Abstract
This study determined the methylation status of cellular retinoic acid-binding protein (CRABP) gene promoters and associated them with demographic characteristics, habits, and the presence of human papilloma virus (HPV) in patients with cervical cancer (CC), low and high squamous intraepithelial lesions, and no intraepithelial lesion. Women (n = 158) were selected from the Colposcopy Clinic of Sanitary Jurisdiction II in Ciudad Juarez, Chihuahua, Mexico. Demographic characteristics and habit information were collected. Cervical biopsy and endocervical scraping were used to determine methylation in promoter regions by methylation-specific polymerase chain reaction technique. We found hemi-methylation patterns in the promoter regions of CRABP1 and CRABP2; there was 28.5% hemi-methylation in CRABP1 and 7.0% in that of CRABP2. Methylation in CRABP1 was associated with age (≥35 years, P = 0.002), family history of cancer (P = 0.032), the presence of HPV-16 (P = 0.013), and no alcohol intake (P = 0.035). These epigenetic changes could be involved in the CC process, and CRABP1 has the potential to be a predictive molecular marker of retinoid therapy response.
Keywords: cervical cancer, squamous intraepithelial lesions, methylation, CRABP
Introduction
Cervical cancer (CC) is the second leading cause of death from malignancy in women in Mexico and the fourth in the world.1 Over the years, CC has been studied to define specific characteristics involved in the cancer process to determine the best time for interventions. One of the processes that is currently being studied is the process of DNA methylation and its involvement in cancer treatment. During the cancer process, promoter methylation profiles of tumor suppressor genes are commonly methylated, creating a mechanism for the promotion and development of cancer.2–4 In CC, it is known that the presence of human papilloma virus (HPV) oncoproteins, such as E6 and E7, increases DNA methyltransferase activity and causes global methylation.4 Nevertheless, other factors may impinge on this process.
Epidemiological and lifestyle factors are implicated in methylation, such as age,5,6 obesity,7 smoking and alcohol intake,8,9 physical activity,10 epigenomic inheritance,11 and circulating estrogens.12,13 However, there are no studies that have evaluated the relationship between lifestyle factors and the methylation processes in CC.
DNA methylation has been useful in identifying the presence of a tumor, as well as determining its status, subtype, and responsiveness to specific therapies such as retinol.14 In CC, it has been shown that there are epigenetic alterations in genes related to retinol metabolism, such as the retinoic acid receptor (RAR) and the cellular retinol-binding protein (CRBP1).15,16 Nevertheless, it has not been reported whether methylation status can occur in cellular retinoic acid-binding protein-1 (CRABP1) and 2 (CRABP2) and silence their gene expression.
Retinoic acid (RA) or vitamin A is a metabolite that has an effect on embryonic development, cell growth, differentiation, and apoptosis.17 These effects are regulated by CRABPs that are related to RA transport within the cell. There are two isoforms, CRABP1 that is expressed in almost all tissues and CRABP2 that is expressed in the skin, nervous system, breast, uterus, and ovary.18 Both proteins protect amphipathic molecules of RA from oxidative degradations and they also control the availability of retinoids in several metabolic processes. The presence of RA is essential for cell cycle regulation blocking the carcinogenesis process.
Therefore, if methylation in the promoter region of these genes exists, the retinol metabolism could change and affect retinol treatment in CC patients, a commonly used therapy for this cancer. Moreover, knowing other personal factors that promote methylation may allow future interventions for risk populations. The aim of this study is to determine the methylation status of CRABP genes and its association with the evolution of the type of squamous intraepithelial lesion (SIL) and CC, as well as the relation with risk factors such as demographic characteristics, habits, and the presence of HPV.
