Abstract
Mycobacterium tuberculosis (MTB) is a specific aerobic bacterium, but can survive under hypoxic conditions, such as those in lung cheese necrosis, granulomas, or macrophages. It is not clear whether the drug sensitivity and growth characteristics of MTB under hypoxic conditions are different from those under aerobic conditions. In this study, we examined the drug resistance and growth characteristics of MTB clinical isolates by a large sample of in vitro drug susceptibility tests, using an automatic growth instrument. Under hypoxic conditions, variance in drug resistance was observed in nearly one-third of the MTB strains and was defined as MTB strains with changed drug sensitivity (MTB-CDS). Among these strains, resistance in a considerable proportion of clinical strains was significantly increased, and some strains emerged as multi-drug resistant. Growth test results revealed a high growth rate and large survival number in macrophages under hypoxia in MTB-CDS. According to the results of fluorescence quantitative PCR, the expression of some genes, including RegX3 (involving RIF resistance), Rv0194 (efflux pump gene), four genes related to transcription regulation (KstR, DosR, Rv0081 and WhiB3) and gene related to translation regulation (DATIN), were upregulated significantly under hypoxic conditions compared to that under aerobic conditions (p < 0.05). Thus, we concluded that some MTB clinical isolates can survive under hypoxic conditions and their resistance could change. As for poor clinical outcomes in patients, based on routine drug susceptibility testing, drug susceptibility tests for tuberculosis under hypoxic conditions should also be recommended. However, the detailed mechanisms of the effect of hypoxia on drug sensitivity and growth characteristics of MTB clinical isolates still requires further study.
Introduction
Tuberculosis (TB) is a serious health problem, and China is one of the countries with high burden of TB [1,2]. One of causes for the difficulty in controlling tuberculosis is TB drug resistance [3–6]. In recent years, there has been a rising trend in the number of multi-drug resistant Mycobacterium tuberculosis (MTB) clinical isolates [5–9].
Currently, to guide the clinically rational use of drugs, routine in vitro drug susceptibility tests for TB are commonly conducted in many laboratories [10–13]. However, despite therapy based on the test results, there are still some cases of treatment failure [14]. We speculate that one of the causes for this may be related to changes in MTB resistance under hypoxia.
The routine TB drug susceptibility tests in vitro are performed under aerobic conditions (21% of oxygen from the atmosphere). However, in vivo conditions such as granuloma, caseous necrosis tissue, or macrophages are hypoxic in nature [15–17]. If MTB resides in these tissues, the features of its biological metabolism might change [15–18], which may lead to variation in its resistance and growth characteristics. To identify whether the drug sensitivity and growth characteristics of MTB under hypoxic conditions are different from those under aerobic conditions, and to explore the related mechanisms, we conducted the following related research, using a large sample of MTB clinical isolates.
Methods and Materials
Ethics statement
This study was approved by the Shanghai Pulmonary Hospital Affiliated to Tongji University School of Medicine Ethics Committee. Subjects were treated in accordance with the Helsinki Declaration on the participation of human subjects in medical research. Written informed consent was obtained from each participant.
Collection of strains and drug susceptibility testing
Two hundred and forty-five MTB clinical isolates were obtained from the Department of Clinical Laboratory Medicine at Shanghai Pulmonary Hospital between 2013 and 2015 (S1 Fig). Routine drug susceptibility tests were performed using a commercial microplate kit (Yibaishi Biotech, Inc., Shen-zhen, China) on the BACTECT 960 System (Becton Dickinson, Franklin Lakes, NJ) under conventional aerobic conditions, and hypoxic drug susceptibility testing was conducted under hypoxic conditions by covering 50 μl of paraffin oil. In this study, susceptibility to first-line drugs like isoniazid (INH), ethambutol (EMB), rifampicin (RMP), and streptomycin (SM) was analyzed. The minimum inhibitory concentrations (MICs) were recorded. The standard laboratory strain H37Rv was purchased from the Chinese National Institute for Food and Drug Control.
Establishment of the hypoxia model and analysis of growth characteristics of MTB
Aerobic cultures (NRP-2) by Wayne's method were used to establish the hypoxia model described in this report [15,19]. Population growth curves were determined by a Bioscreen Growth Curve Instrument (Bioscreen C, Helsinki, Finland), using a honeycomb plate with 100 wells (Bioscreen C, Helsinki, Finland). Briefly, 300 μl of 7H9 medium was added to each well, and cultured with shaking at 37°C. The optical density was measured as absorbance at 600 nm after every 2 hours. Hypoxia conditions were established by covering 50 μl of paraffin oil. If both growth curves under the aerobic and hypoxic conditions were completely in accord with the Webster's model, the model of hypoxia was considered successful. The growth characteristics analysis was performed under aerobic as well as hypoxic conditions.
