Summary
Following injury, adult skeletal muscle undergoes a well-coordinated sequence of molecular and physiological events to promote repair and regeneration. However, a thorough understanding of the in vivo epigenomic and transcriptional mechanisms that control these reparative events is lacking. To address this, we monitored the in vivo dynamics of three histone modifications and coding and noncoding RNA expression throughout the regenerative process in a mouse model of traumatic muscle injury. We first illustrate how both coding and noncoding RNAs in tissues and sorted satellite cells are modified and regulated during various stages after trauma. Next, we use chromatin immunoprecipitation followed by sequencing to evaluate the chromatin state of cis-regulatory elements (promoters and enhancers) and view how these elements evolve and influence various muscle repair and regeneration transcriptional programs. These results provide a comprehensive view of the central factors that regulate muscle regeneration and underscore the multiple levels through which both transcriptional and epigenetic patterns are regulated to enact appropriate repair and regeneration.
Highlights
-
•
Temporal coding and noncoding transcriptomes during the muscle regenerative process
-
•
Transcriptomes of satellite cells during activation, proliferation, differentiation
-
•
Multidimensional chromatin maps of injured and noninjured muscle tissues
-
•
Impact of epigenetic regulation and gene expression on muscle regeneration
In this article, Aguilar and colleagues temporally characterize the in vivo dynamics of chromatin modifications and the coding and noncoding transcriptional landscape in a mouse model of traumatic muscle injury to provide a comprehensive view of the central factors that regulate muscle regeneration.
Introduction
Adult skeletal muscle is a postmitotic organ that coordinates movement and constantly grows and adapts by remodeling its structure and metabolism. After insult or injury, adult skeletal muscle enables repair and regeneration of existing fibers and formation of new fibers through a population of stem cells that reside underneath the basal lamina called satellite cells (SCs) (Scharner and Zammit, 2011). The SCs reside in a specialized niche and change their quiescent complexion after injury, whereby SCs activate, proliferate, differentiate into myoblasts, and fuse to form myofibers (Bentzinger et al., 2013, Kuang et al., 2008). Each step is affected by various environmental signals and communication with infiltrating and resident cells (Aurora and Olson, 2014, Burzyn et al., 2013, Heredia et al., 2013). Multiple repair and regeneration subprocesses accomplished by SCs (and other cell types) after muscle injury are orchestrated by distinct epigenetic (Asp et al., 2011, Brancaccio and Palacios, 2015), transcriptional (Braun and Gautel, 2011, Buckingham and Rigby, 2014) and post-transcriptional events. However, the integrative dynamics of transcriptional networks and regulatory epigenetic switches at genome-wide levels have only been partly characterized in vivo (Liu et al., 2013, Giordani and Puri, 2013) and, as such, our understanding of the molecular processes involved in muscle regeneration have been limited.
Herein, the in vivo evolution of coding and noncoding expression and three different chromatin modifications (H3K4me3, H3K4me1, and H3K27ac) were profiled across nine time points (t = 3 hr to 672 hr) from an injured and uninjured contralateral tibialis anterior (TA) muscle. The generated genomic maps were then contrasted against myogenic transcription factors (MyoD and MyoG) genomic binding data (Cao et al., 2010, The Mouse ENCODE Consortium, 2014) to determine shared and distinguishing signatures at cis-regulatory elements during different stages after injury. The dynamic levels of numerous coding and noncoding transcripts, chromatin state transitions, and differential binding at transcription factor (TF) motifs were integrated and assessed to construct a comprehensive view of the key transcriptional and chromatin factors that influence and modulate in vivo muscle repair and regeneration dynamics.
Results
Severe Muscle Trauma Induces Extensive Transcriptional and Post-transcriptional Regulation in Both Coding and Noncoding Transcripts
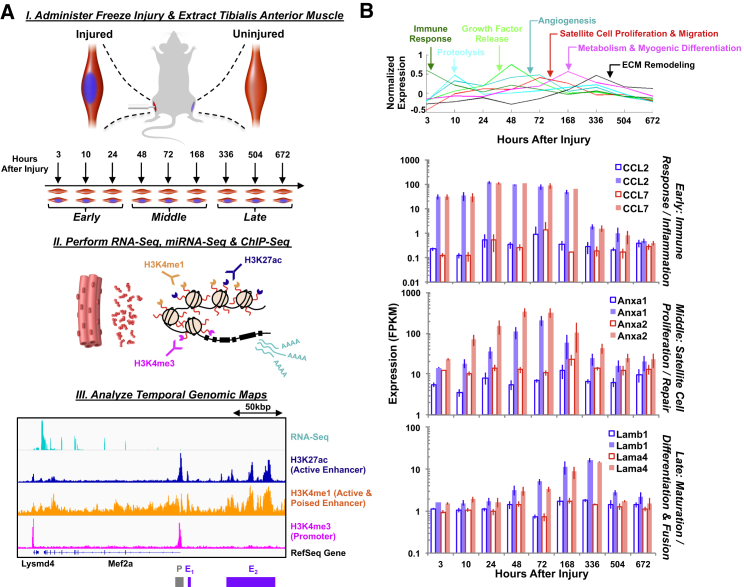
To gain insights into the healing process after administration of trauma, we performed expression profiling by RNA sequencing (RNA-seq) and small RNA-seq (miRNA-seq) (Aguilar et al., 2015) for both the injured and contralateral tissues for multiple time points (Figure 1), and lumped the datasets into three key stages (early, 3–24 hr; middle, 48–168 hr; late, 336–672 hr). Hierarchical clustering of the RNA-seq data through time revealed clusters up- and downregulated that were associated with different stages of muscle repair and regeneration (Figure 1B). For example, chemokine ligands 2 and 7 (Ccl2 and Ccl7), which are important for the recruitment of various immune cells to the injured muscle, peaked in expression in the early period (at 24 hr after injury). Annexins 1 and 2 (Anxa1 and Anxa2), cellular membrane binding proteins that are important for promoting migration of SCs, demonstrated a different expression profile and peaked in the middle period (48–72 hr). Laminin subunits β-1 and α-4 (Lamb1 and Lama4), which are cellular adhesion molecules present in the basement membrane, exhibited a peak in differential expression in the late period (336 hr). Pathway analysis of the time-clustered RNA-seq data showed an initial burst of proinflammatory and immune-response transcripts in the early period, followed by activation, proliferation, and differentiation of myogenic precursors and extracellular matrix (ECM) remodeling in the middle and late time periods, which was consistent with previous studies of muscle tissue injury and in line with a productive healing process (Tidball, 2005, Warren et al., 2007).
Figure 1.
Experimental Overview for Profiling Molecular Mechanisms Governing In Vivo Tibialis Anterior Muscle Regeneration after Severe Trauma
(A) Schematic diagram of injury model and process flow for chromatin and transcript extraction. A representative example of the Mef2a gene at 3 hr post injury is shown where the promoter (labeled P in gray) and enhancer regions (labeled E1 and E2 in purple) are depicted.
(B) (Top) Line plots of hierarchically clustered RNA-seq data through time revealed clusters up- and downregulated at different time periods that were associated with different stages of muscle repair and regeneration. (Bottom) Bar graphs of gene expression values of six different genes corresponding to different stages of the muscle regeneration process through time from left to right. Error bars represent 1 SD.
