Summary
Poly(A) tail length and mRNA deadenylation play important roles in gene regulation. However, how they regulate embryonic development and pluripotent cell fate is not fully understood. Here we present evidence that CNOT3-dependent mRNA deadenylation governs the pluripotent state. We show that CNOT3, a component of the Ccr4-Not deadenylase complex, is required for mouse epiblast maintenance. It is highly expressed in blastocysts and its deletion leads to peri-implantation lethality. The epiblast cells in Cnot3 deletion embryos are quickly lost during diapause and fail to outgrow in culture. Mechanistically, CNOT3 C terminus is required for its interaction with the complex and its function in embryonic stem cells (ESCs). Furthermore, Cnot3 deletion results in increases in the poly(A) tail lengths, half-lives, and steady-state levels of differentiation gene mRNAs. The half-lives of CNOT3 target mRNAs are shorter in ESCs and become longer during normal differentiation. Together, we propose that CNOT3 maintains the pluripotent state by promoting differentiation gene mRNA deadenylation and degradation, and we identify poly(A) tail-length regulation as a post-transcriptional mechanism that controls pluripotency.
Keywords: pluripotent state, pre-implantation development, embryonic stem cell, mRNA deadenylation
Highlights
-
•
CNOT3 is required for mouse epiblast maintenance during early development
-
•
CNOT3 C-terminal domain is necessary for the maintenance of the pluripotent state
-
•
CNOT3 promotes differentiation gene mRNA deadenylation and degradation
-
•
mRNA poly(A) tail regulation plays a critical role in pluripotency
In this article, Hu and colleagues showed that CNOT3, a component of the Ccr4-Not deadenylase complex, is required for the maintenance of the pluripotent state in mouse epiblast and embryonic stem cells. They found that CNOT3 promotes differentiation gene mRNA deadenylation and degradation, and post-transcriptionally regulates the gene expression program in pluripotent cells.
Introduction
Pluripotency is defined as the ability of a single cell to give rise to all the cell types formed by the three germ layers (Posfai et al., 2014). It is a unique property of the epiblast cells in early embryos, and can also be captured in embryonic stem cells (ESCs) in culture (Boroviak and Nichols, 2014, Martello and Smith, 2014). ESCs provide an invaluable platform from which to investigate the molecular mechanisms that regulate pluripotency. It has been shown that the pluripotent state in ESCs is controlled by a combination of signal-transduction pathways, transcription factors, epigenetic modifiers, RNA binding proteins, and regulatory RNA molecules (Dejosez and Zwaka, 2012, Hackett and Surani, 2014, Martello and Smith, 2014). Although extensive research has focused on the signaling, transcriptional, and epigenetic regulation in ESCs (Ng and Surani, 2011, Wang et al., 2014, Young, 2011), how post-transcriptional mechanisms can influence the ESC gene expression program and the pluripotent state has only begun to be revealed. Indeed, recent studies showed that post-transcriptional regulation, such as alternative splicing (Gabut et al., 2011), alternative polyadenylation (Lackford et al., 2014), RNA export (Wang et al., 2013), and RNA modification (Geula et al., 2015), plays critical roles in ESC maintenance and pluripotency (Wright and Ciosk, 2013, Ye and Blelloch, 2014).
Most eukaryotic mRNAs are polyadenylated, and poly(A) tail lengths are important for post-transcriptional gene regulation (Eckmann et al., 2011, Norbury, 2013). It has been shown that mRNA deadenylation is a main determinant of mRNA poly(A) tail length, and can influence mRNA half-life and/or translation efficiency in different cellular contexts (Eckmann et al., 2011, Norbury, 2013). A recent study showed that the poly(A) tail length appears to have a more profound impact on mRNA stability than mRNA translation in cells with active transcription (Subtelny et al., 2014).
To investigate whether and how mRNA poly(A) tail length and deadenylation regulates embryonic development and pluripotent cell fate, we focused on the Ccr4-Not complex. Ccr4-Not is the main mRNA deadenylase complex in eukaryotic cells that shortens mRNA poly(A) tails (Collart and Panasenko, 2012, Shirai et al., 2014, Xu et al., 2014). It has been implicated in various physiological and developmental processes, such as spermatogenesis (Berthet et al., 2004), heart development (Neely et al., 2010), energy metabolism (Morita et al., 2011), B cell differentiation (Inoue et al., 2015), osteoporosis (Watanabe et al., 2014), reprogramming (Kamon et al., 2014), and ESC self-renewal (Hu et al., 2009, Zheng et al., 2012). However, its role in early development and the mechanism by which it regulates pluripotency remain to be fully elucidated. In this study, we focus on the CNOT3 subunit in the complex and present evidence that the poly(A) tail-length regulation by CNOT3 serves as a post-transcriptional regulatory mechanism governing early development and ESC maintenance.
Results
Cnot3 Expression Is Upregulated in the Blastocysts
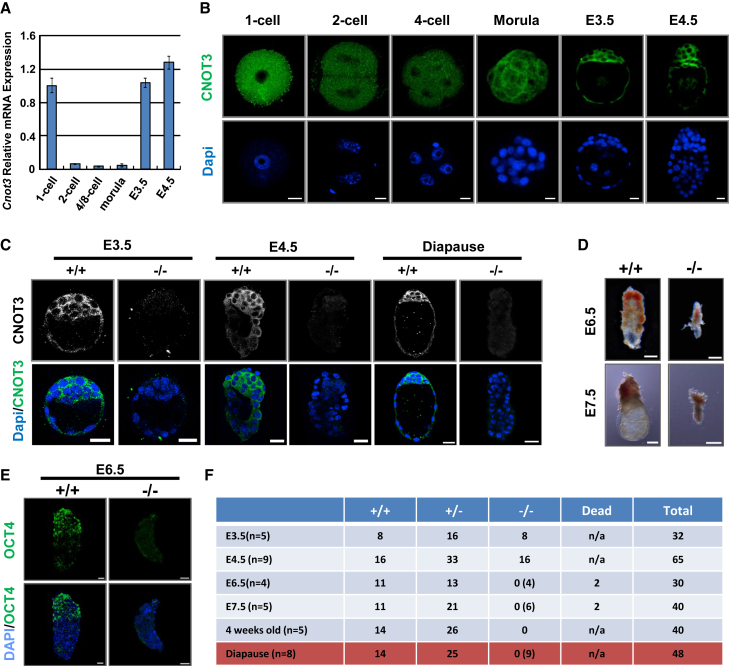
The Ccr4-Not complex is the main deadenylase complex in eukaryotic cells and regulates mRNA poly(A) tail length. To test the roles of Ccr4-Not and mRNA poly(A) tail length in mouse embryonic development, we focused on the Cnot3 subunit because its silencing resulted in prominent phenotypic and gene expression changes in ESCs (Zheng et al., 2012). We first examined Cnot3 expression during pre-implantation development. By qRT-PCR, we found that Cnot3 mRNA level is high in one-cell embryos, presumably from maternal expression, and is elevated again in blastocysts during pre-implantation development (Figure 1A). Immunofluorescence staining showed that Cnot3 protein expression is in agreement with the above pattern (Figure 1B). Furthermore, Cnot3 is enriched in the inner cell mass at the blastocyst stage. It predominantly localizes in the cytoplasm (Figure 1B), consistent with the notion that it is a part of the Ccr4-Not complex that regulates mRNAs.
Figure 1.
Cnot3 Is Required for Early Embryonic Development
(A and B) Cnot3 expression in pre-implantation embryos. Expression was determined by qRT-PCR and plotted as mean ± SEM from three independent experiments (A) and immunofluorescence staining (B). Scale bar, 20 μm.