Enumerating MTB clinical strains in macrophages
Clinical isolates of MTB were cultured under both aerobic and hypoxic conditions. MTB isolates grown to the logarithmic phase were used to infect THP1 macrophages. At different time points after infection, the infected cells were washed twice with PBS and lysed for 10 minutes using Triton X-100. Subsequently, the lysed mixture was inoculated onto 7H9 solid plates and cultured at 37°C for 4 weeks. Finally, MTB clones were counted.
Effect of hypoxia on gene expression in MTB clinical isolates
After reviewing previous studies [11,15–43], the relevant regulatory genes for efflux pumps and transcription factors likely to affect drug resistance in MTB (Table 1) were selected and their expression was quantified by RT-PCR. Briefly, 25 ml of culture was used to extract RNA upon cell lysis via the TRIzol bead beater method and phenol extraction [35]. RNA concentrations were determined using a Nanodrop 2000 (NanoDrop Technologies). RNA was treated with DNase as previously described [35] and reverse-transcribed using the High Capacity RNA-to-cDNA kit (Applied Biosystems) following the manufacturers’ instructions. RT PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems), as previously described [35]. PCR primers for the above genes are specified in S1 Table. The tests in this study were performed on the 7500 Fast Dx Real-Time PCR System (Applied Biosystems). The reaction conditions were as follows: 95°C for 60 s, 40 cycles of 95°C for 5 s, 62°C for 8 s, and 72°C for 20 s, followed by a melt curve analysis.
Table 1. The characteristics of the testing genes in this study.
| Category | Gene name | Rv number | Description |
|---|---|---|---|
| INH regulation gene | FbpC | Rv0129c | Diacylglycerol acyltransferase/mycolyltransferase Ag85C |
| INH regulation gene | EfpA | Rv2846c | MFS-type transporter EfpA |
| INH regulation gene | FadE23 | Rv3140 | Acyl-CoA dehydrogenase FadE23 |
| RIF regulation gene | RegX3 | Rv0491 | Two component sensory transduction protein RegX |
| RIF regulation gene | SigF | Rv3286c | RNA polymerase sigma factor SigF |
| RIF regulation gene | MprA | Rv0981 | Two-component response regulator MrpA |
| Efflux pump regulation gene | Rv0194 | Rv0194 | Multidrug ABC transporter ATPase/permease |
| Efflux pump regulation gene | WhiB7 | Rv3197A | Transcriptional regulator WhiB7 |
| Efflux pump regulation gene | Rv2136c | Rv2136c | Undecaprenyl-diphosphatase |
| Efflux pump regulation gene | Rv1410c | Rv1410c | Aminoglycosides/tetracycline-transport integral membrane protein |
| Transcript regulation gene | Rv0081 | Rv0081 | HTH-type transcriptional regulator |
| Transcript regulation gene | Rv0324 | Rv0324 | Transcriptional regulator |
| Transcript regulation gene | LipY | Rv3097c | Triacylglycerol lipase Lip |
| Transcript regulation gene | LipX | Rv1169c | Lipase LipX |
| Transcript regulation gene | Pks3 | Rv1180 | Polyketide beta-ketoacyl synthase |
| Transcript regulation gene | MutA | Rv1492 | Methylmalonyl-CoA mutase small subunit |
| Transcript regulation gene | AccA3 | Rv3285 | Bifunctional protein acetyl-/propionyl-CoA carboxylase subunit alpha AccA |
| Transcript regulation gene | WhiB3 | Rv3416 | Redox-responsive transcriptional regulator WhiB3 |
| Transcript regulation gene | KstR | Rv3574 | HTH-type transcriptional regulator KstR |
| Transcript regulation gene | Lsr2 | Rv3597c | Iron-regulated H-NS-like protein |
| Transcript regulation gene | DevR | Rv3133c | Two component transcriptional regulator dosR |
| Transcript regulation gene | IciA | Rv1985c | HTH-type transcriptional regulator |
| Transcript regulation gene | DATIN | Rv0079 | Dormancy Associated Translation Inhibitor |
Statistical analysis
SPSS software (SPSS Inc., Chicago, IL, USA) was used for the statistical analysis of results. Chi-square test, non-parametric test, and one-way analysis of variance (ANOVA) were performed to analyze the different data in this study. A p-value less than 0.05 was considered statistically significant.
Results
Drug resistance of clinical strains under hypoxic conditions
In this study, the MICs of 245 MTB clinical isolates determined by the microplate method were found to be identical with those obtained by the BACTECT 960 System (S2 Table).
Addition of liquid paraffin to the microplate allowed the formation of a curve similar to that in the Wayne’s aerobic model, as determined by OD (S2 Fig). Therefore, construction of a hypoxic growth model in the microplate was successful.