Materials and Methods
Tissue collection
A total of 158 women were selected from the Colposcopy Clinic of Sanitary Jurisdiction II in Ciudad Juarez, Chihuahua, Mexico. Patients were selected by colposcopic and histopathological evaluation, and each patient then signed a consent form for the study. The cervix sample of each patient was obtained by biopsy and by endocervical scraping for the control group. Samples were distributed as low squamous intraepithelial lesion (LSIL; n = 42), high squamous intraepithelial lesion (HSIL; n = 69), and CC (n = 25). Women who showed no intraepithelial lesion (NIL) (n = 22) were selected as control group. All tissue samples were stored in 50 µL of RNAlater® at −20°C (Invitrogen). The ethics committee of Universidad Autónoma de Ciudad Juárez approved this study (CBE.ICB/004.01–14). This research complied with the principles of the Declaration of Helsinki. Patients diagnosed with CC by histopathological evaluation were not on any treatment at that moment. Some demographic characteristics and habit information have been reported to influence the methylation process.6 Therefore, we collected information about age, family history of cancer, hormonal contraceptive use, smoking, and alcohol intake from patient interviews.
DNA extraction and HPV genotyping method
DNA of tissue samples was extracted by the phenol–chloroform–isoamyl alcohol-adapted technique. Before DNA extraction, tissue was treated with 500 µL of lysis buffer (0.2 M Tris–HCL pH 8; 10 mM ethylenediaminetetraacetic acid pH 8; 0.5 M NaCl; 1% sodium dodecyl sulfate) and 2.5 µL of proteinase K (20 mg/mL) and incubated for 30 minutes at 56°C. Then, the phenol–chloroform–isoamyl alcohol technique was used.19 HPV genotyping was determined by polymerase chain reaction (PCR). PCR conditions and primer sequences used have been reported elsewhere.20,21
Bisulfite treatment and methylation-specific PCR
Extracted DNA was treated with bisulfite using the DNA Methylation-Gold Kit (Zymo Research Corp.) and following the manufacturer protocol. After bisulfite treatment, modified DNA was used as a template for the methylation-specific PCR (MSP) technique. For PCR amplification, 50 ng of bisulfite-modified DNA was added to a final volume of 25 µL PCR mix containing 12.5 µL GoTaq® Green Master Mix (Promega), 1 µL of forward primer, and 1 µL of reverse primer (4 µM for CRABP1 and 20 µM for CRABP2). Primer sequences are shown in Table 1. The unmethylated and methylated regions of CRABP1 (−193 to +19 bp) and CRABP2 (−265 to −179 bp) were determined in typical PCR conditions. Annealing temperature for methylated and unmethylated CRABP1 primers was 60°C. For CRABP2, the annealing temperature was 60°C for methylated and 65°C for unmethylated primers. PCR products were loaded on 2.0% agarose gels, stained with ethidium bromide, and visualized under ultraviolet illumination.
Table 1.
Primer sequences for CRABP1 and CRABP2.
| 5′ → 3′ | AMPLICON SIZE (bp) | |||
|---|---|---|---|---|
| CRABP1* | Methylated | Fw | GGAGGTTTTTTAGTTGGAGAGC | 212 |
| Rv | CTCGCAAAACGAAAACTAACG | |||
| Unmethylated | Fw | GAGGTTTTTTAGTTGGAGAGTGG | 211 | |
| Rv | AACTCACAAAACAAAAACTAACACT | |||
| CRABP2** | Methylated | Fw | CGTTTTCGCGGAGAGCGCG | 87 |
| Rv | AACCGAAATAACCTTCTCCTACGC | |||
| Unmethylated | Fw | TTTGTTTTTGTGGAGAGTGTGA | 86 | |
| Rv | TCCAAAATAACCTTCTCCTACACT |
Statistical analyses
Comparisons for statistical significance were analyzed using SPSS 15.0 software (SPSS Inc.). A two-proportion z-test was used to analyze the proportions of methylation status among groups. Association between methylation status and personal characteristics of groups was analyzed using χ2 or Fisher’s exact test, as appropriate. Correlations between the process of cancer (NIL to CC) and the presence of methylation were analyzed with Spearman’s correlation coefficient (Rho). All P-values represent two-tailed tests and were considered significant at 0.05.
