ABSTRACT
In order to achieve accurate chromosome segregation, eukaryotic cells undergo a dramatic change in morphology to obtain a spherical shape during mitosis. Interphase cells communicate directly with each other by exchanging ions and small molecules via gap junctions, which have important roles in controlling cell growth and differentiation. As cells round up during mitosis, the gap junctional communication between mitotic cells and adjacent interphase cells ceases. Whether mitotic cells use alternative mechanisms for mediating direct cell-cell communication during rounding is currently unknown. Here, we have studied the mechanisms involved in the remodeling of gap junctions during mitosis. We further demonstrate that mitotic cells are able to form actin-based plasma membrane bridges with adjacent cells during rounding. These structures, termed “mitotic nanotubes,” were found to be involved in mediating the transport of cytoplasm, including Rab11-positive vesicles, between mitotic cells and adjacent cells. Moreover, a subpool of the gap-junction channel protein connexin43 localized in these intercellular bridges during mitosis. Collectively, the data provide new insights into the mechanisms involved in the remodeling of gap junctions during mitosis and identify actin-based plasma membrane bridges as a novel means of communication between mitotic cells and adjacent cells during rounding.
KEYWORDS: actin, cell junctions, connexin, gap junction, mitosis, tunneling nanotubes
Introduction
A characteristic of eukaryotic cells is to round up during mitosis.1 A round morphology is important to ensure proper spindle formation, to distribute components to the daughter cells appropriately, and to establish correct intracellular gradients of signaling molecules.1 Rounding up is followed by the ingression of the cytokinetic furrow at the cellular equator and subsequent membrane abscission at the cytokinetic midbody that separates the two daughter cells. The cell rounding that occurs during mitosis is associated with a major decrease in the cell surface area, which reaches a minimum at metaphase.2 This decrease has been proposed to be due to increased numbers of microvilli, blebs, and folds on the cell surface of the mitotic cell.3,4 Others have reported that the reduction of the cell surface area could be due to continuous membrane internalization accompanied by shutdown of membrane recycling, causing an accumulation of membrane components in intracellular compartments.2 The cell surface increases again with the onset of anaphase when the intracellular reservoir of the membrane components is recycled.2 The reduction in cell surface area during mitosis is associated with a decrease in cell volume.5-7 This cellular condensation occurs as a result of extrusion of osmotically active chloride via voltage-gated channels, resulting in an outward osmotic pressure in dividing cells.8 After reaching a minimum at metaphase, the cell volume increases again, and toward the end of cytokinesis, the combined volume of the two daughter cells equals that of their mother cell during prophase.5-7
As cells round up during mitosis, they remain anchored to the substratum by forming actin-based retraction fibers.9,10 However, the mechanism by which mitotic cells maintain contact and communicate with adjacent cells during rounding is less well understood. Cell-cell adhesion is generally considered to be retained during mitosis, as tight junctions, adherens junctions and desmosomes have been shown to persist throughout cell division.11 In contrast to the other types of intercellular junctions, gap junctions are subjected to a major remodeling during mitosis, resulting in the loss of gap junctional intercellular communication.12-17 The gap junctions consist of arrays of intercellular channels that enable adjacent cells to directly exchange ions, secondary messengers, and other small molecules (<1 kDa). This form of cell-cell communication has important roles in cell growth control and differentiation, in embryogenesis, and in the maintenance of tissue homeostasis.18 The constituents of the gap junction channels are a family of transmembrane proteins called connexins. The human proteome consists of 21 known connexin proteins, of which the most ubiquitously expressed is connexin43 (Cx43).19 Germ-line mutations of connexins are linked to deafness, peripheral neuropathies, skin disorders, cataracts, and cardiovascular diseases.20,21 Moreover, connexins are often downregulated during cancer pathogenesis, resulting in loss of intercellular communication via gap junctions.22,23
In addition to communicating via gap junctions, neighboring cells are also able to form thin membranous extensions that provide a pathway for direct exchange of cytoplasm and for electrical coupling.24-26 These structures, termed tunneling nanotubes, have been identified in a variety of cell types in vitro, including epithelial cells, fibroblasts, neurons, and immune cells.24-26 There is also increasing evidence that tunneling nanotubes exist in vivo. For instance, structures resembling tunneling nanotubes have been identified in solid tumors obtained from patients with malignant pleural mesothelioma 27 and in MHC class II+ cells in the mouse cornea.28 Tunneling nanotubes are considered to have important roles in development and immunity, as well as in pathogen transfer.24 Interestingly, recent studies have demonstrated a close functional interplay between the gap junctions and tunneling nanotubes.29-32 Cx43 has been shown to localize in tunneling nanotubes, where it has essential roles in mediating the electrical coupling between cells via the tunneling nanotubes.31,32
Here, we show that although gap junctions are lost as cells round up during mitosis, the mitotic cells are able to communicate with adjacent cells by forming actin-based intercellular bridges. We demonstrate that such bridges, termed “mitotic nanotubes,” are involved in mediating the intercellular transfer of cytoplasm, including Rab11-positive vesicles, between mitotic cells and adjacent cells. We further show that a subpool of Cx43 localizes in these actin-based intercellular bridges during mitotic rounding.
Results
A Cx43 subpool is subjected to increased endocytosis during mitosis
As a first approach to study the mechanisms involved in the remodeling of gap junctions during mitosis, we analyzed the subcellular localization of Cx43 during mitosis in IAR20 cells, which express high levels of endogenous Cx43 that forms functional gap junctions.33 As determined by fluorescence confocal microscopy, a subpool of Cx43 was found to be subjected to relocalization from the plasma membrane to intracellular vesicular structures, in accordance with previous studies in other cell lines (Fig. 1A).12,16,17,34 The internalized Cx43 was found to partly colocalize with the early endosomal marker EEA1, in line with previous observations in other cell lines (Fig. 1B).12 A quantitative analysis revealed that the level of colocalization between Cx43 and EEA1 started to increase in the early phases of mitosis and reached its peak at anaphase (Fig. 1C). Super-resolution microscopy confirmed that Cx43-positive intracellular vesicles in mitotic cells partly colocalized with EEA1 (Fig. 1D; Fig. S1). These data suggest that a subpool of Cx43 undergoes increased endocytosis and trafficking to early endosomes during mitosis in IAR20 cells.
Figure 1.
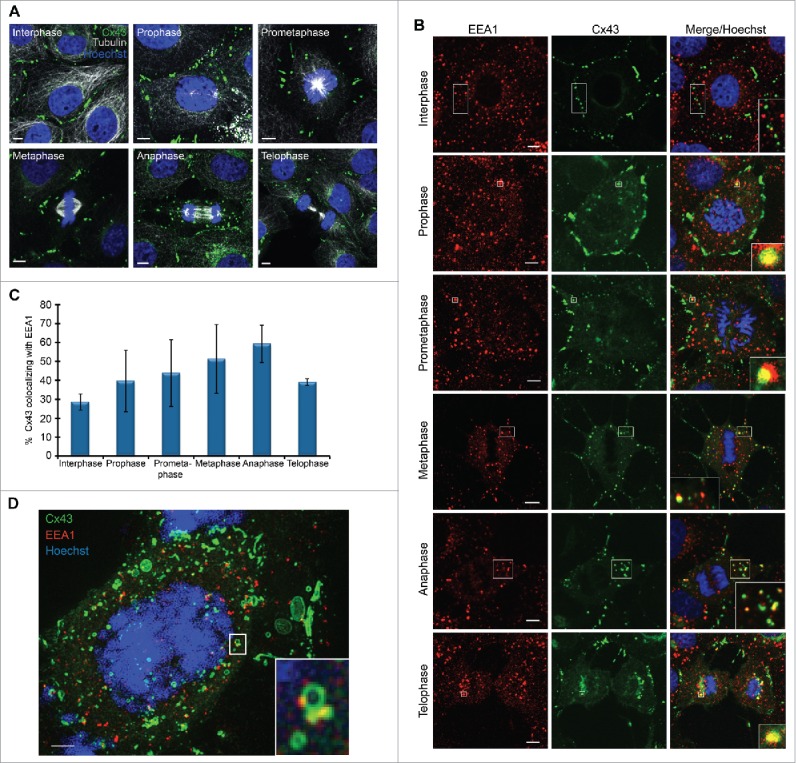
A subpool of Cx43 undergoes increased endocytosis during mitosis. IAR20 cells were fixed and stained with (A) anti-Cx43 (green) and anti-tubulin (white) or (B) anti-Cx43 (green) and anti-EEA1 (red) antibodies. Cells were then visualized by fluorescence confocal microscopy, and representative images of single confocal planes showing the subcellular localization of Cx43 in the various mitotic phases were acquired using fluorescence confocal microscopy. The nuclei were stained with Hoechst (blue). Cell-cycle stages were defined by DNA staining with Hoechst. Scale bars, 5 µm. Inserts in (B) show enlarged views of subcellular structures displaying colocalization between Cx43 and EEA1. Scale bars, 5 µm. (C) The colocalization between Cx43 and EEA1 in interphase cells and in cells in the various mitotic phases was quantified in z-stacks obtained by confocal microscopy, using the IMARIS software. Values shown are the mean ± SD of three independent experiments. (D) The subcellular localization of Cx43 and EEA1 in mitotic cells was analyzed by SIM. A max projection of 57 z-stacks obtained by SIM revealed a large pool of Cx43-positive intracellular vesicles in mitotic cells. Insert shows an enlarged view of an apparent fusion between Cx43-positive vesicles (green) and early endosomes (red). Examples of z-positions from the max projection in which Cx43 and EEA1 colocalize are shown in Fig. S1. Scale bar, 2 µm.