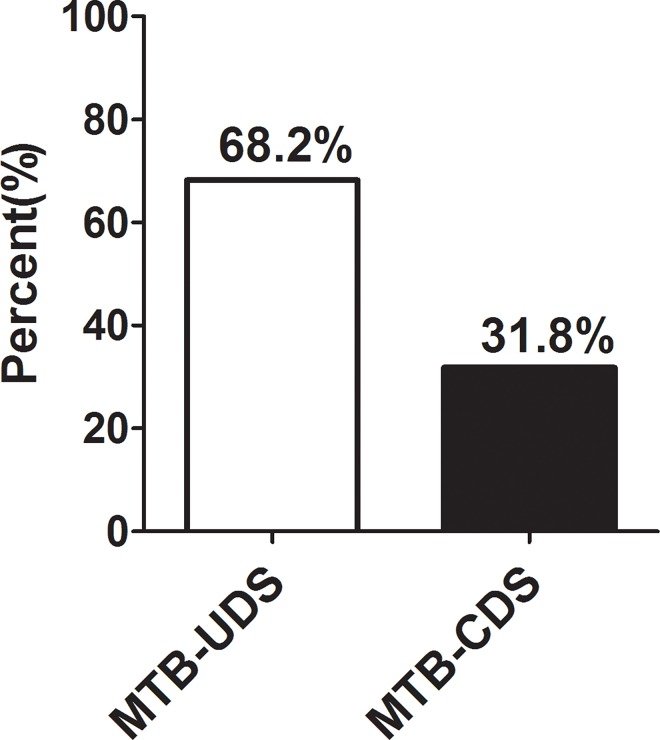
Upon analyzing the MICs of 245 clinical isolates, 68.2% (167/245) were determined as MTB strains of unchanged drug sensitivity (MTB-UDS), whereas others (31.8%, 78/245) belonged to MTB strains of changed drug sensitivity (MTB-CDS; Fig 1).
Fig 1. Comparison of the rates of changed MTB strains of drug sensitivity and unchanged MTB strains of drug sensitivity in 245 strains clinical isolates under hypoxia.
MTB-UDS: changed MTB strains of drug sensitivity; MTB-CDS: unchanged MTB strains of drug sensitivity.
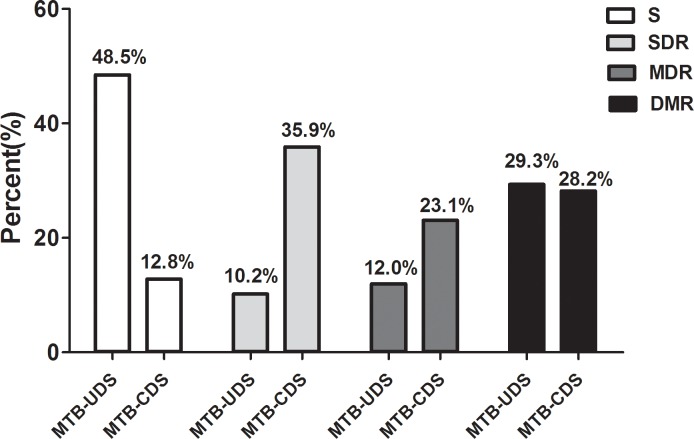
The stratified analysis of the drug susceptibility test result showed that the overall sensitivity in MTB-CDS was significantly lower than that in MTB-UDS (12.8% vs. 48.5%, p < 0.05). Resistance to single drugs in MTB-CDS was also significantly higher than that in MTB-CDS (35.9% vs. 10.2%, p < 0.05); multi-drug resistance in MTB-CDS was also significantly higher than that in MTB-UDS (23.1% vs. 12%, p < 0.05; Fig 2).
Fig 2. Comparison of drug resistances of changed MTB strains of drug sensitivity and unchanged MTB strains of drug sensitivity.
MTB-UDS: changed MTB strains of drug sensitivity under hypoxia; MTB-CDS: unchanged MTB strains of drug sensitivity under hypoxia; S: sensitive; SDR: single drug resistance; MDR: multi-drug resistance; DMR: drug multi-resistance.
Overall, under hypoxic conditions, the rate of MTB clinical isolates for which the MICs of first line drugs were increased was 38.5% (30/78), whereas that for which the MICs were decreased was 23% (18/78; S3 Fig).
