Abstract
Background
The potential clinical impact of KRAS and epidermal growth factor receptor (EGFR) mutations has been investigated in lung adenocarcinomas; however, their prognostic value remains controversial. In our study, we sought to investigate the prognostic significance of driver mutations using a large cohort of early-stage lung adenocarcinomas.
Methods
We reviewed patients with pathologic early-stage, lymph node-negative, solitary lung adenocarcinoma who had undergone surgical resection (1995–2005; stage I/II = 463/19). Tumors were classified according to the IASLC/ATS/ERS classification and genotyped by Sequenom MassARRAY system and polymerase chain reaction-based assays. In stage I disease, the Kaplan-Meier method and cumulative incidence of recurrence (CIR) analyses were used to estimate the probability of overall survival (OS) and recurrence, respectively.
Results
Of all, 129 (27%) patients had mutations in KRAS, 86 (18%) in EGFR, 8 (2%) in BRAF, 8 (2%) in PIK3CA, 4 (1%) in NRAS, and 1 (0.2%) in AKT1. EGFR L858R mutation correlated with lepidic predominant histology (P = 0.006) while exon 19 deletion correlated with acinar predominant histology (P < 0.001). EGFR mutations were not detected in invasive mucinous adenocarcinomas (P = 0.033). The 5-year OS of patients with KRAS mutant tumors was significantly worse (n = 124; 5-year OS, 63%) than those with KRAS wild-type (n = 339; 77%; P < 0.001). In solid predominant tumors, KRAS mutations correlated with worse OS (P = 0.008) and increased risk of recurrence (P = 0.005). On multivariate analysis, KRAS mutation was an independent prognosticator of OS in all patients (hazard ratio, 1.87; P < 0.001) and recurrence in solid predominant tumors (hazard ratio, 4.73; P = 0.012).
Conclusion
In patients with resected stage I lung adenocarcinomas, KRAS mutation was an independent prognostic factor for OS and recurrence, especially in solid predominant tumors.
Keywords: Adenocarcinoma, lung, KRAS, epidermal growth factor receptor, prognosis
INTRODUCTION
Recent advances in thoracic medical oncology have focused on the identification of driver mutations and the development of molecular-targeted therapy in patients with non-small cell lung cancer (NSCLC). Tumors with driver mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR), which occurs primarily in adenocarcinomas, show higher sensitivity to EGFR tyrosine kinase inhibitors (TKIs), erlotinib, and gefitinib in NSCLC patients.1–5 More recently recognized anaplastic lymphoma kinase (ALK) rearrangements also predicted a higher response rate to the targeted agent (crizotinib).6, 7
The 2011 international multidisciplinary histologic classification proposed by the International Association for the Study of Lung Cancer (IASLC), American Thoracic Society (ATS), and European Respiratory Society (ERS)8 demonstrates the prognostic significance of the predominant histologic subtype and has been validated in large independent cohorts (>400 patients) across multiple countries.9–12 Moreover, EGFR and KRAS mutations that correlate with predominant histologic subtypes, according to this classification, have been identified.11–14 However, correlations with other rare mutations (such as BRAF, PIK3CA, and NRAS) and each predominant histologic subtype have not been thoroughly investigated and there is little data suggesting driver mutation status correlates with prognosis, within a single histologic subtype or within a specific tumor grade.
In NSCLCs, although the potential clinical impact of KRAS and EGFR mutations has been investigated, their prognostic significance remains controversial, specifically in early-stage disease.15–25 In our study, we sought to investigate the prognostic significance of driver mutations (mainly in KRAS and EGFR) using a large cohort of resected early-stage lung adenocarcinomas and analyze the molecular (KRAS, EGFR, and other rare gene mutation) correlations with histologic subtypes based on the 2011 IASLC/ATS/ERS classification, which is currently published in the 2015 World Health Organization Classification of Tumours of the Lung.26
MATERIALS AND METHODS
Patients
This retrospective study (WA0269-08) was approved by the Institutional Review Board at Memorial Sloan Kettering Cancer Center (MSK). We reviewed patients with pathologic stage I–II, lymph node-negative, solitary lung adenocarcinomas who had undergone surgical resection at MSK between 1995 and 2005. Of all, only 4 (0.8%) patients received adjuvant chemotherapy. Tumor slides and blocks were available for review and molecular analyses from 482 patients (stage IA [n = 316]; stage IB [n = 147]; and stage II [n = 19]). Clinical data were collected from our prospectively maintained database. Disease stage was assigned by the seventh edition of the American Joint Committee on Cancer TNM Staging Manual.27 According to the sixth edition of TNM classification, all patients in this cohort had stage I disease with no lymph node metastasis. By applying the seventh edition of TNM classification, however, a minority of cases were reclassified as stage II tumors (n = 19). Subsets of these cases have been used in our previous publications.28–33 However, there is no overlap of patients with our recent paper focusing on EGFR and KRAS mutations in lung adenocarcinoma.14
Histologic evaluation
All available hematoxylin and eosin (H&E)-stained slides were reviewed by two pathologists (K.K. and W.D.T.), both of whom were blinded to patient clinical outcomes, using an Olympus BX51 microscope (Olympus, Tokyo, Japan) with a standard 22-mm diameter eyepiece. Tumors were classified according to the IASLC/ATS/ERS classification8 and were grouped into 3 architectural grades according to histologic subtype—low-grade (adenocarcinoma in situ, minimally invasive adenocarcinoma or lepidic predominant), intermediate-grade (papillary predominant or acinar predominant), and high-grade (micropapillary predominant, solid predominant, invasive mucinous, or colloid predominant).10, 29
Percentage of cribriform pattern—which our group has recently published as a distinct histologic pattern in acinar predominant subtype with poor prognosis in stage I lung adenocarcinoma was also recorded in 5% increments and designations of cribriform-predominant subtype were made using criteria similar to the IASLC/ATS/ERS classification.32 The signet-ring cell feature is characterized by abundant intracellular mucin and a crescentic nucleus displaced toward one end of the cell, and it represents a cytologic change that can occur in multiple histologic subtypes of invasive adenocarcinoma (acinar, papillary, micropapillary, and solid predominant). Percentage of signet-ring cell feature was recorded regardless of the histologic subtype of each tumor; this feature was recorded as being present when any percentage was found.
Nuclear atypia was identified in the area with highest degree of atypia and was graded as follows: mild (uniform nuclei in size and shape), moderate (intermediate size nuclei with slight irregularity), and severe (enlarged nuclei in varying degrees and some nuclei at least twice as large as others).31, 34 Mitoses were evaluated in 50 high-power fields (HPFs) of ×400 magnification (0.237 mm2 field) in areas with the highest mitotic activity and were counted as the average number of mitotic figures per 10 HPFs.31, 34 Visceral pleural invasion, lymphovascular invasion, and tumor necrosis were also investigated.
