, as a result, less FMN/FAD is available for the formation of active flavoholoenzymes (Table 1). RoF competes with riboflavin for binding to Lmo1329, which further reduces FMN/FAD levels. In addition to FMN riboswitches and flavin-metabolizing enzymes, flavoproteins are likely targets for flavin analogs, and we propose that one (or several) of the 34 flavoproteins annotated for L. monocytogenes (28) is less active or completely inactive in the presence of either RoFMN or RoFAD as was reported for other bacterial enzymes (5, 6, 36). The identification of the main flavoprotein target(s) in L. monocytogenes might be rewarding (and is under way) since such a protein might be inhibited by a nonflavin compound and may constitute a completely novel bacterial drug target.
Our results with AF show a slightly different picture. Growth of L. monocytogenes was not inhibited at 100 μM AF (Fig. 1), although FMN/FAD levels were drastically reduced in cell extracts of AF-treated L. monocytogenes (as was the case for RoF-treated cells). Rli96-controlled expression of lmo1945 was neither affected in vivo nor in vitro by AF/AFMN. These findings might indicate that the lack of FMN and the reduced levels of FAD are not inhibitory to L. monocytogenes, which would be in line with the long-known fact that B. subtilis strains deficient in the bifunctional flavokinase/FAD synthetase RibC (residual activity, 1%) do not show any signs of growth reduction, at least under laboratory conditions (26). On the other hand, AFMN and AFAD are more hydrophilic than RoFMN/RoFAD and may not bind as tightly to flavoproteins (41, 42), which may explain why reduced FMN/FAD levels (in case of AF treatment) are tolerated by L. monocytogenes.
Riboflavin analogs and/or their degradation products have the potential to negatively interfere with human metabolism (18). The synthesis of RoFMN and AFMN by human hepatocyte cell extracts suggests that cofactor analogs are generated by humans in vivo. For d-amino acid oxidase (EC 1.4.3.3) from the mammal Sus scrofa, it was shown that RoFAD is an inactive cofactor (36), and we expect that human flavoproteins are also inactivated upon combining with RoFMN, RoFAD, AFMN, or AFAD.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the state of Baden-Württemberg (Germany) (Kooperatives Promotionskolleg Krankheitsmodelle und Wirkstoffe) and the research training group NANOKAT (FKZ 0316052A) of the German Federal Ministry of Education and Research (BMBF).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00388-16.
REFERENCES
- 1.Fischer M, Bacher A. 2005. Biosynthesis of flavocoenzymes. Nat Prod Rep 22:324–350. doi: 10.1039/b210142b. [DOI] [PubMed] [Google Scholar]
- 2.Gutiérrez-Preciado A, Torres AG, Merino E, Bonomi HR, Goldbaum FA, Garcia-Angulo VA. 2015. Extensive identification of bacterial riboflavin transporters and their distribution across bacterial species. PLoS One 10:e0126124. doi: 10.1371/journal.pone.0126124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eitinger T, Rodionov DA, Grote M, Schneider E. 2011. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol Rev 35:3–67. doi: 10.1111/j.1574-6976.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- 4.Karpowich NK, Wang DN. 2013. Assembly and mechanism of a group II ECF transporter. Proc Natl Acad Sci U S A 110:2534–2539. doi: 10.1073/pnas.1217361110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langer S, Hashimoto M, Hobl B, Mathes T, Mack M. 2013. Flavoproteins are potential targets for the antibiotic roseoflavin in Escherichia coli. J Bacteriol 195:4037–4045. doi: 10.1128/JB.00646-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langer S, Nakanishi S, Mathes T, Knaus T, Binter A, Macheroux P, Mase T, Miyakawa T, Tanokura M, Mack M. 2013. The flavoenzyme azobenzene reductase AzoR from Escherichia coli binds roseoflavin mononucleotide (RoFMN) with high affinity and is less active in its RoFMN form. Biochemistry 52:4288–4295. doi: 10.1021/bi400348d. [DOI] [PubMed] [Google Scholar]