Growth characteristics of MTB clinical isolates under hypoxic conditions
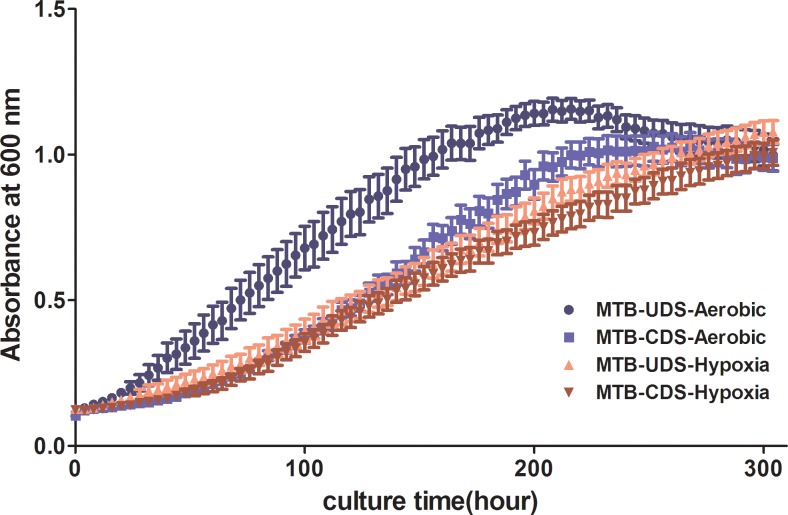
Based on the growth curve analysis, the growth of MTB-CDS was slower that of MTB-UDS under aerobic conditions; whereas under hypoxic conditions, the growth curves of MTB-CDS nearly coincided with those of MTB-UDS, (Fig 3) indicating that MTB-CDS can suitably grow under hypoxia.
Fig 3. Growth curves of MTB clinical isolates under aerobic and hypoxic conditions.
MTB-UDS: changed MTB strains of drug sensitivity; MTB-CDS: unchanged MTB strains of drug sensitivity.
Analysis of survival ability of MTB clinical strains in macrophages under hypoxic conditions
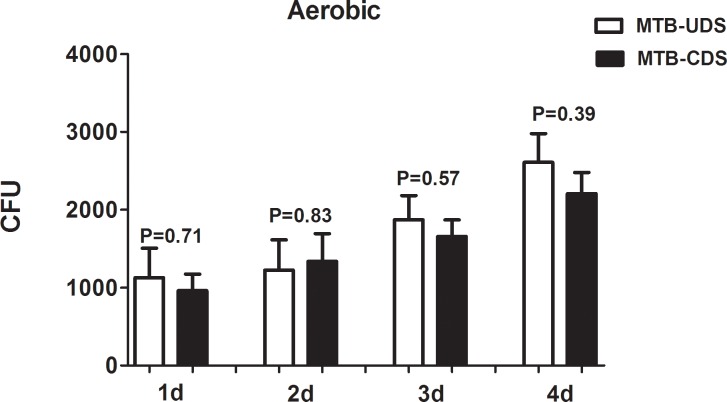
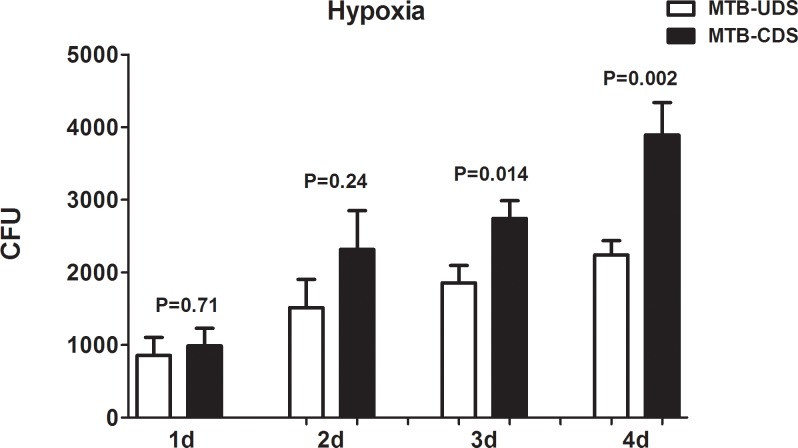
Examination of the MTB survival ability in macrophages at different times under aerobic conditions showed that the difference in survival between the MTB-CDS and MTB-UDS with increasing time was not statistically significant (p > 0.05; Fig 4); whereas under hypoxic conditions, the survival of the MTB-CDS was significantly higher than that of MTB-UDS (p < 0.05; Fig 5).
Fig 4. Survivability of changed MTB strains of drug sensitivity and unchanged MTB strains of drug sensitivity in macrophage cell under aerobic conditions.
MTB-UDS: changed MTB strains of drug sensitivity; MTB-CDS: unchanged MTB strains of drug sensitivity.
Fig 5. Survivability of changed MTB strains of drug sensitivity and unchanged MTB strains of drug sensitivity in macrophage cell under hypoxic conditions.
Gene expression potentially related to resistance in MTB
According to the results of fluorescence quantitative PCR, the expression of some genes, including RegX3 (involving RIF resistance), Rv0194 (efflux pump gene), four genes related to transcription regulation (KstR, DosR, Rv0081 and WhiB3) and gene related to translation regulation (DATIN), was significantly upregulated under hypoxic conditions compared to that under aerobic conditions (p < 0.05; S4, S5, S6 and S7 Figs).