The results of thyroid transcription factor-1 (TTF-1) immunohistochemistry, on the basis of tissue microarray analysis, were obtained from our previous study and any immunoreactivity for TTF-1 was considered positive.30
Mutation analysis
In each case, H&E-stained slides from formalin-fixed paraffin-embedded tumor blocks were reviewed to identify and circle the tumor area, thereby ensuring >50% tumor content in tumor blocks. Ten unstained sections (10-μm) were cut from tumor blocks for molecular analysis. When the tumor content was <50%, macrodissection was performed using the blade tip to scrape off the selected tumor areas on 10 corresponding 10-μm unstained sections on the slides. Genomic DNA was extracted using the DNeasy Tissue Kit (Qiagen, Valencia, CA).35
Tumors were genotyped using the Sequenom MassARRAY system (Sequenom, San Diego, CA), just as in our previous publications.35, 36 Amplification and extension primers were designed using Sequenom Assay Designer v3.1 software to target the driver mutations in 8 oncogenes: EGFR, KRAS, BRAF, PIK3CA, NRAS, AKT1, ERBB2/HER2, and MAP2K1/MEK1 (for a total of 92 nonsynonymous mutations).36 Allele-specific single base extension products were quantitatively analyzed using matrix-assisted laser desorption/ionization-time of flight/mass spectrometry on the Sequenom MassArray Spectrometer. Automatically generated genotype calls were confirmed with manual review of the spectra.35 Additionally, EGFR exon 19 deletion was detected via length analysis of fluorescently labeled polymerase chain reaction products.37
Immunohistochemistry and scoring of ALK by using tissue microarrays
Formalin-fixed, paraffin-embedded tumor specimens were used for tissue microarray construction. Briefly, 6 representative tumor areas were marked on H&E-stained slides and cylindrical 0.6-mm tissue cores were arrayed from the corresponding paraffin blocks into a recipient block using an automated tissue arrayer (ATA-27; Beecher Instruments, Sun Prairie, WI), resulting in 15 tissue microarray blocks. From each tissue microarray block, 4-μm-thick paraffin sections were prepared for immunohistochemical analysis. In total, 471 cases with adequate cores were available for immunohistochemical analysis.
We briefly deparaffinized 4-μm sections from the tissue microarray blocks in xylene and dehydrated in graded alcohols. The standard avidin-biotin complex peroxidase technique was used for immunohistochemical staining of anti-ALK antibodies (clone 5A4; Adcam; diluted at 1:30). Sections were stained using a Ventana Discovery XT Automated Immunohistochemical Stainer (Ventana, Tucson, AZ), in accordance with the manufacturer guidelines. Diaminobenzidine was used as the chromogen and hematoxylin was used as the nuclear counterstain. Positive control tissues were stained in parallel with the study cases.
ALK expression was recorded as intensity of tumor cells with cytoplasmic-positive immunostaining in each tumor core. The intensity of staining was scored as 0 (no staining), 1 (faint cytoplasmic staining), 2 (moderate granular cytoplasmic staining), and 3 (strong granular cytoplasmic staining).14, 38–40 Average intensity score of tumor cores was considered indicative of ALK expression for each patient. According to the intensity score, ALK expression was divided into two groups—negative (score of 0–1) and positive (score > 1).14, 39, 40
Statistical analysis
In the entire cohort (n = 482), associations between variables were analyzed using the Fisher’s exact test for categorical variables and the Wilcoxon test for continuous variables. We investigated the prognostic significance (for survival and recurrence) of each factor only in patients with stage I disease (n = 463). Overall survival (OS) was defined as time from surgery to death or last follow-up, and was estimated using the Kaplan-Meier method. Associations between factors and OS were analyzed using the log-rank test and the Cox proportional hazards regression model. Cumulative incidence of recurrence (CIR) analysis—where death from any causes other than recurrence was considered a competing event—was used to estimate probability of recurrence.41 Follow-up duration was calculated from date of surgery to date of first recurrence, death from any cause, or last follow-up. Differences in CIR between groups were assessed using the Gray method for univariate analyses and the Fine-Gray method for multivariate analyses after adjustment for important potential confounders.42
All P-values were determined using two-tailed statistical analyses and P < 0.05 was considered statistically significant. Statistical analyses were conducted using SAS v9.2 (SAS Institute, Cary, NC) and R (R Development Core Team, 2010), including the “survival” and “cmprsk” packages.
RESULTS
Patient demographics and their associations with EGFR and KRAS mutations
Patient clinicopathologic factors are summarized in Table 1. Of all (n = 482), median patient age was 69 years (range, 33–89 years) and most patients were women (n = 304). During the study period in stage I disease, 76 (16%) patients experienced recurrence, 164 (35%) died from any cause, and median follow-up period for patients without recurrence was 56.8 months (range: 0.3–160.0 months).
Table 1.
Clinicopathologic associations with KRAS and EGFR mutations
| Characteristic | Total, N |
KRAS, N (%)
|
P |
EGFR, N (%)
|
P | ||
|---|---|---|---|---|---|---|---|
| Wild-type | Mutant | Wild-type | Mutant | ||||
| Age, years | 0.35 | 0.74 | |||||
| Median | 69 | 69 | 68 | 69 | 69 | ||
| Range | 33–89 | 33–89 | 50–85 | 33–89 | 37–88 | ||
| Sex | 0.67 | 0.019 | |||||
| Female | 304 | 225 (74) | 79 (26) | 240 (79) | 64 (21) | ||
| Male | 178 | 128 (72) | 50 (28) | 156 (88) | 22 (12) | ||
| Smoking status | <0.001 | <0.001 | |||||
| Never | 77 | 72 (94) | 5 (6) | 40 (52) | 37 (48) | ||
| Former/Current | 405 | 281 (69) | 124 (31) | 356 (88) | 49 (12) | ||
| Total tumor size, cm | 0.87 | 0.95 | |||||
| Median | 2.1 | 2.1 | 2.0 | 2.1 | 2.0 | ||
| Range | 0.3–14 | 0.3–14 | 0.5–7.2 | 0.3–14 | 0.6–6.0 | ||
| Invasive tumor size, cm | 0.65 | 0.063 | |||||
| Median | 1.8 | 1.8 | 1.8 | 1.8 | 1.6 | ||
| Range | 0.1–9.8 | 0.1–9.8 | 0.1–7.0 | 0.1–9.8 | 0.1–5.4 | ||
| Pleural invasion | 0.43 | 0.88 | |||||
| Absent | 390 | 289 (74) | 101 (26) | 321 (82) | 69 (18) | ||
| Present | 92 | 64 (70) | 28 (30) | 75 (82) | 17 (18) | ||
| Lymphatic invasion | 0.99 | 0.79 | |||||
| Absent | 351 | 257 (73) | 94 (27) | 287 (82) | 64 (18) | ||
| Present | 131 | 96 (73) | 35 (27) | 109 (83) | 22 (17) | ||
| Vascular invasion | 0.82 | 0.15 | |||||
| Absent | 343 | 250 (73) | 93 (27) | 276 (80) | 67 (20) | ||
| Present | 139 | 103 (74) | 36 (26) | 120 (86) | 19 (14) | ||
| Necrosis | 0.90 | 0.014 | |||||
| Absent | 393 | 287 (73) | 106 (27) | 315 (80) | 78 (20) | ||
| Present | 89 | 66 (74) | 23 (26) | 81 (91) | 8 (9) | ||
| Nuclear atypia | 0.71 | 0.13 | |||||
| Mild | 241 | 173 (72) | 68 (28) | 192 (80) | 49 (20) | ||
| Moderate | 129 | 95 (74) | 34 (26) | 105 (81) | 24 (19) | ||
| Severe | 112 | 85 (76) | 27 (24) | 99 (88) | 13 (12) | ||
| Mitosis | 0.24 | <0.001 | |||||
| Median | 2 | 2 | 3 | 3 | 1 | ||
| Range | 0–43 | 0–43 | 0–33 | 0–43 | 0–35 | ||
| TTF-1 expression | 0.57 | 0.004 | |||||
| Negative | 39 | 27 (69) | 12 (31) | 38 (97) | 1 (3) | ||
| Positive | 425 | 315 (74) | 110 (26) | 340 (80) | 85 (20) | ||
Significant P-values are shown in bold.
EGFR, epidermal growth factor receptor
EGFR mutation was positively associated with female sex (P = 0.019) while KRAS mutation was not associated with patient gender (P = 0.67). KRAS mutation was more frequently identified in ever smokers than in never smokers (P < 0.001). KRAS transversion mutations were also more frequently identified in ever smokers (P < 0.001) while KRAS transition mutations were not associated with smoking (P = 0.34). EGFR mutation was more frequently identified in never smokers (P < 0.001) and TTF-1 positive tumors (P = 0.004), and negatively associated with presence of tumor necrosis (P = 0.014) and mitotic count (P < 0.001). Both EGFR exon 21 L858R mutations and exon 19 deletions were associated with a history of never smoking (P < 0.001 and P < 0.001, respectively).
