Abstract
Background
The presence of hepatitis B virus (HBV) resistance mutations upon initiation or during antiretroviral therapy (ART) in HIV co-infected patients is an important determinant of treatment response. The main objective of the study was to determine the prevalence of HBV resistance mutations in antiretroviral treatment-naïve and treatment-experienced HBV-HIV co-infected Ghanaian patients with detectable HBV viremia.
Methods
HBV-HIV co-infected patients who were ART-naïve or had received at least 9 months of lamivudine (3TC)-containing ART were enrolled in a cross-sectional study. Demographic and clinical data were collected and HBV DNA quantified. Partial HBV sequences were amplified by PCR and sequenced bi-directionally to obtain a 2.1-2.2 kb fragment for phylogenetic analysis of HBV genotypes and evaluation of drug resistance mutations.
Results
Of the 100 HBV-HIV co-infected study patients, 75 were successfully PCR-amplified, and 63 were successfully sequenced. Of these 63 patients, 27 (42.9%) were ART-experienced, and 58 (92.1%) had HBV genotype E. No resistance mutations were observed in the 36 ART-naïve patients, while 21 (77.8%) of 27 treatment-experienced patients had resistance mutations. All patients with resistance mutations had no tenofovir (TDF) in their regimens, and 80% of them had HIV RNA < 40 copies/mL. The 3TC resistance mutations rtL180M and rtM204V were observed in 10 (47.6%) of the 21 patients, while 5 patients (23.8%) had rtV173, rtL80I, and rtM204V mutations.
Discussion
A high proportion of HBV-HIV co-infected patients with detectable viremia on 3TC-containing ART had resistance mutations despite good ART adherence as determined by HIV RNA suppression. This study emphasizes the need for dual therapy as part of a fully suppressive ART in all HBV-HIV co-infected patients in Ghana.
Keywords: Hepatitis B virus, HIV, co-infection, Ghana, Africa, genotype, drug resistance
Introduction
There are more than 350 million hepatitis B virus (HBV) carriers in the world with the majority living in low- and middle-income countries [1]. HBV DNA levels are associated with risk of disease progression and development of cirrhosis, hepatic decompensation, and hepatocellular carcinoma (HCC) [2-4]. An estimated 10% of HIV-infected persons worldwide are chronically infected with HBV, with the highest rates (up to 25%) occurring in sub-Saharan Africa and Asia [5, 6]. HIV co-infection is a major cause of morbidity and mortality of HBV infection as it is associated with an increased risk of progression to liver cirrhosis and HCC [3]. Effective antiviral therapy is indicated to reduce the viral replication and risk of HBV progression [4]. Major advancements in the treatment of chronic HBV have been made during the last decade with the development of nucleoside reverse transcriptase inhibitors (NRTIs) with anti-HBV activity such as L-nucleosides (lamivudine (3TC) and telbivudine), alkyl phosphonates (adefovir dipivoxil and tenofovir disoproxil fumarate), or D-cyclopentanes (entecavir) [7].
Inadequate suppression of HBV replication by antivirals under drug pressure may result in the development of resistant mutations within the conserved region of the polymerase gene known as the (YMDD) motif [8]. Until recently, 3TC was utilized as the only active drug in ART regimens in sub-Saharan African as part of HIV treatment regimens. However, it has been shown that HBV resistance to 3TC occurs rapidly through the rtM204V/I substitutions [9]. Other studies have reported primary 3TC resistance due to rt181T/S substitutions [10]. HBV mutations also occur in the absence of 3TC therapy and are thought to occur naturally in HBV carriers who have not been treated with the drug, implying that pre-existing mutations may be transmitted or contribute toward early emergence of 3TC resistance during therapy [11, 12].
In the current study, we investigated the prevalence and pattern of HBV resistance mutations in ART-naïve and ART-experienced HBV-HIV co-infected Ghanaian patients on 3TC-containing regimens who had detectable viremia > 400 IU/mL.
Methods
Patient Population
In a previous prospective cross-sectional study [13], serum samples from 235 HBV-HIV co-infected patients were collected at the Fevers Unit of the Korle-Bu Teaching Hospital in Accra, Ghana between 2012 and 2014. HBsAg positivity was confirmed serologically using a third generation ELISA Surase B 96 (TMB) (General Biologicals Corp, Hsinchu, Taiwan). Plasma HBV DNA levels were determined using fully automated COBAS® TaqMan® Analyzer (Roche Diagnostics GmbH, Mannheim, Germany). The lower limit of detection of the Roche TaqMan assay was 20 IU/mL with a linear range of 20-170,000,000 IU/mL.
Of the original 235 samples, 100 samples with the highest HBV viral loads (>400 IU/mL) were evaluated in the current analysis. Of these, 57 patients were ART-naïve, while 43 patients had received at least 9 months of 3TC-containing ART. HBV DNA was extracted from 500 uL of patient serum using the QIAamp UltraSens Virus Kit (QIAGEN, Valencia, CA, USA).