Results
Methylation and hemi-methylation of CRABPs
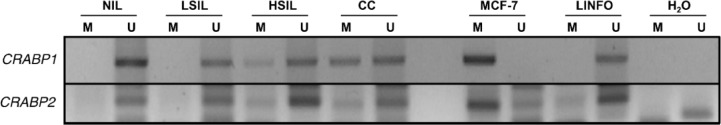
The epigenetic modifications were evaluated in Mexican population distributed in three different groups, according to the grade of SIL or CC. The DNA was modified by bisulfite treatment, and PCR test was performed to determine the methylation status of CRABP1 and CRABP2. The amplification products are shown in Figure 1. Amplification results show hemimethylation (methylation and unmethylation) patterns in the promoter regions of CRABP1 and CRABP2, compared with the control cell line MCF-7. According to the results, 28.5% (45/158) of the samples showed methylation in the promoter region of CRABP1, 7.0% (11/158) in that of CRABP2, and only 2.5% (4/158) methylation in both genes at once (as shown in Table 2). Analysis indicated that CRABP1 methylation is significantly associated with CC (χ2 = 19.7, P < 0.001) and with increases in the degree of injury (rho = 0.290, P < 0.001). Analysis of proportions showed significant differences between CC and the other groups (NIL, P = 0.004; LSIL, P < 0.001; HSIL, P = 0.001). In contrast, CRABP2 methylation was observed to be higher in women with CC but significantly different in women with HSIL (P = 0.047). The linear correlation of CRABP2 methylation and degree of injury showed no statistical significance (rho = 0.145, P = 0.069).
Figure 1.
MSP amplification products of representative samples from each group. Patients with CC (16) and HSIL (18) showed hemi-methylation (methylation and unmethylation) patterns in CRABP1, and CRABP2 hemi-methylation was observed in CC (5) and HSIL (3). MCF-7 cell lines were used as methylated (M) positive controls and LINFO (lymphocytes) as unmethylated (U) positive controls.
Table 2.
Percentage of patients with presence of hemi-methylation in CRABP1 and CRABP2 gene promoters.
|
CRABP1 HEMI-METHYLATION |
CRABP2 HEMI-METHYLATION |
|||
|---|---|---|---|---|
| % (n) | rho (P)§ | % (n) | rho (P)§ | |
| NIL | 18.2 (4/22)* | 0.290 (<0.001) | 4.6 (1/22) | 0.145 (0.069) |
| LSIL | 16.7 (7/42)* | 4.8 (2/42) | ||
| HSIL | 26.1 (18/69)* | 4.6 (3/69)* | ||
| CC | 64.0 (16/25) | 20.0 (5/25) | ||
Notes:
Statistically significant Spearman’s correlation coefficient (rho) (P < 0.05).
Analysis of proportions showing significant differences between CC and the other groups (P < 0.05).
Risk factor associated with methylation profiles
Important factors involved in CC and methylation were considered in this study (age, family history of cancer, hormonal contraceptive use, HPV infection, and habits as alcohol and smoke). The statistical multivariate analysis shows in Table 3 an association analysis that determines whether the presence of methylation is related to certain personal characteristics. Results of methylation in the promoter of CRABP1, adjusted by diagnosis, were found to be 3.6-fold increased when age was ≥35 years (95% confidence interval [95% CI] = 1.58–8.16), 2.2-fold increased with family history of cancer (95% CI = 1.05–4.64), and 2.9-fold increased with the presence of HPV-16 (95% CI = 1.24–6.73). In contrast, methylation of CRABP1 was found to be 0.4-fold decreased in the presence of alcohol consumption (95% CI = 0.18–0.95). The presence of methylation of CRABP2 was not statistically significantly associated with any personal characteristics.
Table 3.
Association between promoter hemi-methylation of CRABPs and personal characteristic variables.