As for correlations between smoking history and predominant histologic subtypes, solid predominant tumors were more frequently observed in ever smokers than in never smokers (14% vs. 3%; P = 0.002). Acinar predominant tumors were more frequently observed in never smokers than in ever-smokers (57% vs. 42%; P = 0.012). Lepidic predominant tumors were more frequently identified in never smokers and former smokers than in current smokers (8% vs. 8% vs. 1%); the difference was not statistically significant (P = 0.11).
Driver mutation profiles according to predominant histologic subtypes
Details of molecular results are summarized in Table 2. There were 129 (27%) patients that had mutations in KRAS, 86 (18%) in EGFR, 8 (2%) in BRAF, 8 (2%) in PIK3CA, 4 (1%) in NRAS, and 1 (0.2%) in AKT1. No tumors had mutations in ERBB2/HER2 and MAP2K1/MEK1. Among KRAS-mutant tumors, 110 were transversion mutations and 19 were transition mutations. Among EGFR-mutant tumors, 42 were exon 21 L858R mutations and 39 were exon 19 deletions. Among PIK3CA-mutant tumors, 2 cases coexisted with KRAS mutations and 2 with EGFR mutations.
Table 2.
Summary of driver mutation types
| Driver mutation | Total (%) | Type | N (%) |
|---|---|---|---|
| KRAS | 129 (27) | G12C | 57 (44) |
| G12V | 22 (17) | ||
| G12D | 15 (12) | ||
| G12A | 14 (11) | ||
| G12F | 2 (2) | ||
| G12R | 1 (1) | ||
| G12S | 1 (1) | ||
| G13C | 8 (6) | ||
| G13D | 1 (1) | ||
| Q61H | 4 (3) | ||
| Q61L | 2 (2) | ||
| Q61R | 1 (1) | ||
| A146T | 1 (1) | ||
| EGFR | 86 (18) | L858R | 42 (49) |
| Exon 19 del. | 39 (45) | ||
| L861Q | 2 (2) | ||
| S768I | 2 (2) | ||
| G719A | 1 (1) | ||
| BRAF | 8 (2) | V600E | 6 (75) |
| D594G | 2 (25) | ||
| PIK3CA* | 8 (2) | E542K | 4 (50) |
| C420K | 1 (13) | ||
| E545K | 1 (13) | ||
| H1047R | 1 (13) | ||
| R88Q | 1 (13) | ||
| NRAS | 4 (1) | Q61L | 2 (50) |
| Q61K | 1 (25) | ||
| G31R | 1 (25) | ||
| AKT1 | 1 (0.2) | E17K | 1 (100) |
Among PIK3CA mutated tumors, 2 coexisted with KRAS mutation and 2 with EGFR mutation.
EGFR, epidermal growth factor receptor
Distribution of driver mutations according to histologic subtypes is summarized in Table 3. KRAS mutation was identified in tumors with all histologic subtypes. EGFR mutation was identified in tumors with all histologic subtypes except invasive mucinous adenocarcinomas and colloid predominant tumors. Invasive mucinous adenocarcinomas and colloid predominant tumors harbored only KRAS mutations.
Table 3.
Driver mutation distribution according to the predominant histologic subtype
| Histologic subtype | Total N (%) | Driver mutation, N (%)
|
||||||
|---|---|---|---|---|---|---|---|---|
| KRAS | EGFR | BRAF | PIK3CA* | NRAS | AKT1 | All (−) | ||
| MIA, nonmucinous | 7 (1) | 1 (1) | 2 (2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (2) |
| MIA, mucinous | 1 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0) |
| Lepidic predominant | 26 (5) | 6 (5) | 8 (9) | 1 (13) | 1 (13) | 0 (0) | 0 (0) | 10 (4) |
| Acinar predominant | 212 (44) | 50 (39) | 54 (63) | 2 (25) | 5 (63) | 3 (75) | 1 (100) | 101 (40) |
| Papillary predominant | 135 (28) | 40 (31) | 18 (21) | 4 (50) | 1 (13) | 0 (0) | 0 (0) | 72 (29) |
| Micropapillary predominant | 14 (3) | 6 (5) | 1 (1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7 (3) |
| Solid predominant | 60 (12) | 16 (12) | 3 (3) | 1 (13) | 1 (13) | 1 (25) | 0 (0) | 38 (15) |
| Invasive mucinous | 20 (4) | 7 (5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 13 (5) |
| Colloid predominant | 7 (1) | 3 (2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (2) |
| Total | 482 (100) | 129 (100) | 86 (100) | 8 (100) | 8 (100) | 4 (100) | 1 (100) | 250 (100) |
Among PIK3CA mutated tumors, 2 coexisted with KRAS mutation and 2 with EGFR mutation.
EGFR, epidermal growth factor receptor
KRAS and EGFR mutation associations with predominant histologic subtypes
KRAS and EGFR mutation associations with histologic predominant subtypes are summarized in Table 4. KRAS mutation was not significantly associated with any histologic subtype, including invasive mucinous adenocarcinoma. EGFR mutation was more likely to be identified in lepidic predominant tumors (29%) than non-lepidic predominant tumors (17%); this difference was only a trend and not statistically significant (P = 0.10). However, EGFR L858R mutation was more frequently identified in lepidic predominant tumors (24%) than non-lepidic predominant tumors (8%; P = 0.006). EGFR mutation was more frequently identified in acinar predominant tumors (25%) than non-acinar predominant tumors (12%; P < 0.001). EGFR exon 19 deletion was more frequently identified in acinar predominant tumors (13%) than non-acinar predominant tumors (4%: P < 0.001) while EGFR L858R mutation was not associated with acinar predominant pattern (P = 0.26). EGFR mutation was less frequently identified in solid predominant tumors (5%) than non-solid predominant tumors (20%; P = 0.004), and was not detected in invasive mucinous adenocarcinomas (P = 0.033).
Table 4.
KRAS and EGFR mutation associations with the histologic predominant subtypes
| Histologic subtypes |
KRAS
|
P |
EGFR
|
P |
EGFR L858R
|
P |
EGFR exon 19
|
P | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild-type | Mutant | Wild-type | Mutant | Wild-type | Mutant | Wild-type | Mutant | |||||
| N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | |||||
| Lepidic | 0.55 | 0.1 | 0.006 | 1.00 | ||||||||
| Predominant* | 27 (79) | 7 (21) | 24 (71) | 10 (29) | 26 (76) | 8 (24) | 32 (94) | 2 (6) | ||||
| Non-predominant | 326 (73) | 122 (27) | 372 (83) | 76 (17) | 414 (92) | 34 (8) | 411 (92) | 37 (8) | ||||
| Acinar | 0.18 | <0.001 | 0.26 | <0.001 | ||||||||
| Predominant | 162 (76) | 50 (24) | 158 (75) | 54 (25) | 190 (90) | 22 (10) | 184 (87) | 28 (13) | ||||
| Non-predominant | 191 (71) | 79 (29) | 238 (88) | 32 (12) | 250 (93) | 20 (7) | 259 (96) | 11 (4) | ||||
| Papillary | 0.42 | 0.11 | 0.59 | 0.35 | ||||||||
| Predominant | 95 (70) | 40 (30) | 117 (87) | 18 (13) | 125 (93) | 10 (7) | 127 (94) | 8 (6) | ||||
| Non-predominant | 258 (74) | 89 (26) | 279 (80) | 68 (20) | 315 (91) | 32 (9) | 316 (91) | 31 (9) | ||||
| Micropapillary | 0.22 | 0.48 | 1.00 | 0.62 | ||||||||
| Predominant | 8 (57) | 6 (43) | 13 (93) | 1 (7) | 13 (93) | 1 (7) | 14 (100) | 0 (0) | ||||
| Non-predominant | 345 (74) | 123 (26) | 383 (82) | 85 (18) | 427 (91) | 41 (9) | 429 (92) | 39 (8) | ||||
| Solid pattern | 1.00 | 0.004 | 0.046 | 0.071 | ||||||||
| Predominant | 44 (73) | 16 (27) | 57 (95) | 3 (5) | 59 (98) | 1 (2) | 59 (98) | 1 (2) | ||||
| Non-predominant | 309 (73) | 113 (27) | 339 (80) | 83 (20) | 381 (90) | 41 (10) | 384 (91) | 38 (9) | ||||
| Invasive mucinous | 0.44 | 0.033 | 0.24 | 0.39 | ||||||||
| Invasive mucinous | 13 (65) | 7 (35) | 20 (100) | 0 (0) | 20 (100) | 0 (0) | 20 (100) | 0 (0) | ||||
| Non-mucinous | 340 (74) | 122 (26) | 376 (81) | 86 (19) | 420 (91) | 42 (9) | 423 (92) | 39 (8) | ||||
Significant P-values are shown in bold.
