Abstract
Introduction
Endoplasmic reticulum aminopeptidase 1 (ERAP1) protein is highly polymorphic with numerous missense amino acid variants. We sought to determine the naturally occurring ERAP1 protein allotypes and their contribution to Behçet’s disease.
Methods
Genotypes of all reported missense ERAP1 gene variants with 1000 Genomes EUR super-population frequency greater than 1% were determined in 1,900 Behçet’s disease cases and 1,779 controls from Turkey. ERAP1 protein allotypes and their contributions to Behçet’s disease risk were determined by haplotype identification and disease association analyses.
Results
One ERAP1 protein allotype with 5 non-ancestral amino acids was recessively associated with disease (P = 3.13 × 10−6, odds ratio 2.55, 95% CI 1.70 to 3.82). The ERAP1 association was absent in individuals who lacked HLA-B*51. Individuals who carry HLA-B*51 and who are also homozygous for the haplotype had an increased disease odds compared with those with neither risk factor (P = 4.80 × 10−20, odds ratio 10.96, 95% CI 5.91 to 20.32).
Discussion
The Behçet’s disease-associated ERAP1 protein allotype was previously shown to have poor peptide trimming activity. Combined with its requirement for HLA-B*51, these data suggest that a hypoactive ERAP1 allotype contributes to Behçet’s disease risk by altering the peptides available for binding to HLA-B*51.
INTRODUCTION
The endoplasmic reticulum aminopeptidase-1 (ERAP1) protein trims intracellular proteasome-processed peptides prior to their loading onto nascent class I HLA molecules in the endoplasmic reticulum. Peptides that are efficiently bound by the class I HLA molecules are transported to and displayed on the surface of nearly all cell types, where they play an important role in immune surveillance and in the function of cytotoxic T and natural killer cells. Variants of the ERAP1 gene have been associated with three polygenic inflammatory diseases with strong class I HLA associations. In all three diseases the ERAP1 association is found only among individuals carrying the disease-associated HLA type, HLA-B*27 in ankylosing spondylitis,1 HLA-Cw6 in psoriasis,2 and HLA-B*51 in Behçet’s disease.3 Interestingly, the ERAP1 variant (p.Arg725Gln) that is associated with Behçet’s disease risk is protective for both ankylosing spondylitis and psoriasis. Variants that influence ERAP1 activity and peptide specificity are likely to influence the ER-peptidome by producing and or destroying peptides that can be efficiently bound by disease-specific HLA class I molecules.
The ERAP1 protein is highly polymorphic with 10 missense amino acid variants reported with greater than 1% minor allele frequency in the 1000 Genomes Project EUR super-population. The enzymatic activity and peptide specificity of the ERAP1 protein is likely dependent on its complete combination of variant amino acids, i.e., its protein allotype, but ERAP1 disease associations have been reported for individual variants or for haplotypes composed of only two to five variants.4-7 Furthermore, ERAP1 activity and specificity in individuals may ultimately be dependent upon allotype combinations as individuals carry a pair of codominantly expressed haplotypes.8 To determine the common protein allotypes present in the Turkish population and evaluate their contributions to Behçet’s disease risk, we genotyped ERAP1 coding region SNPs, imputed additional marker genotypes, and estimated coding variant haplotypes in 1,900 Behçet’s disease cases and 1,779 controls from Turkey. We found a single ERAP1 allotype with a large contribution to disease risk in HLA-B*51 carriers.
MATERIALS AND METHODS
Patients and controls
1,900 unrelated Behçet’s disease cases and 1,779 unrelated controls (from our previous Turkish GWAS and Turkish replication collections3) that passed stringent quality controls applied after genotyping with the Illumina Immunochip were included in the study. For further information and patient characteristics see online supplementary text and Table S1.
SNP genotypes and imputation
973 Immunochip SNP markers from the ERAP1 genomic region (hg build 19, chr5:95,970,970 – 96,427,776) that passed stringent quality controls were used as input for regional imputation with IMPUTE2 software9 with the 1000 Genomes Project phase 1 integrated dataset haplotypes as the reference. Imputed markers with info-score > 0.8 and predicted genotypes with probability > 0.9 were included.
Analysis of ERAP1 coding haplotypes
Genotypes of 10 missense SNPs were used for prediction of coding haplotypes with SNP & Variation Suite 8.4 (SVS) [Golden Helix, Bozeman, Montana].
