ABSTRACT
The transcription factor Pax7 plays a key role during embryonic myogenesis and in adult organisms in that it sustains the proper function of satellite cells, which serve as adult skeletal muscle stem cells. Recently we have shown that lack of Pax7 does not prevent the myogenic differentiation of pluripotent stem cells. In the current work we show that the absence of functional Pax7 in differentiating embryonic stem cells modulates cell cycle facilitating their proliferation. Surprisingly, deregulation of Pax7 function also positively impacts at the proliferation of mouse embryonic fibroblasts. Such phenotypes seem to be executed by modulating the expression of positive cell cycle regulators, such as cyclin E.
KEYWORDS: Cell cycle, differentiation, ESCs, MEFs, mouse, Pax7
Introduction
Embryonic Stem Cells (ESCs) are pluripotent cells that when cultured under appropriate conditions retain the ability to self-renew their population as well as to differentiate into any given tissue. This characteristics makes them a perfect tool in developmental biology and regenerative medicine studies. Developmental biology takes advantage of the methods of ESC genetic modifications and their in vitro as well as in vivo differentiation. Importantly, analyzes of differentiating ESCs are the prerequisite step for the development of therapies involving stem cells which could be used as a treatment for many degenerative diseases, such as muscular dystrophies. In this context the role of crucial regulators of differentiation as well as impact of the culture conditions are essential issues in the studies involving ESCs.1,2
Many studies focusing at the mechanisms of ESC myogenic differentiation took advantage of genetically modified ESCs, such as those lacking functional genes encoding myogenic regulatory factors (MRFs), e.g. myogenin,3 or structural proteins, e.g., desmin.4 Such approach allowed to prove that these genes are essential for myogenic differentiation of ESCs. Our own study showed that myogenic differentiation of ESCs can occur without functional Pax7 gene,5 i.e. crucial regulator of both embryonic myogenesis and maintenance of satellite cells in adult skeletal muscles.6 In the same study we showed that differentiation of ESCs lacking functional Pax7 resulted in the higher number of myoblasts, as compared to wild-type cells. Our observation suggested that in differentiating ESCs Pax7 acts as a cell cycle regulator.
In adult organisms Pax7 is involved in the regulation of the balance between self-renewal and differentiation of the activated satellite cells.7 It is expressed in proliferating myoblasts and downregulated when they differentiate into myotubes.8 Overexpression of Pax7 increases the proliferation of in vitro cultured myoblasts.9 However, other data documented that overexpression of Pax7 in MM14 myoblasts inhibits the cell cycle.10 Pax7 was shown to induce the expression of genes such as Inhibitor of differentiation 3 (Id3),11 known to prevent MyoD expression and to inhibit apoptosis in quiescent satellite cells.10,12 Myoblasts lacking MyoD, similarly to quiescent satellite cells, express anti-apoptotic factors, such as Bcl-2 and Bcl-xl.13 Interestingly, expression of dominant negative form of Pax7 resulted in the increased proportion of myoblasts in S phase. However, at the same time the number of cells per colony of in vitro cultured primary myoblasts decreased suggesting that in the absence of Pax7 G1 cells are lost most probably via apoptosis.14 Importantly, in the absence of functional Pax7 gene the number of satellite cells decreases dramatically after birth in mouse muscles.14,15 Taking together, the influence of Pax7 on the regulation of proliferation and apoptosis of satellite cells and myoblasts is unquestionable. However, its participation in the cell cycle regulation is still less understood when compared to such myogenic regulators like for example MyoD.
MyoD was shown to induce expression of cell cycle suppressor gene encoding pRb protein.16 Active form of pRb results in the dissociation of MyoD from histone deacetylase Hdac-1 what induces expression of its target genes,17 such as the one encoding cell cycle inhibitor p21cip1.18 Interestingly, MyoD acting together with pRb decreases expression of cyclin D1, another positive cell cycle regulator, preventing cell proliferation.19 Myogenic differentiation is also associated with the increase in the levels of other cell cycle inhibitors – p27cip2 and p57kip2 20 (for the review see ref.21).
The role of Pax7 in ESCs was studied by silencing its expression using siRNA what led to the decrease in the levels of mRNAs encoding MyoD, Myf-5, and desmin.22 However, in differentiating ESCs lacking functional Pax7 gene expression of these and other factors, e.g. Pax3, M-cadherin or MyHC, was not affected.5 Interestingly, in these mutant cells the levels of microRNAs, such as miR-133a was modified, suggesting that the regulation of ESC proliferation and/or differentiation may occur at the posttranscriptional level. Importantly, ESCs lacking Pax7 were able to turn into myoblasts and initiate myotube formation in EB outgrowths.5 These observations were consistent with the data showing that mice lacking functional Pax7 do form skeletal muscles, although, of lower mass and containing limited number of satellite cells.8,23 However, the role of Pax7 in the regulation of proliferation and apoptosis of ESCs induced to undergo myogenic differentiation in vitro was not studied. For this reason, we took advantage of cells in that function of Pax7 was ablated. Since cell cycle machinery is specifically adjusted in ESCs (for the review see ref.21,24) we also investigated the influence of Pax7 ablation on cell cycle in well-characterized, “standard” cells, i.e., mouse embryonic fibroblasts (MEFs). By doing so we were able to compare the impact of the Pax7 at the proliferation and apoptosis of differentiating stem cells as well as, non-myogenic cells, non stem cells, i.e. fibroblasts. The role of Pax7 in MEFs has not been studied and documented so far.
Results
Transcription profiling of cells lacking functional Pax7
ESCs lacking functional Pax7 (Pax7ko) are able to undergo myogenic differentiation, i.e., form myoblasts as well as initiate formation of myotubes, similarly to control, i.e. Pax7wt cells.5 In our previous study to induce ESC differentiation we generated embryoid bodies (EBs), i.e., tridimensional structures recapitulating early stages of embryonic development.25,26 By using this technique, we showed that cultures of Pax7ko ESCs resulted in the higher number of myoblasts as compared to control ones. To trace the background of this phenomenon we analyzed the expression of genes regulating myogenic differentiation. Microarray analysis comparing Pax7wt and Pax7ko ESCs did not reveal any significant differences in the levels of mRNAs encoding myogenic factors, such as MyoD, Myf5 or myogenin. This comparison, however, revealed many differentially expressed genes.5 Among them were transcripts encoding cell cycle regulators which upregulation could result in the generation of higher numbers of muscle progenitor cells, and thus, higher number of myoblasts. For this reason, we focused at the cell cycle progression in differentiating ESCs. In addition, we decided to assess the function of Pax7 in mouse embryonic fibroblasts (MEFs) serving as an example of non-myogenic and non-stem cells, in that the function of Pax7 was not reported so far. However, this protein was shown by us to be present in asynchronously proliferating wild-type MEFs (Fig. 1A and B). To our surprise performed analyzes revealed Pax7-related phenotype in case of both, ESCs and also MEFs.
Figure 1.
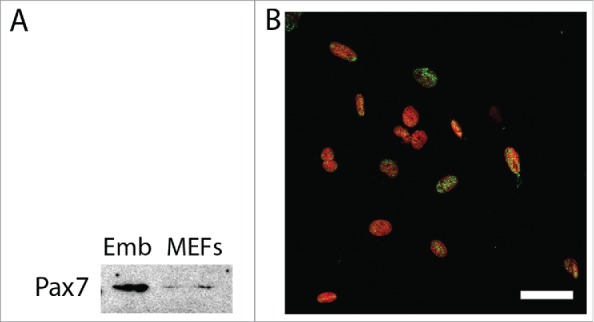
Pax7 in mouse embryonic fibroblasts. (A). Western blotting analysis of Pax7 in whole 13.5-day-old embryo (Emb) and asynchronously dividing MEFs; (B). Localization of Pax7 (green) and nuclei (red) in MEFs; bar 50 µm.