Including minimally invasive and lepidic predominant adenocarcinoma.
Tumors were classified into 3 groups according to percentage of each histologic pattern (0–19%, 20–49% and ≥50%), as previously reported by our group,13 and their associations with KRAS and EGFR mutations were analyzed. KRAS mutation was less frequently identified in tumors with 20–49% and ≥50% lepidic pattern than in those with 0–19% lepidic pattern (frequency of KRAS mutation, 20%, 23%, and 30%, respectively), even though this difference was not statistically significant (P = 0.11). However, incremental increases in the amount of other histologic patterns were not associated with KRAS mutation. Incremental increases in the amount of lepidic and acinar patterns (0–19%, 20–49% and ≥50%) were positively associated with frequency of EGFR mutation (frequency of EGFR mutation by lepidic pattern, 12%, 30%, and 33%, respectively, P < 0.001; frequency of EGFR mutation by acinar pattern, 10%, 14%, and 25%, respectively, P = 0.002). By contrast, incremental increase in the amount of solid pattern (0–19%, 20–49% and ≥50%) was inversely associated with frequency of EGFR mutation (frequency of EGFR mutation, 22%, 9%, and 6%, respectively, P = 0.001). Thirty cribriform predominant tumors were identified, and among them, 9 tumors had KRAS mutation and 2 had EGFR mutation. Presence of signet ring cell features and cribriform predominant pattern were not associated with either KRAS or EGFR mutations (data not shown).
OS analysis by driver mutations in patients with stage I disease
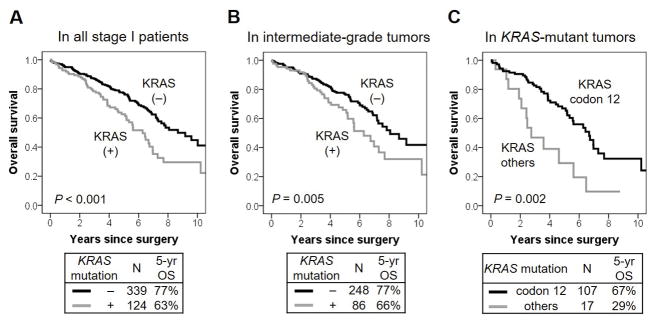
Clinicopathologic associations with OS in patients with stage I disease are summarized in Table 5. In limited resection group, 54 patients (72%) underwent lymph node dissection or sampling while, in lobectomy group, all patients underwent lymph node dissection or sampling. Patients with KRAS-mutant tumors was significantly worse 5-year OS (n = 124; 5-year OS, 63%) than those with KRAS wild-type tumors (n = 339; 77%; P < 0.001) (Fig. 1A). We then analyzed the prognostic value of KRAS mutation in subgroups according to each histologic subtype. In architecturally intermediate-grade tumors (acinar predominant and papillary predominant subtypes), 5-year OS of patients with KRAS-mutant tumors was significantly worse (n = 86; 5-year OS, 66%) than those with KRAS wild-type tumors (n = 248; 77%; P = 0.005) (Fig. 1B). In solid predominant tumors, 5-year OS of patients with KRAS-mutant tumors was significantly worse (n = 16; 5-year OS, 43%) than those with KRAS wild-type tumors (n = 42; 77%; P = 0.008) (Fig. 2A). According to the codon of KRAS mutations, 5-year OS of patients with KRAS codon 12 mutated tumors were significantly better (n = 107; 5-year OS, 67%) than those with other KRAS-mutated tumor types (n = 17; 29%; P = 0.002) (Fig. 1C). Patients with KRAS codon 13 mutated and codon 61 mutated tumors had 5-year OS of 19% and 43%, respectively; these results were based on a small number of patients (n = 9 and n = 7, respectively). Type of KRAS mutation (transversion vs. transition) was not associated with OS (P = 0.69). On multivariate analysis of OS, KRAS mutation remained a significant prognostic factor after adjustment with other prognostic factors (hazard ratio [HR] = 1.87; 95% confidence interval [CI], 1.36–2.58; P < 0.001) (Table 6A).
Table 5.
Clinicopathologic associations with overall survival and disease recurrence in stage I patients
| Characteristic | N | 5-year OS | P | 5-year CIR | P |
|---|---|---|---|---|---|
| Age, years | <0.001 | 0.48 | |||
| ≤65 | 166 | 80% | 19% | ||
| >65 | 297 | 69% | 16% | ||
| Sex | <0.001 | 0.005 | |||
| Female | 294 | 80% | 13% | ||
| Male | 169 | 61% | 24% | ||
| Smoking status | 0.12 | 0.15 | |||
| Never | 72 | 83% | 11% | ||
| Former/Current | 391 | 71% | 18% | ||
| Surgery | <0.001 | 0.013 | |||
| Lobectomy | 388 | 77% | 15% | ||
| Limited resection | 75 | 56% | 27% | ||
| Pathologic stage | 0.010 | <0.001 | |||
| IA | 316 | 76% | 13% | ||
| IB | 147 | 67% | 26% | ||
| Architectural grade | 0.36 | 0.002 | |||
| Low | 34 | 78% | 7% | ||
| Intermediate | 334 | 74% | 16% | ||
| High | 95 | 68% | 25% | ||
| Pleural invasion | 0.045 | 0.065 | |||
| Absence | 376 | 75% | 16% | ||
| Presence | 87 | 67% | 23% | ||
| Lymphatic invasion | 0.018 | 0.008 | |||
| Absence | 341 | 76% | 14% | ||
| Presence | 122 | 65% | 24% | ||
| Vascular invasion | 0.002 | 0.007 | |||
| Absence | 334 | 76% | 14% | ||
| Presence | 129 | 67% | 25% | ||
| Necrosis | <0.001 | <0.001 | |||
| Absence | 384 | 77% | 12% | ||
| Presence | 79 | 54% | 41% |
Significant P-values are shown in bold.
OS, overall survival; CIR, cumulative incidence of recurrence
Figure 1. KRAS mutation associations with overall survival (OS).
(A) 5-year OS of patients with KRAS-mutant tumors was significantly worse (n = 124; 5-year OS, 63%) than those with KRAS wild-type tumors (n = 339; 77%; P < 0.001). (B) In architecturally intermediate-grade tumors (acinar predominant and papillary predominant subtypes), 5-year OS of patients with KRAS-mutant tumors was significantly worse (n = 86; 5-year OS, 66%) than those with KRAS wild-type tumors (n = 248; 77%; P = 0.005). (C) 5-year OS of patient with KRAS codon 12 mutated tumors were significantly better (n = 107; 5-year OS, 67%) than those with other KRAS-mutant tumors (n = 17; 29%; P = 0.002)
Figure 2. KRAS mutation associations with overall survival (OS) and cumulative incidence of recurrence (CIR) in solid predominant tumors.