Discussion
Previous studies have shown that MTB can survive under hypoxic conditions [23,44,45]. In this study, MICs of 245 MTB clinical isolates, determined by the microplate method, were found to be identical with those obtained from the BACTECT 960 System under aerobic conditions. The Wayne model is currently recognized as a reliable in vitro model for simulating in vivo environments such as MTB dormancy or hypoxic environments. In this study, we observed almost coinciding curves upon comparing the growth curve of the MTB culture systems with H37Rv, which was cultured under both aerobic and anaerobic environments. Therefore, the growth model of hypoxia was successfully established. By analyzing the MICs of 245 clinical isolates, we found that about one-third of the MTB strains were MTB-CDS, whereas the others belonged to MTB-UDS.
Further stratified analysis of the results of drug sensitivity testing showed that the overall sensitivity of MTB-CDS was significantly lower than that of MTB-UDS, and the resistance to single drugs and multiple drugs in MTB-CDS were both significantly higher than those in MTB-UDS. Overall, under hypoxic conditions, the proportion of MTB clinical isolates for which the MICs of first line drugs were increased was 38.5%.
Therefore, we can conclude that there was a significant change in drug resistance of some MTB strains under hypoxic conditions. Considering that routine drug susceptibility testing is unable to completely reflect the actual resistance in MTB, we suggest that drug susceptibility tests should also be conducted under hypoxic conditions to exclude hypoxia resistance in MTB, especially for cases showing initial therapeutic failure.
Based on the growth curve analysis, the growth of MTB-CDS was slower than that of MTB-UDS under aerobic conditions, whereas under hypoxic conditions, the growth curves of MTB-CDS were nearly similar to those of MTB-UDS, indicating that MTB-CDS are more suitable for survival under hypoxia.
By determining the survival of MTB in macrophages at different times, we found that the difference in survival under aerobic conditions was not statistically significant between the MTB-UDS and MTB-CDS with increasing time; however, under hypoxic conditions, the survival of MTB-UDS was significantly higher than that of MTB-CDS. This further proves that MTB-CDS can survive under hypoxic conditions.
A previous study has shown that the dormancy survival regulon (DosR regulon) is chiefly responsible for encoding the dormancy related functions of MTB, and appears to be involved in translation regulation through the interaction of its product with bacterial ribosomal subunits [46]. One of the DosR regulon proteins, DATIN, encoded by the gene Rv0079, can stimulate macrophages and peripheral blood mononuclear cells to secrete important cytokines, which may be significant in granuloma formation and maintenance [47]. Additionally, the expression of DATIN in M. bovis BCG was found to be upregulated under pH stress and micro-aerobic conditions [47]. This indicates that MTB survival under hypoxia is closely associated with gene regulation, and some relevant regulatory genes related to drug resistance, such as efflux pumps and transcription factors, are likely involved in MTB drug resistance under hypoxia. According to the results of fluorescence quantitative PCR, the expression of some genes, including RegX3 (involving RIF resistance), Rv0194 (efflux pump gene), four genes related to transcription regulation (KstR, DosR, Rv0081 and WhiB3) and gene related to translation regulation (DATIN), was significantly upregulated under hypoxic conditions compared to that under aerobic conditions. As for other genes such as IciA, we didn’t found difference in gene expression. However, the previous study showed that, IciA has a bearing on the functional role of the important regulator of MTB chromosomal replication in the context of latency [48]. According to the report, synergy between the N-terminal and C-terminal domains of HupB is essential for its DNA-binding ability, and to modulate the topological features of DNA, which has implications for processes such as DNA compaction, gene regulation, homologous recombination, and DNA repair [49]. Thus, we believe that the mechanisms of hypoxia resistance in MTB clinical isolates are associated with the expression of some genes. Moreover, we speculate that the difference in the expression of some genes could result in better survival of MTB-CDS under hypoxia compared to that of MTB-CDS.
Efflux pumps are thought to have emerged for the expulsion of noxious substances from the bacterial cell, thus allowing its survival; increased expression of efflux pumps is associated with resistance to alien substances, including drugs [50]. These pumps are often called multi-drug efflux pumps [50]. In addition, the expression of drug resistance in MTB is linked with some transcription regulation genes. However, the exact mechanisms involved in hypoxia drug resistance of MTB remain unknown.
In summary, in our study, some MTB clinical isolates showed very good adaptability under hypoxic conditions, better survival in macrophages, and resistance to some anti-tuberculosis drugs. This resulted in incorrect results from routine MTB drug susceptibility testing; hence, we recommend that drug susceptibility tests should be simultaneously conducted under hypoxic conditions, especially for cases with initial therapeutic failure. The mechanisms underlying the effect of hypoxia on drug sensitivity and growth characteristics of MTB still require further study.