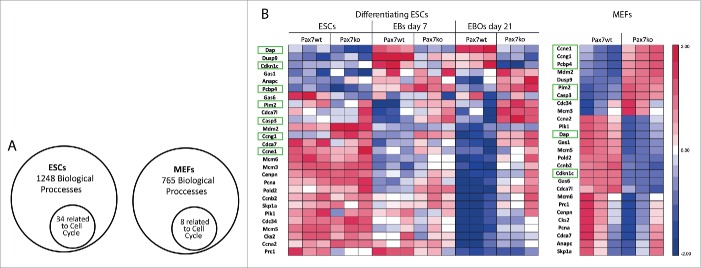
First, we focused at the global gene expression profiles of undifferentiated ESCs, EBs at day 7 of culture, and EB outgrowths (EBOs) at day 21 of culture (Fig. 2), as well as in asynchronously proliferating MEFs. For each cell type we analyzed one Pax7wt as well as one Pax7ko line and compared RNA samples obtained from 3 independent experiments. Performed comparisons revealed many differences in ESC (database access available in NCBI's Gene Expression Omnibus GSE66483) and MEF transcriptomes (database access available in NCBI's Gene Expression Omnibus GSE80658). As mentioned above, among the genes which expression differed between cells expressing and lacking functional Pax7 we identified ones encoding proteins controlling cell cycle. Gene Ontology analysis of significantly expressed genes showed that in differentiating ESCs out of 1248 biological processes 34 were related to the cell cycle (Fig. 3A, Table S1). The same analysis done for asynchronously dividing MEFs revealed that 8 processes out of 765 that were singled out were related to the cell cycle (Fig. 3A, Table S2). Enrichment score included into the supplementary tables represents the level of significance, i.e. the higher the enrichment score, the more given GO functional group is over-represented in the gene list.
Figure 2.
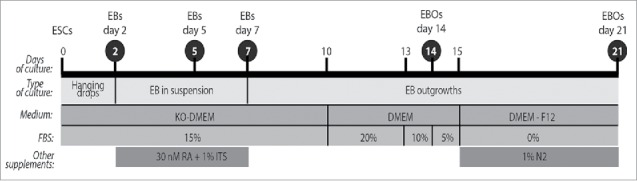
Schematic diagram of the protocol used to induce the differentiation of ESCs (see Materials and Methods); EBs – embryoid bodies, EBOs – embryoid body outgrowths; time points when samples were collected for analysis are indicated in black circles.
Figure 3.
Analysis of expression of genes involved in regulation of cell cycle in both Pax7ko and Pax7wt ESCs and MEFs. (A). Gene Ontology analysis of significantly expressed genes in differentiating ESCs and asynchronously proliferating MEFs showing proportion of cell cycle related processes among all others. List of the processes is presented in Tables S1 and S2; (B). Microarray analysis of the expression levels of transcripts that encoded cell cycle-associated proteins in undifferentiated ESCs and in EBs at day 7 and EBOs at day 21 of differentiation (left diagram) and asynchronously proliferating MEFs (right diagram). For each cell type and genotype one cell line was analyzed. Each group included triplicate measurements. Blue color indicates low expression and red color indicates high expression levels of transcripts. Genes which levels change similarly in differentiating ESCs and MEFs were marked in green. All genes shown were selected from the database complete list presented in Czerwinska et al.5 and Table S3.
In pluripotent Pax7wt and Pax7ko ESCs, as well as in cells analyzed at day 7 of differentiation, the levels of the majority of the cell cycle related transcripts did not significantly differ. However, some of the genes were upregulated in Pax7ko cells. These were Ccng1 encoding cyclin G1, which is a direct target of p53, also involved in the regulation of differentiation. The most profound differences, however, were observed at day 21 of differentiation, i.e., when the cells cease to proliferate (Fig. 3B). At this time point Pax7ko cells were characterized by lower levels of such transcripts as Cdkn1c encoding Cdk inhibitor – p57kip2, Dusp9 encoding phosphatase inactivating MAP kinase and impacting proliferation, and Dap (Death associated protein) which acts as a positive mediator of cell death.27 Levels of several mRNAs encoding positive regulators of cell cycle were higher in Pax7ko ESCs. Among them were Polo-like kinase (Plk1), cyclins E1 (Ccne1), A2 (Ccna2), B2 (Ccnb2), proteins of Mcm (minichromosome maintenance deficient) family, subunit of DNA polimerase delta (Pold2), and subunit of anaphase promoting complex (Anapc1) (Fig. 3B).
Similar analysis carried out for asynchronously proliferating MEFs, which we chosen as control non-myogenic cells, delivered surprising results. During embryonic development Pax7 was never showed to be involved in the function of cells other than neural and myogenic ones. In adults, except satellite cells, it was also detected in testes.28,29 Microarray analyses performed by us showed that in MEFs lacking functional Pax7 the levels of many transcripts differed as compared to Pax7wt cells. Thus, Pax7ko MEFs showed, similarly to Pax7ko ESCs, lower levels of Cdkn1c and Dap transcripts (Fig. 3B). Next, the levels of mRNAs encoding Ccng1 and Ccne1 were higher in Pax7ko MEFs, again similarly to ESCs of the same genotype (21 day of culture). Such data suggested increased proliferation ability of Pax7ko cells. However, at the same time we noticed that in the absence of functional Pax7 both ESCs and MEFs upregulated Casp3 and Pcbp4, i.e. the genes encoding factors involved in the execution of apoptosis. Selected transcripts were further analyzed using RT-qPCR in two Pax7wt and two Pax7ko ESC lines as well as in three Pax7wt and three Pax7ko MEF lines.
The levels of mRNAs encoding Ccnd1, Ccne1, Ccna2, Ccnb2, Cdkn1a, Cdkn1b, Cdkn1c, as well as Plk1, Cdc34, and Mcm5 were compared in non-differentiated ESCs, EBs at day 2, 7, as well as EB outgrowths at day 21 of culture (Fig. 4A). In undifferentiated ESCs only the expression of Mcm5 differed between Pax7wt and Pax7ko cells – it was significantly higher in mutant cells. Next, Pax7ko ESCs expressed significantly higher level of mRNA encoding cyclin E1 (Ccne1), as compared to wild-type control, but only at day 2 of differentiation. Western blot analyzes confirmed that level of cyclin E was the highest in EBs at day 2 of differentiation, then dropped at day 5 and 14. In fact, at 14 day of differentiation it was absent in Pax7wt cells while still detectable in Pax7ko ones. However, the differences between Pax7wt and Pax7ko cells were rather subtle (Fig. 4B). The levels of the other genes analyzed by RT-qPCR did not significantly differ between the cells of different genotypes.
Figure 4.
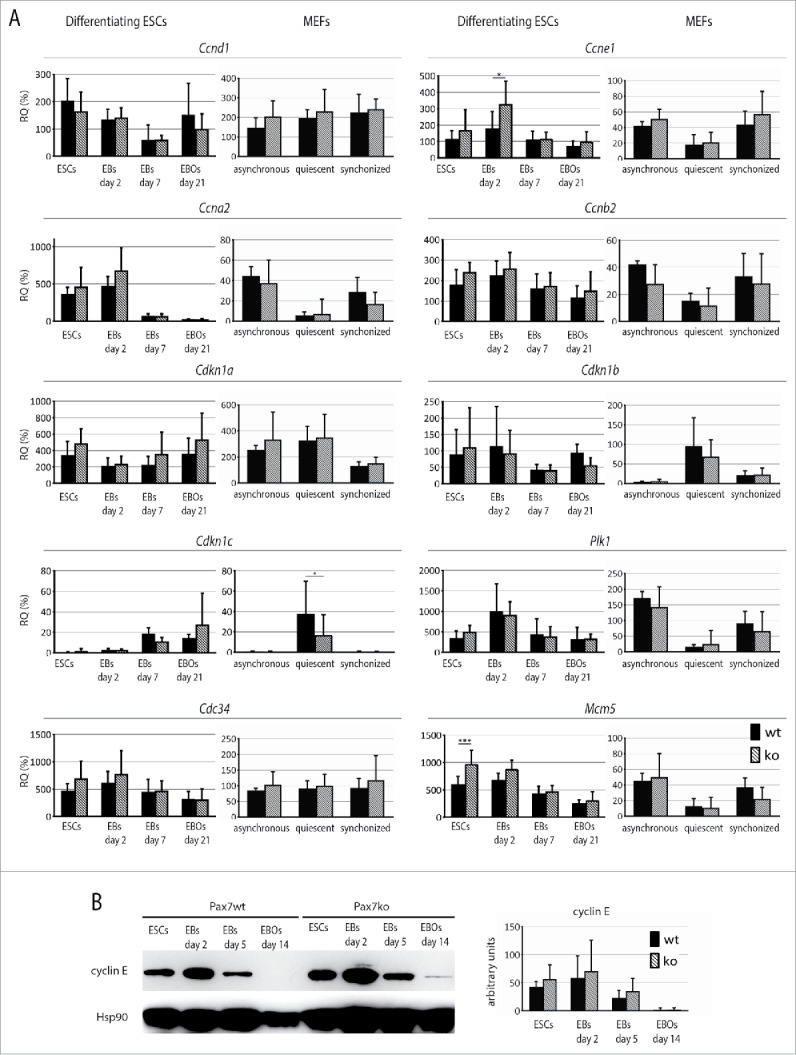
Analysis of factors involved in regulation of cell cycle in both Pax7ko and Pax7wt ESCs and MEFs. (A). RT-qPCR analysis of Ccnd1, Ccne1, Ccna2, Ccnb2, Cdkn1a, Cdkn1b, Cdkn1c, Plk1, Cdc34, and Mcm5 transcript levels in undifferentiated ESCs, EBs at day 2, 7 and EB outgrowths at day 21 of differentiation, as well as asynchronously dividing, quiescent and synchronously cycling MEFs. Data are shown as CT values, which were normalized against those of Actb mRNA level. Data are represented as the percentage of expression observed in mouse embryos at day 13.5 of development. For each genotype 2 ESC and 3 MEF lines were analyzed, graphs represent the mean values. P-values: *<0.05, ** <0.01, *** <0.001. (B). Western blotting analysis of cyclin E1 levels in ESCs, EBs at day 2 and 5, and EB outgrowths at day 14 of differentiation. Probing with anti-Hsp90 antibody was used as a loading control. Graph represents optical density of cyclin E bands compared to density of Hsp90 bands (optical density of Hsp90 was taken as 100 arbitrary units).