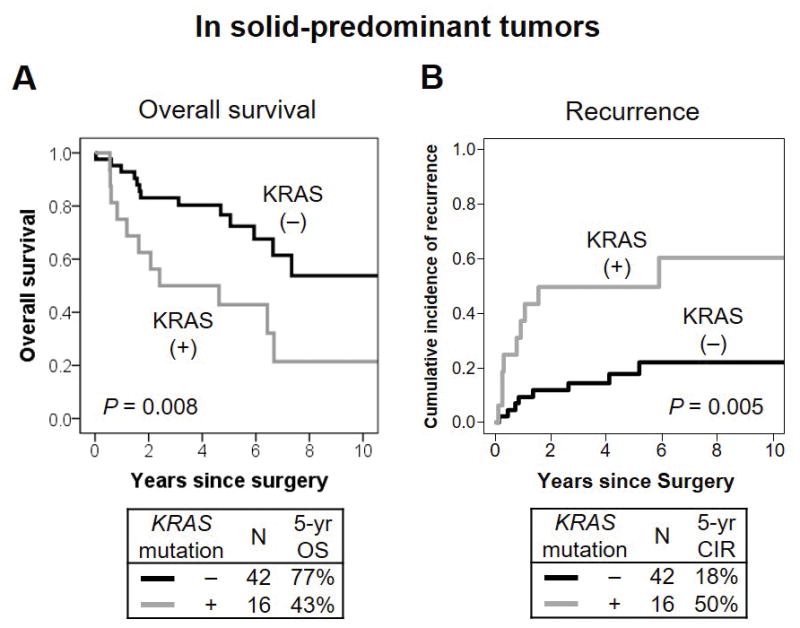
(A) In solid predominant tumors, 5-year OS of patients with KRAS-mutant tumors was significantly worse (n = 16; 5-year OS, 43%) than those with KRAS wild-type (n = 42; 77%; P = 0.008). (B) In solid predominant tumors, 5-year CIR of patients with KRAS-mutant tumors was significantly higher (5-year OS, 50%) than those with KRAS wild-type tumors (18%; P = 0.005).
Table 6.
Multivariate analyses
| (A) For overall survival | ||||
|---|---|---|---|---|
|
| ||||
| Variables | HR | 95% CI | P | |
| Age, years | >65 vs. ≤65 | 1.78 | 1.24–2.55 | 0.002 |
| Sex | male vs. female | 1.73 | 1.26–2.36 | 0.001 |
| Surgery | limited resection vs. lobectomy | 2.31 | 1.62–3.29 | < 0.001 |
| Pathologic stage | IB vs. IA | 1.40 | 1.02–1.93 | 0.039 |
| Vascular invasion | positive vs. negative | 1.45 | 1.04–2.01 | 0.027 |
| KRAS mutation | mutant vs. wild-type | 1.87 | 1.36–2.58 | < 0.001 |
| (B) For disease recurrence | ||||
|---|---|---|---|---|
|
| ||||
| Variables | HR | 95% CI | P | |
| Sex | male vs. female | 1.63 | 0.98–2.71 | 0.058 |
| Smoking | ever vs. never | 1.31 | 0.61–2.81 | 0.49 |
| Necrosis | present vs. absent | 1.27 | 1.14–1.41 | < 0.001 |
| Pathologic stage | IB vs. IA | 1.81 | 1.1–2.97 | 0.019 |
| KRAS mutation | mutant vs. wild-type in solid predominant tumors | 4.73 | 1.41–15.9 | 0.012 |
| mutant vs. wild-type in non-solid predominant tumors | 0.91 | 0.49–1.66 | 0.75 | |
Significant P-values are shown in bold.
HR, hazard ratio; CI, confidence interval
Patients with EGFR-mutant tumors trended with better OS (n = 85; 5-year OS, 86%) than those with EGFR wild-type tumors (n = 378; 70%; P = 0.055) (Fig. 3A). Type of EGFR mutations (exon 21 L858R mutation vs. exon 19 deletion) was not associated with OS (P = 0.64) (Fig. 3B).
Figure 3. EGFR mutation associations with overall survival (OS).
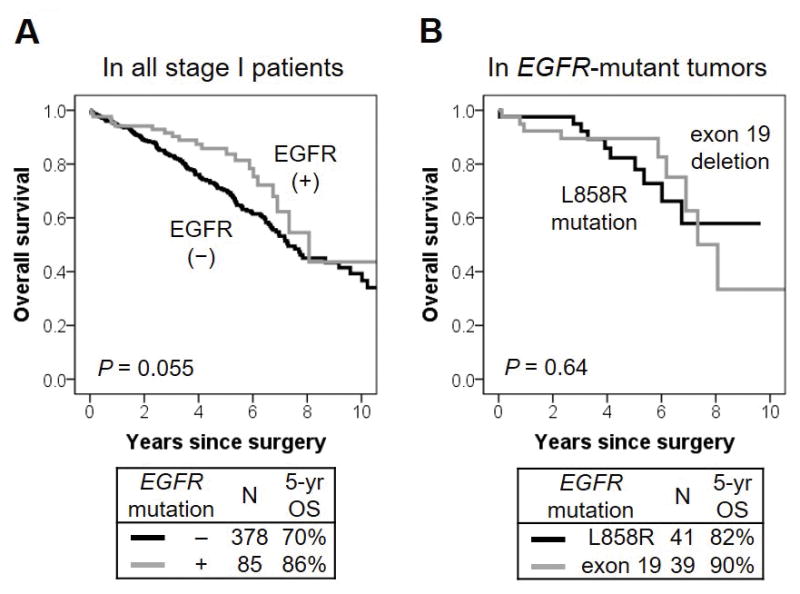
(A) Patients with EGFR-mutant tumors trended with better OS (n = 85; 5-year OS, 86%) than those with EGFR wild-type tumors (n = 378; 70%; P = 0.055). (B) Type of EGFR mutation (exon 21 L858R mutation vs. exon 19 deletion) was not associated with OS (P = 0.64).
Patients with BRAF-mutant tumors were likely to have worse prognosis (5-year OS, 50%) than those with BRAF wild-type tumors (73%); this difference was based on a small number of BRAF-mutant tumors (n = 6) and was not statistically significant (P = 0.19) (Fig. 4). PIK3CA mutation was not associated with OS (P = 0.66). NRAS and AKT1 mutations were not applicable for OS survival because of a small number of patients in these groups.
Figure 4. BRAF mutation associations with overall survival (OS).
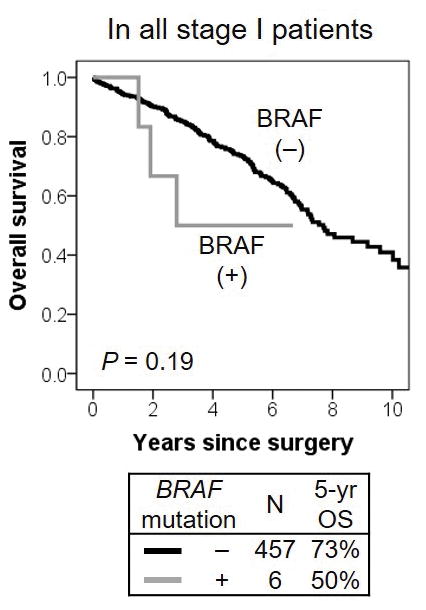
Patients with BRAF-mutant tumors were likely to have worse prognosis (5-year OS, 50%) than those with BRAF wild-type tumors (73%); although, this difference was based on a small number of BRAF-mutant tumors (n = 6) and was not statistically significant (P = 0.19).