(C) Immunofluorescence staining of CNOT3 in WT and Cnot3 deletion embryos at the indicated developmental stages. Scale bars, 20 μm.
(D) Morphology of WT and Cnot3 deletion embryos at E6.5 and E7.5. Scale bars, 100 μm.
(E) Morphology and OCT4 expression of Cnot3 deletion embryo at E6.5. Scale bars, 20 μm.
(F) Numbers and genotypes of embryos collected at the indicated developmental stages. Numbers of morphologically abnormal embryos are listed in parentheses.
Cnot3 Is Required for Epiblast Maintenance
To test its role in embryonic development, we generated a Cnot3 conditional deletion mouse model by conventional gene targeting (Figures S1A–S1D). We confirmed the successful depletion of the Cnot3 protein in the null embryos by immunofluorescence staining (Figure 1C). Because Cnot3 is required for ESC maintenance, we hypothesized that it may play important roles in the specification or maintenance of the epiblast. Consistent with the hypothesis, we found that Cnot3 deletion resulted in early embryonic lethality, as we were not able to recover any viable null pups or embryos with normal morphology at embryonic day 6.5 (E6.5) to E7.5 (Figures 1D–1F, S1F, and S2A). At E3.5 and E4.5, Cnot3 deletion embryos appear normal and were recovered at a Mendelian ratio (Figure 1F). Furthermore, the expression pattern of the epiblast (Nanog), trophectoderm (Cdx2), and primitive endoderm markers (Gata4, Gata6, Pdgfra) was comparable between the null and wild-type (WT) embryos (Figures 2A and S2B). Thus, Cnot3 may not be required for the formation of the blastocysts and the specification of the epiblast lineage, although we cannot rule out the possible contribution from maternal expression (Figures 1A and 1B).
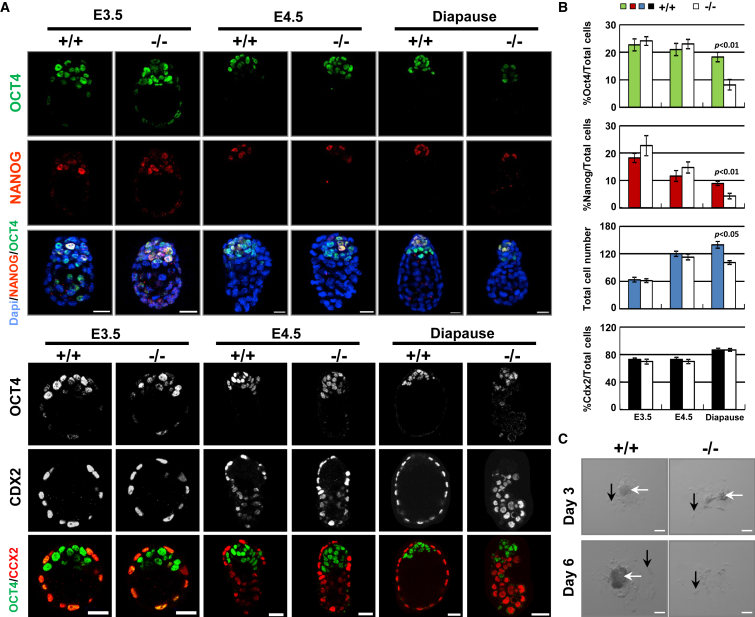
Figure 2.
Cnot3 Deletion Impairs Epiblast Maintenance
(A) Immunofluorescence staining of epiblast markers OCT4, NANOG, and trophectoderm marker CDX2 in WT and Cnot3 deletion embryos. Scale bars, 20 μm.
(B) Total cell number and percentage of OCT4-, NANOG-, or CDX2-positive cells in WT and Cnot3 deletion embryos. Values were plotted as mean ± SEM from three independent experiments.
(C) Epiblast cell outgrowth from WT and Cnot3 deletion blastocysts. White arrows, epiblast cells; black arrows, trophectoderm cells. Scale bars, 20 μm.
To further test the role of Cnot3 in the maintenance of the epiblast, we used the embryonic diapause model. During diapause, the embryos are arrested in utero at the late blastocyst stage and the pluripotent state is maintained in the epiblast cells for an extended period of time (Fenelon et al., 2014). We found that Cnot3 was clearly required for the maintenance of the blastocysts during diapause, as the deletion embryos show significant compromise in morphology and reduction in size (Figures 1F and 2A; Movie S1). Quantitatively, Cnot3 deletion led to a decrease in the total cell number in the embryos. More importantly, it led to a reduction in the percentage of cells expressing epiblast markers Oct4 and Nanog, but not those expressing the trophectoderm marker Cdx2 (Figures 2A and 2B), suggesting that epiblast cells were lost in the null embryos. To further support these findings, we carried out epiblast outgrowth studies. As expected, epiblast cells from Cnot3 null blastocysts failed to expand and grow into colonies, while trophectoderm cells continued to survive (Figure 2C). Together, our data support the notion that Cnot3 is required for the maintenance of the pluripotent epiblast cells in vivo.
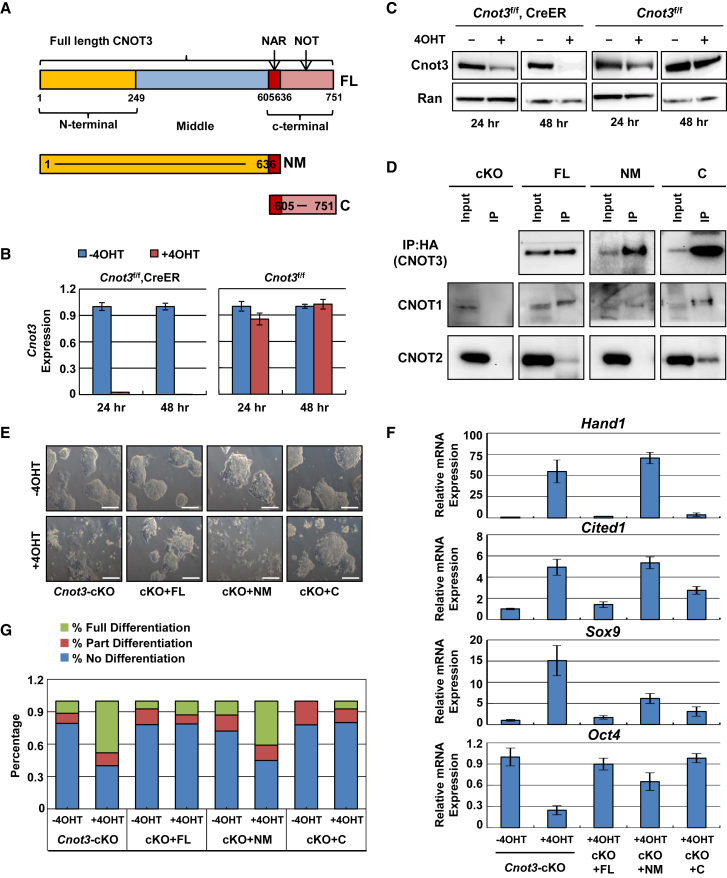
CNOT3 C-Terminal Domain Is Required for ESC Maintenance
To understand how CNOT3 regulates the pluripotent state, we carried out structure-function analysis to determine the functional domain(s) in Cnot3. Based on sequence and structural information, the Cnot3 protein can be divided into the N-terminal, middle (NM), and C-terminal domains (Figure 3A) (Boland et al., 2013). We generated Cnot3 conditional knockout (cKO) ESCs in which Cnot3 mRNA and protein can be quickly depleted upon tamoxifen treatment (Figures S3A, 3B, and 3C). Cnot3 deletion led to ESC differentiation as indicated by obvious changes in cellular morphology and lineage marker expression (Figures S3B and S3C). Importantly, its deletion induced upregulation of differentiation gene markers even in the 2i + LIF medium that supports the ground state (Figure S3D). However, it did not induce obvious changes in caspase-3 cleavage (Figures S3E and S3F), suggesting that CNOT3 does not directly regulate apoptosis. In the Cnot3 cKO ESCs, we overexpressed CNOT3 full-length (FL), N-terminal and NM, and C-terminal domains (Figure S4A). Consistent with previous results from other cell types (Boland et al., 2013), we found that the C-terminal domain in CNOT3 is responsible for its interaction with CNOT1 and CNOT2 in the Ccr4-Not complex in ESCs (Figure 3D, and whole blot images in S4B). More importantly, the overexpression of either the CNOT3 full-length or C-terminal domain led to normal morphology and marker expression after the depletion of the endogenous CNOT3 by tamoxifen treatment (Figures 3E and 3F). Furthermore, the overexpression sustained the formation of normal ESC colonies at clonal density (Figures 3G and S4C). In contrast, overexpression of the CNOT3 N-terminal and NM domains (Figure S4A) had no effect (Figures 3E–3G), even though this fragment retained the interaction with Cnot1 (Figure 3D). Therefore, our results showed that the CNOT3 C-terminal domain is required for the maintenance of the pluripotent state, possibly via the interaction with CNOT1, CNOT2, and other unknown factors.
