When normal cells are challenged with genotoxic agents, tumor suppressor p53 curbs the growth of damaged cells by regulating the expression of genes that promote apoptosis, cell cycle arrest and DNA repair. More enigmatic is how p53 functions to prevent the formation of tumors under normal conditions. Through complex strategies cancer cells evade these gatekeeping mechanisms, which results in sustained proliferation and growth. Since p53 is integral to inhibiting tumourigenesis it is not surprising that this gene is mutated in ∼50% of all cancers. Studies investigating the tumor suppressive functions of p53 have primarily focused on its classical roles in cell death and cell division.1 The mechanisms by which the p53 family, including p63 and p73, regulate genome stability are less understood.
With an evolutionary history dating over 1 billion years, it is likely that the ability of p53 to prevent tumor formation in the soma of complex multicellular organisms occurred later.2 The presence of p53-like proteins in amoeba and choanoflagellates indicates that this family predated multicellularity, but their biological functions are not well characterized. The most ancient role described for a p53 protein is in sea anemone, where it promotes apoptosis of germ cells damaged by UV irradiation. Similarly, p53-like proteins in Drosophila melanogaster and Caenorhabditis elegans activate programmed cell death in the reproductive tissues in response to genotoxic stress. So, what can be learned by studying p53 proteins in simpler organisms that is relevant to its tumor suppressive function in vertebrates? We discovered a previously unknown role for the p53-like protein CEP-1 in ensuring meiotic fidelity in the germline of C. elegans.3 This raises the question of whether the p53 family employ similar mechanisms in somatic tissues of vertebrates.
In more complex organisms, the roles in germline protection were retained by p63, while p53 and p73 execute similar gatekeeping functions in the soma to prevent tumor formation (Fig. 1). The C. elegans germline is an excellent model for understanding general mechanisms that govern genome stability. To maintain organismal fitness it is critical that errors acquired during meiosis, or from exposure to genotoxic agents, are not propagated to the next generation. Therefore, it is tempting to speculate that the tumor suppressive mechanisms carried out by p53 in the soma may have originated from an ancestral role in protecting germline integrity.
Figure 1.
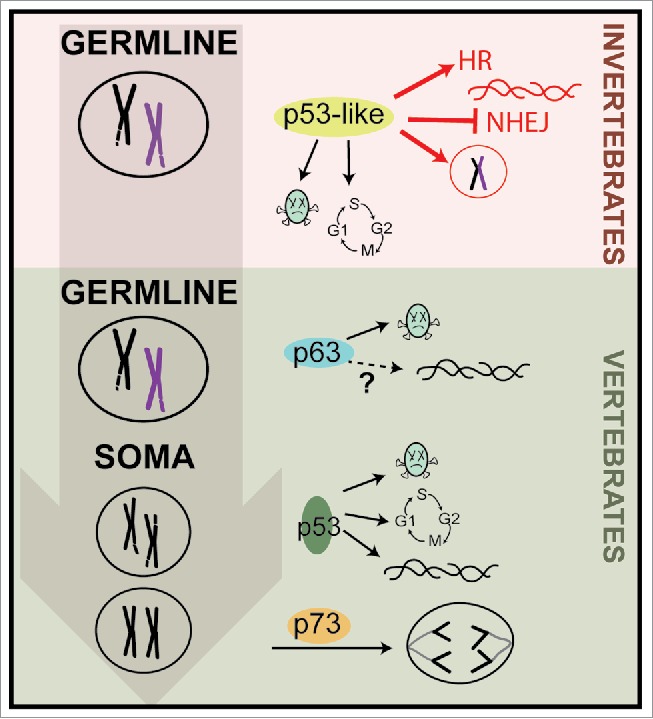
The p53 family of proteins regulate genome fidelity: from germline to soma. We propose that the tumor suppressive functions of p53 in vertebrate organisms originated from its ancient roles in germline protection, such as apoptosis, cell cycle and DNA repair. Highlighted in red are the novel activities we uncovered for CEP-1 in regulating meiotic fidelity in the C. elegans germline.