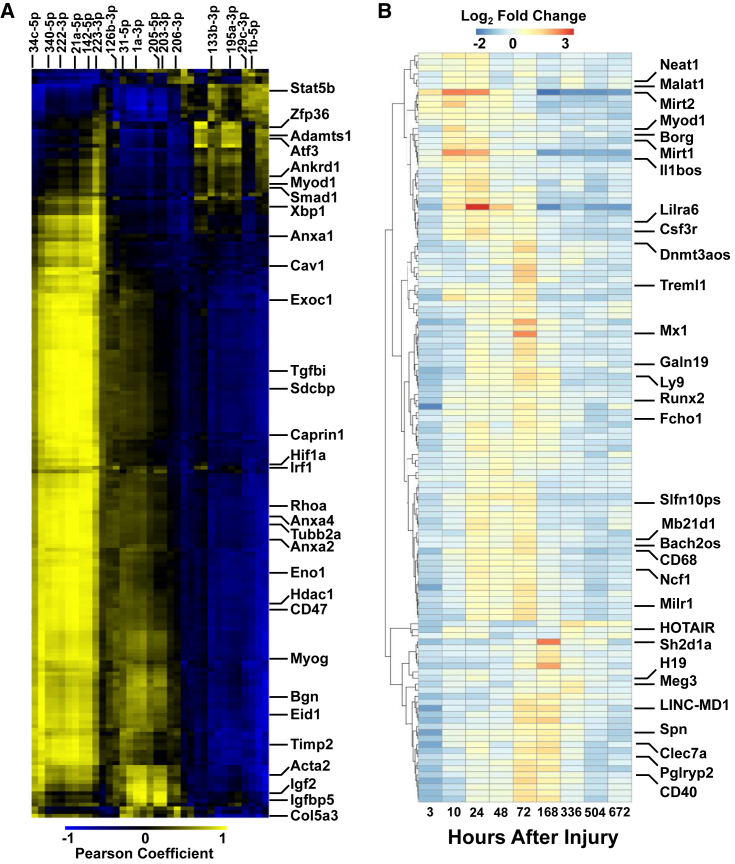
Regulation of muscle regeneration has also recently been shown to occur post-transcriptionally, where microRNAs (miRNAs) bind to the 3′ UTR of mRNAs and inhibit translation (Chen et al., 2005, Kim et al., 2006). Small RNA-seq was performed at multiple time points from each stage and 841 miRNAs were detected. Of these, 143 miRNAs showed dynamic behavior and reinforced many of the results observed from the RNA-seq datasets, where early upregulated miRNAs were associated with inflammation and immune system programs and middle- and late-stage upregulated miRNAs were associated with muscle repair and regeneration (Figure S1). For example, in the early period the inflammatory miR-223 was upregulated whereas its expression subsided in the middle and late periods. In the middle period, miR126b (associated with angiogenic and chemokine signaling, Zhang et al., 2013), the miR-29 family (regulator of fibrosis and collagen expression), miR-21 (regulator of phosphatidylinositol 3-kinase [PI3K]/Akt signaling), and miR-31 and miR-206 (differentiating myoblasts) were upregulated, which was consistent with pathways observed to promote myogenic differentiation during the middle period (Cacchiarelli et al., 2010, Cacchiarelli et al., 2011). To gain insights into the consequences of differential expression of these dynamic miRNAs, we assessed pairwise correlations in expression patterns with their 3′ UTR mRNA targets. We observed multiple miRNA-mRNA targets that were reciprocally regulated during the time course, and hierarchical clustering of the correlations revealed three clusters (Figure 2A). The first cluster contained positively correlated miRNA-mRNA targets that were significantly upregulated in the early period (miR-709, miR-6538, miR-378 family with genes Atf3, Stat5b, Fas, and Btg2) and could be ascribed to functional classes related to immune regulation and DNA-damage cellular response. The second cluster was upregulated in the middle and late periods and was associated with myogenic growth and development. For example, we detected upregulation of miR-206, miR-205, miR-203, miR-1a, miR182, and miR-31 along with several target genes (Igf2, Igfbp5) that were previously identified as critical regulators of SC differentiation (Liu et al., 2012). The third cluster possessed the largest number of positively correlated miRNA-mRNA targets and was also upregulated in the middle period. This cluster was linked to transforming growth factor β (TGF-β) signaling and ECM and cytoskeletal remodeling, and contained miR-18a, miR-21, miR-335, miR34c, and miR-142 along with target genes Tgfbi, Anxa1, Sdcbp, Cav1, and Timp2.
Figure 2.
Dynamics of Noncoding RNAs after Severe Muscle Trauma
(A) Cross-correlation analysis of dynamic miRNAs and their mRNA targets. The heatmap is plotted as the Pearson correlation coefficient of the expression of 35 dynamic miRNAs (x axis) against the expression of their 200 predicted mRNA targets (y axis). A positive correlation coefficient is labeled in yellow and a negative correlation coefficient in blue.
(B) Expression heatmap of 124 lncRNAs through time from left to right. Heatmap is depicted as fold change of injured versus uninjured.
Long noncoding RNAs (lncRNAs) are a new class of transcripts that have recently been discovered and play diverse regulatory roles in muscle differentiation and disease (Neguembor et al., 2014, Cesana et al., 2011, Gong et al., 2015). Previously identified lncRNAs were intersected with the RNA-seq datasets and 2,444 lncRNAs were detected, with 124 lncRNAs exhibiting dynamic behavior. Figure 2B shows the differentially expressed lncRNAs clustered into three groups, and similar to the mRNA and miRNA datasets, we view upregulated lncRNAs involved in inflammatory and immune-related processes (Csf1r, Il1bos, Ly9, Treml1, CD68, and Ncf1) in the early time points followed by increases in expression of lncRNAs that were previously shown to be critical for myogenic differentiation (H19 and linc-MD1) in the middle and late periods (Dey et al., 2014). Uniquely, in the early period we also detected upregulated lncRNAs that were found in other injured muscle tissues (myocardial infarction-associated transcripts Mirt1 and Mirt2), suggesting a common regulatory scheme (Zangrado et al., 2014).
Temporal Transcriptional Signatures of Satellite Cells Reflect a Dynamically Changing Tissue Composition
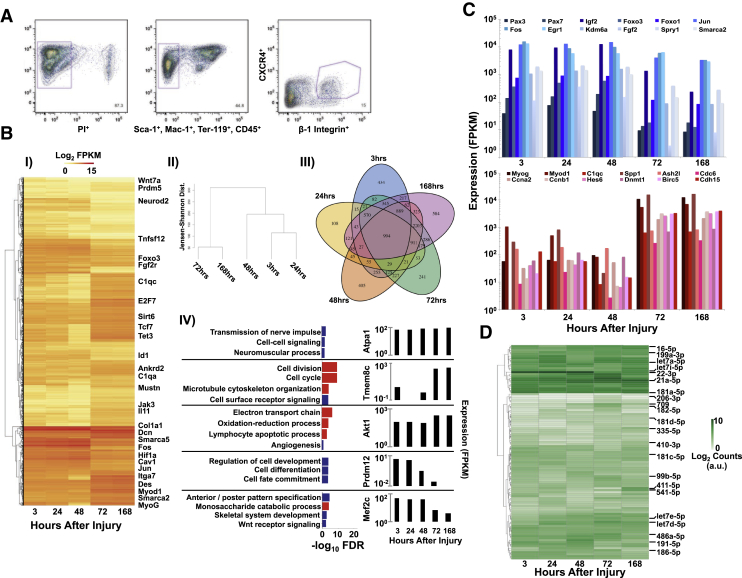
SCs from injured tissues were isolated using flow cytometry (Sca-1−, CD45−, Mac-1−, Ter-119−, β1-integrin+, Cxcr4+; Figure 3A) at time points associated with activation, proliferation, and differentiation (3, 24, 48, 72, and 168 hr), and RNA-seq and miRNA-seq was performed. A total of 17,636 mRNAs were detected and 9,953 genes were differentially expressed at one or more time points (Figure 3B). Of the time points sampled, 168 hr possessed the highest number of genes that were differentially expressed (584) compared with 434 genes for 3 hr, 108 for 24 hr, 405 for 48 hr, and 241 for 72 hr. These datasets were compared with previously published microarray datasets (Liu et al., 2013, Pallafacchina et al., 2010, Farina et al., 2012) of isolated SCs and excellent agreement was observed. Hierarchical clustering of the datasets through time revealed five clusters, and gene ontology (GO) analysis of the differentially expressed genes within each cluster illustrated significant pathway perturbations (Figure 3B). A set of genes that remained highly expressed over the time course were in the first cluster and were associated with ATP synthesis, ion channel activity, and regulation of glucose and insulin. The second and third clusters were low in expression from 3 to 48 hr and upregulated from 72 to 168 hr (Figure 3C) and contained multiple genes associated with myogenic differentiation (MyoD1, MyoG, Des, Ash2l, Hes6), cell-cycle regulation (Ccna2, Ccnb1, Dnmt1, Birc5), cytoskeletal proteins (Cdh15, Tmem8c/Myomaker), and mitochondrial metabolism. The fourth and fifth cluster displayed a relatively high level of expression from 3 to 48 hr that decreased from 72 to 168 hr and were associated with development, and contained markers of quiescent SCs (Pax7, Foxo3; Figure 3C).