PCR Amplification and Sequencing
A 2.1-2.2 kb fragment of the polymerase open reading frame – containing the entire reverse-transcriptase (RT) region – was amplified for each sample with one of two sets of primers (A or B) as described in Table 1. The PicoMaxx High Fidelity PCR system (Agilent, Santa Clara, CA, USA) was used for amplification with primer set A. For any sample that did not amplify, a second PCR was attempted with the same primer set. If the re-amplification was negative again, amplification was attempted with primer set B. Amplification conditions were as follows: 94°C for 2 minutes initially, 40 cycles of 94°C for 40 seconds, 60°C for 90 seconds, and 68°C for three minutes, 2 minutes at 68°C every ten cycles, and ending with 68°C for 8 minutes. PCR products were separated with gel electrophoresis, purified with the QIAquick PCR Purification Kit (QIAGEN, Valencia, CA, USA), and sequenced bi-directionally using the primers described in Table 1.
Table 1. Primers for PCR and sequencing.
| Set | Direction | Primer Name | Primer Sequence | Binding Site (from EcoRI site) | Fragment Length | Number of Samples |
|---|---|---|---|---|---|---|
| PCR Primers | ||||||
| A | Forward | CoreF | 5′-GTGTGGATTCGCACTCCT-3′ | 2269-2287 | 2.1 kb | 65 |
| Reverse | Werle AS | 5′-CGTCAGCAAACACTTGGC-3′ | 1175-1192 | |||
| B | Forward | P6 | 5′-GGGCAGGTCCCCTAGAAGA AGAACT-3′ | 2363-2386 | 2.2 kb | 10 |
| Reverse | FengR | 5′-CCGATGAGCTTTGCTCCAGACC-3′ | 1328-1307 | |||
|
| ||||||
| Sequencing Primers | ||||||
| Reverse | Werle AS | 5′-CGTCAGCAAACACTTGGC-3′ | 1175-1192 | |||
| Forward | P7 | 5′-GTGGGTCACCATATTCTTGGG-3′ | 2820-2840 | |||
| Forward | P9 | 5′-ATTCCTATGGGAGTGGGCCTCAG-3′ | 633-655 | |||
| Reverse | P15 | 5′-ATAACTGAGAGCCAAACAGTGGG-3′ | 718-740 | |||
Phylogenetic Analysis
Sequences were assembled using CodonCode Aligner (CodonCode Corporation, Centerville, MA, USA). All alignments were performed using the neighbor-joining method implemented in Clustal X and trimmed to include only the RT-region (1.04 kb) in which HBV drug resistance mutations are found. The statistical robustness and reliability of the branching order within each phylogenetic tree were confirmed by bootstrap analysis using 1,000 replicates. Additional phylogenetic inference was performed using a Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in the BEAST v1.8.0 program under an uncorrelated lognormal relaxed molecular clock and the general time-reversible model with nucleotide site heterogeneity estimated using a gamma distribution. The MCMC analysis was run for a chain length of 50,000,000 with sampling every 5,000th generation. Results were visualized in Tracer v1.5 to confirm chain convergence, and the effective sample size (ESS) was calculated for each parameter. All ESS values were >200 indicating sufficient sampling. The maximum clade credibility tree was selected from the posterior tree distribution after a 10% burn-in using TreeAnnotator v1.8.0. Patient sequences have been submitted to GenBank with the accession numbers KU711604-KU711666.
Mutation Identification
BioEdit (Ibis Biosciences, Carlsbad, CA, USA) was used to translate the aligned patient nucleotide sequences. The sequences were examined for known vaccine escape mutations (sG145R/A, sP142S, sI/T126A/N/I/S, sQ129H/R, sM133L, sD144A/E, sP120S/E, sK141E, sP134I, and sT116N), immunoprophylaxis escape mutations (sI110L, sI126T, sT131N, sM133T, sS143T, sC149R, and sN204S), mutations associated with occult HBV infection (nucleotide changes A233G and G418T), and diagnostic failure mutations (sT123A/N, sK122I, sT131I, and sK141E) [14-39]. Sequences were examined for known HBV drug resistance mutations (Table 2).
Table 2. HBV drug resistance mutations.