HLA-B*51 imputation
HLA-type imputation was performed with SNP2HLA software with Immunochip HLA region marker genotypes and a reference of 5,225 individuals collected, HLA-typed, and SNP genotyped by the Type I Diabetes Genetics Consortium.10 In 2213 samples typed for HLA-B*51, there were 24 individuals with discordant imputed types (98.9% concordance rate).
Association testing of ERAP1 coding haplotypes with Behçet’s disease
Associations of the ERAP1 coding haplotypes with disease were evaluated by Pearson’s chi squared test in SVS or the exact unconditional chi squared test11 in StatXact11 software (Cytel, Cambridge, Massachusetts) under a recessive model and odds ratios were calculated under a recessive model using SVS. The recessive model was applied because the previously reported single marker associations were found only with the recessive model. Two risk factor analyses (HLA-B*51 and homozygosity for the disease-associated haplotype) were evaluated by 2 × 2 contingency table odds ratios comparing the frequency in cases with controls of the single-risk factor or two-risk factor groups relative to the frequency of individuals with neither risk factor. Significance thresholds for P-values were 0.05 divided by the number of comparisons made (Bonferroni correction).
Molecular modeling of ERAP1
The structure of ERAP1 was evaluated using protein structure summary 3MDJ and the UCSF Chimera package http://www.cgl.ucsf.edu/chimera. Polymorphic amino acid positions (Table 1) were altered to demonstrate the ERAP1 molecules encoded by Hap1 and Hap10.
Table 1.
Common ERAP1 coding variant haplotypes/allotypes and homozygous association with Behçet’s disease in 1,876 cases and 1,761 controls from Turkey.
|
Coding
haplo- typea |
Amino acid position
|
Homozyg
hap freq cases n (%) |
Homozyg
hap freq ctrls n (%) |
Recessive
model P-value |
Homozyg hap
odds ratio (95% CI) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 56 | 127 | 276 | 346 | 349 | 528 | 575 | 725 | 730 | |||||
| Hap1 | Ileb | Glu | Pro | Ile | Gly | Met | Lys | Asp | Arg | Gln | 26 (1.4) | 14 (0.8) | 0.088 | 1.75 (0.91-3.37) |
|
| ||||||||||||||
| Hap2 | Thr | Glu | Arg | Ile | Gly | Met | Lys | Asp | Arg | Gln | 43 (2.3) | 22 (1.2) | 0.018 | 1.85 (1.10-3.11) |
|
| ||||||||||||||
| Hap3 | Thr | Glu | Arg | Ile | Gly | Met | Lys | Asp | Arg | Glu | 46 (2.5) | 34 (1.9) | 0.284 | 1.28 (0.81-2.00) |
|
| ||||||||||||||
| Hap5 | Thr | Glu | Arg | Ile | Asp | Met | Arg | Asp | Arg | Glu | 20 (1.1) | 13 (0.7) | 0.297 | 1.45 (0.72-2.92) |
|
| ||||||||||||||
| Hap6 | Thr | Glu | Pro | Ile | Gly | Met | Arg | Asp | Arg | Glu | 34 (1.8) | 21 (1.2) | 0.126 | 1.53 (0.88-2.65) |
|
| ||||||||||||||
| Hap7 | Thr | Lys | Pro | Ile | Gly | Met | Arg | Asp | Arg | Glu | 1 (0.06) | 2 (0.11) | 0.684c | 0.47 (0.04-5.18) |
|
| ||||||||||||||
| Hap8 | Thr | Glu | Pro | Met | Gly | Met | Arg | Asp | Arg | Glu | 98 (5.2) | 94 (5.3) | 0.888 | 0.98 (0.73-1.31) |
|
| ||||||||||||||
| Hap10 | Thr | Glu | Pro | Ile | Gly | Val | Arg | Asn | Gln | Glu | 87 (4.6) | 33 (1.9) | 3.12E-06** | 2.55 (1.70 -3.82) |
Haplotype numbers according to Ombrello et al, 2015. These 8 haplotypes represent all the coding haplotypes with frequency greater than 1% frequency in the Turkish controls and account for 98.8% of the haplotypes predicted with probability greater than 0.8.
Italic indicates the non-ancestral amino acid; the ancestral amino acid is the amino acid found in the chimpanzee sequence.