Figure 3.
CNOT3 C-Terminal Domain Is Required for ESC Maintenance
(A) Domain structure of mouse CNOT3.
(B and C) Induction of Cnot3 deletion in Cnot3 cKO ESCs. Cells were treated with or without 4-OHT, and Cnot3 expression was determined by qRT-PCR (B) and western blot (C) at the indicated time points. Values were plotted as mean ± SEM from three independent experiments.
(D) Interaction between CNOT3 fragments and CNOT1 or CNOT2. HA-tagged CNOT3 fragments were expressed in Cnot3 cKO ESCs and affinity purified by HA beads. Co-purified endogenous CNOT1 and CNOT2 were detected by western blot. Whole images of the same blots are shown in Figure S4B.
(E–G) Rescue of the deletion phenotype by the overexpression of CNOT3 domains. Cnot3 cKO ESCs expressing various CNOT3 fragments were treated with or without 4-OHT. Changes in cellular morphology (E; scale bars, 200 μm), marker expression (F), and colony formation (G) were determined by imaging, qRT-PCR, and alkaline phosphatase staining, respectively. For qRT-PCR, relative expression was normalized by Actin and plotted as mean ± SEM from three independent experiments.
CNOT3 Negatively Regulates Differentiation Gene Expression
Because the Ccr4-Not complex regulates mRNA deadenylation, we hypothesize that CNOT3 may regulate the gene expression program in pluripotent cells post-transcriptionally. Indeed, immunofluorescence staining showed that CNOT3 localizes predominantly in the cytosol in both epiblast cells in vivo and ESCs in vitro (Figures 1B and S5A). Moreover, CNOT3 remained in the cytosol in ESCs even after nuclear export was blocked by leptomycin B treatment. These results are consistent with the idea that CNOT3 likely acts on mRNAs in the cytosol.
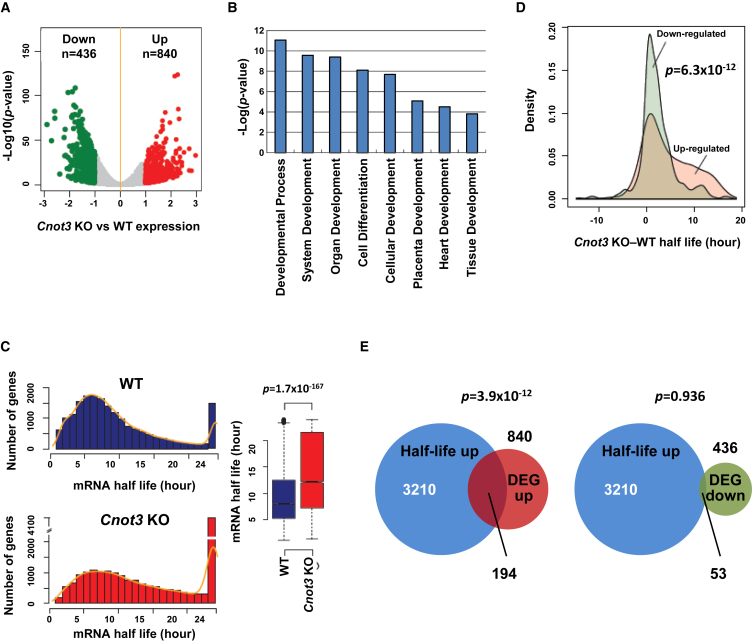
Gene expression analysis showed that Cnot3 deletion led to the upregulation of 840 genes and downregulation of 436 genes at the mRNA level (Figure 4A and Table S1; fold change >2, false discovery rate [FDR] <0.05). Gene ontology (GO) analysis showed that genes upregulated after Cnot3 deletion are highly enriched for those that are involved in normal differentiation and development (Figure 4B). Together, these results support the notion that CNOT3 inhibits differentiation and development of gene expression.
Figure 4.
Cnot3 Deletion Leads to Increases in mRNA Half-Life and Steady-State Level
(A) Gene expression changes after Cnot3 deletion in ESCs. Cnot3 cKO ESCs were treated with (KO) or without 4-OHT (WT). Cells were collected 72 hr after treatment and total RNAs were prepared for RNA-seq.
(B) GO analysis of upregulated genes after Cnot3 deletion. Only selected GO categories were plotted. For complete list of enriched GO categories, see Table S2.
(C and D) Increase in mRNA half-life in a subset of genes after Cnot3 deletion. Cnot3 cKO ESCs were treated with (KO) or without 4-OHT (WT). Actinomycin D was added 48 hr after treatment, and cells were collected at 0, 4, and 8 hr after actinomycin D addition for RNA-seq. (C) Frequency distribution and box plot for mRNA half-life in WT or KO samples. (D) Changes in mRNA half-life for genes down- or upregulated after Cnot3 deletion.
(E) Venn diagrams showing the overlaps between genes with extended mRNA half-lives and those that are down- or upregulated after Cnot3 deletion.
CNOT3 Promotes mRNA Degradation
To determine its role in the gene expression program in pluripotent cells, we first tested whether Cnot3 deletion has an impact on mRNA translation. We carried out polysome fractionation (Sampath et al., 2011) in both control and tamoxifen-treated Cnot3 cKO ESCs, and isolated polysome-associated RNAs. By RNA sequencing (RNA-seq), we found that Cnot3 deletion resulted in very similar changes in total RNAs versus polysome-associated RNAs (Figure S5B), suggesting that Cnot3 deletion did not have a significant impact on mRNA translation.
Next, we tested whether Cnot3 deletion affects mRNA stability. We inhibited transcription in the control and tamoxifen-treated Cnot3 cKO ESCs with actinomycin D, and determined the decay rate of all transcripts by carrying out RNA-seq at defined time points. We found that Cnot3 deletion led to an increase in mRNA half-life (Figure 4C). Importantly, while the increase in half-life (fold change >2, FDR <0.05; see Experimental Procedures for details) was detected for many mRNAs that were upregulated after Cnot3 deletion, there was little change in half-life for those mRNAs that were downregulated (Figure 4D). Indeed, 194 mRNAs show increases in both stability and expression (Figure 4E, p < 3.9 × 10−12) after Cnot3 deletion, which are likely direct targets of CNOT3. In contrast, there is no significant overlap between mRNAs with extended half-life and those with reduced expression. Notably, CNOT3 was recently suggested to regulate cell death-inducing genes in mouse embryonic fibroblasts (MEFs) (Suzuki et al., 2015). However, CNOT3 does not appear to regulate such cell death-inducing genes in ESCs, and there is no significant overlap between the 194 CNOT3 target genes in ESCs and those determined by similar methods in MEFs (Figure S5C). Together, these results suggest that CNOT3 specifically promotes the degradation of a subset of mRNAs in ESCs.