The molecular mechanisms involved in the endocytosis of gap junctions during mitosis have not been characterized. Moreover, whether the increased endocytosis of Cx43 during mitosis is a prerequisite for the remodeling of gap junctions during mitosis is currently unknown. We have previously demonstrated that the E3 ubiquitin ligase SMAD ubiquitination regulatory factor-2 (SMURF2) controls the endocytosis of Cx43 gap junctions under basal conditions and in response to activation of protein kinase C (PKC).35 In mitotic IAR20 cells, SMURF2 was found to partly colocalize with Cx43 gap junctions at the plasma membrane and in intracellular vesicles (Fig. 2A). To determine whether SMURF2 is involved in the remodeling of Cx43 gap junctions during mitosis, we depleted SMURF2 by using small interfering RNA (siRNA). In these experiments, the SMURF2 protein level was reduced by approximately 70% compared with cells transfected with a control siRNA sequence (Fig. 2B and Fykerud et al.35). Depletion of SMURF2 resulted in an overall increase in Cx43 protein levels, in accordance with our previous study (Fig. 2B and Fykerud et al.35). Importantly, depletion of SMURF2 strongly counteracted the remodeling of Cx43 gap junctions during mitosis (Fig. 2C). While Cx43 was found to gradually undergo endocytosis during mitosis in control siRNA-transfected cells, most Cx43 localized at the plasma membrane and formed gap junctions at cell-cell borders in all phases of mitosis in cells depleted of SMURF2 (Fig. 2C). To quantify the effect of SMURF2 depletion on the relocalization of Cx43 during mitosis, we determined the level of Cx43 staining at the plasma membrane as a percentage of overall Cx43 staining in anaphase cells. In control cells, approximately 40% of Cx43 was found to localize at the plasma membrane during anaphase (Fig. 2D). In contrast, cells depleted of SMURF2 expressed approximately 70% of Cx43 at the plasma membrane during anaphase (Fig. 2D). As control, the tight junction protein occludin did not appear to undergo major relocalization from the plasma membrane to endocytic compartments in mitotic cells (Fig. 2C). Collectively, these data indicate that the remodeling of the Cx43 gap junction during mitosis in IAR20 cells is regulated by SMURF2 and that increased Cx43 endocytosis is a prerequisite for gap junction remodeling during mitosis.
Figure 2.
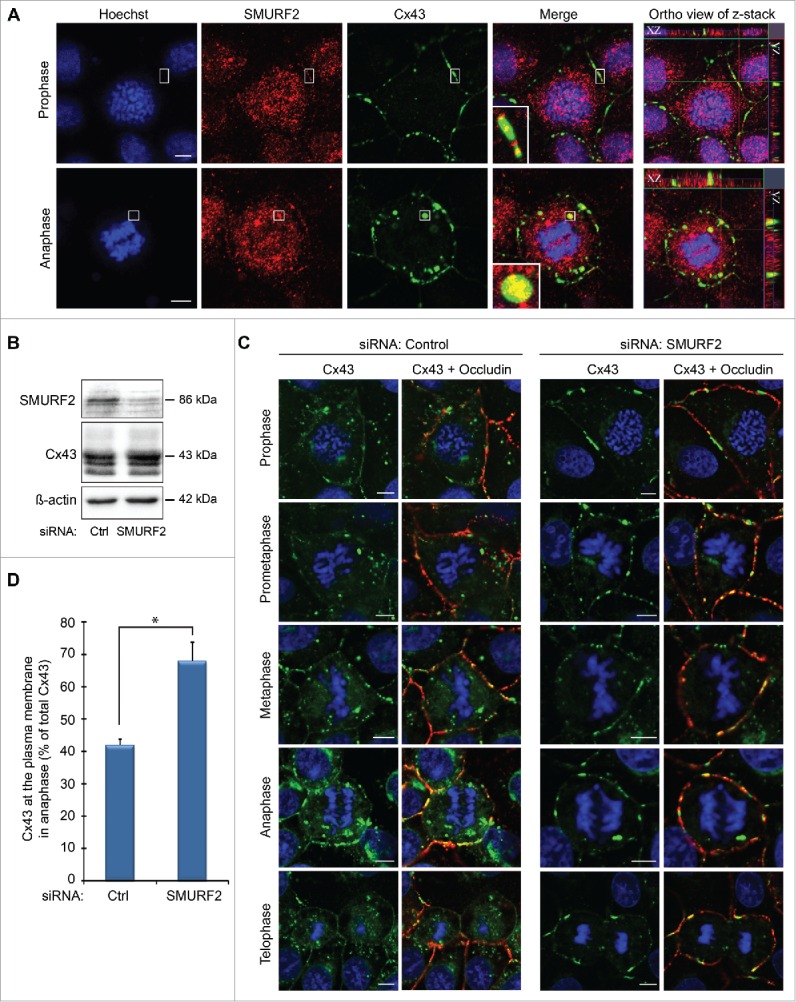
SMURF2 regulates Cx43 gap junction remodelling during mitosis. (A) IAR20 cells were fixed, co-stained against Cx43 (green) and SMURF2 (red), and representative images of single confocal planes were acquired using fluorescence confocal microscopy. Cell-cycle stages were defined by DNA staining with Hoechst. Scale bars, 5 µm. (B) IAR20 cells were either transfected with non-targeting siRNA as a control or with siRNA sequences against SMURF2. After 48 hours of transfection, cell lysates were prepared and equal amounts of total cell protein were subjected to SDS-PAGE. Cx43 and SMURF2 were detected by western blotting, using anti-Cx43 and anti-SMURF2 antibodies, respectively. The blots were stripped and reprobed with anti-β-actin antibodies. (C) IAR20 cells were either transfected with non-targeting siRNA as a control or with siRNA sequences against SMURF2. After 48 hours of transfection, the cells were fixed, co-stained against Cx43 (green) and occludin (red), and representative images of single confocal planes were acquired using fluorescence confocal microscopy. The mitotic phases were determined based on DNA staining with Hoechst. Scale bars, 5 µm. (D) To quantify the percentage Cx43 in the plasma membrane compared to total Cx43, 4 to 8 cells in anaphase from 3 independent experiments were analyzed in control siRNA-transfected cells and SMURF2 siRNA-transfected cells. Values shown are the mean ± SEM *P < 0.001.
A second Cx43 subpool localizes in intercellular plasma membrane bridges during mitosis
Next, mitotic IAR20 cells were studied by live-cell differential interference contrast (DIC) imaging. This analysis revealed that mitotic cells often formed intercellular bridges with adjacent interphase cells as the mitotic cells rounded up (Fig. 3A; Movie 1 and Fig. S2; Movie S1). Such intercellular bridges were also observed when fixed IAR20 cells were analyzed by fluorescence confocal microscopy (Fig. 3B). Furthermore, we observed that a subpool of Cx43 in IAR20 cells did not undergo endocytosis during mitosis. Rather, this Cx43 pool localized in the plasma membrane bridges formed between the mitotic cells and their adjacent interphase cells, as defined by DIC imaging (Fig. 3B). To obtain a better understanding of how the distribution of Cx43 changes throughout mitosis, we performed time-lapse fluorescence microscopy of IAR20 cells cotransfected with Cx43-EGFP and tomato-EEA1-CT in order to define early endosomes. In accordance with the data obtained in fixed IAR20 cells (Fig. 1), subdomains of Cx43-EGFP-positive gap junctions were found to bud into the mitotic cell during mitosis (Fig. 3C; Movie 2). Sometimes, the internalized Cx43-enriched vesicles were found to undergo fusion with early endosomes (Fig. 3C; Movie 2). Over the course of 60 minutes, the level of Cx43-EGFP-positive gap junctions between mitotic cells and adjacent cells was gradually decreased, whereas the intensity of Cx43-EGFP in intracellular vesicular compartments increased (Fig. 3C; Movie 2). Moreover, in accordance with our observations in fixed cells (Fig. 3B), a subpool of Cx43 was found to not undergo endocytosis during mitosis. Rather, this protein pool appeared to remain localized in plasma membrane bridges formed between mitotic cells and neighboring cells (Fig. 3C; Movie 2). After the completion of mitosis, this pool of Cx43 appeared to be able to rapidly reassemble into gap junctions (Fig. 3C; Movie 2). To further characterize the formation of the intercellular plasma membrane bridges formed during mitosis, IAR20 cells transfected with Cx43-EGFP were analyzed by live-cell fluorescence/DIC imaging (Fig. 3D; Movie 3). This analysis confirmed that a subpool of Cx43 localizes in intercellular bridges between mitotic cells and adjacent interphase cells that are formed as the mitotic cells round up (Fig. 3D; Movie 3). In these bridges, Cx43 was sometimes found to localize in vesicular structures, which appeared to be transported to the mitotic cell (Fig. 3D; Movie 3). The analysis further suggested that these bridges are formed during the dislodgement of the mitotic cell and its adjacent cells as the mitotic cell rounds up. Following mitotic exit, gap junctions were found to be gradually established in the two daughter cells (Fig. 3D; Movie 3). Collectively, these observations suggest that Cx43 is subjected to two distinct fates during mitosis. One Cx43 pool undergoes endocytosis and trafficking to early endosomes, whereas a second pool remains localized in plasma membrane bridges that are formed between mitotic cells and their neighboring interphase cells during mitotic rounding.
Figure 3.