| Location | Adefovir | Lamivudine | Tenofovir | Telbivudine | Entecavir |
|---|---|---|---|---|---|
| rtL80V | - | X | - | - | - |
| rtL80I | - | X | - | - | - |
|
| |||||
| rtV84M | X | - | - | - | - |
|
| |||||
| rtS85A | X | - | - | - | - |
|
| |||||
| rtI169T | - | X | - | - | X |
|
| |||||
| rtV173L | - | X* | - | - | X |
|
| |||||
| rtL180M | - | X* | - | X | X |
|
| |||||
| rtA181T | X | X | X | X | - |
| rtA181V | X | X | X | X | - |
|
| |||||
| rtT184S | - | - | - | - | X |
| rtT184A | - | - | - | - | X |
| rtT184I | - | - | - | - | X |
| rtT184L | - | - | - | - | X |
| rtT184C | - | - | - | - | X |
| rtT184M | - | - | - | - | X |
| rtT184G | - | - | - | - | X |
| rtT184S | - | X | - | - | X |
| rtS184G | - | - | - | - | X |
|
| |||||
| rtA194T | - | - | X | - | - |
|
| |||||
| rtS202G | - | X | - | - | X |
| rtS202I | - | - | - | - | X |
|
| |||||
| rtM204V | - | X* | X | X | X |
| rtM204I | - | X | X | X | - |
| rtM204S | - | X | - | - | - |
|
| |||||
| rtV214A | X | X | X | - | - |
| rtV214E | X | - | - | - | - |
|
| |||||
| rtQ215S | X | X | X | - | - |
|
| |||||
| rtN236T | X | X | X | - | - |
|
| |||||
| rtN238D | X | X | - | - | - |
|
| |||||
| rtM250V | - | - | - | - | X |
Results
Of the 235 HIV-infected patients who were HBsAg-positive, 138 (58.7%) and 101 (43.0%) patients had detectable HBV DNA level above 19 and 400 IU/mL, respectively [13]. Of those with HBV DNA > 400 IU/mL, 100 (99.0%) were included in the current study. PCR amplification was successful for 75 of 100 patient samples using one of the two primer sets, with 63 of the 75 samples providing high quality sequence data for further analysis. The demographic and clinical characteristics of the HBV-HIV co-infected patients in whom sequencing was successful (N = 63) were more likely than those from whom PCR or sequencing failed (N = 37) to have higher median HBV DNA levels (53,870,705 vs. 2,953 IU/mL, P < 0.001), be HBeAg positive (61.9% vs. 21.6%, P < 0.001), be HBeAb negative (55.6% vs. 18.9%, P < 0.001) and to have a trend toward longer median duration of therapy (5.6 vs. 4.1 years, P = 0.087). There were no differences in median age, BMI, CD4 count, ART status, or WHO disease stage.
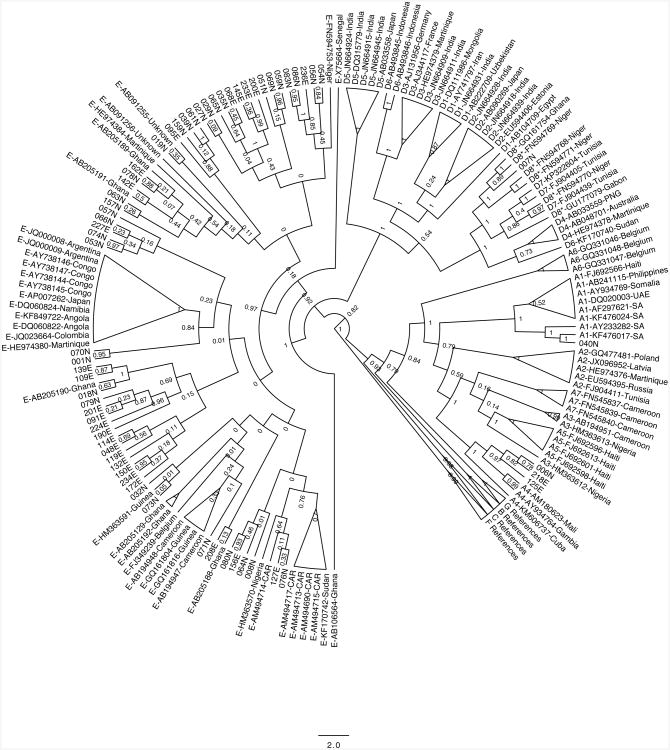
Of the 63 study patients evaluated here, 36 (57.1%) were treatment-naïve and 27 (42.9%) had received a median (range) of 5.6 (1.6 – 9.1) years of 3TC-containing drug therapy. Of the 27 treatment-experienced patients, 2 (7.4%) were on drug therapy containing TDF. Fifty-eight (92.1%) individuals were infected with HBV genotype E, 3 (4.8%) with genotype A4, and 1 (1.6%) each with genotype A1 and genotype D8 as shown in Figure 1.
Figure 1.
Phylogenetic tree for 63 patient samples from the current study. Patient IDs are the numbers formatted as XXXN or XXXE (where XXX is the patient identified). Reference sequences are denoted by their HBV gentoype followed by their accession number and a 3 letter abbreviation corresponding to the country of origin.
Vaccine escape mutations were found in 7 (11.1%) patients including 040N (sG145R), 159N (sQ129H), 007N (sQ129R), 054N (sD144A), 127E (sD144E), 006N (sD144E), and 119E (sD144E). Immunoprophylaxis escape mutations were observed in all 63 (100%) patients. All patients had the N204S mutation, and all but patient 132E had the sI126T mutation. Four (6.35%) patients – 006N, 040N, 125E, and 218E – had both sT131N and sS143T mutations. Additional immunoprophylaxis mutations were observed in patients 224E (sI110L), 080N (sT131N), and 132E (sM133T). No mutations associated with occult HBV infection or diagnostic failure were present.