Cnot3-Dependent Differentiation Gene mRNA Degradation Plays an Important Role in the Maintenance of the Pluripotent State
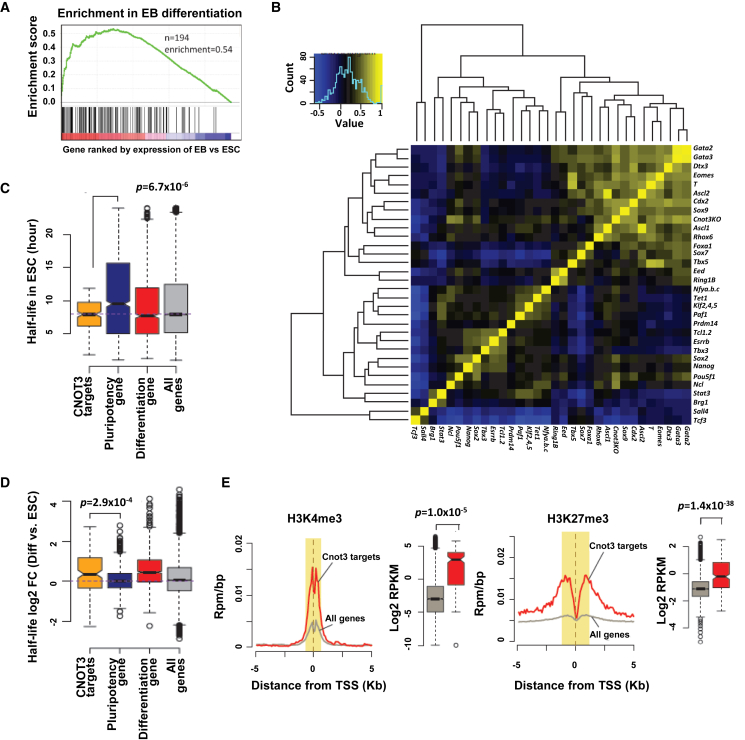
Gene set enrichment analysis (GSEA) showed that the 194 CNOT3 target genes are highly enriched for those that are upregulated during normal ESC differentiation (Figure 5A), suggesting that CNOT3 promotes differentiation gene mRNA degradation. To further test this idea, we compared the gene expression changes caused by Cnot3 deletion with those caused by the depletion of pluripotency genes and overexpression of differentiation genes in ESCs. Unsupervised hierarchical clustering analysis revealed that the pluripotency and differentiation genes clustered separately and formed two distinct modules. Consistent with our hypothesis, Cnot3 deletion clustered with the overexpression of differentiation genes (Figure 5B), including those that are likely its direct targets such as Foxa1 and Sox9 (see below).
Figure 5.
Cnot3 Deletion Impairs Differentiation Gene Degradation
(A) GSEA showing the enrichment for differentiation genes in the 194 CNOT3 target genes from Figure 2E.
(B) Hierarchical clustering analysis showing the similarity in gene expression profiles after depletion of pluripotency genes and overexpression of differentiation genes (see Table S3 for gene expression datasets used in this plot).
(C) Box plot for the half-lives of CNOT3 target genes, pluripotency genes, differentiation genes, and all genes in ESCs.
(D) Box plot showing the changes in half-lives for CNOT3 target genes, pluripotency genes, differentiation genes, and all genes in ESCs during differentiation.
(E) Metagene analysis and box plots (based on yellow highlighted regions in the metagene analysis) for H3K4me3 and H3K27me3 occupancy at the transcription start sites (TSS) of the CNOT3 target genes and all genes.
Intriguingly, the CNOT3 target genes showed shorter half-life compared with pluripotency genes in ESCs (Figure 5C), and their half-lives were extended during normal differentiation (Figure 5D). Indeed, the half-lives of differentiation gene mRNAs in general are shorter in ESCs and become longer during differentiation, while pluripotency gene mRNAs do not show the same trend. These results suggest that CNOT3-dependent regulation of differentiation gene mRNA stability may play an important role in the maintenance of the pluripotent state. Finally, we found that the CNOT3 target genes tend to be bivalently modified by H3K4me3 and H3K27me3 near their transcription start sites (Figure 5E), suggesting that CNOT3 further dampens the expression of poised developmental genes in pluripotent cells at the post-transcriptional level.
Cnot3 Promotes Differentiation Gene mRNA Deadenylation
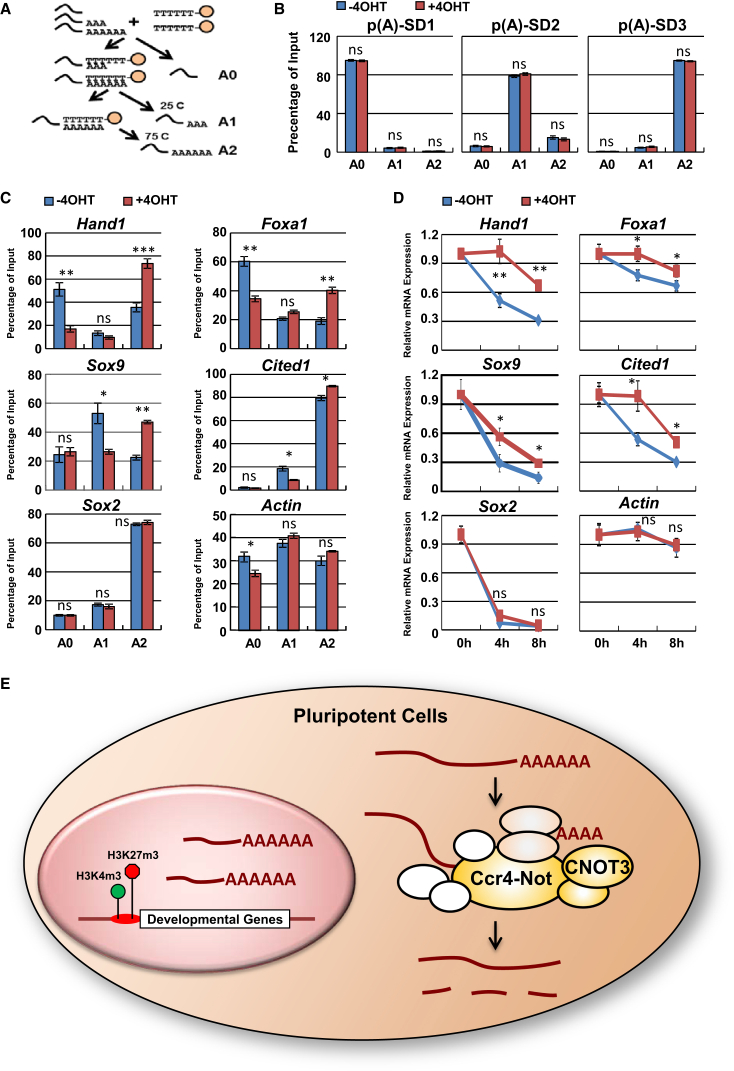
To test whether CNOT3 may regulate the poly(A) tail length of its target mRNAs, we first examined global polyadenylation in WT and KO ESCs. We found that Cnot3 deletion led to a subtle increase in global polyadenylation (Figure S5D), suggesting that only a subset of mRNAs were affected. To further test the impact of Cnot3 deletion on specific mRNAs, we fractionated total RNAs from control or tamoxifen-treated Cnot3 cKO ESCs based on poly(A) tail length using oligo(dT) beads (Meijer and de Moor, 2011) (Figure 6A). To assess the fractionation efficiency, we added in vitro synthesized mRNA standards with defined poly(A) tail lengths (Figure S5E) to both WT and KO total RNAs before the fractionation. By qRT-PCR, we found that the added poly(A) tail standards were recovered from the different fractions with expected efficiency, confirming the validity of the procedure (Figure 6B). Using this method, we tested the impact of Cnot3 deletion on poly(A) tails of several genes, including the housekeeping gene Actin, the pluripotency genes Sox2, Oct4, Nanog, Esrrb, Rex1, and the differentiation genes Cited1, Foxa1, Hand1, and Sox9. We found that Cnot3 deletion led to an increase in mRNA poly(A) tail length in differentiation genes Cited1, Foxa1, Hand1, and Sox9, without affecting the poly(A) tail length of housekeeping or pluripotency genes (Figures 6C and S5F). Interestingly, poly(A) tails appear to be longer in pluripotency than differentiation gene mRNAs (Figures 6C and S5F), consistent with our finding that pluripotency gene mRNAs tend to have longer half-lives (Figure 5C).