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(XLS)
(XLS)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project was funded by the National Basic Research Program of China (973 Program, no. 2012CB518706), the National Natural Science Foundation of China (no. 81201253), and Research Grant from Shanghai Hospital Development Center (no. SHDC12015910). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dodd PJ, Gardiner E, Coghlan R, Seddon JA (2014) Burden of childhood tuberculosis in 22 high-burden countries: a mathematical modelling study. Lancet Glob Health 2: e453–459. 10.1016/S2214-109X(14)70245-1 [DOI] [PubMed] [Google Scholar]
- 2.Li L, Duanmu HJ (2004) [The epidemic of childhood tuberculosis in China]. Zhonghua Yi Xue Za Zhi 84: 1678–1680. [PubMed] [Google Scholar]
- 3.Chen J, Li NX, Wan KL, Yang GJ, Wang Q (2007) [Analysis on risk factors of drug resistance for tuberculosis in Sichuan and Anhui Provinces]. Sichuan Da Xue Xue Bao Yi Xue Ban 38: 135–137. [PubMed] [Google Scholar]
- 4.He GX, Zhao YL, Jiang GL, Liu YH, Xia H, Wang SF, et al. (2008) Prevalence of tuberculosis drug resistance in 10 provinces of China. BMC Infect Dis 8: 166 10.1186/1471-2334-8-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Zhang YY, Gui XH, Yuan ZA, Pan QC, Mei J, et al. (2012) [Prevalence and risk factors on the resistance related to second-line drugs among multi-drug resistant tuberculosis cases in Shanghai, China]. Zhonghua Liu Xing Bing Xue Za Zhi 33: 796–798. [PubMed] [Google Scholar]
- 6.Xiao HP (2010) [The prevalence of drug-resistance tuberculosis in China and the chemotherapy strategies]. Zhonghua Jie He He Hu Xi Za Zhi 33: 481–482. [PubMed] [Google Scholar]
- 7.Abdul-Aziz AA, Elhassan MM, Abdulsalam SA, Mohammed EO, Hamid ME (2013) Multi-drug resistance tuberculosis (MDR-TB) in Kassala State, Eastern Sudan. Trop Doct 43: 66–70. 10.1177/0049475513490421 [DOI] [PubMed] [Google Scholar]
- 8.Liu CH, Li HM, Li L, Hu YL, Wang Q, Yang N, et al. (2011) Anti-tuberculosis drug resistance patterns and trends in a tuberculosis referral hospital, 1997–2009. Epidemiol Infect 139: 1909–1918. 10.1017/S0950268810003158 [DOI] [PubMed] [Google Scholar]
- 9.Mesfin YM, Hailemariam D, Biadgilign S, Kibret KT (2014) Association between HIV/AIDS and multi-drug resistance tuberculosis: a systematic review and meta-analysis. PLoS One 9: e82235 10.1371/journal.pone.0082235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrade BH, Greco DB, Oliveira MT, Lacerda NP, Correa Rde A (2013) Contributions of culture and antimicrobial susceptibility tests to the retreatment of patients with pulmonary tuberculosis. Rev Soc Bras Med Trop 46: 441–446. 10.1590/0037-8682-0047-2013 [DOI] [PubMed] [Google Scholar]
- 11.Kharatmal S, Jhamb SS, Singh PP (2009) Evaluation of BACTEC 460 TB system for rapid in vitro screening of drugs against latent state Mycobacterium tuberculosis H37Rv under hypoxia conditions. J Microbiol Methods 78: 161–164. 10.1016/j.mimet.2009.05.008 [DOI] [PubMed] [Google Scholar]
- 12.Coronel J, Roper MH, Herrera C, Bonilla C, Jave O, Gianella C, et al. (2014) Validation of microscopic observation drug susceptibility testing for rapid, direct rifampicin and isoniazid drug susceptibility testing in patients receiving tuberculosis treatment. Clin Microbiol Infect 20: 536–541. 10.1111/1469-0691.12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aziz MA, Wright A, Laszlo A, De Muynck A, Portaels F, Van Deun A, et al. (2006) Epidemiology of antituberculosis drug resistance (the Global Project on Anti-tuberculosis Drug Resistance Surveillance): an updated analysis. Lancet 368: 2142–2154. 10.1016/S0140-6736(06)69863-2 [DOI] [PubMed] [Google Scholar]
- 14.Hofman S, Segers MM, Ghimire S, Bolhuis MS, Sturkenboom MG, Van Soolingen D, et al. (2016) Emerging drugs and alternative possibilities in the treatment of tuberculosis. Expert Opin Emerg Drugs 21: 103–116. 10.1517/14728214.2016.1151000 [DOI] [PubMed] [Google Scholar]
- 15.Devasundaram S, Raja A (2016) Variable transcriptional adaptation between the laboratory (H37Rv) and clinical strains (S7 and S10) of Mycobacterium tuberculosis under hypoxia. Infect Genet Evol 40: 21–28. 10.1016/j.meegid.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 16.Qualls JE, Murray PJ (2016) Immunometabolism within the tuberculosis granuloma: amino acids, hypoxia, and cellular respiration. Semin Immunopathol 38: 139–152. 10.1007/s00281-015-0534-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zenk SF, Vollmer M, Schercher E, Kallert S, Kubis J, Stenger S (2015) Hypoxia promotes Mycobacterium tuberculosis-specific up-regulation of granulysin in human T cells. Med Microbiol Immunol. [DOI] [PubMed] [Google Scholar]