Next, we analyzed 3 populations of MEFs: asynchronously dividing, serum starved, i.e., quiescent cells, as well as synchronized by serum-starvation proliferating ones. By doing so we hoped to highlight the differences detectable in microarray analyzes of transcriptomes of asynchronously dividing cells. Synchronously dividing Pax7ko MEFs were characterized by higher levels of Ccne1 and Mcm5 transcripts, what resembled the phenotype of ESCs of the same genotype (Fig. 4A and B). Importantly, the expression of genes encoding Cdk inhibitor, i.e. Cdk1c was lower in serum-starved Pax7ko MEFs, as compared to Pax7wt ones.
Proliferation of cells lacking functional Pax7
mRNA analyses showed that Pax7ko ESCs express higher levels of G1 phase cyclins and lower levels of Cdk inhibitors. Thus, we examined the proliferation and cell cycle profiles of ESCs and MEFs (3 cell lines for each genotype). We analyzed non-differentiating ESCs and EBs at day 2 and 5, as well as EB outgrowths at day 14 of culture. Proportion of undifferentiated ESCs expressing Ki-67, i.e., marker of cycling cells,30 was the same in case of Pax7wt and Pax7ko ESC lines, i.e. reached 93% (Fig. 5A and B). Interestingly, EBs generated from Pax7ko ESCs were larger than those obtained from Pax7wt ESCs, both at day 2 and 5 of culture. By disaggregating EBs and counting cells we showed that Pax7ko EBs were composed of more cells than Pax7wt EBs (Fig. 5C). At day 5 of culture the difference was significant (p = 0.0297). On the other hand, at day 14 of culture number of proliferating cells, i.e., expressing Ki-67, was significantly higher (p < 0.0001) in Pax7wt, as compared to Pax7ko EB outgrowths (24% vs. 16%, Fig. 5A and B). This phenomenon most probably reflects the fact that the Pax7ko cells more readily start to differentiate as compared to wild-type ones.5
Figure 5.
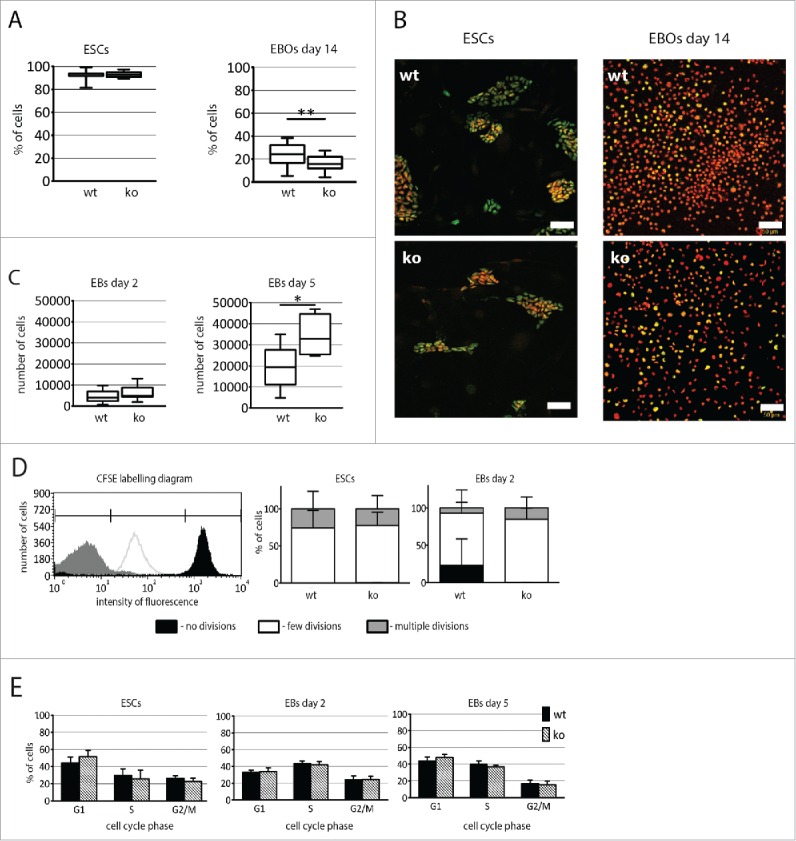
Analysis of proliferation of both Pax7ko and Pax7wt ESCs. (A). The proportion of cells (median values) expressing Ki-67 in ESCs colonies (left diagram) and in EB outgrowths at 14 day of differentiation (right diagram). P-value: **<0.01. (B). Localization of Ki-67 (green) and nuclei (red) in ESCs (left panel) and EB outgrowths at 14 day (right panel); bar 50 µm. (C). The number of cells (median values) per EB at 2 (left diagram) and 5 (right diagram) days of culture. P-value: * <0.05. (D). The proportion of ESCs and cells building EBs retaining fluorescence 48 h after pulse labeling with CFSE. Schematic diagram (left panel) showing the intensity of fluorescence of cells that did not divide (black), underwent few divisions (white), and cells that were nontreated with CFSE and represent ones that divided vigorously, underwent multiple divisions and lost CFSE labeling (gray). Proportion of ESCs and cells building EBs characterized by various fluorescence intensity (right panel). (E). The proportion of cells in G1, S, and G2/M phase of cell cycle among undifferentiated ESCs and cells in EBs at 2 and 5 d of culture. For each genotype 3 cell lines were analyzed, graphs represent the mean values.
Assessment of the proliferation dynamics was done using CFSE labeling method which relies on the pulse labeling of cells with fluorescent dye, i.e. CFSE, followed by FACS analyzes after further in vitro culture. Since each cell division leads to the dilution of incorporated CFSE proliferating cells are characterized by the decreasing fluorescence intensity. Thus, cells that did not divide appear as highly fluorescent (Fig. 5D, marked in black), those that underwent few divisions are characterized by decreased fluorescence (Fig. 5D, marked in white), and finally in those intensively dividing fluorescence is undetectable (Fig. 5D, marked in gray). Using such method we showed that pluripotent Pax7wt and Pax7ko ESCs did not differ in their proliferation rate during 48 h of culture. However, Pax7ko cells building EBs divided more vigorously, as compared to Pax7wt ones (Fig. 5D).
Analysis of cell cycle profile with propidium iodide showed that the highest proportion of cells in S-phase was observed at day 2 of differentiation. At day 5 of differentiation the proportion of Pax7wt cells in G1 increased and the population of cells in G2/M dropped. Importantly, the proportion of cells in various cell cycle phases did not differ between Pax7wt and Pax7ko ESCs at all analyzed stages of differentiation (Fig. 5E).