- 7.Pedrolli DB, Matern A, Wang J, Ester M, Siedler K, Breaker R, Mack M. 2012. A highly specialized flavin mononucleotide riboswitch responds differently to similar ligands and confers roseoflavin resistance to Streptomyces davawensis. Nucleic Acids Res 40:8662–8673. doi: 10.1093/nar/gks616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kearney EB, Goldenberg J, Lipsick J, Perl M. 1979. Flavokinase and FAD synthetase from Bacillus subtilis specific for reduced flavins. J Biol Chem 254:9551–9557. [PubMed] [Google Scholar]
- 9.Mashhadi Z, Xu H, Grochowski LL, White RH. 2010. Archaeal RibL: a new FAD synthetase that is air sensitive. Biochemistry 49:8748–8755. doi: 10.1021/bi100817q. [DOI] [PubMed] [Google Scholar]
- 10.Fraaije MW, Mattevi A. 2000. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem Sci 25:126–132. doi: 10.1016/S0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 11.Macheroux P, Kappes B, Ealick SE. 2011. Flavogenomics—a genomic and structural view of flavin-dependent proteins. FEBS J 278:2625–2634. doi: 10.1111/j.1742-4658.2011.08202.x. [DOI] [PubMed] [Google Scholar]
- 12.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otani S, Takatsu M, Nakano M, Kasai S, Miura R. 1974. Lett: Roseoflavin, a new antimicrobial pigment from Streptomyces. J Antibiot (Tokyo) 27:88–89. doi: 10.7164/antibiotics.27.88. [DOI] [PubMed] [Google Scholar]
- 14.Schwarz J, Konjik V, Jankowitsch F, Sandhoff R, Mack M. 2016. Identification of the key enzyme of roseoflavin biosynthesis. Angew Chem Int Ed Engl 55:6103–6106. doi: 10.1002/anie.201600581. [DOI] [PubMed] [Google Scholar]
- 15.Jankowitsch F, Kuhm C, Kellner R, Kalinowski J, Pelzer S, Macheroux P, Mack M. 2011. A novel N,N-8-amino-8-demethyl-d-riboflavin dimethyltransferase (RosA) catalyzing the two terminal steps of roseoflavin biosynthesis in Streptomyces davawensis. J Biol Chem 286:38275–38285. doi: 10.1074/jbc.M111.292300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tongsook C, Uhl MK, Jankowitsch F, Mack M, Gruber K, Macheroux P. 2016. Structural and kinetic studies on RosA, the enzyme catalysing the methylation of 8-demethyl-8-amino-d-riboflavin to the antibiotic roseoflavin. FEBS J 283:1531−1549. doi: 10.1111/febs.13690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansjö M, Johansson J. 2011. The riboflavin analog roseoflavin targets an FMN-riboswitch and blocks Listeria monocytogenes growth, but also stimulates virulence gene-expression and infection. RNA Biol 8:674–680. doi: 10.4161/rna.8.4.15586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedrolli DB, Nakanishi S, Barile M, Mansurova M, Carmona EC, Lux A, Gartner W, Mack M. 2011. The antibiotics roseoflavin and 8-demethyl-8-amino-riboflavin from Streptomyces davawensis are metabolized by human flavokinase and human FAD synthetase. Biochem Pharmacol 82:1853–1859. doi: 10.1016/j.bcp.2011.08.029. [DOI] [PubMed] [Google Scholar]
- 19.Vogl C, Grill S, Schilling O, Stulke J, Mack M, Stolz J. 2007. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J Bacteriol 189:7367–7375. doi: 10.1128/JB.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goulas T, Cuppari A, Garcia-Castellanos R, Snipas S, Glockshuber R, Arolas JL, Gomis-Ruth FX. 2014. The pCri System: a vector collection for recombinant protein expression and purification. PLoS One 9:e112643. doi: 10.1371/journal.pone.0112643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ott E, Stolz J, Lehmann M, Mack M. 2009. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol 6:276–280. doi: 10.4161/rna.6.3.8342. [DOI] [PubMed] [Google Scholar]
- 22.Phan-Thanh L, Gormon T. 1997. A chemically defined minimal medium for the optimal culture of Listeria. Int J Food Microbiol 35:91–95. doi: 10.1016/S0168-1605(96)01205-6. [DOI] [PubMed] [Google Scholar]
- 23.Pedrolli DB, Mack M. 2014. Bacterial flavin mononucleotide riboswitches as targets for flavin analogs. Methods Mol Biol 1103:165–176. doi: 10.1007/978-1-62703-730-3_13. [DOI] [PubMed] [Google Scholar]