| CRABP1, n (%) | CRABP2, n (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| HM | U | OR* (95% CI) | P | HM | U | OR* (95% CI) | P | |
| Age | ||||||||
| ≥35 years | 28 (42.4) | 38 (57.6) | 3.6 | 0.002 | 7 (10.6) | 59 (89.4) | 2.3 | 0.238 |
| <35 years | 17 (18.5) | 75 (81.5) | (1.58–8.16) | 4 (4.3) | 88 (95.7) | (0.58–9.52) | ||
| Family history of cancer | ||||||||
| Positive | 23 (39.7) | 35 (60.3) | 2.2 | 0.032 | 3 (5.2) | 55 (94.8) | 0.5 | 0.392 |
| Negative | 22 (22.0) | 78 (78.0) | (1.05–4.64) | 8 (8.0) | 92 (92.0) | (0.12–2.23) | ||
| Hormonal contraceptive use | ||||||||
| Positive | 13 (35.1) | 24 (64.9) | 1.4 | 0.406 | 2 (5.4) | 35 (94.6) | 0.6 | 0.578 |
| Negative | 32 (26.5) | 89 (73.5) | (0.61–3.28) | 9 (7.4) | 112 (92.6) | (0.12–3.18) | ||
| HPV§ | ||||||||
| Positive | 37 (90.2) | 88 (83.8) | 1.8 | 0.434 | 10 (90.9) | 115 (85.2) | 1.7 | 1.000 |
| Negative | 4 (9.8) | 17 (16.2) | (0.53–7.77) | 1 (9.1) | 20 (14.8) | (0.22–79.2) | ||
| HPV 16§§ | ||||||||
| Positive | 22 (44.0) | 28 (56.0) | 2.9 | 0.013 | 7 (14.0) | 43 (86.0) | 3.4 | 0.081 |
| Negative | 15 (20.0) | 60 (80.0) | (1.24–6.73) | 3 (4.0) | 72 (96.0) | (0.79–14.9) | ||
| Smoke | ||||||||
| Positive | 8 (22.9) | 27 (77.1) | 0.6 | 0.273 | 2 (5.7) | 33 (94.3) | 0.65 | 0.618 |
| Negative | 37 (30.1) | 86 (69.9) | (0.21–1.53) | 9 (7.3) | 114 (92.7) | (0.13–3.37) | ||
| Alcohol | ||||||||
| Positive | 11 (25.0) | 50 (81.9) | 0.4 | 0.035 | 2 (3.3) | 59 (96.7) | 0.3 | 0.181 |
| Negative | 33 (34.4) | 63 (65.6) | (0.18–0.95) | 9 (9.4) | 87 (90.6) | (0.07–1.71) | ||
Notes:
Analysis of odds ratio (OR) is adjusted with diagnostics.
Analysis of 146 patients.
Analysis of positive HPV (n = 125). Bold values show statistical significance (P ≤ 0.05).
Abbreviations: HM, hemi-methylated; U, unmethylated.
Discussion
Retinoids are commonly used as a chemopreventative and a chemotherapeutic agent for cancer.17 Nevertheless, the effects of retinoids may be altered by epigenetic changes in CC. Mendoza et al16 showed that CRBP1 is methylated in this cancer and that the gene expression is reduced. Additionally, the presence of RARβ2 methylation increases from low grade to invasive in CC patients.15 Therefore, metabolism of all retinols has the potential to be changed and generate a resistance to retinoid therapy.22
The aim of this study was to determine whether the methylation status was present in CRABP1 and CRABP2 gene promoters in CC. First, the proportion of patients with the presence of methylation in CRABP1 was statistically greater in CC patients than in SIL and NIL patients. This result is similar to that reported in another study.23 CRABP2 methylation studies have also described different types and samples of cancer line cells and cancer tissue.24 In this study, the CC group showed methylation in the promoter region of CRABP2, but there was no significant difference when compared with LSIL and NIL. In addition, the proportion of patients with methylation of CRABP2 was less than that of CRABP1. This shows that the process of methylation may be different for the two CRABPs. This study analyzed the association of personal characteristic variables that could contribute to methylation in CRABPs. The results showed a statistical association between methylation and age, family history of cancer, and HPV-16 genotype infection. Researchers have reported that the presence of global methylation can increase in older people, and this is known as age-related methylation.5 A family history of cancer may have a genetic influence on methylation,25 which may contribute to methylation in other chromosome regions, such as the CRABP1 gene promoter. Nevertheless, this result must be studied in more depth to find a correlation between both variables. On the other hand, it is well known that high-risk HPV oncoproteins, such as E6 and E7, increase DNA methyltransferase activity and cause global methylation. In addition, host cells increase the methylation process by regulating regions of E6 and E7 oncogenes as a defense mechanism, which may also be affecting other regions.26 Therefore, this study suggests that the methylation pattern of CRABP1 is changed by personal characteristics and HPV-16 infection. Alcohol consumption has a positive effect on the absence of methylation of CRABP1, and studies have reported that it has an influence on methylation development, especially hypomethylation. Alcohol alters DNA transmethylation and homocysteine metabolism by enzymatic inhibition.27 However, this study suggests that more analysis is needed on the effects of alcohol on the methylation process.