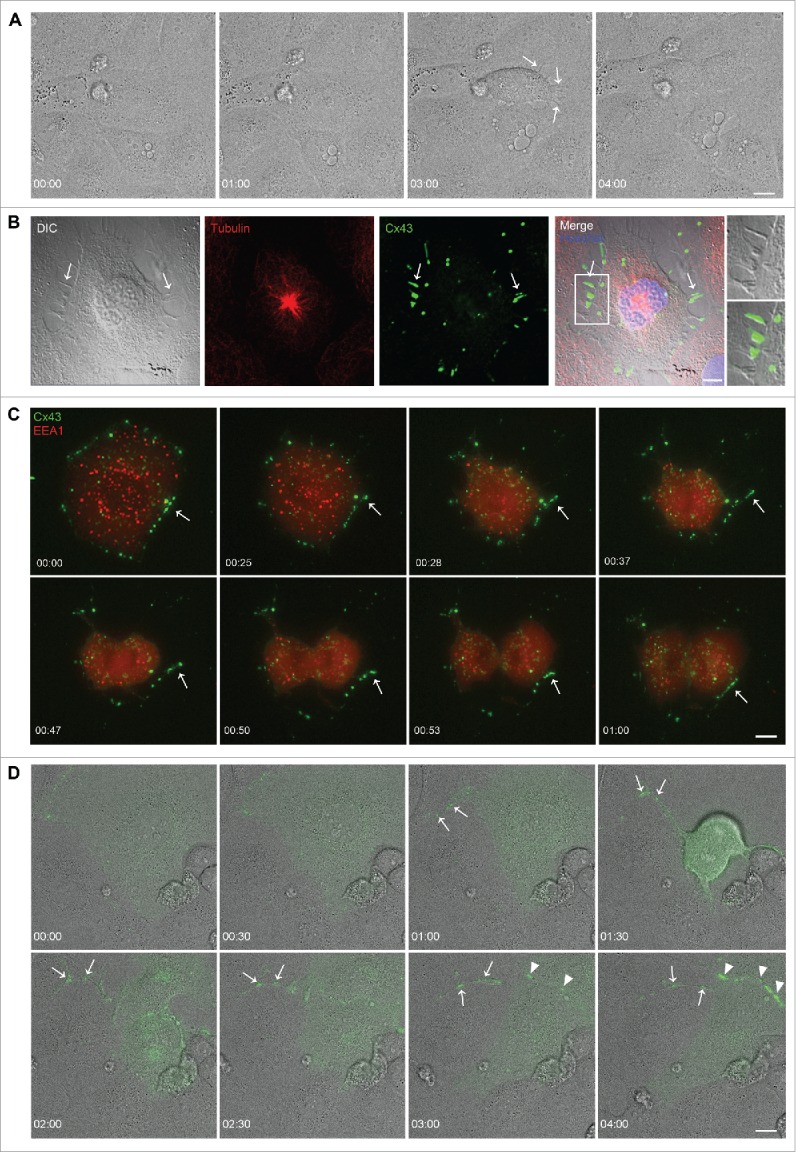
A subpool of Cx43 localizes in plasma membrane bridges between mitotic cells and adjacent cells. (A) IAR20 cells were subjected to DIC live cell imaging, and images were collected every minute over a period of 5.5 hours. Arrows indicate plasma membrane bridges that are formed between a mitotic cell and adjacent interphase cells as the mitotic cell rounds up. Scale bar, 10 µm. For the corresponding time-lapse movie, see Movie 1. (B) IAR20 cells were fixed, stained against tubulin (red) and Cx43 (green), and representative images of single confocal planes were acquired using fluorescence confocal microscopy. Arrows indicate plasma membrane bridges between a mitotic cell and an adjacent cell, as detected by DIC. Insets show enlarged view of representative membrane bridges. Scale bar, 10 µm. (C) IAR20 cells co-expressing Cx43-EGFP and Tomato-EEA1-CT were subjected to live cell imaging, and images were collected every minute over a 60 minutes period. Representative images of single confocal planes at various time points as indicated are shown. Arrows indicates a Cx43 pool that appears to localize in a plasma membrane bridge formed between a mitotic cell and an adjacent cell, which appears to rapidly reassemble into gap junction-like structures in the plasma membrane after the completion of mitosis. Scale bar, 10 µm. For the corresponding time-lapse movie, see Movie 2. (D) IAR20 cells transfected with Cx43-EGFP were subjected to live cell imaging including DIC. Images were collected every minute over a period of 5.5 hours. Representative images of single confocal planes at various time points as indicated are shown. Arrows indicates a Cx43 pool that localizes in a plasma membrane bridge formed between a mitotic cell and an adjacent cell. Arrowheads indicate gap junctions that are formed between the two daughter cells following mitotic exit. Scale bar, 10 µm. For the corresponding time-lapse movie, see Movie 3.
In order to study whether this localization of Cx43 in intercellular plasma membrane bridges during mitosis is specific for IAR20 cells or may also occur in other cell types, we studied HeLa cells stably transfected with Cx43 (HeLa-Cx43). Similar to our observations in IAR20 cells, Cx43 was found to be subjected to two distinct fates during mitosis in the HeLa-Cx43 cells. One Cx43 pool was subjected to endocytosis followed by trafficking to early endosomes (Fig. S3A), whereas a second pool remained localized in intercellular bridges that formed between the mitotic cell and adjacent cells as the mitotic cell rounded up (Fig. S3B).
The plasma membrane bridges formed between mitotic cells and adjacent cells are actin based
Next, we aimed to characterize the molecular composition of the Cx43-positive plasma membrane bridges that formed between mitotic cells and their adjacent cells. Morphologically, these structures resembled tunneling nanotubes, which are recently discovered conduits for long-distance communication between interphase cells.24-26 Since Cx43 has previously been shown to localize in tunneling nanotubes in interphase cells,31 we asked whether the Cx43-positive intercellular bridges that formed between mitotic cells and the adjacent cells displayed molecular features previously described for tunneling nanotubes. In human macrophages, two classes of membrane nanotubes have been observed: thin nanotubes that contain only F-actin and thicker nanotubes that contain both F-actin and microtubules (7). Others have reported that tunneling nanotubes contain F-actin, but not microtubules (8). To characterize the observed intercellular bridges in mitotic cells, we stained HeLa-Cx43 cells against tubulin and F-actin by using anti-tubulin antibodies and fluorophore-conjugated phalloidin, respectively. The Cx43-positive intercellular bridges that formed between mitotic cells and adjacent cells during mitotic rounding were found to be positive for F-actin (Fig. 4A; Fig. S4A). In contrast, microtubules were not detected in these structures (Fig. 3B; Fig. S3B). Thus, the plasma membrane bridges that formed between mitotic cells and their neighboring cells resemble the tunneling nanotubes in that they contain F-actin. Notably, they were also sometimes found to be devoid of Cx43 staining, suggesting that the presence of Cx43 is not a prerequisite for their formation (Fig. 4A, B).
Figure 4.
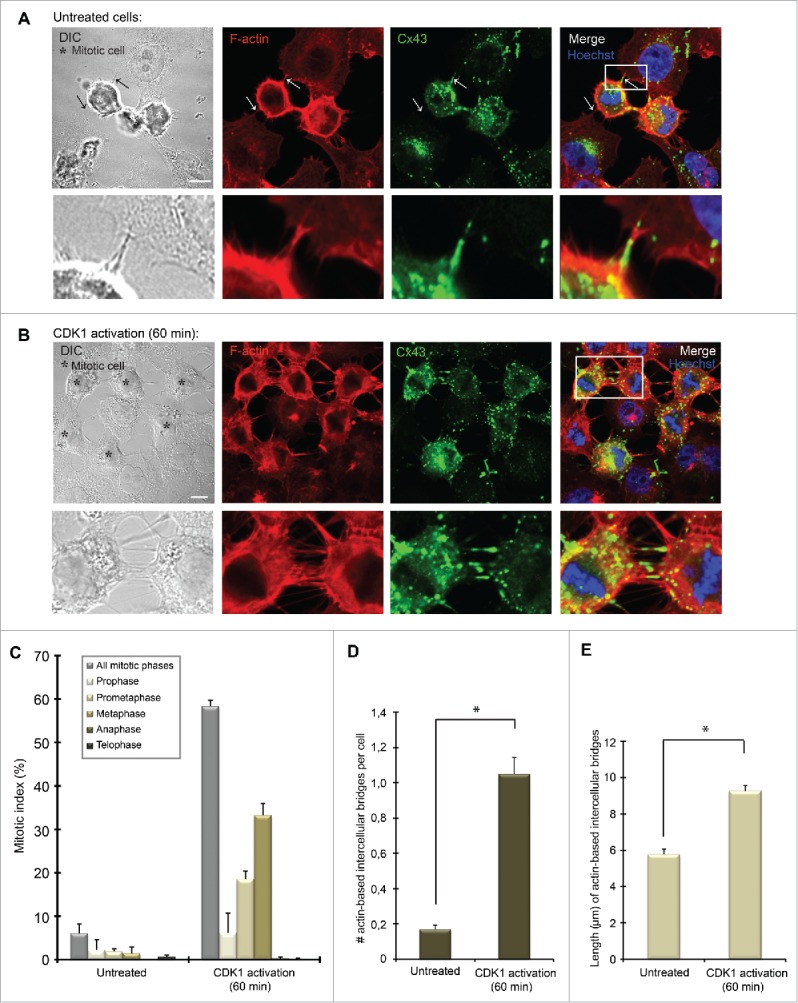
Characterization of plasma membrane bridges between mitotic cells and adjacent cells in non-synchronized and synchronized HeLa cells. Hela-Cx43 cells were (A) left untreated or (B) treated with RO-3306 (10 µM) for 18 hours followed by wash-out of RO-3306 and incubation in normal cell culture medium for 60 minutes. The cells were then fixed and stained against Cx43 using anti-Cx43 antibodies (green) and against F-actin using fluorophore-conjugated Phalloidin (red). The nuclei were stained using Hoechst. Representative images of single confocal planes acquired using fluorescence confocal microscopy are shown. Insets show enlarged view of representative intercellular membrane bridges. The arrow to the right in A indicates an intercellular plasma membrane bridge containing Cx43, whereas the arrow to the left indicates an intercellular plasma membrane bridge negative for Cx43. Scale bars, 10 µm. (C) The graph shows the mitotic index of control Hela-Cx43 cells and cells subjected to RO-3306 treatment for 18 hours followed by wash-out of the drug and incubation for 60 minutes in normal cell culture medium. The mitotic cells were identified and categorized according to mitotic phases on the basis of DNA staining with Hoechst, using fluorescence confocal microscopy. The mitotic index was calculated as the ratio of the number of cells undergoing mitosis to the total number of all cells (n = 628 for untreated cells, n = 614 for cells subjected to CDK1 activation for 1 hour). The number (D) and average length (E) of actin-based intercellular bridges between cells (including both interphase cells and mitotic cells) were quantified in untreated cells (n = 485) and in cells synchronized in mitosis by RO-3306 treatment (n = 259), using fluorescence confocal microscopy. Values shown are the mean ± SEM from three independent experiments. *P < 0.01.
In order to facilitate the study of the actin-based plasma membrane bridges that formed between mitotic cells and neighboring cells, we chemically induced enrichment of cells in mitosis. To this end, HeLa-Cx43 cells were synchronized at the G2/M border by treating the cells with the selective cyclin-dependent kinase 1 (CDK1) inhibitor RO-3306 for 18 hours.36 Cells were then allowed to enter mitosis in a synchronized manner by washing out RO-3306. As expected, following the release of RO-3306, Cx43 was found to undergo increased endocytosis, resulting in a gradual loss of gap junctions at the plasma membrane (Fig. 4B). Moreover, a subpool of Cx43 was found to localize in actin-based plasma membrane bridges between mitotic cells and their neighboring cells in a manner that was similar to that in asynchronous cells (Fig. 4B). Under conditions in which cells were synchronized in mitosis, these bridges were also often found to connect several mitotic cells together (Fig. 4B; Fig. S4A). Notably, actin-based plasma membrane bridges between mitotic cells and adjacent cells were also found in HeLa cells not transfected with Cx43, in accordance with the notion that the presence of Cx43 is not a prerequisite for the formation of these structures (Fig. S4A).