Analysis of CFSE labeling revealed that population of asynchronously dividing Pax7ko MEFs contained more dividing cells (Fig. 6A). After 72 h of culture following CFSE pulse fewer Pax7ko MEFs did not divide, as compared to Pax7wt ones. To more precisely follow the cell cycle progression we analyzed serum starved and synchronously dividing MEFs. Proportion of Ki-67 expressing, i.e., cycling cells, in population of serum-starved MEFs was higher in the population of Pax7wt MEFs, as compared to Pax7ko ones (30% vs. 21%). However, this difference was not statistically significant due to high deviation of obtained results (Fig. 6B). Interestingly, analysis of synchronously proliferating MEFs (i.e. 18 h after serum stimulation) revealed that significantly more Pax7ko cells (p = 0.0127) reentered the cell cycle and expressed Ki-67, as compared to Pax7wt ones (55% vs. 44%) (Fig. 6 B and C). Proportion of G1, S, and G2/M cells among serum-starved MEFs did not differ between Pax7wt and Pax7ko cell lines (Fig. 6D). Serum re-activation increased the proportion of MEFs in S-phase to 9% and 11% in Pax7wt and Pax7ko MEF populations, respectively. Interestingly, the proportion of cells in G1 was significantly (p < 0.0001) lower in the population of Pax7ko than in Pax7wt MEFs (32% vs. 45%). Consequently, the proportion of G2/M cells was significantly higher (p < 0.0001) in case of Pax7ko as compared to Pax7wt MEFs (57% vs. 46%) (Fig. 6D). High fraction of G1 and low fraction of S phase suggested that these cells were in the process of cell cycle re-entry.
Figure 6.
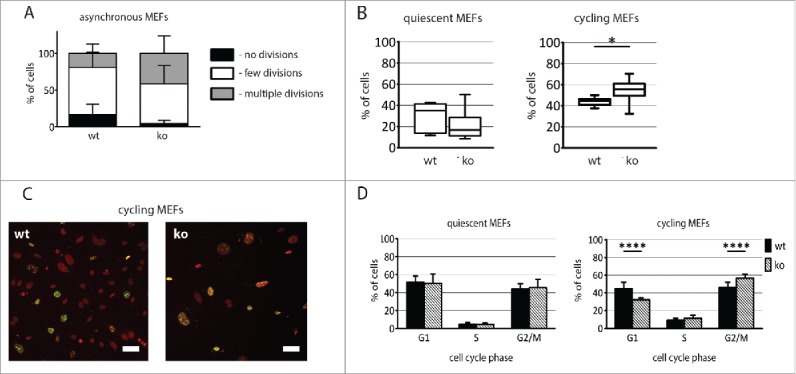
Analysis of proliferation of both Pax7ko and Pax7wt MEFs. (A). Proportion of MEFs characterized by various CFSE fluorescence intensity. (B). The proportion of cells (median values) expressing Ki-67 in quiescent (left diagram) and cycling (right diagram) MEFs. P-value: *<0.05. (C). Localization of Ki-67 (green) and nuclei (red) in cycling MEFs; bar 50 µm. (D). The proportion of cells in G1, S, and G2/M phase of cell cycle among quiescent and cycling MEFs. For each genotype 3 cell lines were analyzed, graphs represent the mean values. P-value: **** <0.0001.
Apoptosis of cells lacking functional Pax7
To evaluate apoptosis rate of ESCs and MEFs lacking functional Pax7 we established the proportion of the cells expressing phosphorylated isoform of histone H2A.X (γ-H2A.X), i.e., the marker of apoptotic cells.31 Again we analyzed 3 lines of ESCs and MEFs for each genotype.
We did not evaluate the proportion of cells expressing γ-H2A.X in undifferentiated ESCs as this isoform is abundant in such cells and its presence does not correlate with apoptosis.32,33 Interestingly, at 14 day of differentiation the fraction of apoptotic cells was significantly higher (p = 0.0004) in Pax7wt EB outgrowths, as compared to Pax7ko ones (11% vs. 7%, Fig. 7A and B). The ratio of apoptotic MEFs was similar in both Pax7wt and Pax7ko serum-starved MEFs, reaching 14% (Fig. 7C and D) and did not change after serum activation, i.e. 14% of Pax7wt and 18% of Pax7ko MEFs expressed γ-H2A.X (Fig. 7C and D).
Figure 7.
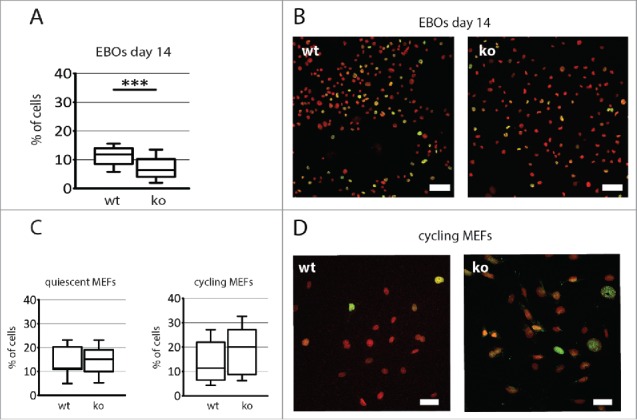
Analysis of apoptosis rate of both Pax7ko and Pax7wt ESCs and MEFs. (A). The proportion of cells expressing γ-H2A.X in EB outgrowths at 14 day of differentiation. P-value: ***<0.001. (B). Localization of γ-H2A.X (green) and nuclei (red) in EB outgrowths at 14 day of in vitro culture; bar 50 µm. (C). The proportion of cells expressing γ-H2A.X in quiescent (left diagram) and cycling (right diagram) MEFs. (D). Localization of γ-H2A.X (green) and nuclei (red) in cycling MEFs; bar 50 µm. For each cell type and genotype 3 cell lines were analyzed, graphs represent the median values.
Discussion
Cell cycle progression is regulated by the universal factors that operate in every dividing cell, regardless of its type and origin (for the review see refs.21,34,35). However, “standard” cell cycle machinery is custom-tuned in many types of cells, such as pluripotent, differentiating or aging ones. Among the cells which mechanisms regulating proliferation are not “custom-modified” are mouse embryonic fibroblasts which for this reason are used in numerous studies focusing on the cell cycle regulation. On the other hand, embryonic stem cells are the example of unique cells which cell cycle regulation is specially adjusted (for the review see ref.24). These cells are characterized by the ability to self-renew and by rapid cell cycles during which function of pRb-Cdk/cyclin D/p16 axis is restricted.36,37 Expression of cyclins D is low and Cdk4 activity is either faint, or if detectable, as in case of Cdk4-cyclin D3 complex, resistant to p16ink4 inhibition.36,38,39 Such restrictions result in the constitutive activation of E2F transcription factors and, as a result, in constant expression of its target genes, such as cyclin E.40 Among the factors which follow “standard” cell cycle pattern are for example cyclin A,41 cyclin B1 or Cdk1.40 Induction of ESC differentiation leads to the resurrection of canonical mechanisms, i.e., increase in the cyclins D levels, Cdk4 activity, and in consequence oscillatory regulation of pRb and E2F activity.42,43 As differentiation progresses it is also accompanied by other changes, such as rise in the levels of Cdk inhibitors, i.e. p21cip1, p27cip2, and p57kip2, leading to the quiescence in case of terminally differentiated cells.