Figure 6.
Cnot3 Deletion Increases Differentiation Gene mRNA Poly(A) Tail Lengths
(A) Schematic drawing for mRNA fractionation based on poly(A) tail length.
(B) Validation of the oligo(dT) fractionation method. Cnot3 cKO ESCs were treated with (KO) or without 4-OHT (WT) for 48 hr. The poly(A) standards were mixed with total RNAs extracted from the cells, and RNAs were fractionated by oligo(dT) beads. The distribution of each standard in each fraction (A0, A1, A2) was determined by qRT-PCR and plotted as mean ± SEM from three independent experiments.
(C) Measurements of poly(A) tail length for the indicated genes by oligo(dT) fractionation.
(D) Examination of mRNA stability for the indicated genes. Cnot3 cKO ESCs were treated with (KO) or without 4-OHT (WT) for 48 hr. Actinomycin D was added to the cells, and mRNA level was measured by qRT-PCR at the indicated time points. Relative expression values were plotted as mean ± SEM from three independent experiments.
(E) Proposed model. CNOT3-dependent differentiation gene mRNA deadenylation and degradation plays a critical role in the maintenance of the pluripotent state.
∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, non-significant.
Consistent with the above data, qRT-PCR showed that while Cnot3 deletion led to increases in the stability of Hand1, Foxa1, Sox9, and Cited1 mRNAs, it had no impact on the stability of Actin or Sox2 mRNAs (Figure 6D). Furthermore, Cnot3 deletion led to an upregulation of the steady-state level of the differentiation gene mRNAs, but not that of Actin or Sox2 (Figure S3C). As we have previously shown that CNOT1, CNOT2, and CNOT3 from the Ccr4-Not complex are all required for ESC maintenance (Zheng et al., 2012), we also tested the effect of Cnot1 and Cnot2 silencing on the half-life of the differentiation gene transcripts. Unlike Cnot3 deletion, Cnot1 and Cnot2 silencing by small interfering RNAs showed very little impact (Figure S5G), suggesting that Cnot3 may play a unique role in post-transcriptional gene regulation in ESCs. This is consistent with our previous finding that Cnot3 silencing resulted in the most prominent phenotypic and gene expression changes in ESCs (Zheng et al., 2012), and is also consistent with the notion that individual subunits in the Ccr4-Not complex may differentially regulate its function (Azzouz et al., 2009). Finally, overexpression of Hand1, Foxa1, Sox9, and Cited1 induced ESC differentiation based on changes in cellular morphology (Figure S5H). Taken together, we propose that CNOT3 promotes the deadenylation and degradation of the differentiation gene mRNAs to maintain the gene expression program in pluripotent cells (Figure 6E).
Discussion
In pluripotent cells, the delicate balance between self-renewal and differentiation is tightly governed by a complex gene expression program. While transcription may function as an on-off switch to activate pluripotency genes and repress differentiation genes, post-transcriptional regulations can provide extra layers of control to refine the transcriptional output. In this study, we show that CNOT3, possibly via the Ccr4-Not complex, maintains pluripotency by promoting the deadenylation and degradation of differentiation gene mRNAs. Our finding reveals a previously uncharacterized mechanism in the maintenance of pluripotency, and provides additional evidence that post-transcriptional regulation is an integral part of the pluripotency regulatory network.
The length of poly(A) tails plays an important role in controlling mRNA degradation and translational silencing, and is therefore dynamically regulated during development and disease (Eckmann et al., 2011, Norbury, 2013). A recent study showed that poly(A) tail length predominantly correlated with mRNA stability in transcriptionally active cells. Furthermore it showed that, during embryonic development, poly(A) tail length initially regulated translational efficiency and gradually shifted to regulate mRNA stability as zygotic transcription started (Subtelny et al., 2014). Our results are consistent with the above observations, and suggest that poly(A) tail-length regulation by Ccr4-Not may start to affect mRNA stability as early as at the blastocyst stage in mammals.
The Ccr4-Not complex is one of the main deadenylases in mammalian cells that remove poly(A) tails. It can catalyze deadenylation in both gene- and context-specific manners to allow control of poly(A) tail lengths (Collart and Panasenko, 2012, Shirai et al., 2014, Xu et al., 2014). It has been proposed that different RNA binding proteins and/or different subunits in the Ccr4-Not complex may mediate the recruitment and regulation of different target mRNAs. For example, in germ cell development, NANOS2 and NANOS3 can interact with different Ccr4-Not subunits to regulate different mRNA targets (Suzuki et al., 2014). In ESCs, PUM1 facilitates the exit from the pluripotent state by binding to pluripotency gene mRNAs and promoting their degradation (Leeb et al., 2014), presumably via the interaction with CNOT7/8 in the Ccr4-Not complex (Van Etten et al., 2012). Thus, we propose that there may exist specific RNA binding proteins that facilitate the regulation of differentiation gene mRNAs by CNOT3 and Ccr4-Not in pluripotent cells. This is consistent with the “RNA Regulon” hypothesis, which suggests that mRNAs encoded by functionally related genes may be coordinately regulated by specific RNA processing machineries (Keene, 2007). Along similar lines, it was recently reported that m6A mRNA methylation reduces pluripotency gene mRNA stability to facilitate the exit from the pluripotent state in ESCs and that m6A depletion led to post-implantation lethality (Geula et al., 2015). Although it is not clear whether Ccr4-Not is involved in the m6A-mediated mRNA regulation, these and our results highlight the critical role of post-transcriptional regulation in the maintenance of the pluripotent state both in vitro and in vivo.
Finally, we found that CNOT3 target gene mRNAs have shorter half-lives in ESCs and acquire longer half-lives during differentiation. This result suggested that mRNA stability may indeed be a critical regulatory step, and that CNOT3, as well as Ccr4-Not, plays an important role in such a regulation to control pluripotent cell-fate specification. In addition, we found that CNOT3 target genes possess both active and repressive histone markers at their promoters. Promoter regions with bivalent histone modifications were initially discovered in ESCs at developmental genes, and were later shown to be present in epiblast cells during mouse development (Voigt et al., 2013). These bivalent genes are transcriptionally poised and can be activated upon suitable developmental cues to facilitate the exit from pluripotency. We propose that in addition to the transcriptional repression, differentiation genes may be further silenced by the Ccr4-Not complex at the mRNA level. Regulation of mRNA poly(A) tail length and stability may provide a quick and potentially reversible means to influence differentiation gene expression. Thus, by controlling the timely expression of differentiation genes, CNOT3 plays an essential role in maintaining the responsiveness of pluripotent cells to developmental signals.