HBV drug resistance mutations were found in 21 (77.8%) of 27 treatment-experienced patients and in 0 (0%) of the 36 treatment-naïve patients. Overall, 21 (33.3%) of the patients in whom sequencing was successful had HBV resistance mutations. There was no statistically significant difference in age, gender, duration of therapy, HBV DNA titer or CD4+ count between the treatment-experienced patients with or without resistance, although there was a trend toward HBeAg-positivity (P=0.056) in ART-experienced patients having resistance mutants. Of the 27 treatment-experienced patients in the current study, 18 (67%) had fully suppressed HIV-1 RNA < 40 copies/mL. Paradoxically, median HBV DNA levels was significantly higher in the patient with suppressed HIV RNA compared to those with detectable viral loads (68,967,835 vs. 3,581,236, P = 0.025). In addition, the patient with HBV resistance mutations were more likely than those without resistance mutation to have suppressed HIV RNA < 40 copies/ML (80.0% vs. 16.7%, P = 0.008).
The resistance mutations pattern is shown in Table 3. Twenty (95.2%) of 21 patients with identified resistance mutations were infected with genotype E, while 1 patient was infected with subgenotype A4. All 21 patients with identified resistance mutations had at least one mutation that confers clinical resistance to both 3TC and telbivudine. No mutations associated with adefovir resistance were observed.
Table 3. HBV drug resistance pattern among 21 HIV-HBV co-infected patients.
| Pattern | Number | Percent |
|---|---|---|
| rtL180M, rtM204V | 10 | 47.6% |
| rtV173L, rtL180M, rtM204V | 5 | 23.8% |
| rtV173L, rtL180M, rtM204I | 1 | 4.76% |
| rtL80I, rtL180M, rtM204V | 2 | 9.52% |
| rtL80I, rtM204I | 1 | 4.76% |
| rtL180M, rtM204I | 1 | 4.76% |
| rtL180M | 1 | 4.76% |
Discussion
Hepatitis B virus infection is a major concern in sub-Saharan Africa; however, only limited data are available on circulating genotypes, vaccine escape mutations, and the presence of drug resistance mutations from this region. The patients in the current study were enrolled previously in a study to investigate the factors associated with HBV viremia in ART-naïve and ART-experienced individuals. Overall, 89% of ART-naïve and 42.1% of ART-experienced patients had detectable HBV DNA > 20 IU/mL [13]. In the current study, the presence of drug resistance mutations in the patients with the highest level of viremia with HBV DNA > 400 IU/mL (N = 100) was investigated. No HBV resistance mutations were observed among the ART-naïve HBV-HIV co-infected patients; however, a high proportion (78%) of ART-experienced patients with viremia had mutations that confer clinical resistance to 3TC. In addition, the vast majority of those with resistance mutations (95%) had HBV genotype E, and vaccine escape mutations were detected in both ART-naïve and ART-experienced patients.
HBV has been classified into 10 genotypes (A-J) with distinct geographical distributions [40]. Along with ART regimen, HBV genotype is a major factor impacting the course of disease and treatment response in both mono- and co-infected patients [41]. In the current study, 20 (95.2%) of 21 patients with identified resistance mutations were infected with genotype E, while 1 patient was infected with genotype A4. Our results are consistent with those of a study in Kumasi, Ghana, in which the investigators demonstrated the predominance of HBV genotypes E (95.3%) and only 4.7% of patients having genotype A [2]. By contrast, a multi-center chronic hepatitis B genotypic resistance study in Europe showed that the most common genotypes were genotype D (63.1%) followed by genotype A (26.3%) [42]. The study in Kumasi found the overall prevalence of major drug resistance mutations in reverse transcriptase (RT) to be 4.6% among 86 HBsAg-positive HIV-infected patients with detectable HBV viremia [2]. The 3TC mutations rtV173L, rtL180M, and rtM204V were observed in 3 patients, although ART information was not reported [2].
The present study found the overall prevalence of drug resistance mutations to be 33.3%; however, HBV resistance mutations were detected exclusively in ART-experienced patients. Previous studies have shown that the resistance mutations present in these patients were associated with 3TC, TDF, telbivudine, and entecavir; nonetheless, it should be noted that the only HBV-active agent in widespread use in Ghana is 3TC [43-46]. The 3TC mutations rtL80I, rtV173L, rtL180M, rtM204V, and/or rtM204I were observed in 21 patients. This was comparable to a study in Kumasi, Ghana which demonstrated 31 of 44 (70.5%) 3TC-experienced HBV-HIV co-infected patients harbored one or more of the 3TC resistant mutations after 45 months of therapy. Overall, 31 (29.2%) out of 106 in the 3TC-experienced group had resistance mutants in the above-mentioned study [47]. Although there were differences in genotype, the dominant pattern of resistance among treatment-experienced patients with mutated strains, (rtM204V + rtL180M), was similar between the European multi-center study (29%) [42] and the present study in Ghana (47.6%).
In our study, twenty (95.2%) of the 21 patients had at least one mutation that may confer clinical resistance to both telbivudine (rtM204V/I) and 3TC (rt180M + rtM204V/I, rtV173L + rtL180M + rtM204V/I, and rtL80I + rtM204I). However, all resistance mutations found were likely due to 3TC cross-resistance as it was the only HBV-active drug in first- and second-line ART for these patients in the treatment-experienced group (Table 4). None of the patients with 3TC+TDF combination ART in the treatment-experienced group showed evidence of resistant mutations.