Due to too few instances for the asymptotic chi squared test, this was computed using the exact unconditional chi squared test.
Significant after Bonferroni correction, P < 3.12E-03 (0.05/[8 haplotypes × 2 models]).
RESULTS
Of the 10 common ERAP1 missense variants identified in the 1000 Genomes Project EUR super-population (online supplementary Table S2), 8 were genotyped successfully on the Immunochip, and 2 (rs2287987/Glu56Lys and rs3734016/Met349Val) were successfully imputed. All 10 missense variants had minor allele frequencies greater than 1% in the Turkish control population (online supplementary Table S2). Strong linkage disequilibrium (LD) was found among these 10 variants (online supplementary Figure S1). Haplotype prediction in the 3,679 Turkish samples identified a pair of haplotypes with probability greater than 0.8 in 3,637 of the samples (98.9%). Eight of the 10 haplotypes reported with greater than 1% frequency in HapMap CEU samples12 were found at greater than 1% frequency and these 8 haplotypes accounted for 98.8% of all haplotypes identified in the Turkish control population (online supplementary Table S3). Results of recessive genotypic association tests for all the imputed SNPs are shown in online supplementary Figure S2.
Association testing under the recessive model revealed that only Hap10 of ERAP1, which constitutes 14.3% of all coding region haplotypes and bears 5 non-ancestral alleles, Met349Val, Lys528Arg, Asp575Asn, Arg725Gln, and Gln730Glu (where the first amino acid is the ancestral amino acid, defined as the one present in chimpanzees), was significantly associated with Behçet’s disease (Table 1). Similar to the reported association of individual ERAP1 SNPs with Behçet’s disease,3 this haplotypic association was only significant under the recessive model (non-significant associations based on haplotype frequency are shown in online supplementary Table S3). Thus, homozygosity for the ERAP1 Hap10 haplotype or for any of the SNPs that specifically tag Hap10 is a risk factor for Behçet’s disease.
A combinatorial analysis integrating the presence of HLA-B*51 and homozygosity for ERAP1 Hap10 demonstrated a strong interaction between these two Behçet’s disease risk factors (Table 2). Compared with individuals with neither risk factor, individuals homozygous for Hap10 but without HLA-B*51 have no detectable increase in disease odds. Individuals who carry HLA-B*51 but are not homozygous for Hap10 have a 3.6 fold increased disease odds. However, individuals who carry HLA-B*51 and are also homozygous for ERAP1 Hap10 had a 10.96 fold increased disease odds (Table 2). We found a similar disease odds in individuals heterozygous for HLA-B*51 (odds ratio 10.07, 95%CI 5.41 to 18.74) compared with the individuals homozygous or heterozygous for HLA-B-*51.
Table 2.
Two risk factor analysis for Behçet’s disease in 1,876 cases and 1,761 controls from Turkey.
| HLA-B*51/Homozygous Hap10 | Number of cases n (%) |
Number of controls n (%) |
Odds Ratio | (95% CI) | P-value |
|---|---|---|---|---|---|
| −/− | 659 (35.1) | 1171 (66.5) | 1.00 | reference | reference |
|
| |||||
| −/+ | 13 (0.7) | 21 (1.2) | 1.10 | (0.55 to2.21) | 7.90 × 10−01 |
|
| |||||
| +/− | 1130 (60.2) | 557 (31.6) | 3.60 | (3.14 to 4.14) | 2.87 × 10−75 ** |
|
| |||||
| +/+ | 74 (3.9) | 12 (0.7) | 10.96 | (5.91 to 20.32) | 4.80 × 10−20 ** |
Significant after Bonferroni correction, P < 0.0167 (0.05/3 groups with one or more risk factors).
DISCUSSION
Our haplotype analysis of ERAP1 missense variants identified 8 ERAP1 protein allotypes with greater than 1% frequency in the Turkish population. Only one of these haplotypes, Hap10, was associated with Behçet’s disease risk. This association was only detected under the recessive model, and moreover it only influenced disease risk in individuals who also carried the Behçet’s disease-associated HLA type, HLA-B*51. Individuals carrying HLA-B*51 who are also homozygous for Hap10 have a nearly 11 fold increased disease odds compared with individuals with neither genetic risk factor. Although homozygosity for Hap10 has a large effect size, particularly in HLA-B*51 carriers, it does not make a large contribution to the overall population risk because of its low frequency. Hap10 represents 14.3% of the ERAP1 gene coding region haplotypes in the Turkish population and therefore, as expected, only about 2% of controls were homozygous for Hap10. Despite the low contribution to the overall population risk, the large effect size conferred by the combination of both risk factors suggests an important mechanism by which their combination contributes to Behçet’s disease risk.