Experimental Procedures
Antibodies
Mouse anti-CNOT3 (H00004849-M01, Abnova), goat anti-OCT3/4 (SC-8628, Santa Cruz Biotechnology), mouse anti-OCT3/4 (SC-5279, Santa Cruz), rabbit anti-Cleaved Caspase-3 (ASP175, Cell Signaling Technology), goat anti-GATA6 (AF1700, R&D Systems), rat anti-PDGFRA (14-1401, eBioscience), goat anti-GATA4 (SC-1237, Santa Cruz), rabbit anti-NANOG (RCAB002P-F, Cosmo Bio), mouse anti-CDX2 (CDX-88, Biogenex) and mouse anti-Cyclin B1 (AS4135, Cell Signaling).
Donkey anti-goat-493 (Nl003, R&D), donkey anti-mouse-594 (A21203, Life Technologies), donkey anti-rabbit-647 (A31573, Life Technologies), goat anti-mouse-594 (A11005, Life Technologies), goat anti-mouse-488 (A11001, Life Technologies), goat anti-rabbit-488 (A11008, Life Technologies), goat anti-rabbit-594 (A11012, Life Technologies), goat anti-mouse-657 (A11078, Life Technologies), rabbit anti-goat-488 (A11078, Life Technologies), and goat anti-rat (A11006, Life Technologies).
Generation of Cnot3 Knockout Mice
Cnot3 conditional deletion (cKO) mice were generated by conventional gene targeting (Figure S1A). The knockout construct was generated by recombineering (Lee and Liu, 2009), linearized, and electroporated into C2 ESCs (Gertsenstein et al., 2010). Correctly targeted clones were verified by both genomic PCR and Southern blot, and injected into C57BL/6 blastocysts. The resulting chimeric mice were crossed with WT C57BL/6 mice to derive the Cnot3tm1(LacZ)Hug F1 mice (Cnot3 LacZ/+). The F1 mice were further bred with Actb:FLPe mice, and backcrossed to the C57BL/6 background to derive the Cnot3tm1.1(flox)Hug mice (Cnot3 flox/+). The Cnot3 flox/+ mice were bred with the Zp3-Cre transgenic mice to drive the Cnot3−/+ mice. Cnot3 null embryos were obtained by breeding the female Cnot3flox/+, Zp3cre mice with the male Cnot3−/+ mice. The genotypes of the animals were determined by Southern blot, genomic PCR (see Table S2 for primer sequences), or qRT-PCR by Transnetyx. All animal research was conducted in accordance with the guidelines of the NIH Animal Care and Use Committee.
Mouse Embryo Collection and Immunofluorescence Analyses
Pre-implantation embryos were flushed out from the oviducts or uterus of pregnant mothers, and implanted E6.5 and E7.5 embryos were surgical dissected from the uterus. Embryonic diapause was induced in pregnant females 3 days after mating by intraperitoneal injection of Nolvadex (pharmaceutical grade tamoxifen, 10 μg in 100 μL of corn oil per mouse) and subcutaneous injection of Depo Provera (pharmaceutical grade medroxyprogesterone, 1 mg in 100 μL of corn oil per mouse) (Buehr and Smith, 2003). Diapause embryos were collected 3 days after the injections.
Embryos were fixed in 4% formaldehyde in PBS and immunofluorescence staining was carried out using standard protocols for whole embryos (Strumpf et al., 2005). Image data were acquired with a Zeiss LSM 780 microscope. Optical sections of 5 μm thickness were collected, and data processed by Imaris software. After imaging, embryos were collected and genotyped by PCR (Ralston et al., 2010).
ESC Derivation and Culture
E14Tg2a (WT) and Cnot3 cKO ESCs (see below) were cultured in gelatin-coated tissue culture plates in DMEM, 10% fetal bovine serum, and 1,000 U/mL LIF, using standard procedures (Zheng et al., 2012, Zheng and Hu, 2012). For inhibition of nuclear export, WT ESCs were treated with 10 nM leptomycin B for 4 hr. For measurement of mRNA stability, WT ESCs were treated with 10 μg/mL actinomycin D (Sigma, A-9415), and cells were harvested at 0, 4, and 8 hr after the treatment.
The Cnot3 flox/+ mice were backcrossed into the 129 strain background and bred with the UBC-Cre/ERT2 transgenic mice. Individual blastocyst embryo was plated, and the resulting ESC lines were genotyped by PCR and Southern blot. Multiple Cnot3flox/flox, UBC-Cre/ERT2 clones were established, and clone 8-4 was used for all the experiments. To induce Cnot3 deletion, we added 4-hydroxytamoxifen (4-OHT, Sigma) to the medium at 0.1 μM.
Cnot3 Domain Cloning and Expression
Mouse CNOT3 fragments corresponding to amino acids 1–751 (full length), 1–636 (NM), and 605–751 (C) were cloned from into pDNR223, sequence verified, and transferred into a gateway lentiviral expression vector PHAGE-EF1a-HA-Puro (Zheng et al., 2012). They were used to package lentiviruses in 293T cells, and the packaged viruses were used to transduce the Cnot3 cKO ESCs to create stable expression clones. Expression of the exogenous CNOT3 fragments was verified by qRT-PCR and western blot. Two to three clones expressing the same fragment were tested and behaved similarly in the rescue of the Cnot3 deletion phenotype, and the results from one clone were included in the figures.
Immunoprecipitation
Cnot3 cKO lines stably expressing hemagglutinin (HA)-tagged CNOT3 domain fragments were lysed in 1% NP-40 buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% NP-40) with EDTA-free protease inhibitors (Roche). Lysates were sonicated, centrifuged, and pre-cleared with protein-A Sepharose beads. HA-tagged fragments were immunoprecipitated with HA-affinity matrix (Roche), and protein was eluted with 2× lithium dodecyl sulfate buffer with 2-mercaptoethanol and boiling. Proteins were resolved on 4%–12% Bis-Tris gels (Life Technologies) and transferred to nitrocellulose membranes. Western blots were conducted using an HA antibody from Cell Signaling and Cnot1, Cnot2 antibodies from Proteintech.
Colony Formation Assay
Cnot3 cKO ESCs stably expressing Cnot3 domain fragments were treated with 4-OHT at 0.1 μM for 48 hr. Cells were replated at 1,000 cells/cm2 in 6-cm plates and allowed to grow for 7 days. The colonies were stained for alkaline phosphatase activity (AP Staining II kit, Stemgent). One hundred colonies were counted and scored as being normal, partially differentiated, or fully differentiated based on intensity of alkaline phosphatase staining and cellular morphology.
qRT-PCR and RNA-Seq
Total RNAs were extracted using the GeneJET RNA Purification Kit (Thermo), and reverse transcribed using the iScript (Bio-Rad, for qRT-PCRs) or the SuperScript II kit (Life Technologies, for poly(A) tail-length determination). qPCRs were performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) on CFX-384 (Bio-Rad).
For RNA-seq, sequencing libraries were prepared using the TruSeq RNA sample preparation kit v2 using the low sample protocol (Illumina). Sequencing was performed on the MiSeq or NextSeq sequencer (Illumina).
Polysome-Associated RNA Purification
Polysome-associated RNAs were purified as previously described (Sampath et al., 2011). In brief, Cnot3 cKO ESCs were treated with or without 4-OHT at 0.1 μM for 72 hr. Cytosolic proteins were extracted and loaded onto linear sucrose gradients of 15%–45% (w/w) and centrifuged for 2.5 hr at 30,000 rpm in an SW-41 rotor (Beckman). The gradients were fractionated into 0.4-mL fractions on a Gradient Master (BioComp) and polysome fractions were pooled. RNAs were extracted from both pooled polysome fractions and total cell lysis and used for RNA-seq.