DNA methylation can be used as a marker to diagnose cancer, evaluate prognosis, or predict a therapy response.28 Considering this, CRABP1 might be an epigenetic marker. The results of this study determined that the CRABP1 gene has epigenetic changes that are in response to personal characteristics of the patient. Consequently, this methylation in the CRABP1 gene promoter may repress gene expression and disturb retinol metabolism. For example, CRABP1 is the protein that regulates cytoplasmic RA concentration and allows the interaction of RA with other proteins.29 The absence of CRABP1 gene expression may alter the correct use of RA and be counterproductive in the use of retinoid treatment.
Conclusion
CRABP1 may be a predictive marker of retinoid therapy response. Nevertheless, we propose to extend this study to determine whether methylation in CRABP1 and the presence of older age, family history of cancer, HPV-16 infection, and alcohol intake could affect the retinoid treatment in CC. Finally, abnormal methylation processes are of recent interest for many researchers who want to generate epigenetic markers for early detection of cancer or therapeutic prognosis. This study showed that CRABP1 may be a marker and an important regulator of the retinol pathway in CC.
Acknowledgments
M.D. CDH and patients who contributed to this study of the Colposcopy Clinic of Sanitary Jurisdiction II in Ciudad Juarez, Chihuahua, Mexico, are acknowledged.
Footnotes
ACADEMIC EDITOR: Christian Bonner, Editor in Chief
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 678 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by CONACyT [Fondo Sectorial de Investigación y Seguridad Social SSA/IMSS/ISSSTE-CONACyT, SALUD-2014-01-233271]. During this work, ALAO and JCSE were recipients of a CONACyT fellowship. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: FJV and MSV. Analyzed the data: ALAO, ADMC. Wrote the first draft of the manuscript: ALAO, FJV and JCSE. Contributed to the writing of the manuscript: ADMC, JALD and CLVR. Agree with manuscript results and conclusions: FJV and MSV. Jointly developed the structure and arguments for the paper: FJV and ALAO. Made critical revisions and approved final version: FJV. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Bruni L, Barrionuevo-Rosas L, Albero G, et al. Human Papillomavirus and Related Diseases in the World. ICO Information Centre on HPV and Cancer (HPV Information Centre); Barcelona, Spain: 2016. [Google Scholar]
- 2.Darroudi F, Bergs JWJ, Bezrookove V, Buist MR, Stalpers LJ, Franken NA. PCC and COBRA-FISH a new tool to characterize primary cervical carcinomas: to assess hall-marks and stage specificity. Cancer Lett. 2010;287(1):67–74. doi: 10.1016/j.canlet.2009.05.034. [DOI] [PubMed] [Google Scholar]
- 3.Vedham V, Verma M. Cancer-associated infectious agents and epigenetic regulation. In: Verma M, editor. Cancer Epigenetics. Risk Assessment, Diagnosis, Treatment, and Prognosis. Vol. 1238. Hatfield, Hertfordshire, UK: Humana Press; 2015. p. 335. [Google Scholar]
- 4.Jiménez-Wences H, Peralta-Zaragoza O, Fernández-Tilapa G. Human papilloma virus, DNA methylation and microRNA expression in cervical cancer. Oncol Rep. 2014;31:2467–2476. doi: 10.3892/or.2014.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Issa J. CpG—island methylation in aging and cancer. Curr Top Microbiol Immunol. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- 6.Lim U, Song M. Dietary and lifestyle factors of DNA methylation. In: Dumitrescu R, Verma M, editors. Cancer Epigenetics: Methods and Protocols. Vol. 863. Springer Science+Business Media; Switzerland: 2012. pp. 359–376. [DOI] [PubMed] [Google Scholar]