Table 4. Treatment regimens and HBV DNA levels for treatment-experienced patients.
| Patient ID | Resistance Mutations Present? | HAART 1 Regimen | HAART 2 Regimen | Duration of HAART (years) | HBV DNA Levels (IU/mL) | HIV-1 RNA (copies/mL) |
|---|---|---|---|---|---|---|
| 048E | Yes | 3TC + AZT + EFV | 7.12 | 49,786,239 | TND | |
| 068E | No | 3TC + TDF + EFV | 2.31 | 1,265 | 318,932 | |
| 091E | Yes | 3TC + AZT + NVP | 2.49 | 20,692,495 | < 40 | |
| 095E | No | 3TC + AZT + EFV | DDI + ABC + LPVr | 8.23 | 6,034,003 | 233,208 |
| 109E | Yes | 3TC + AZT + EFV | 5.68 | 53,870,705 | TND | |
| 114E | Yes | 3TC + AZT + EFV | 2.32 | 170,000,001 | TND | |
| 119E | Yes | 3TC + AZT + EFV | 5.57 | 7,730,963 | TND | |
| 125E | Yes | 3TC + D4T + EFV | 3TC + AZT + EFV | 3.72 | 140,456,193 | TND |
| 127E | No | 3TC + AZT + EFV | 3.39 | 64,830 | 1483 | |
| 132E | Yes | 3TC + AZT + EFV | 1.56 | 821,669 | < 40 | |
| 139E | Yes | 3TC + D4T + EFV | 3TC + AZT + EFV | 7.79 | 172,744 | 283,443 |
| 142E | Yes | 3TC + AZT + EFV | 4.82 | 10,073,311 | TND | |
| 145E | Yes | 3TC + D4T + EFV | 3TC + AZT + EFV | 6.82 | 170,000,001 | TND |
| 150E | Yes | 3TC + AZT + EFV | 2.32 | 170,000,001 | TND | |
| 156E | No | 3TC + AZT + EFV | 3TC + TDF + LPVr | 5.53 | 170,000,001 | < 40 |
| 162E | No | 3TC + AZT + EFV | 6.01 | 170,000,001 | 721 | |
| 172E | Yes | 3TC + D4T + NVP | 3TC + AZT + NVP | 7.00 | 3,581,236 | 2,927 |
| 190E | Yes | 3TC + AZT + EFV | 6.94 | 76,716,146 | TND | |
| 200E | Yes | 3TC + AZT + EFV | 1.79 | 58,688,821 | 174 | |
| 201E | Yes | 3TC + AZT + EFV | 8.68 | 61,219,524 | TND | |
| 208E | No | 3TC + AZT + NVP | 2.26 | 84,536,960 | 188070 | |
| 218E | Yes | 3TC + AZT + EFV | 8.56 | 47,794,228 | TND | |
| 224E | Yes | 3TC + AZT + EFV | 5.23 | 91,534,717 | TND | |
| 227E | Yes | 3TC + AZT + NVP | 5.01 | 143,719,944 | TND | |
| 233E | Yes | 3TC + AZT + EFV | 7.20 | 144,400,877 | TND | |
| 234E | Yes | 3TC + AZT + NVP | 9.05 | 2,357,904 | 1473 | |
| 236E | Yes | 3TC + D4T + NVP | 3TC + AZT + NVP | 7.12 | 16,789,936 | TND |
Key: 3TC = lamivudine, TDF = tenofovir, AZT = zidovudine, NVP = nevirapine, EFV = efavirenz, D4T = stavudine, LPVr = lopinavir/ritonavir, DDI = didanosine, ABC = abacavir, TND = target not detected.
All 63 patient sequences were also screened for known vaccine escape mutations, immunoprophylaxis escape mutations, mutations associated with occult HBV infection, and diagnostic failure mutations. Vaccine escape mutations were found in 11.1% of patients, while 100% of patients had at least one immunoprophylaxis escape mutation (Table 5). No mutations associated with occult infection or diagnostic failure were found.
Limitations to this study should be noted. It was conducted in the main tertiary referral center in Accra serving the majority of the southern half of Ghana. However, results may not be generalizable to the wider Ghanaian population or HIV-negative persons. Furthermore, phylogenetic analysis was restricted by the inability to obtain DNA sequences and HBV genotype data from a subset of HBV DNA positive individuals due to amplification or sequencing failures. Additionally, the analysis of consensus sequence data likely underestimates the true prevalence of drug resistance or vaccine escape mutations if they are present at low levels. Due to the cross-sectional nature of the analysis, the temporal relationship between predictor variables and emergence of HBV drug resistance in treated patients could not be assessed. However, we did not observe a relationship between poor ART adherence as measure by detectable HIV RNA and the level of HBV viremia or presence of HBV resistance mutations. In fact, the patients with suppressed HIV RNA were more likely to have higher HBV DNA levels and an increased number of 3TC resistance mutations. These data suggest that there may be factors other than ART adherence that lead to emergence of 3TC resistance. Thus, prospective studies are necessary to understand the mechanism leading to HBV treatment failure with emergence of drug resistance in some HBV-HIV co-infected patients to guide future therapeutic regimen selection in an HBV-endemic country like Ghana.