The Hap10 allotype bears 5 non-ancestral amino acids (Met349Val, Lys528Arg, Asp575Asn, Arg725Gln, and Gln730Glu), 3 of which (Met349Val, Asp575Asn, and Arg725Gln) are good tags for the haplotype itself. Individual variants encoding these haplotype tagging SNPs,Met349Val (rs2287987), Asp575Asn (rs10050860), and Arg725Gln (rs17482078), were previously reported recessively associated with Behçet’s disease in Turkish3 and Iranian13 studies. Their genotype frequencies were also consistent with recessive association in the Spanish and Chinese populations, but did not reach statistical significance.5, 14
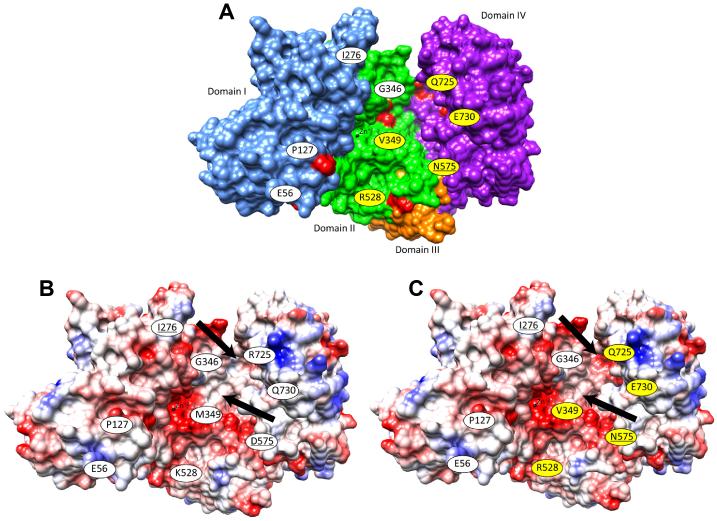
The positions of the ERAP1 variant amino acids and the surface electrostatic potential of of the Hap1 and the Behçet’s disease-associated Hap10 allotypes are shown in structural models of the ERAP1 protein in Figure 1. The altered surface electrostatic potential could result in different characteristics of substrate peptides bound. The Hap10 allotype of the ERAP1 protein was previously found to have poor peptide trimming activity,15-17 thus homozygosity for Hap10 could greatly alter the composition of the peptidome available for binding to HLA-B*51. Recent work by Guasp and colleagues suggests that low ERAP1 activity would lead to a peptidome with low affinity for HLA-B*51, which could contribute to Behçet’s disease risk by enhancing NK cell lytic activity.17
Figure 1.
Behçet's disease-associated form of ERAP1. (A) Surface representation of ERAP1, colored by domain, shows the locations and identities of the common variant amino acid residues (red spheres) in the ERAP1 allotypes, and their proximity to the catalytic Zn2+ atom (black sphere). Models displaying the electrostatic surface potential of Hap1 (B) and the Behçet’s disease-associated Hap10 (C) demonstrate changes in surface potential near to the catalytic site (black arrows). Ancestral alleles are marked with white labels, non-ancestral alleles are marked with yellow labels; labels of residues not visible are underlined. This model was created using 3MDJ and UCSF Chimera software. In (B) and (C), red coloration indicates positive surface charge, blue indicates negative surface charge, and white indicates neutrality.
Thus, the ERAP1 Hap10 allotype could either inefficiently produce disease-protective peptides by failing to trim precursor peptides, or alternatively it could fail to digest/destroy disease-promoting peptides. Identifying the nature and source of such peptides, for example are they self-derived or do they originate in pathogenic or commensal organisms, would be an important step towards elucidating the mechanism by which HLA-B*51 contributes to Behçet’s disease risk.