- 7.Dick KJ, Nelson CP, Tsaprouni L, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 8.Lim U, Flood A, Choi S, et al. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology. 2008;134(1):47–55. doi: 10.1053/j.gastro.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith IM, Mydlarz WK, Mithani SK, Califano JA. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer. 2007;121(8):1724–1728. doi: 10.1002/ijc.22889. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima K, Takeoka M, Mori M, et al. Exercise effects on methylation of ASC gene. Int J Sports Med. 2010;31(9):671–675. doi: 10.1055/s-0029-1246140. [DOI] [PubMed] [Google Scholar]
- 11.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 12.Campesi I, Sanna M, Zinellu A, et al. Oral contraceptives modify DNA methylation and monocyte-derived macrophage function. Biol Sex Differ. 2012;3(4):1–11. doi: 10.1186/2042-6410-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovalchuk O, Tryndyak VP, Montgomery B, et al. Estrogen-induced rat breast carcinogenesis is characterized by alterations in DNA methylation, histone modifications, and aberrant microRNA expression. Cell Cycle. 2007;6(16):2010–2018. doi: 10.4161/cc.6.16.4549. [DOI] [PubMed] [Google Scholar]
- 14.Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: the time is now. Crit Rev Oncol Hematol. 2008;68(1):1–11. doi: 10.1016/j.critrevonc.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Ivanova T, Petrenko A, Gritsko T, et al. Methylation and silencing of the retinoic acid receptor-beta 2 gene in cervical cancer. BMC Cancer. 2002;2:4. doi: 10.1186/1471-2407-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza-Rodriguez M, Arreola H, Valdivia A, et al. Cellular retinol binding protein 1 could be a tumor suppressor gene in cervical cancer. Int J Clin Exp Pathol. 2013;6(9):1817–1825. [PMC free article] [PubMed] [Google Scholar]
- 17.Bushue N, Wan Y-JY. Retinoid pathway and cancer therapeutics. Adv Drug Deliv Rev. 2010;62(13):1285–1298. doi: 10.1016/j.addr.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem J. 2000;348:481–495. [PMC free article] [PubMed] [Google Scholar]
- 19.Ausubel F, Brent R, Kingston RE, et al. Short Protocols in Molecular Biology. 5th ed. New York: John Wiley & Sons; 2002. [Google Scholar]
- 20.Nishiwaki M, Yamamoto T, Tone S, et al. Genotyping of human papilloma-viruses by a novel one-step typing method with multiplex PCR and clinical applications. J Clin Microbiol. 2008;46(4):1161–1168. doi: 10.1128/JCM.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu W, Jiang G, Cruz Y, et al. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J Clin Microbiol. 1997;35(6):1304–1310. doi: 10.1128/jcm.35.6.1304-1310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freemantle SJ, Spinella MJ, Dmitrovsky E. Retinoids in cancer therapy and chemoprevention: promise meets resistance. Oncogene. 2003;22(47):7305–7315. doi: 10.1038/sj.onc.1206936. [DOI] [PubMed] [Google Scholar]
- 23.Wu Q, Lothe RA, Ahlquist T, et al. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol Cancer. 2007;6:45. doi: 10.1186/1476-4598-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calmon MF, Rodrigues RV, Kaneto CM, et al. Epigenetic silencing of CRABP2 and MX1 in head and neck tumors. Neoplasia. 2009;11(12):1329–1339. doi: 10.1593/neo.91110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gertz J, Varley KE, Reddy TE, et al. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet. 2011;7(8):1–10. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgers WA, Blanchon L, Pradhan S, de Launoit Y, Kouzarides T, Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2012;26(11):1650–1655. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varela-Rey M, Woodhoo A, Martinez-Chantar M-L, Mato J, Lu S, Alcohol DNA. methylation, and cancer. Alcohol Res Rev. 2011;35(1):25–36. [PMC free article] [PubMed] [Google Scholar]
- 28.Strathdee G. Epigenetic markers and response to chemotherapy in cancer. Dis Markers. 2007;23(1–2):43–49. doi: 10.1155/2007/610815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfoertner S, Goelden U, Hansen W, et al. Cellular retinoic acid binding protein I: expression and functional influence in renal cell carcinoma. Tumour Biol. 2005;26:313–323. doi: 10.1159/000089262. [DOI] [PubMed] [Google Scholar]