Among the factors involved in cell cycle regulation of differentiating cells are the ones involved in the determination of cell fate. For example, MyoD is well known interactor of pRb making a bridge between cell cycle machinery and myogenic regulators (for review see ref.21). During embryonic myogenesis Pax3 and Pax7 transcription factors play a crucial role in the specification of skeletal muscle precursor cells and also impact at the cell cycle progression. Downregulation of Pax3 in the dermomyotome of chick embryos caused cell cycle arrest of proliferating muscle precursor cells.44 Similarly, lack of Pax3 led to the drop in the number of precursor cells in dermomyotome of mouse embryo.45,46 Pax3 activity is also crucial for the survival of precursor cells migrating from the dermomyotome to the limb buds.46-48 Function of Pax7 in the cell cycle regulation is less obvious, however, chromosomal translocation and fusion of DNA binding domain of either Pax3 or Pax7 genes with transcriptional activation domain of FOXO1a (FKHR) leads to the development of rhabdomyosarcomas.49 Overexpression of Pax3 or Pax7 in in vitro cultured myoblasts increased their proliferation.9 In vitro cultured Pax7-null myoblasts were characterized by decrease in proliferation and overexpression of dominant negative Pax7 significantly increased apoptosis in these cells.14 Next, mice expressing nonfunctional Pax7 developed skeletal muscles, though muscle mass and diameter of myofibers were smaller in comparison to wild-type littermates.8,23 Also in vitro cultured myoblasts lacking functional Pax7 gene form fewer and smaller myotubes.15 What is important, Pax7ko mice die within 2 weeks after birth and during this period the number of satellite cells found in their muscles diminishes rapidly.15,50
Identification of Pax7 transcription factor targets in E16 mouse embryos revealed several genes involved in the regulation of proliferation.51 Among them were genes positively impacting cell proliferation, such as CntfR, receptor of CNTF – a member of IL-6 family, which induces Jak/STAT and Ras/MAPK signaling,52 Camk1d involved in calcium-calmodulin dependent signaling,53 as well as Frk which also regulates cell cycle progression.54 None of these genes was identified in our comparison of differentiating Pax7wt and Pax7ko ESCs or proliferating MEFs. Microarray analyzes focusing at the differentiating ESCs lacking functional Pax7 revealed, however, that they are characterized by upregulated expression of several genes involved in cell cycle regulation, e.g., cell cycle related signaling pathways (Cdca7, Pten, Dusp4, Plk1), G1 and S phase progression (Ccne1, Mcm5, Tfdp1, Anp32b), cell cycle-dependent protein degradation (Skp1a, Huwe1, Cdc34, Trim37, Anapc1), cell division (Ccnb2, Ckap2), or genes involved in the regulation of apoptotic cell death (Casp3, Ddit4, Trp53), and decreased levels of mRNAs encoding apoptosis related genes Tnfsrf21, Dap. Further, RT-qPCR analyzes confirmed that in Pax7ko ESCs Ccne1 gene, encoding cyclin E1 that with Cdk2 regulates G1/S phase transition, was upregulated, but only at the initial stages of in vitro differentiation. Both, cyclin E mRNA and protein decreased at more advanced stages of EBs and EB outgrowths culture. Differences in the levels of other mRNAs analyzed using RT-qPCR technique were noticeable but not statistically significant. Nevertheless, these subtle changes seems to impact at the proliferation of ESCs since in EBs formed from Pax7ko cells we found more cells than in the control ones. At day 5 of culture this difference was significant, suggesting that either more of these cells were cycling or cell cycles were shorter. Next, at day 14 of culture, i.e., when the differentiation is more advanced, Pax7ko EB outgrowths contained less cycling cells, but most probably still expressing higher levels of proliferation-promoting genes, as compared to wild-type control. This may suggest that in the absence of functional Pax7 ESCs progress to the “differentiation state” more readily. Interestingly, at this stage Pax7ko EB outgrowths were also characterized by the lower number of cells undergoing apoptosis what may secure the higher number of myogenic cells that we observed previously.5 This observation is, however, not in line with the already available data showing that muscles of mice lacking functional Pax7 contained apoptotic cells whereas those of wild-type animals did not.14 Notion that satellite cells die in the absence of Pax7 was further supported by the observation that expression of dominant negative Pax7 gene leads to cell death in high proportion of cells (while expression of dominant negative Pax3 has not such effect).14 Thus, it is possible that during ESCs differentiation lack of functional Pax7 does not cause the increase in the apoptosis. Pax7ko cells may execute apoptosis as effectively as wild-type ones but also prematurely exit the cell cycle by forming myotubes, when the Pax7wt cells still proliferate but not differentiate. This is in agreement with our observation that Pax7ko EB outgrowths contained more cells expressing MyoD.5
Surprisingly, results of our analyzes uncovered the differences at the level of cell cycle machinery in MEFs, i.e. the cells in which – to our knowledge – Pax7 function has not been assessed so far. In 2011 Amini-Nik et al.55 documented the involvement of Pax7 expressing cells in the wound repair. However, they suggested that the cells expressing Pax7 originated from satellite cells that adopted fibrotic phenotype. However, Pax7 expression in NIH3T3 fibroblasts was also documented by various vendors distributing anti-Pax7 antibodies (e.g. Abcam, MerckMillipore) what, together with our data, suggests that Pax7 is expressed also in cells other than neuronal or muscle specific satellite cells. In fact, during recent years its presence was also discovered in spermatogonia in adult testes.28,29 The function of Pax7 in MEFs is supported by our results showing that lack of functional Pax7 results in the decrease of proportion of MEFs in G1 phase, increase of proportion of MEFs expressing Ki-67 and being at G2/M transition. In Pax7ko MEFs, similarly to ESCs, we observed upregulation in the level of mRNA encoding cyclin E1, again indicating the relation between Pax7 and the regulation of at least G1/S phase progression. We did not, however, notice any differences in the number of apoptotic cells between Pax7wt and Pax7ko cells. Thus, Pax7 is a factor regulating proliferation of both types of cells studied, i.e., unique mammalian cells such as ESCs and “standard” mammalian cells such as MEFs.
Materials and methods
Animal studies were approved by Local Ethics Committee No. 1 in Warsaw, Poland according to the European Union Directive on the approximation of laws, regulations and administrative provisions of the Member States regarding the protection of animals used for experimental and scientific purposes.56,57 All mice were raised on the premises and were maintained under a 12 hours light/12 hours dark cycle. All analyses were performed at least in 3 independent experiments.
Embryonic stem cells derivation and culture
Embryonic stem cells (ESCs) lacking functional Pax7 were derived, genotyped, and analyzed as described previously.5 Medium for ESCs culture was composed of KnockOut Dulbecco's modified Eagle's Medium (KnockOut DMEM, Gibco) supplemented with 15% heat-inactivated fetal bovine serum (FBS, Performance Plus, Gibco) with addition of nonessential amino acids (0.1 mM, Gibco), L-glutamine (2 mM, Gibco), β-mercaptoethanol (0.1 mM, Sigma-Aldrich), penicillin and streptomycin (5000 units/ml each, Gibco), as well as murine leukemia inhibitory factor (LIF, 1000 IU/ml, ESGRO, Chemicon International). ESCs were cultured on feeder layer, i.e. inactivated mouse embryonic fibroblasts (MEFs) prepared according to Robertson (also see below).58
In vitro differentiation of ESCs
Differentiation of ESCs was carried as described previously.5 Briefly, ESCs cultured at the layer of inactivated MEFs were detached with 0.05% trypsin-EDTA (Gibco). MEFs were separated by pre-plating, i.e., single cell suspension of ESCs and MEFs was seeded onto gelatin-coated dish for 20 min to allow MEFs to attach to the culture dish. Next, medium containing ESCs only was collected and EBs were generated by placing of 800 ESCs in 30 µl drops of culture medium lacking LIF formed at the culture dish covers, i.e. so called hanging drops culture method was used (for the review see refs.26,59). After 2 days of culture EBs were collected from hanging drops and cultured in suspension for further 5 days (Fig. 1). During this period, culture medium was supplemented with retinoic acid (30 nM in dimethylsulfoxide; Sigma-Aldrich) and 1% of supplement consisting of insulin, transferrin, selenite (Gibco). At 7 day of culture EBs were plated onto coverslips (about 20 EBs per coverslip) coated with gelatin (Sigma-Aldrich) to generate EB outgrowths. Starting from day 10 of culture, EB outgrowths were cultured in DMEM containing 20% heat-inactivated FBS, penicillin and streptomycin. Next, the concentration of FBS in culture medium was reduced to 10% (day 13) and 5% (day 14). Starting from day 15 of culture medium composed of DMEM and F12 (1:1) supplemented with 1% N2 (Gibco), penicillin and streptomycin but lacking FBS was used. Culture media were changed every 2–3 days. EBs and EB outgrowths were analyzed at 2, 5, 7, 14, and 21 day of culture (Fig. 2).
Mouse embryonic fibroblasts isolation and culture
Primary MEFs lacking functional Pax7 gene were derived from 13.5-day-old embryos obtained after crossing of F1 (C57Bl/6N × 129Sv) Pax7+/− females with the males of the same genetic background and genotype. Primary MEFs were cultured in DMEM (with 4500 mg/l glucose, Gibco) supplemented with 10% of heat-inactivated FBS (Gibco), penicillin and streptomycin. Genotyping of MEFs was performed as previously described for ESCs.5 Briefly, heads of the embryos that served to isolate MEFs were incubated in 10% Chelex 100 (Bio-Rad) solution in deionized water, in 98°C, for 15 minutes. After centrifugation, supernatant containing DNA was collected and 1 μl of obtained solution was used for PCR analysis using RedTaq Ready Mix (Sigma-Aldrich) and primers according to conditions described previously.23 PCR products were separated in 1.5% agarose gel (Bio-Rad) and visualized with ethidium bromide (1 mg/ml, Sigma-Aldrich). Agarose gels were analyzed with GelDoc 2000 (Bio-Rad) using Quantity One software (Bio-Rad). Wild-type allele was represented by 200 bp and knock-out allele by 600 bp band.23
Mouse embryonic fibroblasts synchronization
MEFs were synchronized by 48-hour-long serum starvation, i.e., culture in DMEM (containing 4500 mg/l glucose, Gibco) supplemented with antibiotics, but lacking FBS. Afterwards, MEFs were either collected for analysis, or cultured in DMEM supplemented with 10% inactivated FBS. After an additional 18 hours of culture MEFs were collected for further analyses.