- 24.Löw C, Jegerschöld C, Kovermann M, Moberg P, Nordlund P. 2012. Optimisation of over-expression in E. coli and biophysical characterisation of human membrane protein synaptogyrin 1. PLoS One 7:e38244. doi: 10.1371/journal.pone.0038244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karpowich NK, Song JM, Cocco N, Wang DN. 2015. ATP binding drives substrate capture in an ECF transporter by a release-and-catch mechanism. Nat Struct Mol Biol 22:565–571. doi: 10.1038/nsmb.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mack M, van Loon AP, Hohmann HP. 1998. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the flavokinase/flavin adenine dinucleotide synthetase encoded by ribC. J Bacteriol 180:950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herguedas B, Martinez-Julvez M, Frago S, Medina M, Hermoso JA. 2010. Oligomeric state in the crystal structure of modular FAD synthetase provides insights into its sequential catalysis in prokaryotes. J Mol Biol 400:218–230. doi: 10.1016/j.jmb.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Barthelmes J, Ebeling C, Chang A, Schomburg I, Schomburg D. 2007. BRENDA, AMENDA and FRENDA: the enzyme information system in 2007. Nucleic Acids Res 35:D511−D514. doi: 10.1093/nar/gkl972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, Archambaud C, Cossart P, Sorek R. 2012. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol 8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedrolli D, Langer S, Hobl B, Schwarz J, Hashimoto M, Mack M. 2015. The ribB FMN riboswitch from Escherichia coli operates at the transcriptional and translational level and regulates riboflavin biosynthesis. FEBS J 282:3230−3242. doi: 10.1111/febs.13226. [DOI] [PubMed] [Google Scholar]
- 32.Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T. 2011. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res 39:4235–4248. doi: 10.1093/nar/gkr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abbas CA, Sibirny AA. 2011. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol Mol Biol Rev 75:321–360. doi: 10.1128/MMBR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodionov DA, Hebbeln P, Eudes A, ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD, Eitinger T. 2009. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jankowitsch F, Schwarz J, Ruckert C, Gust B, Szczepanowski R, Blom J, Pelzer S, Kalinowski J, Mack M. 2012. Genome sequence of the bacterium Streptomyces davawensis JCM 4913 and heterologous production of the unique antibiotic roseoflavin. J Bacteriol 194:6818–6827. doi: 10.1128/JB.01592-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grill S, Busenbender S, Pfeiffer M, Kohler U, Mack M. 2008. The bifunctional flavokinase/flavin adenine dinucleotide synthetase from Streptomyces davawensis produces inactive flavin cofactors and is not involved in resistance to the antibiotic roseoflavin. J Bacteriol 190:1546–1553. doi: 10.1128/JB.01586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yruela I, Arilla-Luna S, Medina M, Contreras-Moreira B. 2010. Evolutionary divergence of chloroplast FAD synthetase proteins. BMC Evol Biol 10:311. doi: 10.1186/1471-2148-10-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pedrolli DB, Kuhm C, Sevin DC, Vockenhuber MP, Sauer U, Suess B, Mack M. 2015. A dual control mechanism synchronizes riboflavin and sulphur metabolism in Bacillus subtilis. Proc Natl Acad Sci U S A 112:14054–14059. doi: 10.1073/pnas.1515024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herguedas B, Lans I, Sebastian M, Hermoso JA, Martinez-Julvez M, Medina M. 2015. Structural insights into the synthesis of FMN in prokaryotic organisms. Acta Crystallogr D Biol Crystallogr 71:2526–2542. doi: 10.1107/S1399004715019641. [DOI] [PubMed] [Google Scholar]
- 40.Akompong T, Ghori N, Haldar K. 2000. In vitro activity of riboflavin against the human malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother 44:88–96. doi: 10.1128/AAC.44.1.88-96.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedrolli DB, Jankowitsch F, Schwarz J, Langer S, Nakanishi S, Frei E, Mack M. 2013. Riboflavin analogs as antiinfectives: occurrence, mode of action, metabolism and resistance. Curr Pharm Des 19:2552–2560. doi: 10.2174/1381612811319140006. [DOI] [PubMed] [Google Scholar]
- 42.Pedrolli DB, Jankowitsch F, Schwarz J, Langer S, Nakanishi S, Mack M. 2014. Natural riboflavin analogs. Methods Mol Biol 1146:41–63. doi: 10.1007/978-1-4939-0452-5_3. [DOI] [PubMed] [Google Scholar]
- 43.Skliarova SA, Kreneva RA, Perumov DA, Mironov AS. 2012. The characterization of internal promoters in the Bacillus subtilis riboflavin biosynthesis operon. Genetika 48:1133–1141. (In Russian.) [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.