- 18.McGillivray A, Golden NA, Kaushal D (2015) The Mycobacterium tuberculosis Clp gene regulator is required for in vitro reactivation from hypoxia-induced dormancy. J Biol Chem 290: 2351–2367. 10.1074/jbc.M114.615534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wayne LG, Hayes LG (1996) An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun 64: 2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreuder LJ, Parish T (2014) Mycobacterium tuberculosis DosR is required for activity of the PmbtB and PmbtI promoters under hypoxia. PLoS One 9: e107283 10.1371/journal.pone.0107283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaur K, Taneja NK, Dhingra S, Tyagi JS (2014) DevR (DosR) mimetic peptides impair transcriptional regulation and survival of Mycobacterium tuberculosis under hypoxia by inhibiting the autokinase activity of DevS sensor kinase. BMC Microbiol 14: 195 10.1186/1471-2180-14-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez Campos GN, Velarde Felix JS, Sandoval Ramirez L, Cazares Salazar S, Corona Nakamura AL, Amaya Tapia G, et al. (2014) Polymorphism in cathelicidin gene (CAMP) that alters Hypoxia-inducible factor (HIF-1alpha::ARNT) binding is not associated with tuberculosis. Int J Immunogenet 41: 54–62. 10.1111/iji.12080 [DOI] [PubMed] [Google Scholar]
- 23.Galagan JE, Minch K, Peterson M, Lyubetskaya A, Azizi E, Sweet L, et al. (2013) The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499: 178–183. 10.1038/nature12337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxena AK, Roy KK, Singh S, Vishnoi SP, Kumar A, Kashyap VK, et al. (2013) Identification and characterisation of small-molecule inhibitors of Rv3097c-encoded lipase (LipY) of Mycobacterium tuberculosis that selectively inhibit growth of bacilli in hypoxia. Int J Antimicrob Agents 42: 27–35. 10.1016/j.ijantimicag.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 25.Eoh H, Rhee KY (2013) Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 110: 6554–6559. 10.1073/pnas.1219375110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phong WY, Lin W, Rao SP, Dick T, Alonso S, Pethe K (2013) Characterization of phosphofructokinase activity in Mycobacterium tuberculosis reveals that a functional glycolytic carbon flow is necessary to limit the accumulation of toxic metabolic intermediates under hypoxia. PLoS One 8: e56037 10.1371/journal.pone.0056037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klinkenberg LG, Karakousis PC (2013) Rv1894c is a novel hypoxia-induced nitronate monooxygenase required for Mycobacterium tuberculosis virulence. J Infect Dis 207: 1525–1534. 10.1093/infdis/jit049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang X, Wallqvist A, Reifman J (2012) Modeling phenotypic metabolic adaptations of Mycobacterium tuberculosis H37Rv under hypoxia. PLoS Comput Biol 8: e1002688 10.1371/journal.pcbi.1002688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickel D, Busch M, Mayer D, Hagemann B, Knoll V, Stenger S (2012) Hypoxia triggers the expression of human beta defensin 2 and antimicrobial activity against Mycobacterium tuberculosis in human macrophages. J Immunol 188: 4001–4007. 10.4049/jimmunol.1100976 [DOI] [PubMed] [Google Scholar]
- 30.Dhingra S, Kaur K, Taneja NK, Tyagi JS (2012) DevR (DosR) binding peptide inhibits adaptation of Mycobacterium tuberculosis under hypoxia. FEMS Microbiol Lett 330: 66–71. 10.1111/j.1574-6968.2012.02534.x [DOI] [PubMed] [Google Scholar]
- 31.Gideon HP, Wilkinson KA, Rustad TR, Oni T, Guio H, Kozak RA, et al. (2010) Hypoxia induces an immunodominant target of tuberculosis specific T cells absent from common BCG vaccines. PLoS Pathog 6: e1001237 10.1371/journal.ppat.1001237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abomoelak B, Hoye EA, Chi J, Marcus SA, Laval F, Bannantine JP, et al. (2009) mosR, a novel transcriptional regulator of hypoxia and virulence in Mycobacterium tuberculosis. J Bacteriol 191: 5941–5952. 10.1128/JB.00778-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontan PA, Voskuil MI, Gomez M, Tan D, Pardini M, Manganelli R, et al. (2009) The Mycobacterium tuberculosis sigma factor sigmaB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J Bacteriol 191: 5628–5633. 10.1128/JB.00510-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rustad TR, Sherrid AM, Minch KJ, Sherman DR (2009) Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol 11: 1151–1159. 10.1111/j.1462-5822.2009.01325.x [DOI] [PubMed] [Google Scholar]