Next, we aimed to obtain a quantitative assessment of the level of the actin-based plasma membrane bridges that formed between mitotic cells and the adjacent cells. One hour following the release of RO-3306, approximately 60% of the cells were found to be in mitosis (Fig. 4C). At this time point, the number of actin-based intercellular plasma membrane bridges per cell in the culture was approximately 6-fold higher than that in untreated cells (Fig. 4D). Moreover, the average length of the actin-based intercellular plasma membrane bridges increased from 5.8 µm in untreated cells to 9.3 µm following the release of RO-3306 for 60 minutes (Fig. 4E). This effect was likely due to the fact that the distance between the cells in the culture increases as more cells enter mitosis. As expected, the average number of actin-based intercellular plasma membrane bridges per mitotic cell was the same in the untreated cells and in the cells exposed to RO-3306 treatment and washout (Fig. S4C). Collectively, these data indicate that increasing the mitotic index in HeLa cells is associated with an increase in the number of actin-based intercellular plasma membrane bridges between cells.
To obtain a better understanding of the localization of Cx43 and F-actin in the actin-based plasma membrane bridges formed between mitotic cells and adjacent cells, we performed super-resolution microscopy. To this end, Hela-Cx43 cells were allowed to enter mitosis in a synchronized manner by using RO-3306 as described above, and the cells were then fixed, stained against Cx43 and F-actin, and analyzed by structured illumination microscopy (SIM). Three-dimensional reconstruction of z-stacks obtained by SIM imaging indicated that the diameter of the actin-based plasma membrane bridges formed between mitotic cells and the adjacent cells varied considerably (Fig. 5A). Sometimes, the actin-based plasma membrane bridges appeared to have diameters of >1 µm as observed by SIM imaging. Such structures seemed to contain bundles of actin, which resembled the parallel actin bundles that support filopodial protrusions (Fig. 5A).37 Notably, actin-based plasma membrane bridges were found to form between mitotic cells and adjacent cells even though the cells were closely localized to each other (Fig. 5B).
Figure 5.
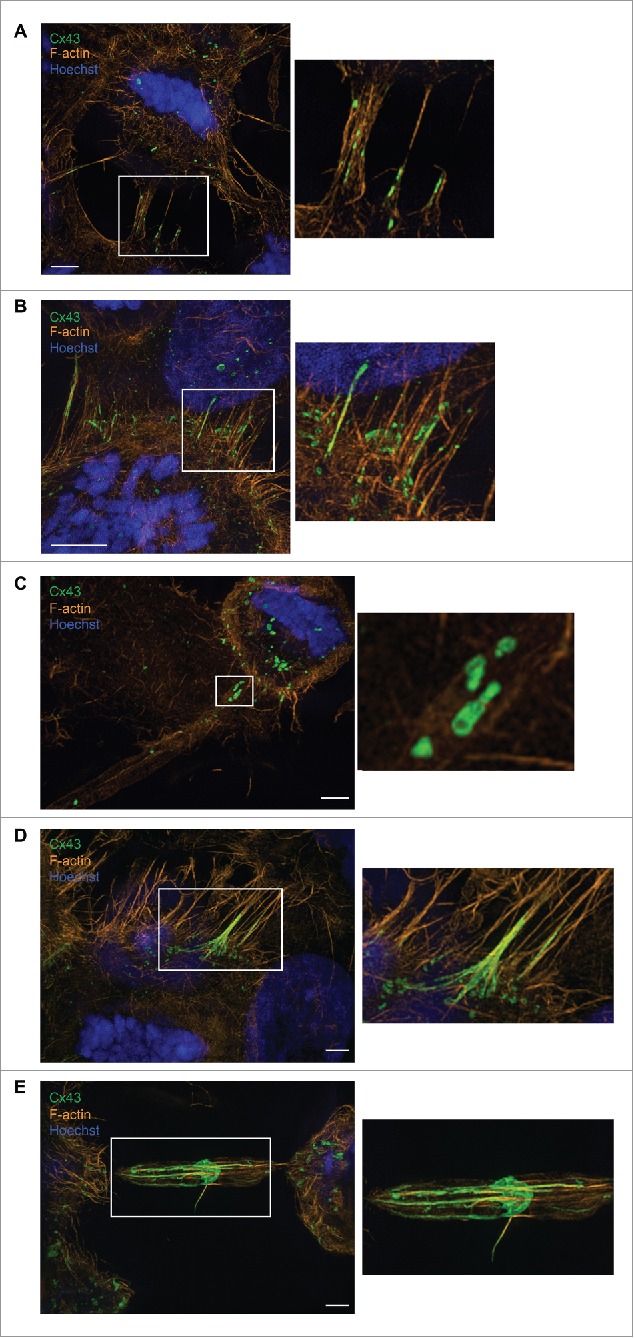
Super-resolution microscopy analysis of actin-based plasma membrane bridges between mitotic cells and adjacent cells. (A-E) Hela-Cx43 cells were treated with 10 µM RO-3306 for 18 hours followed by wash-out of RO-3306 and incubation in normal cell culture medium for 60 minutes. The cells were then fixed and stained against Cx43 using anti-Cx43 antibodies (green) and against F-actin using fluorophore-conjugated Phalloidin (red). The nuclei were stained using Hoechst. The cells were analyzed by SIM, and the images acquired were then subjected to 3-dimensional reconstruction using IMARIS. Scale bars, 3 µm.
SIM imaging further revealed that the actin-based plasma membrane bridges between mitotic cells and adjacent cells contained both Cx43-positive vesicles (Fig. 5A, B, C) as well as elongated gap junction-like structures (Fig. 5B, D, E). Sometimes, Cx43 was found to form gap junction-like structures localized in the center of the tube, with approximately equal distance to the mitotic cell and the adjacent cell (Fig. 5E). These structures had a disc-like shape (Fig. 5E).
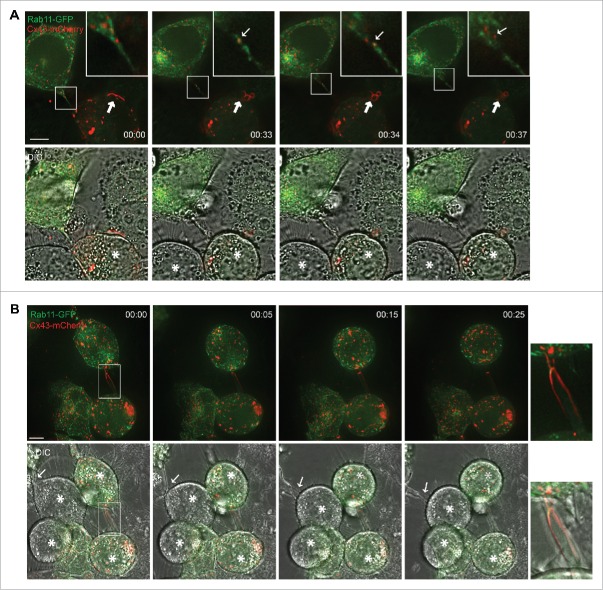
To obtain a better understanding of how the gap junction-like structures within the actin-based bridges between mitotic cells and adjacent cells are formed, we co-seeded HeLa-Cx43 cells transfected with Cx43-EGFP with HeLa-Cx43 cells transfected with Cx43-mCherry. As determined by fluorescence confocal microscopy, the intercellular bridges that formed between mitotic cells and adjacent interphase cells were found to contain both Cx43-EGFP and Cx43-mCherry, as indicated by yellow fluorescence (Fig. 6). These observations suggest that both mitotic cells and adjacent cells contribute to the formation of the gap junction-like structures localizing in the intercellular bridges.
Figure 6.
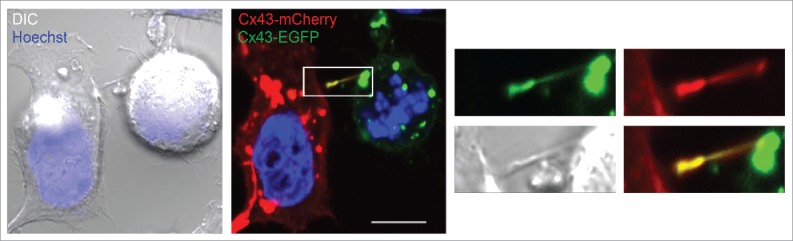
Analysis of Cx43 in actin-based plasma membrane bridges between mitotic cells and adjacent cells. HeLa-Cx43 cells were transiently transfected with either Cx43-EGFP (green) or with Cx43-mCherry (red) for 24 hours. The two cell populations were then trypsinized, mixed and reseeded for an additional 24 hours. The cells were treated with RO-3306 (10 µM) for 18 hours followed by wash-out of RO-3306 and incubation in normal cell culture medium for 60 minutes. The cells were then fixed and the nuclei were stained with Hoechst. The cells were visualized by fluorescence confocal microscopy. Representative images of single confocal planes are shown. Yellow color indicates colocalization between Cx43-EGFP and Cx43-mCherry. Scale bar, 10 µm.
The actin-based plasma membrane bridges between mitotic cells and their non-mitotic neighboring cells mediate intercellular transfer of vesicles
Tunneling nanotubes have previously been shown to be involved in mediating the transport of vesicles and organelles between interphase cells.24-26 Among the vesicles transported via tunneling nanotubes are Rab11-positive vesicles.38 Rab11 is a well-established marker for recycling endosomes.39 To elucidate whether Cx43-positive vesicles and/or Rab11-positive vesicles are trafficked between mitotic cells and their neighboring cells via the actin-based intercellular bridges, HeLa-Cx43 cells were co-transfected with Cx43-mCherry and Rab11-GFP. Cells were then allowed to enter mitosis in a synchronous manner by treating the cells with RO-3306 as described above. Fifteen minutes after the release of RO3306, cells were analyzed by time-lapse fluorescence microscopy. Rab11-positive vesicles were found to be transported between mitotic cells and their neighboring interphase cells via the actin-based plasma membrane bridges (Fig. 7A; Movie 4). Some of these Rab11-positive vesicles were positive for Cx43, suggesting that Cx43 is transported between mitotic cells and neighboring interphase cells through actin-based bridges via recycling endosomes (Fig. 7A; Movie 4).
Figure 7.