Figure 3.
Temporal Coding and Noncoding Transcriptional Signatures of Sorted Satellite Cells Post Trauma Highlights Regenerative Transitions
(A) Representative isolation plot of FACS of satellite cells for Sca-1−, CD45−, Mac-1−, Ter-119−, β1-integrin+, and Cxcr4+. Purple gates indicate subpopulations containing satellite cells and numbers specify percentage of cells within gate.
(B) (I) Expression heatmap of 994 representative differentially expressed genes through time where yellow is low expression and red is high expression. (II) Dendrogram showing global hierarchical clustering of RNA-seq datasets separated by their Jensen-Shannon distance. (III) Venn diagram of unique and overlapping differentially expressed genes for the time points sampled. (IV) Left: bar graphs of false discovery rates for over-represented GO pathways derived from hierarchical clustering. Blue bars are upregulated GO terms and red bars are downregulated GO terms. Right: bar plots of RNA expression profiles of representative genes from each cluster are plotted to the right of each cluster through time from left to right. FPKM, fragments per kilobase of transcript per million mapped reads.
(C) Bar graphs of individual gene expression values through time from left to right.
(D) Expression heatmap of 107 differentially expressed miRNAs observed where white is low expression and green is high expression.
miRNA-seq was performed on the fluorescence-activated cell sorting (FACS)-sorted SCs, and 341 miRNAs were detected for at least one time point (Figure 3D). Of these, 107 miRNAs were differentially expressed (Figure S1) and many of the dynamic miRNAs mirrored expression patterns observed from whole tissues. For example, in the early period miR-22 was downregulated in both SCs and at the tissue level. miR-22 has recently been shown (Lu et al., 2015) to inhibit Hdac4 expression, which is a well-known negative regulator of myogenesis. During this time period, the miR-181 family (modulators of PI3K signaling and metabolic adaption needed for subsequent proliferation) and miR-191 (promotes cell migration via induction of Tgf-β signaling) also showed decreases in expression, which is consistent with transcriptional changes in metabolism, Tgf-β-signaling, and PI3K signaling observed in the tissue at that time. In the middle time periods (≥72 hr), changes in expression of let-7c (SMAD and Tgf-β signaling inhibitor), miR-16 (regulator of proliferation and cell-cycle genes, Liu et al., 2008), miR-148a (modulates fate commitment), miR-182 (repressor of negative regulators of cell-cycle genes), and several other members of the let-7 family (associated with cell cycling and self-renewal) were detected compared with low expression in the earlier time points (3–48 hr). miR-486a and miR-486b were also upregulated at these time points, and were previously shown to potentiate proliferation by targeting p85α, Igf1r, and Igf1, which is consistent with a decrease in expression of these genes at the same time points (72 and 168 hr). Multiple other miRNAs exhibited similar expression profiles (low expression from 3 to 48 hr and high expression from >72 hr) such as 21a, 125a, 127, 199a, 206, 411, and 541, consistent with previous findings for isolated SCs (Arnold et al., 2011).
Acute Muscle Trauma Stimulates Genome-wide Chromatin Remodeling
To further probe the regulation of the coding and noncoding transcriptional programs, we used chromatin immunoprecipitation followed by sequencing (ChIP-seq) to globally map the chromatin state of various cis-regulatory elements at nine time points. Validated antibodies for histone H3 lysine 4 trimethylation (H3K4me3), a modification associated with promoters, and H3K4me1 and H3K27ac, associated with poised and active enhancer regions (Ernst et al., 2011), respectively, were used to enrich chromatin. Each tissue from each time point was immunoprecipitated using the three antibodies, and ChIP-seq maps from the same antibody and time point were merged, resulting in 54 chromatin state maps covering >1.5 billion reads.
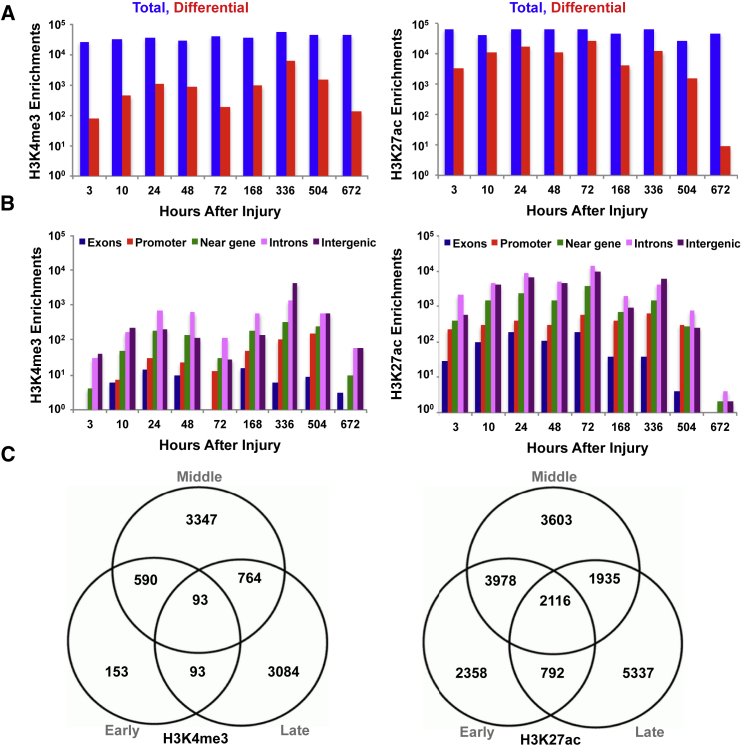
In the early time period 93,149 sites were enriched for H3K4me3 and 104,890 sites for the middle period, followed by 141,948 sites for the late period (Figure 4). Of these, 1,606 were differentially enriched in the early time points relative to the controls, 6,733 in the middle time points, and 6,121 in the late time points. To integrate sites that gained or lost the H3K4me3 modification during the time course with transcriptional activity (fragments per kilobase of transcript per million mapped reads [FPKM] > 1), we merged RNA-seq results with H3K4me3 enrichments. Approximately 58% of sites were found to associate with transcriptional activity (FPKM > 1) and 4,241 sites exhibited dynamics (acquisition or loss of the histone modification) through at least one stage (early to middle, middle to late, early to late).
Figure 4.
Distribution of Enriched Chromatin Sites across the Genome during In Vivo Muscle Regeneration
(A) Total and differential number of enriched sites for H3K4me3 and H3K27ac.
(B) Distribution of peaks across various genomic elements (promoter, intergenic, intron, near-gene: ± 2 kbp from transcriptional start site, exon).
(C) Shared and unique numbers of enriched chromatin sites corresponding to different genomic elements in (B).
In contrast to H3K4me3, which has previously been shown to be largely static during chromatin remodeling events (Garber et al., 2012), H3K4me1 and H3K27ac, which demarcate enhancer elements, were observed to be highly dynamic (Rada-Iglesias et al., 2011). H3K4me1 resides on both poised and active enhancers, whereas H3K27ac marks active sites (Creyghton et al., 2010). In the early time period 93,938 sites enriched for H3K4me1 were identified, followed by 106,353 sites for the middle period and 28,663 sites for the late period. H3K27ac showed similar behavior with 32,285 sites enriched in the early time period, 43,780 sites in the middle period, and 13,748 sites in the late period. The highest number of sites that acquired H3K27ac was found in the 72-hr period (28,178 sites) compared with 11,535 for the 48-hr stage and 4,067 for the 168-hr stage (Figure 4). In contrast to H3K4me3 enrichments, which were primarily found at or near transcriptional start sites, the enhancer marks H3K4me1 and H3K27ac were more broadly distributed across inter- and intragenic loci (Figure 4). Since H3K4me1 demarcates both poised and active enhancers and H3K27ac marks only active enhancers (Ostuni et al., 2013), the total number of enhancer elements (identified by H3K4me1+, H3K27ac+, and H3K4me3−) was determined, and the ratio of the enriched enhancers was quantified as active (H3K4me1+ and H3K27ac+) or poised (H3K4me1+ and H3K27ac−). A wide spectrum of binding patterns was observed for the two enhancer categories across the nine time points, with the majority of differential active enhancers occurring at intragenic and intergenic loci for all time periods. The highest number of differential active enhancers (injured+ and control−) was found for the middle period, with the largest number of active enhancers occurring in the 72-hr period.