- 35.Shi L, Sohaskey CD, North RJ, Gennaro ML (2008) Transcriptional characterization of the antioxidant response of Mycobacterium tuberculosis in vivo and during adaptation to hypoxia in vitro. Tuberculosis (Edinb) 88: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar A, Toledo JC, Patel RP, Lancaster JR Jr., Steyn AJ (2007) Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A 104: 11568–11573. 10.1073/pnas.0705054104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sardiwal S, Kendall SL, Movahedzadeh F, Rison SC, Stoker NG, Djordjevic S (2005) A GAF domain in the hypoxia/NO-inducible Mycobacterium tuberculosis DosS protein binds haem. J Mol Biol 353: 929–936. 10.1016/j.jmb.2005.09.011 [DOI] [PubMed] [Google Scholar]
- 38.Saini DK, Malhotra V, Dey D, Pant N, Das TK, Tyagi JS (2004) DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150: 865–875. 10.1099/mic.0.26218-0 [DOI] [PubMed] [Google Scholar]
- 39.Bagchi G, Mayuri, Tyagi JS (2003) Hypoxia-responsive expression of Mycobacterium tuberculosis Rv3134c and devR promoters in Mycobacterium smegmatis. Microbiology 149: 2303–2305. 10.1099/mic.0.C0120-0 [DOI] [PubMed] [Google Scholar]
- 40.Stenina MA, Voevodin DA, Stakhanov VD, Kisilevich ON, Rozanova GN (2003) Tissue hypoxia and intestinal dysbiosis in children with tuberculosis. Bull Exp Biol Med 135: 178–180. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto T, Nagao K, Okada O, Yasuda J, Tanabe N, Kato K, et al. (1996) [Relation of pulmonary hemodynamics and ventilation to tissue hypoxia during exercise in patients with tuberculosis sequelae]. Kekkaku 71: 331–337. [PubMed] [Google Scholar]
- 42.Sakuma T, Kimura H, Tatsumi K, Okada O, Kato K, Kuriyama T (1995) [Does nocturnal hypoxia relate to acute exacerbation of chronic respiratory failure with right heart failure in late sequelae of pulmonary tuberculosis?]. Kekkaku 70: 1–7. [PubMed] [Google Scholar]
- 43.Fesenko LD (1987) [Characteristics of the systemic hemodynamics and oxygen-transport function of the blood in respiratory hypoxia in pulmonary tuberculosis patients]. Vrach Delo: 57–59. [PubMed] [Google Scholar]
- 44.Dione N, Khelaifia S, La Scola B, Lagier JC, Raoult D (2016) A quasi-universal medium to break the aerobic/anaerobic bacterial culture dichotomy in clinical microbiology. Clin Microbiol Infect 22: 53–58. 10.1016/j.cmi.2015.10.032 [DOI] [PubMed] [Google Scholar]
- 45.Manabe YC, Bishai WR (2000) Latent Mycobacterium tuberculosis-persistence, patience, and winning by waiting. Nat Med 6: 1327–1329. 10.1038/82139 [DOI] [PubMed] [Google Scholar]
- 46.Kumar A, Lewin A, Rani PS, Qureshi IA, Devi S, Majid M, et al. (2013) Dormancy Associated Translation Inhibitor (DATIN/Rv0079) of Mycobacterium tuberculosis interacts with TLR2 and induces proinflammatory cytokine expression. Cytokine 64: 258–264. 10.1016/j.cyto.2013.06.310 [DOI] [PubMed] [Google Scholar]
- 47.Kumar A, Majid M, Kunisch R, Rani PS, Qureshi IA, Lewin A, et al. (2012) Mycobacterium tuberculosis DosR regulon gene Rv0079 encodes a putative, 'dormancy associated translation inhibitor (DATIN)'. PLoS One 7: e38709 10.1371/journal.pone.0038709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Farhana A, Hasnain SE (2009) In-vitro helix opening of M. tuberculosis oriC by DnaA occurs at precise location and is inhibited by IciA like protein. PLoS One 4(1):e4139 10.1371/journal.pone.0004139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharadamma N, Khan K, Kumar S, Patil KN, Hasnain SE, Muniyappa K (2011) Synergy between the N-terminal and C-terminal domains of Mycobacterium tuberculosis HupB is essential for high-affinity binding, DNA supercoiling and inhibition of RecA-promoted strand exchange. FEBS J 278(18):3447–62. 10.1111/j.1742-4658.2011.08267.x [DOI] [PubMed] [Google Scholar]
- 50.Gupta AK, Reddy VP, Lavania M, Chauhan DS, Venkatesan K, Sharma VD, et al. (2010) jefA (Rv2459), a drug efflux gene in Mycobacterium tuberculosis confers resistance to isoniazid & ethambutol. Indian J Med Res 132: 176–188. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.