Vesicles containing Rab11 and Cx43 are transported between mitotic cells and adjacent cells via the actin-based plasma membrane bridges. HeLa-Cx43 cells co-transfected with plasmids encoding Cx43-mCherry (red) and Rab11-GFP (green) were treated with RO-3306 (10 µM) for 18 hours, followed by RO-3306 wash-out and incubation in normal cell culture medium for 15 minutes. The cells were then subjected to live cell imaging, and z-stack images were taken every 50 second over a period of 40 minutes. Representative images of single confocal planes at various time points as indicated are shown. (A) The images demonstrate the trafficking of Rab11- and Cx43-positive vesicles between the mitotic cell and one of its neighboring cells via an actin-based plasma membrane bridge. Insets show vesicle transport in the mitotic nanotube in higher magnification. Small arrow shows a vesicle positive for both Rab11 and Cx43, which is transported between the mitotic cell and an adjacent interphase cell. Mitotic cells are indicated by asterisks. Large arrow indicates a gap junction in the process of being internalized and forming annular gap junctions during cell rounding. Scale bar, 7 µm. For the corresponding time-lapse movie, see Movie 4. (B) The actin-based plasma membrane bridges sometimes contained large gap junction-like structures (insets). Mitotic cells are indicated by asterisks. Arrows indicate the formation of a plasma-membrane bridge between a mitotic cell and an adjacent cell. Scale bar, 7 µm. For the corresponding time-lapse movie, see Movie 5.
In addition to localizing in the actin-based bridges, vesicles containing both Cx43-mCherry and Rab11-GFP were found to localize intracellularly (Fig. 7A; Movie 4). In accordance with the notion that a subpool of Cx43 localizes in recycling endosomes during mitosis, Cx43 was found to partly colocalize with Rab11 in mitotic HeLa-Cx43 cells, as determined by analysis of fixed cells by fluorescence confocal microscopy (Fig. S5). Collectively, these observations suggest that Cx43 may undergo recycling following endocytosis during mitosis, in accordance with the findings of a previous study by Boassa et al.12
The actin-based bridges between mitotic cells and adjacent cells were also found to contain large gap junction-like structures, in accordance with the time-lapse fluorescence microscopy analysis and super-resolution microscopy analysis of IAR20 cells described earlier (Figs. 3C, 7B; Movie 2; Movie 5). These structures possibly act as reservoirs of Cx43 for the rapid reassembly of gap junctions at the mitotic exit. In accordance with the observations in IAR20 cells (Fig 3A; Movie 1, Fig S2; Movie S1 and Fig. 3D; Movie 3), the time-lapse fluorescence microscopy analysis of HeLa-Cx43 cells indicated that the actin-based bridges are formed by the dislodgement of the mitotic cells and its adjacent cells as the mitotic cell rounds up during mitosis (Fig 7B; Movie 5).
Collectively, these observations indicate that the actin-based plasma membrane bridges that are formed between mitotic cells and their adjacent cells during mitotic rounding have certain characteristics in common with tunneling nanotubes that have previously been described between interphase cells. For instance, they contain F-actin and have the ability to mediate the intercellular exchange of Rab11-positive vesicles.24-26 However, whereas tunneling nanotubes are less than 500 nm in diameter,24,25 the membrane tubes between mitotic cells and adjacent cells were observed to sometimes have diameters over 1 μm, as determined by super-resolution microscopy. Moreover, our data suggest that these structures may have unique characteristics and functions specifically related to the mitotic process. Thus, we propose the new term “mitotic nanotubes” to refer to these actin-based plasma membrane bridges between mitotic cells and adjacent cells.
Mitotic nanotubes contain ezrin and cell-cell junction proteins in addition to Cx43
Ezrin forms a complex with radixin and moesin, together called the ERM (ezrin/radixin/moesin) protein family, which participates in cross-linking actin to the plasma membrane and organizing actin filaments in cellular extensions.40 Previous studies have reported that ezrin localizes in tunneling nanotubes.27 As determined by fluorescence confocal microscopy, ezrin was found to localize in the mitotic nanotubes (Fig. S6A). These observations suggest that the ERM complex may have a role in cross-linking the actin filaments to the plasma membrane in these structures.
The above data suggest that mitotic nanotubes, among other functions, may act as plasma membrane reservoirs for gap junctions during mitosis. Notably, these structures were also found to contain the tight junction proteins ZO-1 and occludin, the adherens junction protein β-catenin, and the desmosome protein desmoplakin (Fig. S6B).
Comparison between mitotic nanotubes and retraction fibers
As determined by DIC imaging, the mitotic nanotubes were found to morphologically resemble the retraction fibers, which are actin-based protrusions involved in anchoring mitotic cells to the substratum during rounding (Fig. 8A).9,10 However, in contrast to mitotic nanotubes, retraction fibers did not contain Cx43 (Fig. 8A). We also compared mitotic nanotubes with retraction fibers by 3-dimensional reconstruction of z-stacks from SIM. This analysis revealed that, in contrast to retraction fibers, mitotic nanotubes were not attached to the substratum (Fig. 8B).
Figure 8.
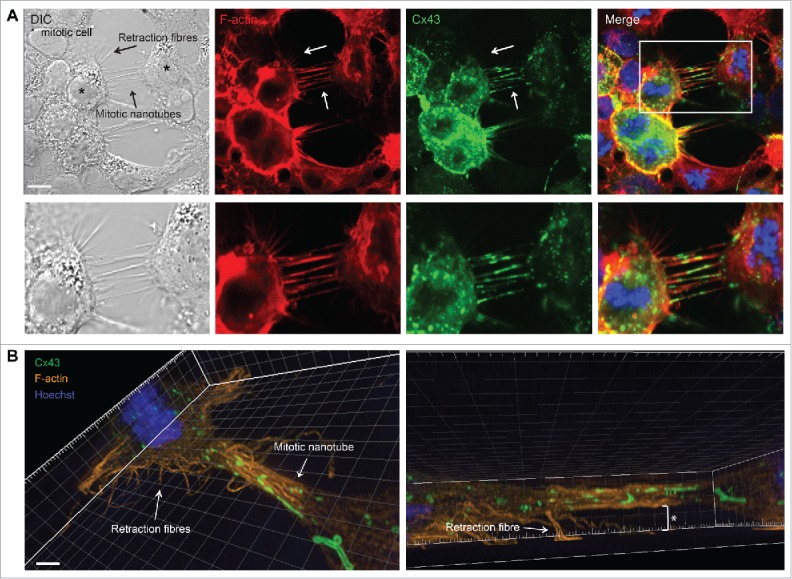
Morphological and molecular comparison between retraction fibers and mitotic nanotubes. Hela-Cx43 cells were treated with RO-3306 (10 µM) for 18 hours followed by wash-out of RO-3306 and incubation in normal cell culture medium for 60 minutes. The cells were then fixed and stained against Cx43 using anti-Cx43 antibodies (green) and F-actin was stained using fluorophore-conjugated Phalloidin (red). The nuclei were stained using Hoechst. (A) The cells were then visualized by fluorescence confocal microscopy. Representative images of single confocal planes are shown. Scale bars, 10 µm. (B) The cells were imaged by SIM, followed by 3-dimensional reconstruction of images using the IMARIS software. The asterisk indicates the distance between the mitotic nanotube and the substratum. Scale bars, 2 µm.
Discussion
The round morphology obtained by cells during mitosis has been shown to be associated with a rapid decrease in cell surface area and volume.2,5-7 However, how mitotic cells communicate with adjacent cells during rounding has remained elusive. In the present study, we demonstrate that mitotic cells are able to communicate with neighboring cells by forming actin-based plasma membrane bridges. The ability of these structures to mediate the exchange of cytoplasmic content between cells along with their high F-actin content is reminiscent of tunneling nanotubes, which have previously been described between interphase cells.24-26 However, our data indicate that these structures may also have unique characteristics and functions specifically related to the mitotic process. Thus, we propose the new term “mitotic nanotubes” to refer to these intercellular structures that are formed during mitosis (Fig. 9).
Figure 9.
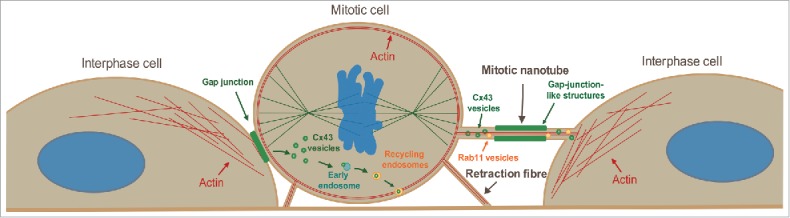
Model on the regulation of Cx43 during mitosis. Based on the data presented in this study, we propose that mitotic cells are able to form actin-based plasma membrane bridges with adjacent cells during rounding, termed mitotic nanotubes. These structures are involved in mediating trafficking of vesicles to and from the mitotic cells and may act as plasma membrane reservoirs during cell rounding. We also propose that Cx43 is subjected to two different fates during mitosis. One subpool undergoes increased endocytosis and subsequent sorting to early endosomes and recycling endosomes. Cx43 may then undergo recycling to the plasma membrane and form de novo functional gap junctions at the mitotic exit. A second pool of Cx43 localizes in the mitotic nanotubes formed between the mitotic cell and its neighboring cells. At the completion of mitosis, both the Cx43 that localizes in the mitotic nanotubes as well as the Cx43 pool that has been internalized may reassemble to form new gap junctions between cells.
Mitotic cell rounding involves major changes in cytoskeleton organization. The microtubules are remodeled in order to generate a mitotic spindle, whereas changes in the actin cytoskeleton drive the rounding and cortical stiffening of mitotic cells.41,42 The actin cytoskeleton is also involved in forming retraction fibers, which anchor the rounded mitotic cells to the substratum.9,10 In the present study, we show that actin is also involved in forming plasma membrane bridges between mitotic cells and adjacent cells and that these structures act as conduits for intercellular communication. In analogy to the role of retraction fibers in anchoring the rounded mitotic cells to the substratum, mitotic nanotubes may participate in anchoring the rounded mitotic cells to their adjacent cells. It is also possible that the actin fibers localized in mitotic nanotubes may be involved in the transport of vesicles between the mitotic cells and their neighboring cells.
Interestingly, early scanning electron microscopy studies on Chinese hamster ovary cells described filopodia-like structures with a diameter of 50–100 nm and a length of several micrometers that appeared to link mitotic cells with other cells.4 However, the molecular composition or functions of these structures were not characterized. Thus, it remains to be determined whether these structures may have molecular and functional similarities to the actin-based plasma membrane bridges that we have characterized in the IAR20 and HeLa cells in the present study.
Our data indicate that the mitotic nanotubes are involved in mediating transfer of Rab11-positive vesicles between mitotic cells and adjacent cells. In future studies it will be interesting to further characterize the type of cargo that is transported to and from the mitotic cells via these structures. Recent studies have shown that tunneling nanotubes are also involved in mediating electrical coupling between non-mitotic cells.31,32 The presence of Cx43 in the tunneling nanotubes was shown to be essential for the ability of the tunneling nanotubes to electrically couple cells.31,32 Here, we have shown that Cx43 was frequently found to localize in the mitotic nanotubes. Whether the mitotic nanotubes may be involved in meditating electrical coupling between mitotic cells and adjacent cells, and the possible role of such coupling in mitosis, remains to be investigated. The Cx43 pool in the mitotic nanotubes was found to localize in vesicular structures, some of which were Rab11 positive, or in gap junction-like structures. Further studies are required in order to precisely define these structures, and to determine whether they are double-membrane structures as in gap junctions, or whether they consist of hemichannels not assembled into gap junctions.