Integrative Analysis of Enriched Enhancers Highlights Transient Recruitment of Different Types of Immune Cells after Muscle Trauma
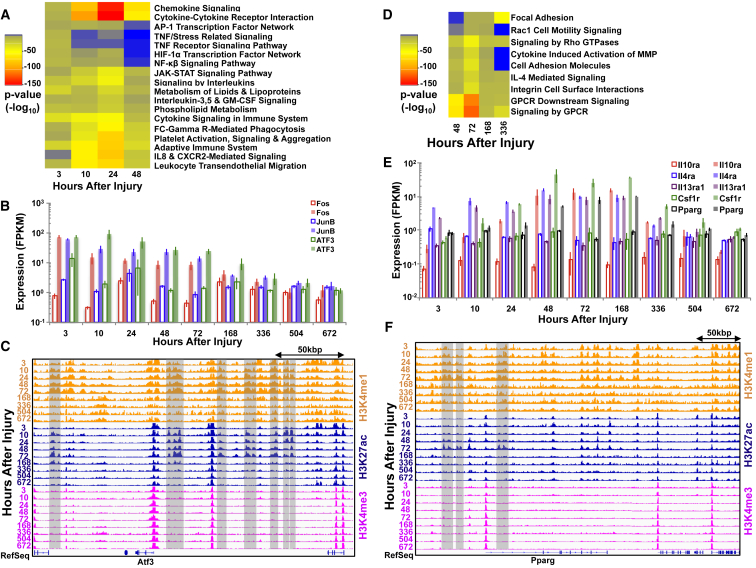
GO term analysis of the enriched differential enhancer peaks for the early period demonstrated over-represented terms, similar to the transcriptional groups (immune response, chemokine and cytokine signaling, inflammation, and metabolism of lipids and lipoproteins; Figure 5A). The result of these chromatin and transcriptional enrichments in the early period is consistent with upregulation of cytokines and chemokines (Il-6 and TNFα), and increases in expression of pleiotropic transcription factors such as components of the activator protein 1 (AP-1) complex (c-Fos, Atf3, JunB), STATs, NF-κβ, EGR, and PU.1 (Figures 5B and S2), which stimulate a permissive chromatin state and have previously been shown to induce SC activation genes such as Myod1, Myf6, and c-Myc (Toth et al., 2011).
Figure 5.
Chromatin Landscapes Are Immediately Modified after Severe Muscle Trauma and Reflect Recruitment of Different Types of Immune Cells
(A) Heatmap of p values for over-represented pathways derived from enriched H3K27ac peaks in the early time period (3–48 hr). AP-1, activator protein 1; CXCR2, C-X-C motif chemokine receptor 2; FC gamma R, Fc-γ receptor; GM-CSF, granulocyte macrophage colony-stimulating factor; HIF-1α, hypoxia-inducible factor 1α; IL8, interleukin-8; NF-κβ, nuclear factor κB; TNF, tumor necrosis factor.
(B) Bar graphs of individual gene expression values through time from left to right of immune-cell related transcripts upregulated in the early period (injured samples are colored and uninjured samples are uncolored). Error bars represent 1 SD.
(C) Normalized ChIP-seq tracks of H3K4me1, H3K27ac, and H3K4me3 profiles around the Atf3 gene. Enriched enhancer regions are highlighted in gray.
(D) Heatmap of over-represented pathways derived from enriched H3K27ac peaks in the middle time period (48–336 hr). GPCR, G-protein-coupled receptor; IL-4, interleukin-4; MMP, matrix metalloprotein.
(E) Histograms of individual gene expression values through time from left to right of anti-inflammatory related transcripts. Interleukin-4 receptor α (Il4ra), interleukin-10 receptor α (Il10ra), and interleukin-13 receptor α 1 (Il13ra1), colony-stimulating factor 1 receptor (Csf1r), and peroxisome proliferator-activated receptor γ (Pparg). Error bars represent 1 SD.
(F) Normalized ChIP-seq tracks of H3K4me1, H3K27ac, and H3K4me3 profiles around the Ppar-γ gene. Enriched enhancer regions are highlighted in gray.
In the middle time period (48–168 hr after injury), multiple genes associated with Tgf-β signaling were differentially expressed, such as several Smads (Figure S3), which have been shown to interact with chromatin remodeling complexes such as histone acetyltransferases p300 and CBP (CREB-binding protein) to induce H3K27 acetylation (Mullen et al., 2011). Consistent with this view, we observed the highest number of sites that acquired H3K27ac in the middle time period (Figure 4). GO annotation of the differential enhancer peaks for the middle period revealed multiple enrichments for anti-inflammatory cytokines, G-protein-coupled receptors, and the Rho family of small GTPases (Figures 5 and S4). This result is in agreement with the peak in expression of SRF target genes (Kuwahara et al., 2005) and anti-inflammatory cytokines interleukin-4, -10, and -13 (Il-4, Il-10, Il-13), which induce M2 macrophage polarization and are essential components for resolution of inflammation and tissue repair (Figure 5). Increases in expression of these factors were also mirrored by upregulation of the macrophage-derived matrix metalloproteinase 12 (Mmp12), which cleaves and inactivates CXC chemokines (Cxcl1, -2, -3, -5, and -8) and monocyte chemotactic proteins (Ccl2, -7, -8, and -13), inhibiting leukocyte flux to the injured site and abrogating the amount of proinflammatory molecules present in the injured tissue (Figure S5).
Chromatin State Transitions Associated with Activation of ECM Repair and Myogenic Regeneration
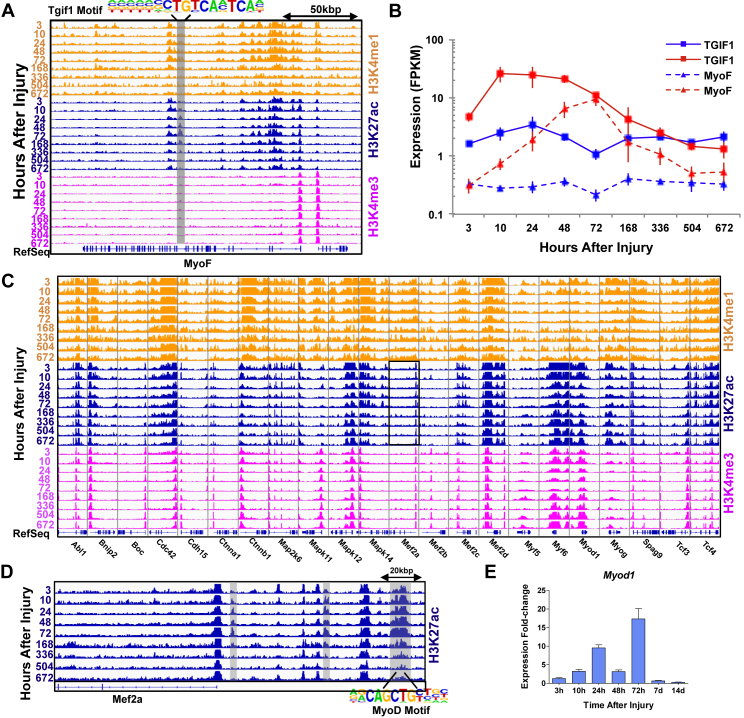
A primary determinant of successful muscle regeneration after injury is ECM remodeling (Calve et al., 2010), which provides a scaffold for proliferating SCs to migrate, differentiate, and fuse in the correct orientation. In the middle period, increases in expression and multiple enhancer enrichments were viewed for genes associated with ECM remodeling, such as components of the basement membrane (myoferlin, laminin, collagen VI genes, annexins Figures 6 and S5), glycoproteins, and Mmps. In addition, during the middle period chromatin remodeling and upregulation of genes associated with muscle development and architecture were also detected (Figure 6).
Figure 6.
Chromatin State Transitions Associated with Activation of Basement Membrane Repair and Myogenic Regeneration
(A) Normalized ChIP-seq tracks of H3K4me1, H3K27ac, and H3K4me3 profiles around the myoferlin (MyoF) gene. Enriched enhancer regions are highlighted in gray and corresponding enriched TF motif is labeled.