Poly(A) Tail-Length Analysis
DNA templates used for poly(A) tail-length standards were cloned by PCR into the PCR-blunt II vector. Poly(A) tail-length standards were generated by first synthesizing mRNAs from the DNA templates by in vitro transcription (RiboMax, Promega) and then adding poly(A) tails using the poly(A) polymerase tailing kit (Epicenter) with different reaction times. Products from the poly(A) tailing reactions were purified by denaturing PAGE and analyzed on Agilent's Bioanalyzer to determine the poly(A) tail lengths.
Separation of mRNAs based on poly(A) tail length was performed based on a published protocol (Meijer and de Moor, 2011) with modifications. In brief, 1 μg of total RNA was first mixed with spike-in standards and then incubated with Oligo-dT Dynabeads (Life Technologies) at room temperature for 10 min. The supernatant (the A0 fraction) was kept, and the elution solution was added to the beads. The elution was carried out at 25°C for the A1 fraction, and then again at 75°C for the A2 fraction. RNAs in each fraction were ethanol precipitated, and reverse transcribed with the SuperScript II kit using random primers (Life Technologies). The percentage of individual transcript in each fraction relative to that in 1 μg of total RNA was determined by qRT-PCR.
Bioinformatics Analysis
Sequenced reads were aligned to the mouse genome (mm9) using the latest version of STAR (Dobin et al., 2013) by allowing a maximum of three mismatches and retaining only unique alignments. Gene expression raw counts were quantified by counting aligned reads using the htseq-count program (Anders et al., 2015) with Ensembl gene annotations. Raw gene counts were subsequently normalized using the DESeq2 R package (Love et al., 2014) for each replicate of the WT and Cnot3 KO comparison experiment; and for each time point (0 hr, 4 hr, and 8 hr) and each replicate of the WT and Cnot3 KO half-life experiment. For the WT and Cnot3 KO comparison experiment, normalized gene counts were used to identify differentially expressed genes using DESeq2. Genes with an FDR smaller than 0.05 and a fold change greater than 2 were deemed as differentially expressed.
For RNA half-life estimation, genes that have fewer than six normalized read counts at all three time points (0, 4, and 8 hr) in either WT or Cnot3 KO were deemed as lowly expressed and were removed. For genes that passed the filtering, their half-lives were estimated as described in Sharova et al. (2009). First, since the same amount of each sample was sequenced for each time point, gene expression at 4 hr and 8 hr was corrected by timing a scaling factor to account for relative increase of stable mRNA due to the degradation of unstable species after block of transcription. Then a linear regression of the form y = a − bt was performed on each gene through the three time points, where y is the log-transformed (base 10) read count, t is the time, b is the slope, a is intercept, and d = b × ln(10) is the instantaneous decay rate. Half-life of each gene was estimated and projected as H = min(24, ln(2)/d) for positive d and h = 24 hr for negative d. Genes that displayed significant difference of half-life in WT and Cnot3 KO were determined by requiring a fold change greater than 2 and an FDR smaller than 0.05, calculated as , where d1 and d2 are decay rates estimated from regression for WT and Cnot3 KO, respectively, and s1 and s2 are standard errors of the estimated decay rates.
Fisher's exact test was performed on overlap of gene sets that were significant in the half-life experiment and in the differential expression experiment, and on overlap of gene sets that are significant in WT versus Cnot3 KO in ESCs and MEFs (Suzuki et al., 2015). GSEA was performed to test for enrichment of gene set against EB differentiation data at day 9 (Hailesellasse Sene et al., 2007). Hierarchical clustering was performed to generate the heatmap profile and clustering global expression change after knockout, knockdown, or overexpression of each given gene (Correa-Cerro et al., 2011, Zheng et al., 2012). ESC-associated genes and differentiation-associated genes were defined by taking the top 5% and the bottom 5% of the genes ranked in Cinghu et al. (2014). Tag densities of H3K4me3 and H3K27me3 were generated by aligning H3K4me3 (Agarwal and Jothi, 2012) and H3K27me3 (Ho et al., 2011) sequencing data to mm9 genome using Bowtie (Langmead et al., 2009) by allowing two mismatches and retaining only unique alignment.
Author Contributions
G.H. and X.Z. conceived the study. X.Z., B.L., L.W., H.L., L.M., Y.W., Y.M., J.F.F., and G.H. carried out the experiments. P.Y. and B.D.B. analyzed the data. G.H., R.J., C.J.W., D.C.F., and Y.J. interpreted the data. G.H. and X.Z. wrote the manuscript.
Acknowledgments
We thank Dr. Anna-Katerina Hadjantonakis for helpful discussions. We thank the NIEHS Animal, Epigenomics, Bioinformatics, Protein Expression, Imaging, and Histology core facility for assistance with various techniques and experiments. This study was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences Z01ES102745 (to G.H.), Z01ES102985 (to C.J.W.), and Z01ES102625 (to R.J.).
Published: October 13, 2016
Footnotes
Supplemental Information includes five figures, four tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.09.007.
Accession Numbers
Sequencing data have been uploaded to the GEO database with the accession number GEO: GSE84953.
Supplemental Information
References
- Agarwal S.K., Jothi R. Genome-wide characterization of menin-dependent H3K4me3 reveals a specific role for menin in the regulation of genes implicated in MEN1-like tumors. PLoS One. 2012;7:e37952. doi: 10.1371/journal.pone.0037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouz N., Panasenko O.O., Deluen C., Hsieh J., Theiler G., Collart M.A. Specific roles for the Ccr4-Not complex subunits in expression of the genome. RNA. 2009;15:377–383. doi: 10.1261/rna.1348209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet C., Morera A.M., Asensio M.J., Chauvin M.A., Morel A.P., Dijoud F., Magaud J.P., Durand P., Rouault J.P. CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol. Cell. Biol. 2004;24:5808–5820. doi: 10.1128/MCB.24.13.5808-5820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland A., Chen Y., Raisch T., Jonas S., Kuzuoglu-Ozturk D., Wohlbold L., Weichenrieder O., Izaurralde E. Structure and assembly of the NOT module of the human CCR4-NOT complex. Nat. Struct. Mol. Biol. 2013;20:1289–1297. doi: 10.1038/nsmb.2681. [DOI] [PubMed] [Google Scholar]
- Boroviak T., Nichols J. The birth of embryonic pluripotency. Philos. Trans. R Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M., Smith A. Genesis of embryonic stem cells. Philos. Trans. R Soc. Lond. B Biol. Sci. 2003;358:1397–1402. doi: 10.1098/rstb.2003.1327. [discussion: 1402] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinghu S., Yellaboina S., Freudenberg J.M., Ghosh S., Zheng X., Oldfield A.J., Lackford B.L., Zaykin D.V., Hu G., Jothi R. Integrative framework for identification of key cell identity genes uncovers determinants of ES cell identity and homeostasis. Proc. Natl. Acad. Sci. USA. 2014;111:E1581–E1590. doi: 10.1073/pnas.1318598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M.A., Panasenko O.O. The Ccr4-not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- Correa-Cerro L.S., Piao Y., Sharov A.A., Nishiyama A., Cadet J.S., Yu H., Sharova L.V., Xin L., Hoang H.G., Thomas M. Generation of mouse ES cell lines engineered for the forced induction of transcription factors. Sci. Rep. 2011;1:167. doi: 10.1038/srep00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejosez M., Zwaka T.P. Pluripotency and nuclear reprogramming. Annu. Rev. Biochem. 2012;81:737–765. doi: 10.1146/annurev-biochem-052709-104948. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann C.R., Rammelt C., Wahle E. Control of poly(A) tail length. Wiley Interdiscip. Rev. RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- Fenelon J.C., Banerjee A., Murphy B.D. Embryonic diapause: development on hold. Int. J. Dev. Biol. 2014;58:163–174. doi: 10.1387/ijdb.140074bm. [DOI] [PubMed] [Google Scholar]