Our data suggest that the Cx43 pool that localizes in mitotic nanotubes may be rapidly reassembled to form new gap junctions at the mitotic exit, raising the possibility that the mitotic nanotubes can act as reservoirs for Cx43 during mitosis. This potential novel mechanism for gap junction remodeling during mitosis could be highly beneficial and energy saving, allowing cells to quickly restore gap junction intercellular communication as soon as they have finished dividing. In contrast to the gap junctions, the tight junctions, adherens junctions and desmosomes have been shown to persist throughout cell division.11 However, the mitotic nanotubes were observed to contain not only Cx43 but also protein constituents of the tight junctions, adherens junctions, and desmosomes, suggesting that mitotic nanotubes may to some extent also act as a reservoir for the other intercellular junctions during mitosis.
Gap junctions are highly dynamic plasma membrane domains, and newly synthesized Cx43 is continuously recruited to the outer edges of gap junction plaques, while the older Cx43 pool is removed from the center of the gap junction plaques by endocytosis.43,44 Gap junction levels can be adjusted by altering the rate of connexin synthesis, the trafficking of connexins to the plasma membrane, the assembly of connexins into gap junctions, or the rate of endocytosis and degradation.45 The endocytosis of gap junctions is a unique process in which both membranes of the junction are internalized into one of the adjacent cells, forming a double-membrane vesicle called an annular gap junction or connexosome.46-51 Following gap junction endocytosis, connexins may be trafficked to and degraded in lysosomes.52,53 Our data show that Cx43 is transported to early endosomes following gap junction endocytosis during mitosis. This observation is in agreement with a previous study by Boassa et al., showing that Cx43 colocalizes with EEA1 in mitotic normal rat kidney (NRK) cells.12 The early endosome is known to act as a major sorting station for endocytosed plasma membrane proteins.54 Hence, the finding that Cx43 localizes to early endosomes in mitotic cells raises the interesting possibility that the early endosomes may be involved in regulating the reestablishment of Cx43 gap junctions after cytokinesis. The results obtained in the present study are in agreement with those of previous studies demonstrating that Cx43 gap junctions resume at the plasma membrane after cytokinesis.12,16,17,34 Boassa et al. have proposed that the reestablishment of Cx43 gap junctions after cytokinesis is due to the recycling of endocytosed Cx43 from intracellular compartments to the plasma membrane.12 This model for regulation of gap junction levels during mitosis would be in accordance with findings from previous studies suggesting that the modulation of endosomal recycling is a general mechanism for regulating the surface expression of plasma membrane proteins and for controlling the plasma membrane area during mitosis.2 The data obtained in the current study are in line with a scenario in which Cx43 undergoes recycling to the plasma membrane to reassemble into gap junctions as the cells exit mitosis. For instance, we have found that Cx43 localizes in intracellular vesicles that contain Rab11, a marker for recycling endosomes. On the basis of the data obtained in the present and previous studies on the mitotic regulation of Cx43, a possible scenario for the remodeling of gap junctions during mitosis could be that Cx43 is subjected to two distinct fates during mitosis, in which one Cx43 pool is internalized and trafficked to early endosomes and recycling endosomes, while a second Cx43 pool may localize in mitotic nanotubes. In this scenario, both Cx43 pools would be able to reassemble into gap junctions after mitotic exit. Further studies are required to precisely define the mechanisms involved in the reestablishment of gap junctions after mitotic exit. It is important to take into consideration that during the reestablishment of gap junctions following mitotic exit, also newly synthesized Cx43 is likely to play an important contribution. The relative importance of Cx43 recycling and new synthesis of Cx43 in the reestablishment of gap junctions following mitotic exit remains to be determined.
We have previously demonstrated that the E3 ubiquitin ligase SMURF2 regulates the endocytosis of Cx43 gap junctions in IAR20 cells under basal conditions and in response to PKC activation.35 In the present study, we found that SMURF2 has a major role in regulating the endocytosis of Cx43 gap junctions during mitosis in IAR20 cells. The data further suggest that endocytosis of Cx43 is essential for the remodeling of gap junctions during mitosis. SMURF2 has previously been shown to regulate the normal progression of the spindle assembly checkpoint (Osmundson et al., 2009). Our findings raise the intriguing scenario that SMURF2 may have a role in coordinating the downregulation of Cx43 gap junction intercellular communication with the control of the mitotic spindle checkpoint and possibly other mitotic events.
In conclusion, this study provide new insights into the mechanisms involved in the remodeling of gap junctions during mitosis, and identifies actin-based plasma membrane bridges as a novel means of communication between mitotic cells and their neighboring cells. Collectively, the data add another layer of complexity to our understanding of how cells communicate with their neighboring cells and modulate their surface area during mitosis.
Materials and methods
Cell culture
The rat liver epithelial cell line IAR20, originally isolated from normal inbred BD-IV rats, was obtained from the International Agency for Research on Cancer (Lyon, France) (14). HeLa cells stably transfected with rat Cx43 (HeLa-Cx43) were a kind gift from Professor Klaus Willecke (University of Bonn, Germany), and have been described previously (15). Cells were grown on 35-mm or 100-mm Petri dishes in Dulbecco´s modified Eagle´s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco BRL Life Technologies, Inchinnan, UK) and l-glutamine (Sigma). Cell cultures were regularly tested for mycoplasma infection using Myco Alert (Lonza, Walkersville, MD) according to the manufacturer's protocol.
Reagents and antibodies
The CDK1 inhibitor IV, RO-3306, was obtained from VWR (Radnor, PA). Anti-Cx43 antibodies were obtained by injecting rabbits with a synthetic peptide consisting of the 20 C-terminal amino acids of Cx43 (dilutions for Western blotting were 1:4000; for immunofluorescence 1:500) (16). Mouse anti-Cx43 antibodies were from Chemicon International, Inc. (Temecula, CA; 1:100). Mouse anti-occludin (33–1500; 1:100) was from Thermo Fisher Scientific (Maltham, MA). Mouse anti-Actin (A2228; 1:5000) antibodies were from Sigma-Aldrich (St. Louis, MO). Mouse anti-ZO-1 (610966; 1:100) antibodies, mouse anti-β-Catenin (610153; 1:100) antibodies, and mouse anti-EEA1 (610456; 1:100) antibodies were all from BD Transduction Laboratories (Franklin Lakes, NJ). Mouse anti-vimentin (3390; 1:100), rabbit anti-ezrin (3145; 1:100), and rabbit anti-moesin (3150; 1:100) were all obtained from Cell Signaling Technology (Danvers, MA). Rabbit anti-SMURF2 (H-50, sc-25511; 1:100) antibodies were obtained from Santa Cruz (Berkely, CA). Rabbit anti-tubulin (EP1332Y; 1:200) antibodies and mouse anti-desmoplakin I+II antibodies (2Q400; 1:100) were both from Abcam (Cambridge, UK). Alexa488-conjugated goat anti-rabbit IgG (A11034; 1:1000) and Alexa555-conjugated goat anti-mouse IgG (A21424; 1:1000) were from Thermo Fisher Scientific (Maltham, MA). Horseradish peroxidase-conjugated Goat anti-rabbit IgG secondary antibodies (170–6515; 1:5000) were from Bio-Rad (Hercules, CA). Horseradish peroxidase-conjugated donkey anti-mouse IgG antibodies (715–035–150; 1:5000) were obtained from Jackson Immunoresearch Laboratories Inc. (West Grove, PA). Hoechst 33342 were from Life Technologies. Alexa Fluor 488 Phalloidin and Alexa Fluor 594 Phalloidin were obtained from Thermo Fisher Scientific (Maltham, MA).
DNA and siRNA transfection
The expression plasmids encoding Cx43-EGFP were a kind gift from Klaus Willecke (University of Bonn, Germany). Cx43-mCherry was a kind gift from Michael Davidson (Addgene plasmid #55023). The Tomato-EEA1-CT construct, made by recombining pDest-Tomato and pEntr-EEA1-CT, was kindly provided by Harald Stenmark (The Norwegian Radium Hospital, Oslo, Norway). Rab11-GFP was a gift from Richard Pagano (Addgene plasmid #12674). Cells were grown on 35-mm Petri dishes and transfected 24 h after seeding using Lipofectamine 2000 reagent Thermo Fisher Scientific (Maltham, MA) according to the recommendations of the manufacturers. Prior to plasmid transfection, the cell culture medium containing FBS and l-glutamine was replaced with DMEM containing only l-glutamine. Five hours after transfection, DMEM containing only l-glutamine was replaced with DMEM containing both FBS and l-glutamine.
The siRNA oligo targeted against SMURF2 (Stealth Select RNAi SMURF2RSS314470) was obtained from Thermo Fisher Scientific (Maltham, MA), and had the following sequence: 5´- GAGAUGAUAUCUACACGUUACAGAU-3´. As siRNA control constructs, Stealth RNAi Negative Control (Medium GC) from Thermo Fisher Scientific (Maltham, MA) was used. siRNA was transfected into cells using Lipofectamine 2000 at a final concentration of 80 nM, according to the manufacturer's procedure. The cell culture medium was replaced with DMEM supplemented with 10% (v/v) FBS 24 hours after transfection. The cells were assayed 48 hours after transfection.
Cell-cycle synchronization
Cell-cycle synchronization at the G2/M phase border was achieved by treating cells with the reversible CDK1 inhibitor RO-3306 (10 µM, Sigma Aldrich, St. Louis, MO) for 18 hours. The cells were then allowed to enter mitosis in a synchronous manner by washing the cells three times with cell culture medium without RO-3306 and incubated for various time points without RO-3306 before assaying.