(B) Line graphs of individual gene expression values of MyoF and associated transcription factor (Tgif1) through time from left to right (injured samples are colored red and uninjured samples are colored blue). Error bars represent 1 SD.
(C) Normalized ChIP-seq tracks of H3K4me1, H3K27ac, and H3K4me3 profiles for a subset of myogenesis genes. The black box outlines the H3K27ac region, which is expanded in (D) and illustrates the Mef2a locus.
(D) Normalized H3K27ac ChIP-seq track highlighting enriched enhancers around the Mef2a gene. The enriched enhancer regions are highlighted in gray and corresponding enriched TF motif (MyoD) is labeled beneath the track.
(E) Bar graph of expression for MyoD. Error bars represent SEM.
To understand this critical step further, we conducted additional analysis of the differential active enhancer sites during the middle period by evaluating the number of enriched active enhancers that overlapped with MyoD and MyoG ChIP-seq data (Cao et al., 2010, The Mouse ENCODE Consortium, 2014, Mousavi et al., 2013). The largest increase in MyoD binding sites at active enhancers was found at 72 hr (Figure S6), which is consistent with the largest increase in expression (Figure 6E) and known association between MyoD and p300/CBP on E-box motifs of target genes during myogenic differentiation (Palacios and Puri, 2006). The greatest increase in MyoG binding sites at active enhancers was viewed at 336 hr, which agrees well with observation of late myogenic differentiation programs in that time period. Compiling the enriched enhancer sites coincident for MyoD or MyoG binding and performing GREAT (genomic regions enhancement of annotations tool) analysis revealed over-represented terms nominally associated with myogenic differentiation such as different types of growth factor signaling (insulin growth factor [Igf], fibroblast growth factor [Fgf], hedgehog), p38 mitogen-activated protein kinase (p38-MAPK), and PI3K signaling (Figure S6 and discussed further below).
Chromatin Regulation of PI3K Pathway Reflects Transition from Myoblast Proliferation and Differentiation
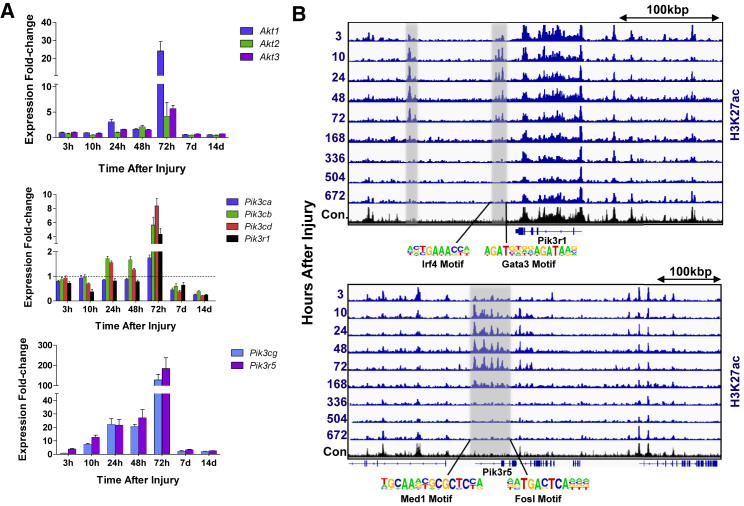
IGF signaling plays a crucial role in muscle repair and regeneration after injury, most notably during the transition between myoblast proliferation and differentiation whereby Igf2 activates the Akt pathway and targets bound by MyoD. In line with this observation, an increase in expression of Igf1 was detected in the early period (Pelosi et al., 2007), which subsided in the middle period when several Igfbps and Igf2 and members of the Akt pathway were observed to be upregulated along with miR-483 (which is embedded in the second intron of Igf2 and is an SRF target) (Figure 7A; Schiaffino and Mammucari, 2011). One component of the IGF pathway, the PI3K component, is essential for skeletal muscle regeneration and is composed of three primary classes (classes I, II, III) that are structurally and functionally distinct. The three class IA PI3K p110 catalytic subunits (α, β, and δ) are expressed in skeletal muscle (Matheny and Adamo, 2010) and of these subunits, p110α and p110β have been shown to positively influence myoblast proliferation and differentiation (Matheny et al., 2012, Matheny et al., 2015). In the early and middle periods following injury, we observed differential enhancer binding and increases in expression of PI3K catalytic subunits p110α (Pik3ca), p110β (Pik3cb), and p110δ (Pik3cd) as well as the PI3K p85α (Pik3r1) regulatory subunit for the injured muscle compared with the contralateral control (Figure 6A). Increases in expression and active enhancers were also viewed for class IB p110γ (Pik3cg) and its regulatory subunit p101 (Pik3r5) during the healing process, which are expressed abundantly in immune cells. TF binding analysis of the enriched enhancer regions for the regulatory subunit p85α showed motifs for signal mediators of Il-4 and the anti-inflammatory response, which is consistent with activation of pathways during this time period (Figure 7B).
Figure 7.
PI3K/Akt Activation Promotes Transition from Proliferation toward Myogenic Differentiation
(A) Bar graphs of individual gene expression values for various components of the PI3K and AKT pathways through time from left to right. Error bars represent SEM.
(B) Normalized ChIP-seq tracks of H3K27ac profiles showing differential enhancer activity of the PI3Kr1, PI3Kr5, and PI3Kr6 loci. Enriched enhancer regions are highlighted in gray and corresponding enriched TF motifs are labeled underneath.
The temporal increases in PI3K expression (and other positive myogenic regulatory pathways) between 24 and 72 hr post injury were also concomitant with increases in expression of negative feedback regulators such as Mg53 (disruptor of upstream IGF signaling via ubiquitin ligases, Yi et al., 2013), Xbp1 (Acosta-Alvear et al., 2007), and Usf1 (which compete with or co-occupy binding sites for MyoD and Mef2), and Id1, Id2, and Snai1 (which reduce the binding affinity of MyoD and Mef2; Soleimani et al., 2012). The negative regulators began to decrease in expression starting at 72 hr (Figure S7), further reinforcing the observation of myogenic differentiation at this time.
Discussion
Muscle recovery after traumatic injuries such as an occupational crush or blast suffered during military combat canonically induces a predisposition for additional injuries and chronic pain owing to incomplete regeneration of the tissue. Our understanding of how different factors translate to influence the chromatin architecture that regulates muscle regeneration has been limited. Herein, we used integrative genomic mapping technologies to profile coding and noncoding expression and the in vivo chromatin state of various cis-regulatory elements, and found successive waves of transcriptional and chromatin changes during the course of healing. To obtain broad, unbiased views of the diverse repair and regeneration pathways utilized by different cell types after trauma, we performed profiling of both whole, unfractionated muscle tissue and FACS-sorted SCs (Liu et al., 2015). Whole-tissue profiling enabled capture of signals emanating from multiple cell types and the capacity to profile multiple chromatin modifications from a single tissue without pooling tissues from multiple animals. The contralateral/uninjured tissue was used as the control for this study; however, recently (Rodgers et al., 2014) it was shown that systemic signals induced from acute trauma stimulate SCs within the contralateral muscle to undergo a transition to an alert state that primes their differentiation potential. Thus, while we normalized changes to the contralateral tissue, the normalization may not be reflective of an unperturbed muscle.
Pathway analysis of the time-clustered whole-tissue RNA-seq data revealed waves of transcription associated with proinflammatory and immune responses in the early period, followed by transcriptional signatures associated with myogenic differentiation and ECM remodeling in the middle and late time periods. Disruption of the muscle integrity by acute trauma also produced noncoding RNA (ncRNA) dynamics very similar to those observed in other muscle myopathies (Neguembor et al., 2014, Cesana et al., 2011, Eisenberg et al., 2007), suggesting the modulation of common molecular pathways. miRanda-mirSVR (Betel et al., 2010) was used to study the interaction of dynamic miRNA-mRNA pairs and whether their expression patterns changed concomitantly with time. We identified 200 miRNA-mRNA mutually dynamic relationships and clustered the pairs into three categories of immune regulation, TGF-β signaling, and ECM and cytoskeletal remodeling followed by myogenic differentiation. These three clusters highlight a temporal regulatory program whereby infiltrating immune cells release signaling molecules that trigger SC activation, followed by a transition whereby the activated progenitors proliferate and are repressed from differentiating by TGF-β signaling and changes to the surrounding matrix. These repressive cues were extended via other regulatory mechanisms (see below) and began to subside at 72 hr when a new class of ncRNAs (miR-206, miR-205, miR-203, miR-1a, miR182, miR-31, H19, and linc-MD1) increased in expression. Similar to the mRNA and miRNA-seq results, three clusters of differentially expressed lncRNAs were detected. Interestingly, H19 (which was upregulated in the middle and late periods) and another lncRNA called lncMyoD (which was upregulated in the early and middle periods) bind mRNA IGF2 binding proteins, suggesting that induction and stabilization of this pathway may be regulated temporally via different combinations of lncRNAs. Since muscle repair and regeneration utilizes many feedforward and feedback loops, tightly regulated expression patterns across multiple levels may facilitate a precise way to prevent extrinsic signal propagation from adjacent tissues that are also regenerating or responding to the injury, reinforce or tune noisy expression patterns, and perhaps facilitate a metabolically efficient mechanism to quickly respond after injury.