Until TDF became available in Ghana in 2012, the recommendation was to use 3TC-containing ART for HBV-HIV co-infected individuals. The most recent HIV treatment guidelines recommend the use of TDF plus 3TC or emtricitabine in patients with HBV-HIV co-infection [48]. However, none of the patients with HBV-resistant mutations were on the recommended dual-anti-HBV therapy at the time of the study. Our data provide evidence in support of the notion that using 3TC as the only anti-viral active against HBV in ART is not an effective treatment plan in co-infected patients. The data also provide evidence that HBsAg testing should be introduced as part of routine HIV care prior to initiation of ART in sub-Saharan Africa and that HBV-HIV co-infected patients should be given 3TC or emtricitabine plus TDF as part of their regimen unless TDF is contraindicated per treatment guidelines [48, 49]. We recommend HBV screening for all treatment-experienced patients who were initiated on a non-TDF containing ART prior to availability of TDF or universal screening in Ghana and intensifying their regimen by substituting the second NRTI with TDF in HBV-HIV co-infected patients. When TDF cannot be used in naïve or treatment-experienced patients with HBV co-infection, fully suppressive ART plus entecavir or telbivudine may be needed as per guidelines [48, 49].
Acknowledgments
We thank the patients for participation in this study. This study was funded by a 2012 International Developmental Grant from the Lifespan/Tufts/Brown CFAR (P30AI042853) and the Brown/Tufts AIDS International Training and Research Program (D43TW000237) to Drs. Archampong and Kwara. Additional support was provided by Brown University – University of Ghana partnership through a USAID/HED grant (Award # AEG-A-00-05-00007). Dr. Kwara received additional support from Fogarty international Center (D43TW010055). We wish to acknowledge MDS Lancet Laboratories, Ghana for performing HBV DNA on study participants. We are grateful for the support of Drs. Michael Stein and Annie Gjelsvik at Brown University for their assistance in developing the study design.
Footnotes
Author Disclosure Statement: The authors have no further disclosures relevant to this article and declare no other conflicts of interest.
References
- 1.Maynard JE. Hepatitis B: global importance and need for control. Vaccine. 1990;8 Suppl:S18–20. doi: 10.1016/0264-410x(90)90209-5. discussion S21-13. [DOI] [PubMed] [Google Scholar]
- 2.Geretti AM, Patel M, Sarfo FS, et al. Detection of highly prevalent hepatitis B virus coinfection among HIV-seropositive persons in Ghana. J Clin Microbiol. 2010;48:3223–3230. doi: 10.1128/JCM.02231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thio CL, Seaberg EC, Skolasky R, Jr, et al. HIV-1, hepatitis B virus, and risk of liver-related mortality in the Multicenter Cohort Study (MACS) Lancet. 2002;360:1921–1926. doi: 10.1016/s0140-6736(02)11913-1. [DOI] [PubMed] [Google Scholar]
- 4.Hafkin JS, Osborn MK, Localio AR, et al. Incidence and risk factors for incomplete HBV DNA suppression in HIV/HBV-co-infected patients initiating tenofovir-based therapy. J Viral Hepat. 2014;21:288–296. doi: 10.1111/jvh.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kourtis AP, Bulterys M, Hu DJ, Jamieson DJ. HIV–BV Coinfection — Global Challenge. N Engl J Med. 2012;366:1749–1752. doi: 10.1056/NEJMp1201796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann CJ, Thio CL. Clinical implications of HIV and hepatitis B co-infection in Asia and Africa. Lancet Infect Dis. 2007;7:402–409. doi: 10.1016/S1473-3099(07)70135-4. [DOI] [PubMed] [Google Scholar]
- 7.Zoulim F. Hepatitis B virus resistance to antiviral drugs: where are we going? Liver Int. 2011;31(1):111–116. doi: 10.1111/j.1478-3231.2010.02399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison TJ. Hepatitis B virus: molecular virology and common mutants. Semin Liver Dis. 2006;26:87–96. doi: 10.1055/s-2006-939754. [DOI] [PubMed] [Google Scholar]
- 9.Allen MI, Deslauriers M, Andrews CW, et al. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 10.Ghany MG, Doo EC. Antiviral resistance and hepatitis B therapy. Hepatology. 2009;49:S174–184. doi: 10.1002/hep.22900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selabe SG, Lukhwareni A, Song E, Leeuw YG, Burnett RJ, Mphahlele MJ. Mutations associated with lamivudine-resistance in therapy-naive hepatitis B virus (HBV) infected patients with and without HIV co-infection: implications for antiretroviral therapy in HBV and HIV co-infected South African patients. J Med Virol. 2007;79:1650–1654. doi: 10.1002/jmv.20974. [DOI] [PubMed] [Google Scholar]