Preparation of feeder cells for ESC culture
Primary MEFs used as a feeder cells were derived from 13.5-day-old embryos obtained after crossing of F1 (C57Bl/6NxCBA/H) mice. Next, confluent MEFs were treated with mitomycin C (10 µg/ml, Sigma-Aldrich) for 2 h, stored in −80°C, and plated on gelatin-coated dishes one day before the plating of ESCs.
Isolation of mRNA and RT-qPCR
Total RNA was extracted from ESCs, EBs, and EB outgrowths using High Pure RNA isolation kit (Roche) and from MEFs using mirVana miRNA Isolation Kit (Ambion), according to the manufacturer's instructions. Reverse transcription was performed using 0.5 µg of RNA and RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to manufacturer's instruction. Next, qPCR analysis was performed using LightCycler 96 instrument (Roche), TaqMan Gene Expression Master Mix (Life Technologies), and specific TaqMan probes: Mm00432359_m1 (Ccnd1), Mm00432367_m1 (Ccne1), Mm00438066_m1 (Ccna2), Mm01171453_m1 (Ccnb2), Mm01243769_m1 (Mcm5), Mm00775040_gH (Cdc34), Mm00440924_g1 (Plk1), Mm04205640_g1 (Cdkn1a), Mm00438168_m1 (Cdkn1b), Mm01272135_g1 (Cdkn1c), Mm01205647_g1 (Actb). Data were collected and analyzed with LightCycler 96 SW1.1 software (Roche). Ddct analysis was performed according to Livak.60
Microarray analysis
For microarray analyses total RNA was isolated from undifferentiated ESCs, EBs (day 7 of culture), EB outgrowths (day 21 of culture) and asynchronously proliferating MEFs. For each type of cell cultures samples were collected from 4 independent experiments carried out at the same time. RNA was isolated using High Pure RNA isolation kit. Next, its integrity was checked with 2100 Bioanalyzer (Agilent Technologies) using RNA 6000 NAno LAb Chip kit (Agilent Technologies). All RNA samples had integrity number above 8.5. For target preparation Ambion® WT Expression Kit (Ambion) was used. Sense-strand cDNA after fragmentation was labeled using GeneChip® WT Terminal Labeling (Affymetrix) and hybridized onto Affymetrix® Mouse Gene 2.1 ST Array Strip, according to the manufacturer's instructions. Following hybridization the arrays were washed and stained using Affymetrix GeneAtlas fluidic station and scanned on Affymetrix GeneAtlas Imaging station. The intensity signals for each of the probe sets were written by into the “.cel files” and were imported into the Partek Genomic Suite v6.6 software with the use of RMA (Robust Multiarray Averaging). Background correction was applied based on the global distribution of the PM (perfect match) probe intensities and the affinity for each of the probes (based on their sequences) was calculated during import. Further, the probe intensities were quantile normalized,61 log2 transformed and median polish summarization to each of the probe sets was applied. Then the quality check was performed, e.g., Principal Component Analysis, in order to identify outliers and artifacts on the microarray. Afterwards the Analysis of Variance (ANOVA) allowed to create lists of significantly and differentially expressed genes between the biological variants. The selected lists were subjected to cluster analysis with the use of unsupervised Hierarchical Clustering algorithm in order to find genes and samples with similar profiles. Clusters were calculated on raw intensities that were standardized, i.e. shifted to mean of 0 and scaled to standard deviation of 1. Further, the color scale among all heat maps was set to the range of −2 to 2 in order to equalize dynamic ranges of all the intensity plots. The data coming from ESCs analyses have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession numbers: GSE66483 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66483) and as a supplementary tables in Czerwinska et al.5 Data coming from MEFs analyses have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE80658 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80658). Gene Ontology analysis was performed with the use of Partek Genomic Suite v6.6 software. Enrichment score (ES) represents the level of significance; the higher the enrichment score, the more a given GO functional group is over-represented in the gene list. The enrichment score was calculated using a chi-square test comparing the proportion of the gene list in a group to the proportion of the background in the group. ES > 1 means that the functional category is overexpressed, ES => 3 represents a significant overexpression (P-val < 0.05).
Propidium iodide staining and flow cytometry analysis
ESCs after pre-plating, EBs collected at day 2 or 5 of culture, and 2 types of MEFs, i.e., serum starved or serum starved for 48 h and then cultured for 18 hours in DMEM containing FBS, were disaggregated using trypsin-EDTA. Next, cells were washed twice in PBS (phosphate-buffered saline) and fixed in 70% cold ethanol (Chempur). After another 2 washes in PBS, cells were suspended in PBS containing 10 μg/ml ribonuclease (Sigma-Aldrich) and 50 μg/ml propidium iodide (PI, Sigma-Aldrich) and incubated in 37°C in darkness for 40 min. Finally, cells were filtered through 35 µm pores and 2–5 × 104 cells were analyzed using FACSCalibur and CellQuest software.
CFSE staining and flow cytometry analysis
CellTrace CFSE Cell Proliferation Kit (Invitrogen) was used for analysis of ESCs and MEFs proliferation. Briefly, ESCs collected after pre-plating or MEFs treated with 0.05% trypsin-EDTA were suspended in 0.1% bovine serum albumin (BSA, Sigma) in PBS containing 20 µM CFSE (carboxyfluorescein diacetate, ester). After 10 min incubation in 37°C, cold appropriate culture medium was added and cells were kept on ice for further 5 min. Afterwards, cells were washed twice in appropriate culture medium and either immediately collected for FACS analysis or subjected to further in vitro culture. ESCs were either cultured (5 × 104/cm2) on feeder layer or in hanging drops to form EBs. MEFs were plated in density of 4 × 104/cm2. ESCs (after 1 or 2 d of culture), EBs (after 1 or 2 d of culture), and MEFs (after 3 d of culture) were trypsynized, washed 2 times in PBS, and fixed in 3% paraformaldehyde (PFA, Sigma) for 10 min. After further 3 washes in PBS cells were analyzed using FL1 detector at FACSCalibur (Becton Dickinson) and CellQuest software (Becton Dickinson). Control cells, that served as negative control, i.e. those that were not stained with CFSE, were characterized by the lowest fluorescence level. They represent cells that underwent many divisions what resulted in the loss of fluorescence. Cells collected for analysis immediately after CFSE staining were positive control as non-divided ones.
Immunolocalization
ESCs, EB outgrowths, and MEFs plated and cultured at coverslips were washed 3 times in PBS and fixed in 3% PFA for 10 min. After another 3 washes in PBS ESCs and EB outgrowths were incubated in 0.5% Triton X100 (Sigma-Aldrich) in PBS for 5 min, and MEFs were incubated in 0.05% Triton X-100 in PBS for 3 min. Next, cells were washed in PBS and incubated in 3% BSA in PBS for 1 h. Afterwards, incubation with primary antibodies solutions was conducted in 4°C overnight. Primary antibodies against Ki-67 (ab15580, Abcam) and γ-H2A.X (ab22551, Abcam), Pax7 (DSHB) were used, diluted 1:500 (Ki-67 and γ-H2A.X) or 1:100 (Pax7) in 0.5% BSA in PBS. Next, cells were washed 3 times in PBS and incubated in solution containing appropriate secondary antibodies conjugated with AlexaFluor594 (Molecular Probes, diluted 1:200) and DRAQ5 (Biostatus, diluted 1:1000) in 0.5% BSA in PBS for 2 h. Specimens were washed 3 times in PBS and mounted using Fluorescent Mounting Medium (Dako). Specificity of primary antibodies was confirmed by incubation of ESCs, EB outgrowths, and MEFs with secondary antibodies only. The specimens were analyzed using Axiovert 100M scanning confocal microscope (Zeiss) equipped with LSM 510 software.