Confocal microscopy
Cells grown in monolayer were fixed with 4% formaldehyde in PBS for 15 minutes at room temperature, rinsed in PBS and permeabilized with 0.1% Triton X-100 for 30 minutes. The cells were then rinsed twice in PBS and incubated with PBS containing 5% (w/v) dry milk and 0.1% Tween for 1 hour. The cells were incubated with primary antibodies overnight, washed with PBS and incubated with Alexa488- and/or Alexa555-conjugated secondary antibodies for 1 hour. The cells were then rinsed in PBS and the nuclei were stained with Hoechst 33342 (10 µg/ml in PBS) prior to mounting with ProLong Gold Antifade Mountant (ThermoFisher Scientific, Maltham, MA). The cells were analyzed with a LSM 710 META confocal microscope (Carl Zeiss Inc., Oberkochen, Germany) equipped with a Plan Apochromat 63× 1.4 NA oil immersion objective (Carl Zeiss Inc., Oberkochen, Germany). Images were acquired with the ZEN 2009 edition software and processed with Adobe Photoshop CS4. The level of co-localization between Cx43 and EEA1 in the various mitotic phases was quantified in z-stacks using the IMARIS software (Bitplane, Andor Technology, Zurich, Switzerland). The level of Cx43 staining in the cell–cell interface as a percentage of the total cellular Cx43 staining in the same focal plane was quantified using ImageJ software, as described previously.35 To quantify the level of mitotic nanotubes in controls HeLa-Cx43 cells and in cells synchronized in mitosis, z-stacks of whole cells were acquired. The length and number of actin-based bridges observed between mitotic cells and adjacent cells was then quantified using the ZEN 2009 edition software.
Live-cell imaging
IAR20 and HeLa-Cx43 cells were seeded in MatTek's glass bottom culture dishes (MatTek Corporations, Ashland, MA) and co-transfected with plasmids 24 hours after seeding. Live-cell imaging was performed 48 hours after transfection. To record the time-lapse movies of the IAR20 cells, a DeltaVision fluorescence microscope (Applied Precision Inc., Issaquah, WA) equipped with Elite TruLight Illumination System, a CoolSNAP HQ2camera and a 60x Plan Apochromat (1.42 NA) lens was used. Images were taken every minute over a period of 60 minutes for up to 20 hours. Time-lapse images were acquired as z-sections, 0.6 µm apart. To increase the mitotic index, the plasmid-transfected HeLa-Cx43 cells were treated with RO-3306 for 18 hours. RO-3306 was then washed out and the cells were incubated in medium without RO-3306 for 15 minutes prior to initiation of live-cell imaging. To record the time-lapse movies of the HeLa-cells, a Deltavision OMX V4 microscope (Applied Precision Inc., Issaquah, WA) equipped with an Olympus ×60 NA 1.42 Plan Apochromat objective, an InSightSSI™ widefield illumination module, and 3 cooled sCMOS cameras was used. Images were taken every 50 second over a period of 40 minutes. Time-lapse images were acquired as 13 z-sections, 0.6 µm apart. Live-cell samples were enclosed in environmental chambers that were maintained at 37°C with 5% CO2 level during imaging. All time-lapse images acquired were deconvolved and processed using the softWoRx software (Applied Precision Inc., Issaquah, WA).
SIM
Hela-Cx43 cells were grown on high precision coverglasses, transfected and treated with RO-3306 for 18 hours followed by wash-out of RO-3306 and incubation in cell culture medium without RO-3306 for 60 minutes. The cells were then fixed with 4% formaldehyde diluted in PBS. For immunostaining, the cells were permeabilized with 0.1% Triton-X-100 in PBS. Immunostaining was done with antibodies diluted in PBS containing 5% (w/v) dry milk and 0.1% Tween, followed by Hoechst staining and washes. The samples were mounted with ProLong Gold Antifade Mountant on object slides.
Three-dimensional structured illumination microscopy was performed on a Deltavision OMX V4 microscope (Applied Precision, Inc., Issaquah, WA) equipped with an Olympus ×60 NA 1.42 Plan Apochromat objective, 405 nm, 445 nm, 488 nm, 568 nm, and 642 nm laserlines, and 3 cooled sCMOS cameras. Z-Stacks covering the whole cell were recorded, with a z-spacing of 125 nm. For each focal plane, 15 raw images (5 phases for 3 different angular orientations of the illumination pattern) were captured. Super-resolution images were reconstructed, aligned, and processed for presentation using Softworx software (Applied Precision, Inc., Issaquah, WA). IMARIS software was used for 3-dimensional reconstruction.
Statistical analysis
For statistical analysis, the independent sample t-test was used to determine significance. P values less than 0.05 were considered significant. Standard errors were calculated in Statistical Package for the Social Sciences (SPSS) (IBM), and graphs were created in Microsoft Excel. The box plot shown in Fig. S4B was made in SPSS. The other graphs were made in Microsoft Excel, which was also used to calculate standard deviations.
Western blotting
Cells grown in 35-mm Petri dishes were washed with PBS and scraped in 300 µl sodium dodecyl sulfate (SDS) electrophoresis sample buffer (10 mM Tris, pH 6.8, 15% w/v glycerol, 3% w/v SDS, 0.01% w/v bromophenol blue and 5% v/v 2-mercaptoethanol). The cell lysates were sonicated and heated for 5 minutes at 95°C. Samples were separated by 8% SDS-PAGE and transferred to nitrocellulose membranes as described (16). The membranes were developed with chemiluminescence using Lumiglo (EMD Millipore, Billerica, MA) or SuperSignal West Dura Extended Duration Substrate Thermo Fisher Scientific (Maltham, MA), and imaged using the ChemiDoc XRS+ System (Bio-Rad, Hercules, CA). Bands were quantified using the Image Lab v2.0 software (Bio-Rad, Hercules, CA).
Supplementary Material
Abbreviations
- CDK1
Cyclin-dependent kinase 1
- Cx43
connexin43
- DIC
differential interference contrast
- EEA1
Early endosome antigen 1
- ERM
ezrin/radixin/moesin
- PKC
protein kinase C
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SiRNA
small interfering RNA
- SMURF2
SMAD ubiquitination regulatory factor-2
- SIM
Structured Illumination Microscopy
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Zeremariam Yohannes for excellent technical support and to Dr. Eva Wenzel (Oslo University Hospital, Oslo, Norway) for excellent technical assistance on the DeltaVision fluorescence microscope.
Funding
This work was supported by the Norwegian Cancer Society [grant number 709125 to E.L.], the KG Jebsen Foundation [to R.A.L], and the Research Council of Norway through its Centres of Excellence funding scheme [project number 179571 to R.A.L, A.B., V.S., and E.L.].
ORCID
Vigdis Sørensen http://orcid.org/0000-0002-1694-890X
References
- [1].Cadart C, Zlotek-Zlotkiewicz E, Le BM, Piel M, Matthews HK. Exploring the function of cell shape and size during mitosis. Dev Cell 2014; 29:159-169; PMID:24780736; http://dx.doi.org/ 10.1016/j.devcel.2014.04.009 [DOI] [PubMed] [Google Scholar]
- [2].Boucrot E, Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proc Natl Acad Sci U S A 2007; 104:7939-7944; PMID:17483462; http://dx.doi.org/ 10.1073/pnas.0702511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Erickson CA, Trinkaus JP. Microvilli and blebs as sources of reserve surface membrane during cell spreading. Exp Cell Res 1976; 99:375-384; PMID:1269533; http://dx.doi.org/ 10.1016/0014-4827(76)90595-4 [DOI] [PubMed] [Google Scholar]
- [4].Porter K, Prescott D, Frye J. Changes in surface morphology of Chinese hamster ovary cells during the cell cycle. J Cell Biol 1973; 57:815-836; PMID:4735453; http://dx.doi.org/ 10.1083/jcb.57.3.815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boucrot E, Kirchhausen T. Mammalian cells change volume during mitosis. PLoS One 2008; 3:e1477; PMID:18213385; http://dx.doi.org/ 10.1371/journal.pone.0001477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Furthauer M, Gonzalez-Gaitan M. Endocytosis and mitosis: a two-way relationship. Cell Cycle 2009; 8:3311-3318; PMID:19770584; http://dx.doi.org/ 10.4161/cc.8.20.9700 [DOI] [PubMed] [Google Scholar]
- [7].Habela CW, Sontheimer H. Cytoplasmic volume condensation is an integral part of mitosis. Cell Cycle 2007; 6:1613-1620; PMID:17581282; http://dx.doi.org/ 10.4161/cc.6.13.4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cuddapah VA, Habela CW, Watkins S, Moore LS, Barclay TT, Sontheimer H. Kinase activation of ClC-3 accelerates cytoplasmic condensation during mitotic cell rounding. Am J Physiol Cell Physiol 2012; 302:C527-C538; PMID:22049206; http://dx.doi.org/ 10.1152/ajpcell.00248.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cramer LP, Mitchison TJ. Investigation of the mechanism of retraction of the cell margin and rearward flow of nodules during mitotic cell rounding. Mol Biol Cell 1997; 8:109-119; PMID:9017599; http://dx.doi.org/ 10.1091/mbc.8.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mitchison TJ. Actin based motility on retraction fibers in mitotic PtK2 cells. Cell Motil Cytoskeleton 1992; 22:135-151; PMID:1633624; http://dx.doi.org/ 10.1002/cm.970220207 [DOI] [PubMed] [Google Scholar]
- [11].Baker J, Garrod D. Epithelial cells retain junctions during mitosis. J Cell Sci 1993; 104 (Pt 2):415-425; PMID:7685036 [DOI] [PubMed] [Google Scholar]
- [12].Boassa D, Solan JL, Papas A, Thornton P, Lampe PD, Sosinsky GE. Trafficking and recycling of the connexin43 gap junction protein during mitosis. Traffic 2010; 11:1471-1486; PMID:20716111; http://dx.doi.org/ 10.1111/j.1600-0854.2010.01109.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kanemitsu MY, Jiang W, Eckhart W. Cdc2-mediated phosphorylation of the gap junction protein, connexin43, during mitosis. Cell Growth Differ 1998; 9:13-21; PMID:9438384 [PubMed] [Google Scholar]