CIR analysis by driver mutations in patients with stage I disease
Clinicopathologic associations with CIR in patients with stage I disease are summarized in Table 5. KRAS mutation (mutant vs. wild-type) was not associated with a risk of recurrence (P = 0.29). In solid predominant tumors, 5-year CIR of patients with KRAS-mutant tumors was significantly higher (5-year CIR, 50%) than those with KRAS wild-type tumors (18%; P = 0.005) (Fig. 2B). However, KRAS mutation in architecturally intermediate-grade tumors was not associated with risk of recurrence (P = 0.85). Patients with KRAS codon 12 mutated tumors were likely to have lower risk of recurrence (5-year CIR, 17%) than those with other KRAS-mutant tumors (31%); this difference was not statistically significant (P = 0.24). Type of KRAS mutation (transversion vs. transition) was not associated with risk of recurrence (P = 0.69). On multivariate analysis of CIR, KRAS mutation in patients with solid predominant tumors remained a significant risk factor for recurrence after adjustment with other prognostic factors (HR = 4.73; 95% CI, 1.41–15.9; P = 0.012) (Table 6B).
Presence of EGFR mutation (mutant vs. wild-type) and their types of mutation (exon 21 L858R mutation vs. exon 19 deletion) were not associated with risk of recurrence (P = 0.74 and P = 0.73, respectively). BRAF and PIK3CA mutations were not associated with risk of recurrence (P = 0.98 and P = 0.21).
Association of ALK expression with predominant histologic subtypes and prognoses (OS and CIR)
ALK expression was positive in 15 (3%) cases. Among them, 2 (13%) cases were classified as lepidic predominant, 6 (40%) as acinar predominant, 5 (33%) as papillary predominant, 1 (7%) as micropapillary predominant, and 1 (7%) as solid predominant. ALK expression was more frequently identified in tumors with signet ring cell features than those without signet ring cell features (19% vs. 2%; P = 0.003). However, ALK expression was not associated with patient gender, age, smoking history, predominant histologic subtype (including cribriform pattern), disease recurrence, and OS (data not shown).
DISCUSSION
We have demonstrated that, in patients with resected early-stage lung adenocarcinomas, KRAS mutation was an independent prognostic factor for OS in all tumors, for disease recurrence in solid predominant tumors. Moreover, EGFR mutations, especially exon 19 deletions, correlated with acinar predominant pattern while EGFR L858R mutation correlated with lepidic predominant pattern.
In our study, KRAS mutation was identified in 27% of cases, which is consistent with the rate in previous studies of lung adenocarcinomas from patients in Western countries.14, 24, 43 Initially, KRAS mutation was thought to be an unfavorable prognostic factor in NSCLC patients, but data regarding its prognostic impact has been contradictory. Several studies demonstrated that KRAS mutation was associated with shorter OS and disease-free survival in patients with NSCLC.17–21 Most studies focused on cohorts with only adenocarcinoma histology, two of which were composed of only stage I patients.17, 18 Two meta-analyses found KRAS mutation to be a poor prognostic marker.15, 16 Huncharek et al. found that in a meta-analysis of 8 studies KRAS mutation was an unfavorable prognosticator of survival in NSCLC with a combined HR of 2.35 (95% CI 1.61–3.22); however, this analysis was not adjusted according to TNM stage.15 Another meta-analysis of 28 studies also demonstrated KRAS mutation was an overall poor prognostic indicator in NSCLC (HR = 1.40, 95% CI, 1.18–1.65) and lung adenocarcinomas (HR = 1.50, 95% CI, 1.26–1.80), but not in squamous cell carcinoma; although, the finding in NSCLC was no longer significant after adjusting for disease stage.16 By contrast, other studies reported no prognostic value of KRAS mutations in patients who had TNM stage I–III disease with adenocarcinoma 23, 25, as well as other types of NSCLC. 22, 24 However, the strength of the conclusions in most of these studies may be limited for several reasons: a) prognostic implication of KRAS mutation was retrospectively investigated using study cohorts that were heterogeneous in tumor histology (including adenocarcinoma and squamous cell carcinoma); b) disease stage (including early-stage and advanced-stage disease); c) treatment (including patients treated with surgery alone, chemotherapy alone, and multimodality therapy); and d) they was statistically analyzed using various end points (death or/and recurrence). In our study and on the basis of a homogeneous cohort of patients with surgically resected early stage lung adenocarcinomas, we identified KRAS mutation to be an independent prognostic factor for OS with a HR of 1.87, after adjustment with important confounders, including patient age, gender, surgical procedure (lobectomy vs. limited resection), pathologic stage (IA vs. IB), and tumoral vascular invasion.
Another possible explanation for the disparate data regarding the prognostic impact of KRAS mutation is that a variety of molecular techniques were used in the previous studies, including direct sequencing, polymerase chain reaction-based assay, and mass spectrometry-based assay (Sequenom). Additionally, some studies focused on the most common types of KRAS mutations in codon 12 but others used methods which could detect mutations in codons 13 and 61.15–25 Mass spectrometry-based assays have been shown to be suitable for a screening method that is more sensitive and broader than direct sequencing.35 In our study, we used a mass spectrometry-based assay that was able to detect KRAS mutations in codons 12, 13, 61, and 146, and we demonstrated that KRAS codon 12 mutated tumors were associated with better prognoses (5-year OS, 67%) than other KRAS-mutant tumors (5-year OS, 19% for codon 13 and 43% for codon 61). By contrast, Villaruz et al. reported that there was a trend toward better OS in patients with KRAS codon 13 mutations than those with KRAS codon 12 mutations in lung adenocarcinomas however, this difference was not statistically significant on multivariate analysis and the study cohort included patients with early-stage disease as well as advanced-stage disease.43 Therefore, the prognostic impact of KRAS-mutant codon types remains unclear because of varying results on the basis of small cohorts that were heterogeneous with regards to patient characteristics. However, presence of KRAS mutation appears to be prognostically significant in patients with surgically resected stage I lung adenocarcinoma.
Although EGFR mutation may be associated with better prognosis specifically in advanced-stage lung adenocarcinoma patients who were treated with TKIs; its prognostic significance remains unclear in early-stage patients.20–24 In our study—which was based on a stage I cohort—patients with EGFR mutation had a tendency to have better OS than those with EGFR wild-type, although, this finding was not statistically significant. Interestingly, a previous study demonstrated that, after treatment with TKI for advanced-stage NSCLC with distant metastases, patients with EGFR exon 19 deletions had significantly longer OS than those with EGFR L858R mutations.44 However, in our present study, the specific type of EGFR mutation (exon 19 deletion vs. L858R mutation) was not associated with prognostic differences in stage I lung adenocarcinomas.
In lung adenocarcinomas, correlations between activating mutations and histologic patterns have been reported. EGFR mutation is more frequently identified in tumors with lepidic pattern (formerly nonmucinous bronchioloalveolar carcinoma [BAC] pattern),5, 45–47 while KRAS mutation is more frequently identified in invasive mucinous adenocarcinoma (formerly called mucinous BAC).48–52 These findings were confirmed when classifying tumors using the 2011 IASLC/ATS/ERS lung adenocarcinoma classification by our group and others. EGFR mutation is frequently identified in non-mucinous lepidic predominant tumors while it is less frequently identified in solid predominant tumors and not detected in invasive mucinous adenocarcinomas.11–14 By contrast, KRAS mutation correlates with invasive mucinous adenocarcinoma and is frequently identified in solid predominant tumors, but it is less frequently identified in non-mucinous lepidic predominant tumors.11–14 In our study, we demonstrated that EGFR mutation was more frequently identified in non-mucinous lepidic predominant tumors (29%) than in non-lepidic tumors (17%), while it was less frequently identified in solid predominant tumors (5%) than in non-solid tumors (20%). Although non-mucinous lepidic predominant tumors have been shown to positively correlate with EGFR mutation, associations between type of EGFR mutation and histologic subtype have not been investigated. Our study demonstrates that EGFR L858R mutations correlate with lepidic predominant subtype while EGFR exon 19 deletions correlate with acinar predominant subtype. Our group has previously reported that solid growth pattern is associated with presence of KRAS mutation using cohorts composed of both early-stage and advanced-stage patients.13, 14 Nevertheless, we did not identify the positive association between solid predominant tumors and KRAS mutations in our early-stage cohort of lung adenocarcinomas but found that, interestingly, KRAS mutation was a strong prognostic factor for OS and recurrence in subgroup analysis of patients with solid predominant tumors. However, this result was based on a small number of tumors with KRAS mutations (n = 16) in the solid predominant group. This should be considered a potential limitation of this finding and warrants further investigation with larger cohorts.