- 12.Bivigou-Mboumba B, Françis-Souquièe S, Deleplancque L, et al. Broad Range of Hepatitis B Virus (HBV) Patterns, Dual Circulation of Quasi-Subgenotype A3 and HBV/E and Heterogeneous HBV Mutations in HIV-Positive Patients in Gabon. PLoS One. 2016;11:e0143869. doi: 10.1371/journal.pone.0143869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Archampong TN, Lartey M, Sagoe KW, et al. Proportion and factors associated with Hepatitis B viremia in antiretroviral treatment naive and experienced HIV co-infected Ghanaian patients. BMC Infect Dis. 2016;16:14. doi: 10.1186/s12879-016-1342-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carman WF, Zanetti AR, Karayiannis P, et al. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 15.Zuckerman JN, Zuckerman AJ. Mutations of the surface protein of hepatitis B virus. Antiviral Res. 2003;60:75–78. doi: 10.1016/j.antiviral.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Hsu HY, Chang MH, Liaw SH, Ni YH, Chen HL. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology. 1999;30:1312–1317. doi: 10.1002/hep.510300511. [DOI] [PubMed] [Google Scholar]
- 17.Francois G, Kew M, Van Damme P, Mphahlele MJ, Meheus A. Mutant hepatitis B viruses: a matter of academic interest only or a problem with far-reaching implications? Vaccine. 2001;19:3799–3815. doi: 10.1016/s0264-410x(01)00108-6. [DOI] [PubMed] [Google Scholar]
- 18.Coleman PF, Chen YC, Mushahwar IK. Immunoassay detection of hepatitis B surface antigen mutants. J Med Virol. 1999;59:19–24. doi: 10.1002/(sici)1096-9071(199909)59:1<19::aid-jmv4>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 19.Ireland JH, O'Donnell B, Basuni AA, et al. Reactivity of 13 in vitro expressed hepatitis B surface antigen variants in 7 commercial diagnostic assays. Hepatology. 2000;31:1176–1182. doi: 10.1053/he.2000.6407. [DOI] [PubMed] [Google Scholar]
- 20.Lee KM, Kim YS, Ko YY, et al. Emergence of vaccine-induced escape mutant of hepatitis B virus with multiple surface gene mutations in a Korean child. J Korean Med Sci. 2001;16:359–362. doi: 10.3346/jkms.2001.16.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seddigh-Tonekaboni S, Lim WL, Young B, et al. Hepatitis B surface antigen variants in vaccinees, blood donors and an interferon-treated patient. J Viral Hepat. 2001;8:154–158. doi: 10.1046/j.1365-2893.2001.00275.x. [DOI] [PubMed] [Google Scholar]
- 22.Carman WF. The clinical significance of surface antigen variants of hepatitis B virus. J Viral Hepat. 1997;4(1):11–20. doi: 10.1111/j.1365-2893.1997.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 23.Hou J, Wang Z, Cheng J, et al. Prevalence of naturally occurring surface gene variants of hepatitis B virus in nonimmunized surface antigen-negative Chinese carriers. Hepatology. 2001;34:1027–1034. doi: 10.1053/jhep.2001.28708. [DOI] [PubMed] [Google Scholar]
- 24.Chen CY, Hsu HY, Liu CC, Chang MH, Ni YH. Stable seroepidemiology of hepatitis B after universal immunization in Taiwan: A 3-year study of national surveillance of primary school students. Vaccine. 2010;28:5605–5608. doi: 10.1016/j.vaccine.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 25.Chong-Jin O, Wei Ning C, Shiuan K, Gek Keow L. Identification of hepatitis B surface antigen variants with alterations outside the “a” determinant in immunized Singapore infants. J Infect Dis. 1999;179:259–263. doi: 10.1086/314553. [DOI] [PubMed] [Google Scholar]
- 26.Ngui SL, O'Connell S, Eglin RP, Heptonstall J, Teo CG. Low detection rate and maternal provenance of hepatitis B virus S gene mutants in cases of failed postnatal immunoprophylaxis in England and Wales. J Infect Dis. 1997;176:1360–1365. doi: 10.1086/514133. [DOI] [PubMed] [Google Scholar]
- 27.He C, Nomura F, Itoga S, Isobe K, Nakai T. Prevalence of vaccine-induced escape mutants of hepatitis B virus in the adult population in China: a prospective study in 176 restaurant employees. J Gastroenterol Hepatol. 2001;16:1373–1377. doi: 10.1046/j.1440-1746.2001.02654.x. [DOI] [PubMed] [Google Scholar]
- 28.Oon CJ, Tan KL, Harrison T, Zuckerman A. Natural history of hepatitis B surface antigen mutants in children. The Lancet. 1996;348:1524. doi: 10.1016/S0140-6736(05)65950-8. [DOI] [PubMed] [Google Scholar]
- 29.Wiseman E, Fraser MA, Holden S, et al. Perinatal transmission of hepatitis B virus: an Australian experience. Med J Aust. 2009;190:489–492. doi: 10.5694/j.1326-5377.2009.tb02524.x. [DOI] [PubMed] [Google Scholar]
- 30.Ghaziasadi A, Alavian SM, Norouzi M, Fazeli Z, Jazayeri SM. Mutational analysis of HBs Ag-positive mothers and their infected children despite immunoprophylaxis. Iran J Allergy Asthma Immunol. 2013;12:352–360. [PubMed] [Google Scholar]