- [14].Lampe PD, Kurata WE, Warn-Cramer BJ, Lau AF. Formation of a distinct connexin43 phosphoisoform in mitotic cells is dependent upon p34cdc2 kinase. J Cell Sci 1998; 111 (Pt 6):833-841; PMID:9472011 [DOI] [PubMed] [Google Scholar]
- [15].Stein LS, Boonstra J, Burghardt RC. Reduced cell-cell communication between mitotic and nonmitotic coupled cells. Exp Cell Res 1992; 198:1-7; PMID:1727042; http://dx.doi.org/ 10.1016/0014-4827(92)90141-T [DOI] [PubMed] [Google Scholar]
- [16].Vanderpuye OA, Bell CL, Murray SA. Redistribution of connexin 43 during cell division. Cell Biol Int 2016; 40(4):387-396; PMID:26724787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xie H, Laird DW, Chang TH, Hu VW. A mitosis-specific phosphorylation of the gap junction protein connexin43 in human vascular cells: biochemical characterization and localization. J Cell Biol 1997; 137:203-210; PMID:9105048; http://dx.doi.org/ 10.1083/jcb.137.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Saez JC, Berthoud VM, Branes MC, Martinez AD, Beyer EC. Plasma membrane channels formed by connexins: their regulation and functions. Physiol Rev 2003; 83:1359-1400; PMID:14506308; http://dx.doi.org/ 10.1152/physrev.00007.2003 [DOI] [PubMed] [Google Scholar]
- [19].Sohl G, Willecke K. An update on connexin genes and their nomenclature in mouse and man. Cell Commun Adhes 2003; 10:173-180; PMID:14681012; http://dx.doi.org/ 10.1080/cac.10.4-6.173.180 [DOI] [PubMed] [Google Scholar]
- [20].Laird DW. Syndromic and non-syndromic disease-linked Cx43 mutations. FEBS Lett 2014; 17;588(8):1339-1348 [DOI] [PubMed] [Google Scholar]
- [21].Wei CJ, Xu X, Lo CW. Connexins and cell signaling in development and disease. Annu Rev Cell Dev Biol 2004; 20:811-838; PMID:15473861; http://dx.doi.org/ 10.1146/annurev.cellbio.19.111301.144309 [DOI] [PubMed] [Google Scholar]
- [22].Mesnil M, Crespin S, Avanzo JL, Zaidan-Dagli ML. Defective gap junctional intercellular communication in the carcinogenic process. Biochim Biophys Acta 2005; 1719:125-145; PMID:16359943; http://dx.doi.org/ 10.1016/j.bbamem.2005.11.004 [DOI] [PubMed] [Google Scholar]
- [23].Naus CC, Laird DW. Implications and challenges of connexin connections to cancer. Nat Rev Cancer 2010; 10:435-441; PMID:20495577; http://dx.doi.org/ 10.1038/nrc2841 [DOI] [PubMed] [Google Scholar]
- [24].Abounit S, Zurzolo C. Wiring through tunneling nanotubes–from electrical signals to organelle transfer. J Cell Sci 2012; 125:1089-1098; PMID:22399801; http://dx.doi.org/ 10.1242/jcs.083279 [DOI] [PubMed] [Google Scholar]
- [25].Abounit S, Delage E, Zurzolo C. Identification and characterization of tunneling nanotubes for intercellular trafficking. Curr Protoc Cell Biol 2015; 67:12; PMID:26061240 [DOI] [PubMed] [Google Scholar]
- [26].Rustom A, Saffrich R, Markovic I, Gerdes HH. Nanotubular highways for intercellular organelle transport. Science 2004; 303:1007-1010; PMID:14963329; http://dx.doi.org/ 10.1126/science.1093133 [DOI] [PubMed] [Google Scholar]
- [27].Lou E, Fujisawa S, Morozov A, Barlas A, Romin Y, Dogan Y, Gholami S, Moreira AL, Manova-Todorova K, Moore MA. Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS One 2012; 7:e33093; PMID:22427958; http://dx.doi.org/ 10.1371/journal.pone.0033093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chinnery HR, Pearlman E, McMenamin PG. Cutting edge: Membrane nanotubes in vivo: a feature of MHC class II+ cells in the mouse cornea. J Immunol 2008; 180:5779-5783; PMID:18424694; http://dx.doi.org/ 10.4049/jimmunol.180.9.5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lou E. Intercellular conduits in tumours: the new social network. Trends Cancer 2016; 2:3-5; PMID:26949744; http://dx.doi.org/ 10.1016/j.trecan.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Osswald M, Jung E, Sahm F, Solecki G, Venkataramani V, Blaes J, Weil S, Horstmann H, Wiestler B, Syed M. Brain tumour cells interconnect to a functional and resistant network. Nature 2015; 528:93-98; PMID:26536111 [DOI] [PubMed] [Google Scholar]
- [31].Wang X, Veruki ML, Bukoreshtliev NV, Hartveit E, Gerdes HH. Animal cells connected by nanotubes can be electrically coupled through interposed gap-junction channels. Proc Natl Acad Sci U S A 2010; 107:17194-17199; PMID:20855598; http://dx.doi.org/ 10.1073/pnas.1006785107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang X, Gerdes HH. Long-distance electrical coupling via tunneling nanotubes. Biochim Biophys Acta 2012; 1818:2082-2086; PMID:21930113; http://dx.doi.org/ 10.1016/j.bbamem.2011.09.002 [DOI] [PubMed] [Google Scholar]
- [33].Leithe E, Rivedal E. Epidermal growth factor regulates ubiquitination, internalization and proteasome-dependent degradation of connexin43. J Cell Sci 2004; 117:1211-1220; PMID:14970263; http://dx.doi.org/ 10.1242/jcs.00951 [DOI] [PubMed] [Google Scholar]
- [34].Lampe PD, TenBroek EM, Burt JM, Kurata WE, Johnson RG, Lau AF. Phosphorylation of connexin43 on serine368 by protein kinase C regulates gap junctional communication. J Cell Biol 2000; 149:1503-1512; PMID:10871288; http://dx.doi.org/ 10.1083/jcb.149.7.1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fykerud TA, Kjenseth A, Schink KO, Sirnes S, Bruun J, Omori Y, Brech A, Rivedal E, Leithe E. Smad ubiquitination regulatory factor-2 controls gap junction intercellular communication by modulating endocytosis and degradation of connexin43. J Cell Sci 2012; 125:3966-3976; PMID:22623726; http://dx.doi.org/ 10.1242/jcs.093500 [DOI] [PubMed] [Google Scholar]
- [36].Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A 2006; 103:10660-10665; PMID:16818887; http://dx.doi.org/ 10.1073/pnas.0600447103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol 2008; 9:446-454; PMID:18464790; http://dx.doi.org/ 10.1038/nrm2406 [DOI] [PubMed] [Google Scholar]
- [38].Burtey A, Wagner M, Hodneland E, Skaftnesmo KO, Schoelermann J, Mondragon IR, Espedal H, Golebiewska A, Niclou SP, Bjerkvig R, et al.. Intercellular transfer of transferrin receptor by a contact-, Rab8-dependent mechanism involving tunneling nanotubes. FASEB J 2015; 29(11):4695-4712 [DOI] [PubMed] [Google Scholar]
- [39].van Ijzendoorn SC. Recycling endosomes. J Cell Sci 2006; 119:1679-1681; PMID:16636069; http://dx.doi.org/ 10.1242/jcs.02948 [DOI] [PubMed] [Google Scholar]
- [40].Neisch AL, Fehon RG. Ezrin, Radixin and Moesin: key regulators of membrane-cortex interactions and signaling. Curr Opin Cell Biol 2011; 23:377-382; PMID:21592758; http://dx.doi.org/ 10.1016/j.ceb.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kunda P, Baum B. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol 2009; 19:174-179; PMID:19285869; http://dx.doi.org/ 10.1016/j.tcb.2009.01.006 [DOI] [PubMed] [Google Scholar]
- [42].Lancaster OM, Baum B. Shaping up to divide: coordinating actin and microtubule cytoskeletal remodelling during mitosis. Semin Cell Dev Biol 2014; 34:109-115; PMID:24607328; http://dx.doi.org/ 10.1016/j.semcdb.2014.02.015 [DOI] [PubMed] [Google Scholar]
- [43].Falk MM, Baker SM, Gumpert AM, Segretain D, Buckheit RW 3rd. Gap junction turnover is achieved by the internalization of small endocytic double-membrane vesicles. Mol Biol Cell 2009; 20:3342-3352; PMID:19458184; http://dx.doi.org/ 10.1091/mbc.E09-04-0288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science 2002; 296:503-507; PMID:11964472; http://dx.doi.org/ 10.1126/science.1068793 [DOI] [PubMed] [Google Scholar]
- [45].Goodenough DA, Paul DL. Gap junctions. Cold Spring Harb Perspect Biol 2009; 1:a002576; PMID:20066080; http://dx.doi.org/ 10.1101/cshperspect.a002576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jordan K, Chodock R, Hand AR, Laird DW. The origin of annular junctions: a mechanism of gap junction internalization. J Cell Sci 2001; 114:763-773; PMID:11171382 [DOI] [PubMed] [Google Scholar]
- [47].Larsen WJ, Hai N. Origin and fate of cytoplasmic gap junctional vesicles in rabbit granulosa cells. Tissue Cell 1978; 10:585-598; PMID:725913 [DOI] [PubMed] [Google Scholar]
- [48].Leithe E, Brech A, Rivedal E. Endocytic processing of connexin43 gap junctions: a morphological study. Biochem J 2006; 393:59-67; PMID:16162097; http://dx.doi.org/ 10.1042/BJ20050674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Murray SA, Nickel BM, Gay VL. Endocytosis of connexin protein in adrenal cells. Endocr Res 2004; 30:647-654; PMID:15666807; http://dx.doi.org/ 10.1081/ERC-200043942 [DOI] [PubMed] [Google Scholar]
- [50].Nickel B, Boller M, Schneider K, Shakespeare T, Gay V, Murray SA. Visualizing the effect of dynamin inhibition on annular gap vesicle formation and fission. J Cell Sci 2013; 126:2607-2616; PMID:23591819; http://dx.doi.org/ 10.1242/jcs.116269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Piehl M, Lehmann C, Gumpert A, Denizot JP, Segretain D, Falk MM. Internalization of large double-membrane intercellular vesicles by a clathrin-dependent endocytic process. Mol Biol Cell 2007; 18:337-347; PMID:17108328; http://dx.doi.org/ 10.1091/mbc.E06-06-0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Laird DW. Life cycle of connexins in health and disease. Biochem J 2006; 394:527-543; PMID:16492141; http://dx.doi.org/ 10.1042/BJ20051922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Leithe E, Sirnes S, Fykerud T, Kjenseth A, Rivedal E. Endocytosis and post-endocytic sorting of connexins. Biochim Biophys Acta 2012; 1818:1870-1879; PMID:21996040; http://dx.doi.org/ 10.1016/j.bbamem.2011.09.029 [DOI] [PubMed] [Google Scholar]
- [54].Gould GW, Lippincott-Schwartz J. New roles for endosomes: from vesicular carriers to multi-purpose platforms. Nat Rev Mol Cell Biol 2009; 10:287-292; PMID:19277045; http://dx.doi.org/ 10.1038/nrm2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.