- Gabut M., Samavarchi-Tehrani P., Wang X., Slobodeniuc V., O'Hanlon D., Sung H.K., Alvarez M., Talukder S., Pan Q., Mazzoni E.O. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Gertsenstein M., Nutter L.M., Reid T., Pereira M., Stanford W.L., Rossant J., Nagy A. Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS One. 2010;5:e11260. doi: 10.1371/journal.pone.0011260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Hackett J.A., Surani M.A. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15:416–430. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]
- Hailesellasse Sene K., Porter C.J., Palidwor G., Perez-Iratxeta C., Muro E.M., Campbell P.A., Rudnicki M.A., Andrade-Navarro M.A. Gene function in early mouse embryonic stem cell differentiation. BMC Genomics. 2007;8:85. doi: 10.1186/1471-2164-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Miller E.L., Ronan J.L., Ho W.Q., Jothi R., Crabtree G.R. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Kim J., Xu Q., Leng Y., Orkin S.H., Elledge S.J. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Morita M., Hijikata A., Fukuda-Yuzawa Y., Adachi S., Isono K., Ikawa T., Kawamoto H., Koseki H., Natsume T. CNOT3 contributes to early B cell development by controlling Igh rearrangement and p53 mRNA stability. J. Exp. Med. 2015;212:1465–1479. doi: 10.1084/jem.20150384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamon M., Katano M., Hiraki-Kamon K., Hishida T., Nakachi Y., Mizuno Y., Okazaki Y., Suzuki A., Hirasaki M., Ueda A. Identification of Ccr4-not complex components as regulators of transition from partial to genuine induced pluripotent stem cells. Stem Cell Dev. 2014;23:2170–2179. doi: 10.1089/scd.2013.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Lackford B., Yao C., Charles G.M., Weng L., Zheng X., Choi E.A., Xie X., Wan J., Xing Y., Freudenberg J.M. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 2014;33:878–889. doi: 10.1002/embj.201386537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Liu P. Construction of gene-targeting vectors by recombineering. Cold Spring Harb. Protoc. 2009;2009 doi: 10.1101/pdb.prot5291. pdb prot5291. [DOI] [PubMed] [Google Scholar]
- Leeb M., Dietmann S., Paramor M., Niwa H., Smith A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell. 2014;14:385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Smith A. The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- Meijer H.A., de Moor C.H. Fractionation of mRNA based on the length of the poly(A) tail. Methods Mol. Biol. 2011;703:123–135. doi: 10.1007/978-1-59745-248-9_9. [DOI] [PubMed] [Google Scholar]
- Morita M., Oike Y., Nagashima T., Kadomatsu T., Tabata M., Suzuki T., Nakamura T., Yoshida N., Okada M., Yamamoto T. Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3(+/-) mice. EMBO J. 2011;30:4678–4691. doi: 10.1038/emboj.2011.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely G.G., Kuba K., Cammarato A., Isobe K., Amann S., Zhang L., Murata M., Elmen L., Gupta V., Arora S. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H.H., Surani M.A. The transcriptional and signalling networks of pluripotency. Nat. Cell Biol. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- Norbury C.J. Cytoplasmic RNA: a case of the tail wagging the dog. Nat. Rev. Mol. Cell Biol. 2013;14:643–653. doi: 10.1038/nrm3645. [DOI] [PubMed] [Google Scholar]
- Posfai E., Tam O.H., Rossant J. Mechanisms of pluripotency in vivo and in vitro. Curr. Top. Dev. Biol. 2014;107:1–37. doi: 10.1016/B978-0-12-416022-4.00001-9. [DOI] [PubMed] [Google Scholar]
- Ralston A., Cox B.J., Nishioka N., Sasaki H., Chea E., Rugg-Gunn P., Guo G., Robson P., Draper J.S., Rossant J. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. 2010;137:395–403. doi: 10.1242/dev.038828. [DOI] [PubMed] [Google Scholar]
- Sampath P., Lee Q.Y., Tanavde V. Identifying translationally regulated genes during stem cell differentiation. Curr. Protoc. Stem Cell Biol. 2011;Chapter 1 doi: 10.1002/9780470151808.sc01b08s18. Unit1B 8. [DOI] [PubMed] [Google Scholar]
- Sharova L.V., Sharov A.A., Nedorezov T., Piao Y., Shaik N., Ko M.S. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai Y.T., Suzuki T., Morita M., Takahashi A., Yamamoto T. Multifunctional roles of the mammalian CCR4-NOT complex in physiological phenomena. Front. Genet. 2014;5:286. doi: 10.3389/fgene.2014.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D., Mao C.A., Yamanaka Y., Ralston A., Chawengsaksophak K., Beck F., Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Subtelny A.O., Eichhorn S.W., Chen G.R., Sive H., Bartel D.P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Niimi Y., Saga Y. Interaction of NANOS2 and NANOS3 with different components of the CNOT complex may contribute to the functional differences in mouse male germ cells. Biol. Open. 2014;3:1207–1216. doi: 10.1242/bio.20149308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Kikuguchi C., Sharma S., Sasaki T., Tokumasu M., Adachi S., Natsume T., Kanegae Y., Yamamoto T. CNOT3 suppression promotes necroptosis by stabilizing mRNAs for cell death-inducing proteins. Sci. Rep. 2015;5:14779. doi: 10.1038/srep14779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J., Schagat T.L., Hrit J., Weidmann C.A., Brumbaugh J., Coon J.J., Goldstrohm A.C. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J. Biol. Chem. 2012;287:36370–36383. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P., Tee W.W., Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Miao Y.L., Zheng X., Lackford B., Zhou B., Han L., Yao C., Ward J.M., Burkholder A., Lipchina I. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell. 2013;13:676–690. doi: 10.1016/j.stem.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Du Y., Ward J.M., Shimbo T., Lackford B., Zheng X., Miao Y.L., Zhou B., Han L., Fargo D.C. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell. 2014;14:575–591. doi: 10.1016/j.stem.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe C., Morita M., Hayata T., Nakamoto T., Kikuguchi C., Li X., Kobayashi Y., Takahashi N., Notomi T., Moriyama K. Stability of mRNA influences osteoporotic bone mass via CNOT3. Proc. Natl. Acad. Sci. USA. 2014;111:2692–2697. doi: 10.1073/pnas.1316932111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J.E., Ciosk R. RNA-based regulation of pluripotency. Trends Genetics. 2013;29:99–107. doi: 10.1016/j.tig.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Xu K., Bai Y., Zhang A., Zhang Q., Bartlam M.G. Insights into the structure and architecture of the CCR4-NOT complex. Front. Genet. 2014;5:137. doi: 10.3389/fgene.2014.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Blelloch R. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell. 2014;15:271–280. doi: 10.1016/j.stem.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Hu G. Oct4GiP reporter assay to study genes that regulate mouse embryonic stem cell maintenance and self-renewal. J. Vis. Exp. 2012;63 doi: 10.3791/3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Dumitru R., Lackford B.L., Freudenberg J.M., Singh A.P., Archer T.K., Jothi R., Hu G. Cnot1, Cnot2, and Cnot3 maintain mouse and human ESC identity and inhibit extraembryonic differentiation. Stem Cells. 2012;30:910–922. doi: 10.1002/stem.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.