Our current and previous studies also confirmed complete lack of EGFR mutations in invasive mucinous adenocarcinoma using two independent large cohorts and performing two different types of mutations analyses (PCR-based assay and mass spectrometry-based assay).14 Additionally, we identified only KRAS mutation in invasive mucinous adenocarcinomas. Interestingly, a recently discovered somatic gene fusion, CD74-NRG1, has been specifically identified in invasive mucinous adenocarcinoma of the lung.53 ALK rearrangement and other rare oncogenic fusion gene were also detected in lung invasive mucinous adenocarcinomas.11, 54 Taking these findings into consideration, only fusion genes and KRAS mutation may be able to act as oncogenic molecular alteration in invasive mucinous adenocarcinomas.
BRAF mutation is very rare (<5% in lung adenocarcinomas) and its clinical impact remains unclear although it may be associated with resistance to EGFR TKIs, micropapillary morphology, and poor prognosis.55–58 Specifically in early-stage lung adenocarcinomas, a prognostic value of BRAF mutation was not investigated. In our study, 5-year OS of patients with BRAF-mutant tumors was lower (50%) than those with BRAF wild-type tumors (73%) in stage I lung adenocarcinomas While the difference was not statistically significant it was based on a small number of patients with BRAF mutations. As for its histologic correlations, we identified BRAF mutation more frequently in papillary predominant tumors but did not detect it in micropapillary predominant tumors. However, all studies, including our own, investigated clinical impacts of BRAF mutation in a small number of patients due to its rarity. Further investigations will be warranted using a larger cohort with BRAF-mutant tumors.
Studies have reported that ALK rearrangement is associated with mucinous features, such as signet-ring cell feature and extracellular mucin, and cribriform pattern in lung adenocarcinoma59–61 Our group has previously reported that evaluating ALK expression via tissue microarray analysis in the independent cohort from our current study, signet ring cell features were associated with positive ALK expression but was not associated with KRAS or EGFR mutation; this finding was validated in our current study.
In conclusion, our study reported that, in early-stage lung adenocarcinomas, KRAS mutation was a strong prognostic factor, especially in patients with solid predominant tumors. Although solid predominant tumors can be classified as high-grade histology with poor prognoses, KRAS mutation may help select for patients with worse prognoses from this group. Additionally, with the exception of invasive mucinous adenocarcinoma and colloid adenocarcinoma which consistently show KRAS rather than EGFR mutations—there do not appear to be specific correlations between histologic subtype and presence of specific driver mutation.
Acknowledgments
We thank Joe Dycoco of the MSK Thoracic Surgery Service, for assisting with the MSK Thoracic Surgery Service’s Lung Cancer Database; and Alex Torres of the MSK Thoracic Surgery Service for his editorial assistance.
FUNDING SUPPORT
This work is supported by grants from the National Institutes of Health (R21 CA164568-01A1, R21 CA164585-01A1, R01 CA136705-06, U54 CA137788, P50 CA086438-13, and P30 CA008748), the U.S. Department of Defense (PR101053 and LC110202), and the Mr. William H. Goodwin and Mrs. Alice Goodwin, the Commonwealth Foundation for Cancer Research, and the Experimental Therapeutics Center of Memorial Sloan Kettering Cancer Center.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
The authors made no disclosures.
References
- 1.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 5.Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 6.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warth A, Muley T, Meister M, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol. 2012;30:1438–1446. doi: 10.1200/JCO.2011.37.2185. [DOI] [PubMed] [Google Scholar]
- 10.Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol. 2011;24:653–664. doi: 10.1038/modpathol.2010.232. [DOI] [PubMed] [Google Scholar]
- 11.Tsuta K, Kawago M, Inoue E, et al. The utility of the proposed IASLC/ATS/ERS lung adenocarcinoma subtypes for disease prognosis and correlation of driver gene alterations. Lung Cancer. 2013;81:371–376. doi: 10.1016/j.lungcan.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Yoshizawa A, Sumiyoshi S, Sonobe M, et al. Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: analysis of 440 Japanese patients. J Thorac Oncol. 2013;8:52–61. doi: 10.1097/JTO.0b013e3182769aa8. [DOI] [PubMed] [Google Scholar]
- 13.Rekhtman N, Ang DC, Riely GJ, Ladanyi M, Moreira AL. KRAS mutations are associated with solid growth pattern and tumor-infiltrating leukocytes in lung adenocarcinoma. Mod Pathol. 2013;26:1307–1319. doi: 10.1038/modpathol.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadota K, Yeh Y-C, D’Angelo SP, et al. Associations between mutations and histologic patterns of mucin in lung adenocarcinoma: Invasive mucinous pattern and extracellular mucin are associated with KRAS mutation. Am J Surg Pathol. 2014;38:1118–1127. doi: 10.1097/PAS.0000000000000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huncharek M, Muscat J, Geschwind JF. K-ras oncogene mutation as a prognostic marker in non-small cell lung cancer: a combined analysis of 881 cases. Carcinogenesis. 1999;20:1507–1510. doi: 10.1093/carcin/20.8.1507. [DOI] [PubMed] [Google Scholar]
- 16.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silini EM, Bosi F, Pellegata NS, et al. K-ras gene mutations: an unfavorable prognostic marker in stage I lung adenocarcinoma. Virchows Arch. 1994;424:367–373. doi: 10.1007/BF00190558. [DOI] [PubMed] [Google Scholar]
- 18.Woo T, Okudela K, Yazawa T, et al. Prognostic value of KRAS mutations and Ki-67 expression in stage I lung adenocarcinomas. Lung Cancer. 2009;65:355–362. doi: 10.1016/j.lungcan.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 19.Grossi F, Loprevite M, Chiaramondia M, et al. Prognostic significance of K-ras, p53, bcl-2, PCNA, CD34 in radically resected non-small cell lung cancers. Eur J Cancer. 2003;39:1242–1250. doi: 10.1016/s0959-8049(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 20.Kim YT, Kim TY, Lee DS, et al. Molecular changes of epidermal growth factor receptor (EGFR) and KRAS and their impact on the clinical outcomes in surgically resected adenocarcinoma of the lung. Lung Cancer. 2008;59:111–118. doi: 10.1016/j.lungcan.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 21.Johnson ML, Sima CS, Chaft J, et al. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2013;119:356–362. doi: 10.1002/cncr.27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosaka T, Yatabe Y, Onozato R, Kuwano H, Mitsudomi T. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol. 2009;4:22–29. doi: 10.1097/JTO.0b013e3181914111. [DOI] [PubMed] [Google Scholar]
- 23.Liu HP, Isaac Wu HD, Chang JW, et al. Prognostic implications of epidermal growth factor receptor and KRAS gene mutations and epidermal growth factor receptor gene copy numbers in patients with surgically resectable non-small cell lung cancer in Taiwan. J Thorac Oncol. 2010;5:1175–1184. doi: 10.1097/JTO.0b013e3181e2f4d6. [DOI] [PubMed] [Google Scholar]
- 24.D’Angelo SP, Janjigian YY, Ahye N, et al. Distinct clinical course of EGFR-mutant resected lung cancers: results of testing of 1118 surgical specimens and effects of adjuvant gefitinib and erlotinib. J Thorac Oncol. 2012;7:1815–1822. doi: 10.1097/JTO.0b013e31826bb7b2. [DOI] [PubMed] [Google Scholar]