Supplementary Material
ACKNOWLEDGMENTS
This study utilized the high-performance computational capabilities of the Helix Systems (http://helix.nih.gov) and the Biowulf cluster (http://hpc.nih.gov) at the National Institutes of Health, Bethesda, MD. Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
FUNDING
This study was supported by the Intramural Research Programs of the National Human Genome Research Institute and the National Institute of Arthritis and Musculoskeletal and Skin Diseases. Dr. Kirino is supported by grants from Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (Grant No. 26713036), the Naito Memorial Foundation, the Kanae Foundation for the Promotion of Medical Science, and Yokohama Foundation for Advancement of Medical Science.
Footnotes
(For a concise report in The Annals of the Rheumatic Diseases)
REFERENCES
- 1.Evans DM, Spencer CC, Pointon JJ, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–7. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strange A, Capon F, Spencer CC, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirino Y, Bertsias G, Ishigatsubo Y, et al. Genome-wide association analysis identifies new susceptibility loci for Behcet's disease and epistasis between HLA-B*51 and ERAP1. Nat Genet. 2013;45:202–7. doi: 10.1038/ng.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettencourt BF, Rocha FL, Alves H, et al. Protective effect of an ERAP1 haplotype in ankylosing spondylitis: investigating non-MHC genes in HLA-B27-positive individuals. Rheumatology (Oxford) 2013;52:2168–76. doi: 10.1093/rheumatology/ket269. [DOI] [PubMed] [Google Scholar]
- 5.Conde-Jaldon M, Montes-Cano MA, Garcia-Lozano JR, et al. Epistatic interaction of ERAP1 and HLA-B in Behcet disease: a replication study in the Spanish population. PLoS One. 2014;9:e102100. doi: 10.1371/journal.pone.0102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadi A, Izac B, Said-Nahal R, et al. Investigating the genetic association between ERAP1 and spondyloarthritis. Ann Rheum Dis. 2013;72:608–13. doi: 10.1136/annrheumdis-2012-201783. [DOI] [PubMed] [Google Scholar]
- 7.Maksymowych WP, Inman RD, Gladman DD, et al. Association of a specific ERAP1/ARTS1 haplotype with disease susceptibility in ankylosing spondylitis. Arthritis Rheum. 2009;60:1317–23. doi: 10.1002/art.24467. [DOI] [PubMed] [Google Scholar]
- 8.Reeves E, Colebatch-Bourn A, Elliott T, et al. Functionally distinct ERAP1 allotype combinations distinguish individuals with Ankylosing Spondylitis. Proc Natl Acad Sci U S A. 2014;111:17594–9. doi: 10.1073/pnas.1408882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–70. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia X, Han B, Onengut-Gumuscu S, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lydersen S, Langaas M, Bakke O. The exact unconditional z-pooled test for equality of two binomial probabilities: optimal choice of the Berger and Boos confidence coefficient. Journal of Statistical Computation and Simulation. 2012;82:1311–1316. [Google Scholar]
- 12.Ombrello MJ, Kastner DL, Remmers EF. Endoplasmic reticulum-associated amino-peptidase 1 and rheumatic disease: genetics. Curr Opin Rheumatol. 2015;27:349–56. doi: 10.1097/BOR.0000000000000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousa I, Shahram F, Francisco D, et al. Brief report: association of CCR1, KLRC4, IL12A-AS1, STAT4, and ERAP1 With Behcet's disease in Iranians. Arthritis Rheumatol. 2015;67:2742–8. doi: 10.1002/art.39240. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Yu H, Zheng M, et al. Association of ERAP1 Gene Polymorphisms With Behcet's Disease in Han Chinese. Invest Ophthalmol Vis Sci. 2015;56:6029–35. doi: 10.1167/iovs.15-17544. [DOI] [PubMed] [Google Scholar]
- 15.Reeves E, Edwards CJ, Elliott T, et al. Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J Immunol. 2013;191:35–43. doi: 10.4049/jimmunol.1300598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Medel N, Sanz-Bravo A, Van Nguyen D, et al. Functional interaction of the ankylosing spondylitis-associated endoplasmic reticulum aminopeptidase 1 polymorphism and HLA-B27 in vivo. Mol Cell Proteomics. 2012;11:1416–29. doi: 10.1074/mcp.M112.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guasp P, Alvarez-Navarro C, Gomez-Molina P, et al. The peptidome of the Behcet's disease-associated HLA-B*51:01 includes two sub-peptidomes differentially shaped by ERAP1. Arthritis Rheumatol. 2015 doi: 10.1002/art.39430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.