The importance of the p53 family in guarding the germline is underscored by observations showing that programmed DNA double strand breaks (DSB) generated by Spo11 results in activation of p53 in the germlines of Drosophila and mice.4 In addition, ablation of the sole p53-like gene in C. elegans, cep-1, results in sex chromosome non-disjunction during meiosis.5 These observations hinted at an active role for p53 proteins in meiosis, but the mechanisms by which they function were unclear. Our recent work sheds light on this elusive role and, for the first time, provides mechanistic insight into how p53-like proteins ensure meiotic fidelity.3 Alone, CEP-1 has a minor role in meiosis but when ablated in combination with the meiotic protein HIM-5, they cause a synthetic lethal phenotype that is in part due to defects in programmed DSB induction and subsequent crossover formation. Our study also reveals that CEP-1 and HIM-5 suppress the error-prone nonhomologous end joining (NHEJ) pathway to ensure that DSBs are repaired by error-free homologous recombination (HR). Surprisingly, CEP-1 likely carries out these DNA repair functions independent of its transcriptional activity.3 Perhaps, these previously undefined roles in meiosis can help explain the reproductive defects observed in p53 mutant mice.6
Our findings in C. elegans also raise the question of whether p53, p63 and/or p73 have similar roles in antagonizing inappropriate activation of NHEJ in the soma of vertebrates. This could explain the differences in the severity of phenotypes in mice where p53 has been knocked out (Trp53−/−) versus mice harboring mutations that specifically disable its apoptotic function (Trp53515C/515C). Unlike Trp53515C/515C mice, Trp53−/− mice develop tumors much earlier in life and exhibit chromosomal abnormalities, which were initially attributed to p53s role in centrosome duplication.7 Alternatively, the chromosome deletions and translocations observed in these mice may be due to a role for p53 in suppressing NHEJ, similar to what we found with CEP-1 in the C. elegans germline. Testing this hypothesis in mouse models will refine our understanding of the tumor suppressive activity of the p53 family of proteins. Another question is whether p53 functions alone or in collaboration with p63 and p73 to carry out these repair activities? Invertebrates with a single p53 family member, like C. elegans, have provided insights that are more difficult to obtain in organisms with 3 p53 family members that share some overlapping functions.
Our work in C. elegans suggests that the tumor suppressive roles of p53 may have stemmed from its ability to maintain germline fidelity by regulating programmed DSB induction and repair pathway choice. Perhaps the “pro-choice” activities of p53 are critical to its maintenance of genome integrity in the soma of vertebrates? Understanding how the p53 family of proteins regulate DNA repair may reveal alternative approaches for treating ∼50% of human tumors that have a mutated p53 gene.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Bieging KT, et al. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer 2014; 14:359-70; PMID:24739573; http://dx.doi.org/ 10.1038/nrc3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Belyi VA, et al. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol 2010; 2:a001198; PMID:20516129; http://dx.doi.org/ 10.1101/cshperspect.a001198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mateo AR, et al. The p53-like Protein CEP-1 Is Required for Meiotic Fidelity in C. elegans. Curr Biol 2016; 26:1148-58; PMID:27151662; http://dx.doi.org/ 10.1016/j.cub.2016.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lu WJ, et al. Meiotic recombination provokes functional activation of the p53 regulatory network. Science 2010; 328:1278-81; PMID:20522776; http://dx.doi.org/ 10.1126/science.1185640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Derry WB, et al. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 2001; 294:591-5; PMID:11557844; http://dx.doi.org/ 10.1126/science.1065486 [DOI] [PubMed] [Google Scholar]
- [6].Rotter V, et al. Mice with reduced levels of p53 protein exhibit the testicular giant-cell degenerative syndrome. Proc Natl Acad Sci U S A 1993; 90:9075-9; PMID:8415656; http://dx.doi.org/ 10.1073/pnas.90.19.9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu G, et al. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet 2004; 36:63-8; PMID:14702042; http://dx.doi.org/ 10.1038/ng1282 [DOI] [PubMed] [Google Scholar]