- 25.Shepherd FA, Domerg C, Hainaut P, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31:2173–2181. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Travis W, Brambilla E, Muller-Hermelink H, Harris C. Tumours of the Lung, Pleura, Thymus, and Heart. IARC; Lyon, France: 2015. World Health Organization Classification of Tumours. Pathology and Genetics. [Google Scholar]
- 27.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. American Joint Committee on Cancer Cancer Staging Manual. 7. New York, NY: Springer; 2009. pp. 253–270. [Google Scholar]
- 28.Suzuki K, Kadota K, Sima CS, et al. Clinical impact of immune microenvironment in stage I lung adenocarcinoma: Tumor interleukin-12 receptor beta2 (IL-12Rbeta2), IL-7R, and stromal FoxP3/CD3 ratio are independent predictors of recurrence. J Clin Oncol. 2013;31:490–498. doi: 10.1200/JCO.2012.45.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadota K, Colovos C, Suzuki K, et al. FDG-PET SUVmax combined with IASLC/ATS/ERS histologic classification improves the prognostic stratification of patients with stage I lung adenocarcinoma. Ann Surg Oncol. 2012;19:3598–3605. doi: 10.1245/s10434-012-2414-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadota K, Nitadori J, Sarkaria IS, et al. Thyroid transcription factor-1 expression is an independent predictor of recurrence and correlates with the IASLC/ATS/ERS histologic classification in patients with stage I lung adenocarcinoma. Cancer. 2013;119:931–938. doi: 10.1002/cncr.27863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadota K, Suzuki K, Kachala SS, et al. A grading system combining architectural features and mitotic count predicts recurrence in stage I lung adenocarcinoma. Mod Pathol. 2012;25:1117–1127. doi: 10.1038/modpathol.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kadota K, Yeh YC, Sima CS, et al. The cribriform pattern identifies a subset of acinar predominant tumors with poor prognosis in patients with stage I lung adenocarcinoma: a conceptual proposal to classify cribriform predominant tumors as a distinct histologic subtype. Mod Pathol. 2014;27:690–700. doi: 10.1038/modpathol.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadota K, Villena-Vargas J, Yoshizawa A, et al. Prognostic significance of adenocarcinoma in situ, minimally invasive adenocarcinoma, and nonmucinous lepidic predominant invasive adenocarcinoma of the lung in patients with stage I disease. Am J Surg Pathol. 2014;38:448–460. doi: 10.1097/PAS.0000000000000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadota K, Suzuki K, Colovos C, et al. A nuclear grading system is a strong predictor of survival in epitheloid diffuse malignant pleural mesothelioma. Mod Pathol. 2012;25:260–271. doi: 10.1038/modpathol.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arcila M, Lau C, Nafa K, Ladanyi M. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn. 2011;13:64–73. doi: 10.1016/j.jmoldx.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rekhtman N, Paik PK, Arcila ME, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res. 2012;18:1167–1176. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLeer-Florin A, Moro-Sibilot D, Melis A, et al. Dual IHC and FISH Testing for ALK Gene Rearrangement in Lung Adenocarcinomas in a Routine Practice: A French Study. J Thorac Oncol. 2012;7:348–354. doi: 10.1097/JTO.0b013e3182381535. [DOI] [PubMed] [Google Scholar]
- 39.Paik JH, Choi C-M, Kim H, et al. Clinicopathologic implication of ALK rearrangement in surgically resected lung cancer: A proposal of diagnostic algorithm for ALK-rearranged adenocarcinoma. Lung Cancer. 2012;76:403–409. doi: 10.1016/j.lungcan.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer: correlation with fluorescence In situ hybridization. J Thorac Oncol. 2011;6:466–472. doi: 10.1097/JTO.0b013e31820b82e8. [DOI] [PubMed] [Google Scholar]
- 41.Dignam JJ, Zhang Q, Kocherginsky M. The use and interpretation of competing risks regression models. Clin Cancer Res. 2012;18:2301–2308. doi: 10.1158/1078-0432.CCR-11-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Annals of Statistics. 1988;16:1141–1154. [Google Scholar]
- 43.Villaruz LC, Socinski MA, Cunningham DE, et al. The prognostic and predictive value of KRAS oncogene substitutions in lung adenocarcinoma. Cancer. 2013;119:2268–2274. doi: 10.1002/cncr.28039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 45.Marchetti A, Martella C, Felicioni L, et al. EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 46.Hsieh RK, Lim KH, Kuo HT, Tzen CY, Huang MJ. Female sex and bronchioloalveolar pathologic subtype predict EGFR mutations in non-small cell lung cancer. Chest. 2005;128:317–321. doi: 10.1378/chest.128.1.317. [DOI] [PubMed] [Google Scholar]
- 47.Blons H, Cote JF, Le Corre D, et al. Epidermal growth factor receptor mutation in lung cancer are linked to bronchioloalveolar differentiation. Am J Surg Pathol. 2006;30:1309–1315. doi: 10.1097/01.pas.0000213285.65907.31. [DOI] [PubMed] [Google Scholar]
- 48.Marchetti A, Buttitta F, Pellegrini S, et al. Bronchioloalveolar lung carcinomas: K-ras mutations are constant events in the mucinous subtype. J Pathol. 1996;179:254–259. doi: 10.1002/(SICI)1096-9896(199607)179:3<254::AID-PATH589>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 49.Finberg KE, Sequist LV, Joshi VA, et al. Mucinous differentiation correlates with absence of EGFR mutation and presence of KRAS mutation in lung adenocarcinomas with bronchioloalveolar features. J Mol Diagn. 2007;9:320–326. doi: 10.2353/jmoldx.2007.060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casali C, Rossi G, Marchioni A, et al. A single institution-based retrospective study of surgically treated bronchioloalveolar adenocarcinoma of the lung: clinicopathologic analysis, molecular features, and possible pitfalls in routine practice. J Thorac Oncol. 2010;5:830–836. doi: 10.1097/jto.0b013e3181d60ff5. [DOI] [PubMed] [Google Scholar]
- 51.Hata A, Katakami N, Fujita S, et al. Frequency of EGFR and KRAS mutations in Japanese patients with lung adenocarcinoma with features of the mucinous subtype of bronchioloalveolar carcinoma. J Thorac Oncol. 2010;5:1197–1200. doi: 10.1097/JTO.0b013e3181e2a2bc. [DOI] [PubMed] [Google Scholar]
- 52.Kakegawa S, Shimizu K, Sugano M, et al. Clinicopathological features of lung adenocarcinoma with KRAS mutations. Cancer. 2011;117:4257–4266. doi: 10.1002/cncr.26010. [DOI] [PubMed] [Google Scholar]
- 53.Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov. 2014;4:415–422. doi: 10.1158/2159-8290.CD-13-0633. [DOI] [PubMed] [Google Scholar]
- 54.Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable Oncogene Fusions in Invasive Mucinous Lung Adenocarcinoma. Clin Cancer Res. 2014 Apr 11; doi: 10.1158/1078-0432.CCR-14-0107. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–2133. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol. 2011;29:3574–3579. doi: 10.1200/JCO.2011.35.9638. [DOI] [PubMed] [Google Scholar]
- 57.Ilie M, Long E, Hofman V, et al. Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann Oncol. 2013;24:742–748. doi: 10.1093/annonc/mds534. [DOI] [PubMed] [Google Scholar]
- 58.Paik PK, Arcila ME, Fara M, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol. 2011;29:2046–2051. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol. 2009;22:508–515. doi: 10.1038/modpathol.2009.2. [DOI] [PubMed] [Google Scholar]
- 60.Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15:5216–5223. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jokoji R, Yamasaki T, Minami S, et al. Combination of morphological feature analysis and immunohistochemistry is useful for screening of EML4-ALK-positive lung adenocarcinoma. J Clin Pathol. 2010;63:1066–1070. doi: 10.1136/jcp.2010.081166. [DOI] [PubMed] [Google Scholar]