- 31.Lee le Y, Aw M, Rauff M, Loh KS, Lim SG, Lee GH. Hepatitis B immunoprophylaxis failure and the presence of hepatitis B surface gene mutants in the affected children. J Med Virol. 2015;87:1344–1350. doi: 10.1002/jmv.24193. [DOI] [PubMed] [Google Scholar]
- 32.Ishigami M, Honda T, Ishizu Y, et al. Frequent incidence of escape mutants after successful hepatitis B vaccine response and stopping of nucleos(t)ide analogues in liver transplant recipients. Liver Transpl. 2014;20:1211–1220. doi: 10.1002/lt.23935. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida EM, Ramji A, Erb SR, et al. De novo acute hepatitis B infection in a previously vaccinated liver transplant recipient due to a strain of HBV with a Met 133 Thr mutation in the “a” determinant. Liver. 2000;20:411–414. doi: 10.1034/j.1600-0676.2000.020005411.x. [DOI] [PubMed] [Google Scholar]
- 34.Bruni R, Prosperi M, Marcantonio C, et al. A computational approach to identify point mutations associated with occult hepatitis B: significant mutations affect coding regions but not regulative elements of HBV. Virol J. 2011;8:394. doi: 10.1186/1743-422X-8-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Servant-Delmas A, Mercier-Darty M, Ly TD, et al. Variable capacity of 13 hepatitis B virus surface antigen assays for the detection of HBsAg mutants in blood samples. J Clin Virol. 2012;53:338–345. doi: 10.1016/j.jcv.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Verheyen J, Neumann-Fraune M, Berg T, Kaiser R, Obermeier M. The detection of HBsAg mutants expressed in vitro using two different quantitative HBsAg assays. J Clin Virol. 2012;54:279–281. doi: 10.1016/j.jcv.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 37.Tian Y, Xu Y, Zhang Z, et al. The amino Acid residues at positions 120 to 123 are crucial for the antigenicity of hepatitis B surface antigen. J Clin Microbiol. 2007;45:2971–2978. doi: 10.1128/JCM.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jongerius JM, WM M, C HT, et al. New hepatitis B virus mutant form in a blood donor that is undetectable in several hepatitis B surface antigen screening assays. Transfusion. 1998;38:56–59. doi: 10.1046/j.1537-2995.1998.38198141499.x. [DOI] [PubMed] [Google Scholar]
- 39.Seddigh-Tonekaboni S, Waters JA, Jeffers S, et al. Effect of variation in the common “a” determinant on the antigenicity of hepatitis B surface antigen. J Med Virol. 2000;60:113–121. doi: 10.1002/(sici)1096-9071(200002)60:2<113::aid-jmv2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 40.Sunbul M. Hepatitis B virus genotypes: Global distribution and clinical importance. World J Gastroenterol. 2014;20:5427–5434. doi: 10.3748/wjg.v20.i18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain MK, Comanor L, White C, et al. Treatment of hepatitis B with lamivudine and tenofovir in HIV/HBV-coinfected patients: factors associated with response. J Viral Hepat. 2007;14:176–182. doi: 10.1111/j.1365-2893.2006.00797.x. [DOI] [PubMed] [Google Scholar]
- 42.Hermans LE, Svicher V, Pas SD, et al. Combined Analysis of the Prevalence of Drug-Resistant Hepatitis B Virus in Antiviral Therapy-Experienced Patients in Europe (CAPRE) J Infect Dis. 2016;213:39–48. doi: 10.1093/infdis/jiv363. [DOI] [PubMed] [Google Scholar]
- 43.Koziel MJ, Peters MG. Viral Hepatitis in HIV Infection. The New England Journal of Medicine. 2007;356:1445–1454. doi: 10.1056/NEJMra065142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Locarnini S, Mason WS. Cellular and virological mechanisms of HBV drug resistance. J Hepatol. 2006;44:422–431. doi: 10.1016/j.jhep.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 45.Michailidis E, Kirby KA, Hachiya A, et al. Antiviral therapies: focus on hepatitis B reverse transcriptase. Int J Biochem Cell Biol. 2012;44:1060–1071. doi: 10.1016/j.biocel.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keeffe EB, Dieterich DT, Pawlotsky JM, Benhamou Y. Chronic hepatitis B: preventing, detecting, and managing viral resistance. Clin Gastroenterol Hepatol. 2008;6:268–274. doi: 10.1016/j.cgh.2007.12.043. [DOI] [PubMed] [Google Scholar]
- 47.Stockdale AJ, Phillips RO, Beloukas A, et al. Liver Fibrosis by Transient Elastography and Virologic Outcomes After Introduction of Tenofovir in Lamivudine-Experienced Adults With HIV and Hepatitis B Virus Coinfection in Ghana. Clinical Infectious Diseases. 2015 doi: 10.1093/cid/civ421. [DOI] [PubMed] [Google Scholar]
- 48.DHHS, editor. Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1 infected adults and adolescents. 2016. [Google Scholar]
- 49.WHO Guidelines for the prevention, care and treatment of persons with chronic hepatitis B infection. 2015 Mar; www.who.int. [PubMed]