RNA-seq of SCs reinforced transcriptional dynamics observed from whole tissues, whereby in the early period, immune-response and chemokine transcripts were upregulated, followed by proliferation and differentiation during the middle time period. Several TFs associated with quiescent cells (Pax3, Pax7, Foxo3, Spry1) and immune response (c-Jun, c-Fos, Atf3, Egr1) exhibited downregulation at the same time that genes associated with cell-cycle progression (Ccna2, Ccnb1, Dnmt1, Birc5), differentiation, and fusion (MyoG, Hes6, Tmem8c, Cdh15) were upregulated (>72 hr). During the same time period, we also observed a switch from high to low expression of lysine demethylase 6A (Kdm6a or Utx), which regulates removal of the repressive H3K27me3chromatin mark (Faralli et al., 2016), to low to high expression of a component of a histone methyltransferase complex (Ash2l). Ash2l is part of the Trithorax complex that specifically methylates the fourth lysine residue of histone H3 (H3K4) and is targeted by Pax7 to activate Myf5 target genes (Almada and Wagers, 2016). Combining these dynamics suggests that in the early time period, upregulated TFs and chromatin remodelers help to establish a more accessible chromatin state at myogenic loci, which is consistent with the increase in enrichments of H3K4me3, H3K4me1, and H3K27ac. Moving into the middle period, as stress-responsive and other immune-activated transcripts decrease in expression, another set of TFs and chromatin remodelers is then upregulated and titrated via miRNAs (see below) to enact appropriate satellite cell proliferation and differentiation. miRNA-seq on the FACS-sorted SCs demonstrated that the programs of activation, proliferation, and subsequent differentiation are tightly controlled. For the time points linked with activation and mobilization (3–48 hr), several miRNAs were downregulated, such as miR-22, which is activated by the transcription factors AP-1 and NF-κB. Since miR-22 inhibits expression of Hdac4 and targets c-myc, which also suppresses MyoD-initiated myogenic differentiation and promotes proliferation, a regulatory program can be inferred whereby stimuli from infiltrating immune cells induce activation of TFs that promote the progenitors to activate, mobilize, and begin to proliferate but are repressed from differentiating. Other post-transcriptional factors such as miR-181, miR-191, and miR-222 appear to reinforce this program in the early time periods and as inflammation and immune-stimulated signatures drop in expression, another program begins around 72 hr via increases of expression of regulators of cell-cycle genes (miR-16, miR-182, miR-486) and myogenic differentiation (let-7 family, miR-206).
The efficacy of myogenic transcription factors to initiate and maintain muscle gene expression programs depends critically on the chromatin state (Blais et al., 2005, Blum et al., 2012) of their targets as well as their interactions with each other (Liu et al., 2014, Molkentin et al., 1995) and other TFs. Quantifying the genome-wide changes in histone modifications revealed that a robust induction of chromatin remodeling occurs at a genome-wide level after muscle trauma, and cis-regulatory elements such as promoters were largely invariant when compared with regions demarcated by enhancer marks (H3K4me1, H3K27ac). Immediately after injury, the enriched enhancer regions were associated with canonical mediators of early stress, immunity, and growth factor responses, which was consistent with the upregulation of cytokines and TFs such as AP-1 (Fos/Jun), SRF, NF-κβ, Egr, and Stats. The detection of these transcripts and enriched enhancer sites was expected since an appropriate inflammatory response is essential for functional recovery after muscle injury. A significant fraction of the enriched chromatin sites correlated with transcriptional dynamics and was most likely attributable to infiltrating monocytes that secrete and respond to cytokines and chemokines. These early immune-related changes were serially followed by changes in expression and chromatin remodeling of genes associated with anti-inflammatory macrophages (Arnold et al., 2007) and activation and proliferation of SCs. During this period, upregulation of Smads and Tgf-β signaling was observed along with increases in expression of TFs such as Tead4 (Benhaddou et al., 2012), Runx1 (Umansky et al., 2015), and MyoD (Mullen et al., 2011). Smads have been shown to interact with chromatin remodeling complexes such as histone acetyltransferases p300 and CBP to induce H3K27 acetylation (Pouponnot et al., 1998), which was consistent with the observation that the highest number of sites that acquired H3K27ac was in the middle time period. In addition, MyoD has been shown to associate with p300 and p300/CBP on E-box motifs of target genes (Puri et al., 1997), and interactions between Smads and Tgf-β signaling have been shown to alter myogenic differentiation kinetics, suggesting a cooperative interaction between these different TFs and chromatin remodeling complexes during this phase of the injury response. Integrating these results reveals a transient shift in the balance of pro- and anti-inflammatory cytokine programs (high expression of Il-6 and low expression of Tgf-β in the early period to low Il-6 expression and high expression of Tgf-β in the middle period), which has previously been shown to promote the recruitment and differentiation of different types of cells that potentiate muscle repair (neutrophils, M1 macrophages, TH17 cells for the early period, and M2 macrophages and Treg cells for the middle period), influences chromatin state and expression programs associated with proliferation and restraint of myogenic differentiation (Dionyssiou et al., 2013).
IGF and PI3K signaling in regenerating muscle was tightly regulated across multiple levels and temporally, especially during the transition between myoblast proliferation and differentiation. In the early period, IGF1 was upregulated and quickly followed by increases in expression and chromatin remodeling of myogenic negative regulators (Mg53, Hdacs, Xbp1, Usf1) that alter how myogenic regulatory factors bind to lineage-specific genes. Starting at 72 hr, IGF1 and negative regulators began to decrease in expression while positive differentiation genes increased in expression, which is consistent with previous observations (Braun and Gautel, 2011, Buckingham and Rigby, 2014, Pelosi et al., 2007, Schiaffino and Mammucari, 2011, Matheny and Adamo, 2010). This switch is consistent with the view that miRs 206, 29, and 1a bind to Hdacs that reduce the binding affinity of MyoD and Mef2, permitting these TFs to initiate myogenic regeneration. Given that IGF-activated Akt1 modulates p300 to associate with MyoD (Serra et al., 2007), it is intriguing to hypothesize how other parallel regulatory schemas of this signaling pathway alter fundamental changes in TF binding and chromatin states that drive muscle regeneration after severe trauma. Collectively, these experiments imply that muscle repair and regeneration uses different sets of transcriptional programs, ncRNAs, and combinations of TFs as well as chromatin remodeling factors to precisely execute stage-specific gene expression programs. We envision that a wider combinatorial interrogation of such a dataset can represent a valuable resource to extend the networks acting in such a complex microenvironment like the cis-regulatory modules engaged by TFs, miRNAs, and lncRNAs.
Experimental Procedures
Animals and Traumatic Injury Model
Mice were cared for in accordance with the Guide for the Care and Use of Laboratory Animals in a facility accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Male C57BL/6J mice (10 weeks of age, 24–27 g) were obtained from The Jackson Laboratory. Prior to administration of the freeze injury, mice were anesthetized, and the TA muscle was exposed via a small incision. Freeze injury was administered to the left hindlimb by applying a 6-mm diameter steel probe (cooled to −70°C) to the belly of the TA muscle (directly below incision site) for 10 s. Following injury, the skin incision was closed using absorbable sutures (Ethicon). Five mice were euthanized at each time point post injury (3, 10, 24, 48, 72, 168, 336, 504, and 672 hr) via CO2 inhalation (2 L/min), thoracotomy, and exsanguination. TA muscles were removed from the injured and contralateral limb and weighed, and approximately half of the tissue was processed further (area delineated by the injury).