Western blotting
Protein extracts were produced by homogenization and lysis of in vitro cultured ESCs, EBs, EB outgrowths, asynchronously dividing MEFs, and whole 13.5-day-old mouse embryos. Briefly, cells and embryos were homogenized and lysed in ELB buffer containing protease inhibitors (Complete, Roche) and obtained lysates were kept on ice for 30 minutes, microcentrifuged and stored at −80°C. Protein concentration was determined by using the Bradford's assay (Sigma). Twenty μg of protein was subjected to 12% SDS-PAGE and Western-blot analysis and probed with antibodies against cyclin E (sc-481, Santa Cruz Biotechnology), Pax7 (ARP32742, Aviva Systems Biology) or Hsp90 (TA500494, Origene). As secondary antibodies goat anti-mouse (170-6516, Bio-Rad) or goat anti-rabbit HRP conjugate (170-6515, Bio-Rad) were used, followed by chemiluminescence detection. Films were photographed and optical density of resulting bands was measured using GelDocXR+ (Bio-Rad) with ImageLab software.
Supplementary Material
Abbreviations
- ESCs
Embryonic Stem Cells
- EBs
Embryoid Bodies
- EBOs
Embryoid Body Outgrowths
- MEFs
Mouse Embryonic Fibroblasts
- CFSE
carboxyfluorescein diacetate, ester
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Analyzes of embryonic stem cells were supported by grant provided by budget funds from National Science Center (Poland) for Areta Czerwinska - 2012/05/N/NZ3/00314 for 2013 – 2015. Analyzes of mouse embryonic fibroblasts were supported by grant provided by budget funds from National Science Centre (Poland) for Iwona Grabowska - N N303 548139 for 2010–2013. Areta Czerwinska was a recipient of EMBO short-term fellowship and a scholarship for PhD students under projects co-financed by European Social Found during realization of this project.
ORCID
Sylwia Gawrzak http://orcid.org/0000-0002-7047-3319
Karolina Archacka http://orcid.org/0000-0002-0274-9117
Marta Koblowska http://orcid.org/0000-0003-4497-6200
References
- [1].Grabowska I, Brzoska E, Gawrysiak A, Streminska W, Moraczewski J, Polanski Z, Hoser G, Kawiak J, Machaj EK, Pojda Z, et al.. Restricted Myogenic Potential of Mesenchymal Stromal Cells Isolated From Umbilical Cord. Cell Transplantation 2012; 21:1711-26; PMID:22525423; http://dx.doi.org/ 10.3727/096368912X640493 [DOI] [PubMed] [Google Scholar]
- [2].Swierczek B, Ciemerych MA, Archacka K. From pluripotency to myogenesis: a multistep process in the dish. J Muscle Res Cell Motil 2015; PMID:26715014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Myer A, Olson EN, Klein WH. MyoD cannot compensate for the absence of myogenin during skeletal muscle differentiation in murine embryonic stem cells. Dev Biol 2001; 229:340-50; PMID:11203698; http://dx.doi.org/ 10.1006/dbio.2000.9985 [DOI] [PubMed] [Google Scholar]
- [4].Weitzer G, Milner DJ, Kim JU, Bradley A, Capetanaki Y. Cytoskeletal control of myogenesis: a desmin null mutation blocks the myogenic pathway during embryonic stem cell differentiation. Dev Biol 1995; 172:422-39; PMID:8612961; http://dx.doi.org/ 10.1006/dbio.1995.8070 [DOI] [PubMed] [Google Scholar]
- [5].Czerwinska AM, Grabowska I, Archacka K, Bem J, Swierczek B, Helinska A, Streminska W, Fogtman A, Iwanicka-Nowicka R, Koblowska M, et al.. Myogenic differentiation of mouse embryonic stem cells that lack a functional Pax7 gene. Stem Cells Dev 2016; 25:285–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 2005; 435:948-53; PMID:15843801; http://dx.doi.org/ 10.1038/nature03594 [DOI] [PubMed] [Google Scholar]
- [7].Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, Beauchamp JR. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci 2006; 119:1824-32; PMID:16608873; http://dx.doi.org/ 10.1242/jcs.02908 [DOI] [PubMed] [Google Scholar]
- [8].Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell 2000; 102:777-86; PMID:11030621; http://dx.doi.org/ 10.1016/S0092-8674(00)00066-0 [DOI] [PubMed] [Google Scholar]
- [9].Collins CA, Gnocchi VF, White RB, Boldrin L, Perez-Ruiz A, Relaix F, Morgan JE, Zammit PS. Integrated functions of Pax3 and Pax7 in the regulation of proliferation, cell size and myogenic differentiation. PLoS One 2009; 4:e4475; PMID:19221588; http://dx.doi.org/ 10.1371/journal.pone.0004475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev Biol 2004; 275:375-88; PMID:15501225; http://dx.doi.org/ 10.1016/j.ydbio.2004.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. Mol Biol Cell 2009; 20:3170-7; PMID:19458195; http://dx.doi.org/ 10.1091/mbc.E08-12-1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Olguin HC, Yang Z, Tapscott SJ, Olwin BB. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol 2007; 177:769-79; PMID:17548510; http://dx.doi.org/ 10.1083/jcb.200608122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hirai H, Verma M, Watanabe S, Tastad C, Asakura Y, Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J Cell Biol 2010; 191:347-65; PMID:20956382; http://dx.doi.org/ 10.1083/jcb.201006025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol 2006; 172:91-102; PMID:16380438; http://dx.doi.org/ 10.1083/jcb.200508044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J 2004; 23:3430-9; PMID:15282552; http://dx.doi.org/ 10.1038/sj.emboj.7600346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Martelli F, Cenciarelli C, Santarelli G, Polikar B, Felsani A, Caruso M. MyoD induces retinoblastoma gene expression during myogenic differentiation. Oncogene 1994; 9:3579-90; PMID:7970718 [PubMed] [Google Scholar]
- [17].Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE, Giordano A, Kedes L, Wang JY, Sartorelli V. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell 2001; 8:885-97; PMID:11684023; http://dx.doi.org/ 10.1016/S1097-2765(01)00373-2 [DOI] [PubMed] [Google Scholar]
- [18].Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 1995; 267:1018-21; PMID:7863327; http://dx.doi.org/ 10.1126/science.7863327 [DOI] [PubMed] [Google Scholar]
- [19].Rajabi HN, Takahashi C, Ewen ME. Retinoblastoma protein and MyoD function together to effect the repression of Fra-1 and in turn cyclin D1 during terminal cell cycle arrest associated with myogenesis. J Biol Chem 2014; 289:23417-27; PMID:25006242; http://dx.doi.org/ 10.1074/jbc.M113.532572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, Baldini A, Harper JW, Elledge SJ. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev 1995; 9:650-62; PMID:7729684; http://dx.doi.org/ 10.1101/gad.9.6.650 [DOI] [PubMed] [Google Scholar]
- [21].Ciemerych MA, Archacka K, Grabowska I, Przewozniak M. Cell cycle regulation during proliferation and differentiation of mammalian muscle precursor cells. Results Probl Cell Differ 2011; 53:473-527; PMID:21630157; http://dx.doi.org/ 10.1007/978-3-642-19065-0_20 [DOI] [PubMed] [Google Scholar]
- [22].Bhagavati S, Song X, Siddiqui MA. RNAi inhibition of Pax3/7 expression leads to markedly decreased expression of muscle determination genes. Mol Cell Biochem 2007; 302:257-62; PMID:17396232; http://dx.doi.org/ 10.1007/s11010-007-9444-3 [DOI] [PubMed] [Google Scholar]
- [23].Mansouri A, Stoykova A, Torres M, Gruss P. Dysgenesis of cephalic neural crest derivatives in Pax7−/− mutant mice. Development 1996; 122:831-8; PMID:8631261 [DOI] [PubMed] [Google Scholar]
- [24].Momčilović O, Navara C, Schatten G. Cell cycle adaptations and maintenance of genomic integrity in embryonic stem cells and induced pluripotent stem cells In: Kubiak JZ, ed. Cell Cycle in Development: Springer-Verlag GmbH, 2011. [DOI] [PubMed] [Google Scholar]
- [25].Desbaillets I, Ziegler U, Groscurth P, Gassmann M. Embryoid bodies: an in vitro model of mouse embryogenesis. Exp Physiol 2000; 85:645-51; PMID:11187960 [PubMed] [Google Scholar]