Chromatin Isolation and Sequencing Library Preparation
Each frozen tissue was thawed for at least 30 min and homogenized (Tissue Ruptor, Qiagen), and the cells lysed for at least 10 min. Next, nuclei were isolated, resuspended, and lysed for at least 10 min. The chromatin was then sheared using a Branson sonifier into fragments of size range 150–700 bp. The sheared chromatin was then incubated overnight at 4°C with validated antibodies (Modified Histone Peptide Array, Active Motif) H3K4me3 (Millipore catalog #07-743), H3K27ac (Active Motif #39133), and H3K4me1 (Abcam #ab8895) using constant agitation, and co-immunoprecipitated using a mixture of Protein A and Protein G Dynabeads (Life Technologies) for 2 hr at 4°C with constant agitation. The immunoprecipitated chromatin was then washed, the crosslinks were reversed and treated with Proteinase K, and DNA was purified. Approximately 10 ng of isolated DNA was end-repaired (End-It DNA End-Repair Kit, Epicenter), extended, and A-tailed (New England Biolabs), and ligated to sequencing adaptor oligos (Illumina). The adaptor-modified library was then amplified by PFU Ultra II Hotstart Master Mix (Agilent) and size-selected to a range of 300–600 bp prior to sequencing. Libraries were pooled and sequenced using an Illumina Genome Analyzer IIx using 44-bp single end reads to achieve approximately 20 M aligned reads per sample.
mRNA and Small RNA Sequencing Library Preparation and qPCR
The extracted tissue was thawed and then homogenized for 30 s (Tissue Ruptor, Qiagen), and total RNA was isolated using the miRNeasy Mini Kit (Qiagen) as per the manufacturer's instructions. Total RNA from sorted SCs were extracted using the miRNeasy Micro Kit (Qiagen), and RNA concentration and integrity for both types of samples (tissues and sorted cells) were measured with a Nanodrop spectrophotometer (Nanodrop 2000c) and Bioanalyzer (Agilent 2100). One microgram of isolated total RNA from tissues was used to produce strand-specific cDNA libraries using the Truseq (Illumina) protocol, as per the manufacturer's instructions. Ten nanograms of isolated total RNA from SCs was used to produce cDNA libraries using the SmartSeq4 protocol (Clontech), as per the manufacturer's instructions. Individual libraries from tissues were pooled and sequenced using 12 lanes of 76-bp paired-end reads on an Illumina Genome Analyzer IIx to an average depth of 50 M reads per library. Individual libraries for the sorted SCs were pooled and sequenced using 76-bp paired-end reads on an Illumina NextSeq to an average depth of 50 M reads per library.
For the production of miRNA-seq libraries 500 ng of total RNA from the tissues and 100 ng of total RNA from the sorted cells was used, and libraries were prepared according to the specifications of the Truseq small-RNA kit (Illumina). Each library was then pooled and sequenced on a MiSeq single-ended 35-bp run (Illumina) to >1 M reads per library.
Acknowledgments
The authors thank Chet Beal for assistance with artwork, Sara Chauvin, Patrick Boyle, Fontina Kelley, and the Broad Institute Genomics Platform for sequencing and technical assistance, Tara Boettcher for assistance with ChIP and sequencing library preparation, Mary Abdalla, Christina Zook, and Alyssa Geddis for technical assistance with PCR, and Darrell O. Ricke and Michael J. Ziller for insightful discussions. This material is based upon work supported under Air Force Contract No. FA8721-05-C-0002 and/or FA8702-15-D-0001. Opinions, interpretations, recommendations, and conclusions are those of the authors and are not necessarily endorsed by the United States Government. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the U.S. Air Force. C.C. was supported by an appointment to the postgraduate research participation program at the US Army Research Institute of Environmental Medicine, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US Army Medical Research and Materiel Command. The views, opinions, and/or findings of this report are those of the authors and should not be construed as an official US Department of the Army position, policy, or decision unless so designated by other official documentation.
Published: October 20, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.09.009.
Supplemental Information
References
- Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N.H., Arias C., Lennon C.J., Kluger Y., Dynlacht B.D. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Aguilar C.A., Shcherbina A., Ricke D.O., Pop R., Carrigan C.T., Gifford C.A., Urso M.L., Kottke M.A., Meissner A. In vivo monitoring of transcriptional dynamics after lower-limb muscle injury enables quantitative classification of healing. Sci. Rep. 2015;5:13885. doi: 10.1038/srep13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almada A.E., Wagers A.J. Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat. Rev. Mol. Cell Biol. 2016;17:267–279. doi: 10.1038/nrm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold L., Henry A., Poron F., Baba-Amer Y., van Rooijen N., Plonquet A., Gherardi R.K., Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into anti-inflammatory macrophages to support myogenesis. J. Exp. Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C.P., Tan R., Zhou B., Yue S.B., Schaffert S., Biggs J.R., Doyonnas R., Lo M.C., Perry J.M., Renault V.M. MicroRNA programs in normal and aberrant stem and progenitor cells. Genome Res. 2011;21:798–810. doi: 10.1101/gr.111385.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J., Bowman C., Kluger Y., Dynlacht B.D. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. USA. 2011;108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurora A.B., Olson E.N. Immune modulation of stem cells and regeneration. Cell Stem Cell. 2014;15:14–25. doi: 10.1016/j.stem.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhaddou A., Keime C., Ye T., Morlon A., Michel I., Jost B., Mengus G., Davidson I. Transcription factor TEAD4 regulates expression of myogenin and the unfolded protein response genes during C2C12 cell differentiation. Cell Death Differ. 2012;19:220–231. doi: 10.1038/cdd.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger C.F., Wang Y.X., Dumont N.A., Rudnicki M.A. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 2013;14:1062–1072. doi: 10.1038/embor.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D., Koppal A., Agius P., Sander C., Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R., Vethanthum V., Bowman C., Rudnicki M., Dynlacht B.D. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev. 2012;26:2763–2779. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio A., Palacios D. Chromatin signaling in muscle stem cells: interpreting the regenerative microenvironment. Front. Aging Neurosci. 2015;7:1–17. doi: 10.3389/fnagi.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011;12:349–361. doi: 10.1038/nrm3118. [DOI] [PubMed] [Google Scholar]
- Buckingham M., Rigby P.W.J. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- Burzyn D., Kuswanto W., Kolodin D., Shadrach J.L., Cerletti M., Jang Y., Sefik E., Tan T.G., Wagers A.J., Benoist C., Mathis D. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–1295. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacchiarelli D., Martone J., Girardi E., Cesana M., Incitti T., Morlando M., Nicoletti C., Santini T., Sthandier O., Barberi L. MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 2010;12:341–351. doi: 10.1016/j.cmet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Cacchiarelli D., Incitti T., Martone J., Cesana M., Cazzella V., Santini T., Sthandier O., Bozzoni I. miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO Rep. 2011;12:136–141. doi: 10.1038/embor.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calve S., Odelberg S.J., Simon H.G. A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev. Biol. 2010;344:259–271. doi: 10.1016/j.ydbio.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., MacQuarrie K.L., Davison J., Morgan M.T., Ruzzo W.L. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2005;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B.K., Pfeifer K., Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionyssiou M.G., Salma J., Bevzyuk M., Zakharyan L., McDermott J.C. Kruppel-like factor 6 (KLF6) promotes cell proliferation in skeletal myoblasts in response to TGF-β/Smad3 signaling. Skeletal Muscle. 2013;3:7. doi: 10.1186/2044-5040-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg I., Eran A., Nishino I., Moggio M., Lamperti C., Amato A.A., Lidov H.G., Kang P.B., North K.N., Mitrani-Rosenbaum S. Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. USA. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faralli H., Wang C., Nakka K., Benyoucef A., Sebastian S., Zhuang L., Chu A., Palii C.G., Liu C., Camellato B. UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J. Clin. Invest. 2016;126:1555–1565. doi: 10.1172/JCI83239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina N.H., Hausberg M., Betta N.D., Pulliam C., Srivastava D., Cornelison D., Olwin B.B. A role for RNA post-transcriptional regulation in satellite cell activation. Skeletal Muscle. 2012;2:21. doi: 10.1186/2044-5040-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M., Yosef N., Goren A., Raychowdhury R., Thielke A., Guttman M., Robinson J., Minie B., Chevrier N., Itzhaki Z. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell. 2012;47:1–13. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordani L., Puri P.L. Epigenetic control of skeletal muscle regeneration: integrating genetic determinants and environmental changes. FEBS J. 2013;280:4014–4025. doi: 10.1111/febs.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Li Z., Ramanujan K., Clay I., Zhang Y., Lemire-Brachat S., Glass D.J. A long noncoding RNA, LincMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell. 2015;34:181–191. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Heredia J.E., Mukundan L., Chen F.M., Mueller A.A., Deo R.C., Locksley R.M., Rando T.A., Chawla A. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 2013;153:376–388. doi: 10.1016/j.cell.2013.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.K., Lee Y.S., Sivaprasad U., Malhotra A., Dutta A. Muscle specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S., Gillespie M.A., Rudnicki M.A. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell. 2008;2:22–31. doi: 10.1016/j.stem.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Kuwahara K., Barrientos T., Pipes G.C.T., Li S., Olson E.N. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol. Cell. Biol. 2005;25:3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Fu H., Sun F., Zhang H., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Williams A.H., Maxeiner J.M., Bezprozvannaya S., Shelton J.M., Richardson J.A., Bassel-Duby R., Olson E.N. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J. Clin. Invest. 2012;122:2054–2065. doi: 10.1172/JCI62656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Cheung T.H., Charville G.W., Hurgo B.M., Leavitt T., Shih J., Brunet A., Rando T.A. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Nelson B.R., Bezprozvannaya S., Shelton J.M., Richardson J.A., Bassel-Duby R., Olson E.N. Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc. Natl. Acad. Sci. USA. 2014;111:4109–4114. doi: 10.1073/pnas.1401732111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Cheung T.H., Charville G.W., Rando T.A. Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc. 2015;10:1612–1624. doi: 10.1038/nprot.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., You R., Yuan X., Yang T., Samuel E.L., Marcano D.C., Sikkema W.K., Tour J.M., Rodriguez A., Kheradmand F., Corry D.B. The microRNA miR-22 inhibits the histone deacetylase HDAC4 to promote TH17 cell-dependent emphysema. Nat. Immuno. 2015;16:1185–1194. doi: 10.1038/ni.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny R.W., Adamo M.L. PI3K p110 alpha and p110 beta have differential effects on Akt activation and protection against oxidative stress-induced apoptosis in myoblasts. Cell Death Differ. 2010;17:677–688. doi: 10.1038/cdd.2009.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny R.W., Lynch C.M., Leandry L.A. Enhanced Akt phosphorylation and myogenic differentiation in PI3K p110beta-deficient myoblasts is mediated by PI3K p110alpha and mTORC2. Growth Factors. 2012;30:367–384. doi: 10.3109/08977194.2012.734507. [DOI] [PubMed] [Google Scholar]
- Matheny R.W., Riddle-Kottke M.A., Leandry L.A., Lynch C.M., Abdalla M.N., Geddis A.V., Piper D.R., Zhao J.J. Role of phosphoinositide 3-OH kinase p110beta in skeletal myogenesis. Mol. Cell. Biol. 2015;35:1182–1196. doi: 10.1128/MCB.00550-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J.D., Black B.L., Martin J.F., Olson E.N. Cooperative activation of muscle gene expression by MEF2 and myogenic BHLH proteins. Cell. 1995;83:2–14. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Mousavi K., Zare H., Dell'orso S., Grontved L., Gutierrez-Cruz G., Derfoul A., Hager G.L., Sartorelli V. eRNAs promoted transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A.C., Orlando D.A., Newman J.J., Lovén J., Kumar R.M., Bilodeau S., Reddy J., Guenther M.G., DeKoter R.P., Young R.A. Master transcription factors determine cell-type-specific responses to TGFβ signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neguembor M.V., Jothi M., Gabellini D. Long noncoding RNAs, emerging players in muscle differentiation and disease. Skeletal Muscle. 2014;4:1–12. doi: 10.1186/2044-5040-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R., Piccolo V., Barozzi I., Polletti S., Termanini A., Bonifacio S., Curina A., Prosperini E., Ghisletti S., Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Palacios D., Puri P.L. The epigenetic network regulating muscle development and regeneration. J. Cell Physiol. 2006;207:1–11. doi: 10.1002/jcp.20489. [DOI] [PubMed] [Google Scholar]
- Pallafacchina G., Francois S., Regnault B., Czarny B., Drive V., Cumano A., Montarras D., Buckingham M. An adult tissue-specific stem cell in its niche: a agene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 2010;4:77–91. doi: 10.1016/j.scr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Pelosi L., Giacinti C., Nardis C., Borsellino G., Rizzuto E., Nicoletti C., Wannenes F., Battistini L., Rosenthal N., Molinaro M., Musarò A. Local expression of IGF-1 accelerates muscle regeneration by rapidly modulating inflammatory cytokines and chemokines. FASEB J. 2007;21:1393–1402. doi: 10.1096/fj.06-7690com. [DOI] [PubMed] [Google Scholar]
- Pouponnot C., Jayaraman L., Massague J. Physical and functional interaction of SMADs and p300/CBP. J. Biol. Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- Puri P.L., Avantaggiati M.L., Balsano C., Sang N., Graessmann A., Giordano A., Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J.T., King K.Y., Brett J.O., Cromie M.J., Charville G.W., Maguire K.K., Brunson C., Mastey N., Liu L., Tsai C.R. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to GAlert. Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharner J., Zammit P.S. The muscle satellite cell at 50: the formative years. Skeletal Muscle. 2011;1:28. doi: 10.1186/2044-5040-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S., Mammucari C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skeletal Muscle. 2011;1:1–14. doi: 10.1186/2044-5040-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra C., Palacios D., Mozzetta C., Forcales S.V., Morantte I., Ripani M., Jones D.R., Du K., Jhala U.S., Simone C., Puri P.L. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol. Cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani V.D., Yin H., Jahani-Asl A., Ming H., Kockx C.E.M., van Ijcken W.F.J., Grosveld F., Rudnicki M.A. Snail regulates Myod binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol. Cell. 2012;47:457–468. doi: 10.1016/j.molcel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Mouse ENCODE Consortium A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball J.G. Inflammatory processes in muscle injury and repair. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R345–R353. doi: 10.1152/ajpregu.00454.2004. [DOI] [PubMed] [Google Scholar]
- Toth K.G., McKay B.R., De Lisio M., Little J.P., Tarnopolsky M.A., Parise G. IL-6 induced Stat3 signalling is associated with the proliferation of human muscle satellite cells following acute muscle damage. PLoS One. 2011;6:e17392. doi: 10.1371/journal.pone.0017392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umansky K.B., Gruenbaum-Cohen Y., Tsoory M., Feldmesser E., Goldenberg D., Brenner O., Groner Y. Runx1 transcription factor is required for myoblast proliferation during muscle regeneration. PLoS Genet. 2015;11:e1005457. doi: 10.1371/journal.pgen.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G.L., Summan M., Gao X., Chapman R., Hulderman T., Simeonova P.P. Mechanisms of skeletal muscle injury and repair revealed by gene expression studies in mouse models. J. Physiol. 2007;582:825–841. doi: 10.1113/jphysiol.2007.132373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J.S., Park J.S., Ham Y.M., Nguyen N., Lee N.R., Hong J., Kim B.W., Lee H., Lee C.S., Jeong B.C. MG53-induced IRS-1 ubiquitination negatively regulates skeletal myogenesis and insulin signalling. Nat. Commun. 2013;4:2354. doi: 10.1038/ncomms3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangrado J., Zhang L., Vausort M., Maskali F., Marie P.Y., Wagner D.R., Devaux Y. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics. 2014;15:460. doi: 10.1186/1471-2164-15-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang P., Sun T., Li D., Xu X., Rui Y., Li C., Chong M., Ibrahim T., Mercatali L. miR-126 and miR-126∗ repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat. Cell Biol. 2013;15:284–294. doi: 10.1038/ncb2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.