- [26].Grabowska I, Archacka K, Czerwinska AM, Krupa M, Ciemerych MA. Mouse and human pluripotent stem cells and the means of their myogenic differentiation. Results Probl Cell Differ 2012; 55:321-56; PMID:22918815; http://dx.doi.org/ 10.1007/978-3-642-30406-4_18 [DOI] [PubMed] [Google Scholar]
- [27].Levy-Strumpf N, Kimchi A. Death associated proteins (DAPs): from gene identification to the analysis of their apoptotic and tumor suppressive functions. Oncogene 1998; 17:3331-40; PMID:9916995; http://dx.doi.org/ 10.1038/sj.onc.1202588 [DOI] [PubMed] [Google Scholar]
- [28].Aloisio GM, Nakada Y, Saatcioglu HD, Pena CG, Baker MD, Tarnawa ED, Mukherjee J, Manjunath H, Bugde A, Sengupta AL, et al.. PAX7 expression defines germline stem cells in the adult testis. J Clin Invest 2014; 124:3929-44; PMID:25133429; http://dx.doi.org/ 10.1172/JCI75943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kumar TR. The quest for male germline stem cell markers: PAX7 gets ID'd. J Clin Invest 2014; 124:4219-22; PMID:25157826; http://dx.doi.org/ 10.1172/JCI77926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol 1984; 133:1710-5; PMID:6206131 [PubMed] [Google Scholar]
- [31].Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273:5858-68; PMID:9488723; http://dx.doi.org/ 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- [32].Banath JP, Banuelos CA, Klokov D, MacPhail SM, Lansdorp PM, Olive PL. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp Cell Res 2009; 315:1505-20; PMID:19154734; http://dx.doi.org/ 10.1016/j.yexcr.2008.12.007 [DOI] [PubMed] [Google Scholar]
- [33].Chuykin IA, Lianguzova MS, Pospelova TV, Pospelov VA. Activation of DNA damage response signaling in mouse embryonic stem cells. Cell Cycle 2008; 7:2922-8; PMID:18787397; http://dx.doi.org/ 10.4161/cc.7.18.6699 [DOI] [PubMed] [Google Scholar]
- [34].Ciemerych MA, Sicinski P. Cell cycle in mouse development. Oncogene 2005; 24:2877-98; PMID:15838522; http://dx.doi.org/ 10.1038/sj.onc.1208608 [DOI] [PubMed] [Google Scholar]
- [35].Gopinathan L, Ratnacaram CK, Kaldis P. Established and novel Cdk/cyclin complexes regulating the cell cycle and development. Results Probl Cell Differ 2011; 53:365-89; PMID:21630153; http://dx.doi.org/ 10.1007/978-3-642-19065-0_16 [DOI] [PubMed] [Google Scholar]
- [36].Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene 1994; 9:809-18; PMID:8108123 [PubMed] [Google Scholar]
- [37].Conklin JF, Sage J. Keeping an eye on retinoblastoma control of human embryonic stem cells. J Cell Biochem 2009; 108:1023-30; PMID:19760644; http://dx.doi.org/ 10.1002/jcb.22342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB, Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene 1996; 12:p309-22. [PubMed] [Google Scholar]
- [39].Faast R, White J, Cartwright P, Crocker L, Sarcevic B, Dalton S. Cdk6-cyclin D3 activity in murine ES cells is resistant to inhibition by p16(INK4a). Oncogene 2004; 23:491-502; PMID:14724578; http://dx.doi.org/ 10.1038/sj.onc.1207133 [DOI] [PubMed] [Google Scholar]
- [40].Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene 2002; 21:8320-33; PMID:12447695; http://dx.doi.org/ 10.1038/sj.onc.1206015 [DOI] [PubMed] [Google Scholar]
- [41].Kalaszczynska I, Geng Y, Iino T, Mizuno S, Choi Y, Kondratiuk I, Silver DP, Wolgemuth DJ, Akashi K, Sicinski P. Cyclin A is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell 2009; 138:352-65; PMID:19592082; http://dx.doi.org/ 10.1016/j.cell.2009.04.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev 2005; 1:131-8; PMID:17142847; http://dx.doi.org/ 10.1385/SCR:1:2:131 [DOI] [PubMed] [Google Scholar]
- [43].White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell 2005; 16:2018-27; PMID:15703208; http://dx.doi.org/ 10.1091/mbc.E04-12-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Amthor H, Christ B, Patel K. A molecular mechanism enabling continuous embryonic muscle growth - a balance between proliferation and differentiation. Development 1999; 126:1041-53; PMID:9927604 [DOI] [PubMed] [Google Scholar]
- [45].Franz T, Kothary R, Surani MA, Halata Z, Grim M. The Splotch mutation interferes with muscle development in the limbs. Anat Embryol (Berl) 1993; 187:153-60; PMID:8238963 [DOI] [PubMed] [Google Scholar]
- [46].Goulding M, Lumsden A, Paquette AJ. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development 1994; 120:957-71; PMID:7600971 [DOI] [PubMed] [Google Scholar]
- [47].Bober E, Franz T, Arnold HH, Gruss P, Tremblay P. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development 1994; 120:603-12; PMID:8162858 [DOI] [PubMed] [Google Scholar]
- [48].Williams BA, Ordahl CP. Pax-3 expression in segmental mesoderm marks early stages in myogenic cell specification. Development 1994; 120:785-96; PMID:7600957 [DOI] [PubMed] [Google Scholar]
- [49].Barr FG. Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene 2001; 20:5736-46; PMID:11607823; http://dx.doi.org/ 10.1038/sj.onc.1204599 [DOI] [PubMed] [Google Scholar]
- [50].Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol 2006; 172:103-13; PMID:16391000; http://dx.doi.org/ 10.1083/jcb.200508001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].White RB, Ziman MR. Genome-wide discovery of Pax7 target genes during development. Physiol Genomics 2008; 33:41-9; PMID:18198279; http://dx.doi.org/ 10.1152/physiolgenomics.00256.2007 [DOI] [PubMed] [Google Scholar]
- [52].Kuroda H, Sugimoto T, Horii Y, Sawada T. Signaling pathway of ciliary neurotrophic factor in neuroblastoma cell lines. Med Pediatr Oncol 2001; 36:118-21; PMID:11464862; http://dx.doi.org/ 10.1002/1096-911X(20010101)36:1%3c118::AID-MPO1028%3e3.0.CO;2-R [DOI] [PubMed] [Google Scholar]
- [53].Verploegen S, Lammers JW, Koenderman L, Coffer PJ. Identification and characterization of CKLiK, a novel granulocyte Ca(++)/calmodulin-dependent kinase. Blood 2000; 96:3215-23; PMID:11050006 [PubMed] [Google Scholar]
- [54].Anneren C, Lindholm CK, Kriz V, Welsh M. The FRK/RAK-SHB signaling cascade: a versatile signal-transduction pathway that regulates cell survival, differentiation and proliferation. Curr Mol Med 2003; 3:313-24; PMID:12776987; http://dx.doi.org/ 10.2174/1566524033479744 [DOI] [PubMed] [Google Scholar]
- [55].Amini-Nik S, Glancy D, Boimer C, Whetstone H, Keller C, Alman BA. Pax7 expressing cells contribute to dermal wound repair, regulating scar size through a beta-catenin mediated process. Stem Cells 2011; 29:1371-9; PMID:21739529 [DOI] [PubMed] [Google Scholar]
- [56].Close B, Banister K, Baumans V, Bernoth EM, Bromage N, Bunyan J, Erhardt W, Flecknell P, Gregory N, Hackbarth H, et al.. Recommendations for euthanasia of experimental animals: Part 1. DGXI of the European Commission. Lab Anim 1996; 30:293-316; PMID:8938617; http://dx.doi.org/ 10.1258/002367796780739871 [DOI] [PubMed] [Google Scholar]
- [57].Close B, Banister K, Baumans V, Bernoth EM, Bromage N, Bunyan J, Erhardt W, Flecknell P, Gregory N, Hackbarth H, et al.. Recommendations for euthanasia of experimental animals: Part 2. DGXT of the European Commission. Lab Anim 1997; 31:1-32; PMID:9121105; http://dx.doi.org/ 10.1258/002367797780600297 [DOI] [PubMed] [Google Scholar]
- [58].Robertson E. Embryo-derived stem cell lines In: Robertson E, ed. Teratocarcinomas and Embryonic Stem Cells: A Practical Approach: Oxford University Press, 1987:71-112. [Google Scholar]
- [59].Wobus AM, Boheler KR. Embryonic stem cells: prospects for developmental biology and cell therapy. Physiol Rev 2005; 85:635-78; PMID:15788707; http://dx.doi.org/ 10.1152/physrev.00054.2003 [DOI] [PubMed] [Google Scholar]
- [60].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001; 25:402-8; PMID:11846609; http://dx.doi.org/ 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- [61].Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003; 19:185-93; PMID:12538238; http://dx.doi.org/ 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.