Summary
Naive and primed pluripotent stem cells (PSCs) and germ cells express the Oct4 gene. The Oct4 gene contains two cis-regulatory elements, the distal enhancer (DE) and proximal enhancer (PE), which differentially control Oct4 expression in a cell-type-specific and stage-specific manner. Here, we generated double transgenic mice carrying both Oct4-ΔPE-GFP and Oct4-ΔDE-tdTomato (RFP), enabling us to simultaneously monitor the activity of DE and PE. Oct4 expression is stage-specifically regulated by DE and PE during embryonic and germ cell development. Using this dual reporter system, we successfully cultured pure populations of naive (GFP+RFP−) and primed (GFP−RFP+) PSCs. We found that GFP+RFP− cells were metastable (not naive) in serum-containing medium; stable naive pluripotent cells were observed in medium containing two inhibitors (Meki and GSKi) but lacked serum. Finally, we suggest that the activity of Oct4 DE and PE is regulated by the repressive histone marks and DNA methylation in a cell-type-specific manner.
Keywords: naive pluripotency, primed pluripotency, Oct4, enhancer
Graphical Abstract
Highlights
-
•
A defined model for Oct4 enhancer activity in the totipotent cycle
-
•
Culturing pure populations of naive and primed PSCs by a double reporter system
-
•
Altering Oct4 enhancer activity in PSCs by changing culture conditions
-
•
Histone modification and DNA methylation regulate Oct4 enhancer activity
Do and colleagues show that Oct4 expression is stage-specifically regulated by enhancer activity during embryonic and germ cell development by using a dual reporter system. Enhancer activity is altered in pluripotent stem cells by culture environment, which is regulated by histone modification and DNA methylation.
Introduction
Pluripotent stem cells (PSCs) can self-renew unlimitedly and have the potential to differentiate into all somatic and germ cell types. PSCs can be derived from the inner cell mass (ICM) of blastocyst or implanted epiblast cells (Hanna et al., 2010, Nichols and Smith, 2009). ICM and epiblast cells are pluripotent cells but not bona fide stem cells in vivo, as they differentiate to eventually establish the three germ layers in the gastrulating embryo. Both cell populations can be grown as PSCs when they are cultured in vitro with stem cell maintenance medium. Interestingly, the ICM of blastocysts forms “naive” pluripotent embryonic stem cells (ESCs) and epiblast cells form “primed” pluripotent epiblast stem cells (EpiSCs) (Hanna et al., 2010). Primed PSCs have limited differentiation potential in vivo; they barely contribute to chimeras by blastocyst injection analysis. Primed PSCs maintain stemness through basic fibroblast growth factor (bFGF) and Activin/Nodal signaling pathways (Brons et al., 2007, Greber et al., 2010, Vallier et al., 2009) but not by STAT3 and bone morphogenetic protein 4 pathways (Nichols and Smith, 2009). These two PSCs exhibit different molecular signatures but still share many important markers. One of the commonly expressed genes in these cells is Oct4, which is a PSC and germ cell marker (Scholer et al., 1990). In addition to maintenance of PSCs, Oct4 alone can transform differentiated cells into PSCs, referred to as induced PSCs (iPSCs) (Kim et al., 2009). The Oct4 gene contains three distinct cis-regulatory elements: the proximal promoter (PP), the distal enhancer (DE), and the proximal enhancer (PE), which differentially control Oct4 expression during embryogenesis (Yeom et al., 1996). Although Oct4 is expressed in both naive and primed PSCs, the regulatory mechanism of Oct4 expression differs between these cell types; Oct4 expression in naive and primed pluripotent cells is differentially controlled by DE and PE, respectively (Brons et al., 2007, Tesar et al., 2007, Yeom et al., 1996). Accordingly, enhancer activity is altered as primed PSCs are converted into naive PSCs through the induction of extrinsic signaling or genetic modification (Bao et al., 2009, Guo et al., 2009, Hanna et al., 2009).
Two recent reports used the Oct4-ΔPE-GFP marker to discriminate naive human PSCs from primed human PSCs (Gafni et al., 2013, Theunissen et al., 2014). However, as shown in flow cytometry data in Theunissen et al. (2014), Oct4-ΔPE-GFP reporter activity is not completely negative (including weak GFP activity) in primed human ESCs. Moreover, the Oct4-ΔPE-GFP+ cells may still include ESCs utilizing Oct4-PE, since a mono-transgenic system cannot discriminate between cells using only Oct4-DE and cells using both Oct4-PE and Oct4-DE, which may constitute an impure population of naive PSCs. Therefore, in this study we established a dual reporter system for naive and primed mouse pluripotent cells, using two fluorescent reporters, GFP and tdTomato (RFP), controlled by the cis-regulatory elements DE and PE, respectively. We found that the expression of Oct4-ΔPE-GFP and Oct4-ΔDE-RFP accurately represents the expression of naive and primed cells during the development of double transgenic mice. Thus, this double transgenic system can reproduce the in vivo Oct4 regulatory system, providing a tool for studying the regulation of naive and primed pluripotency and enabling the separation of pure populations of naive and primed PSCs.
Results
Generation of Dual-Color Fluorescence Transgenic Mice Containing Oct4-ΔPE-GFP and Oct4-ΔDE-RFP
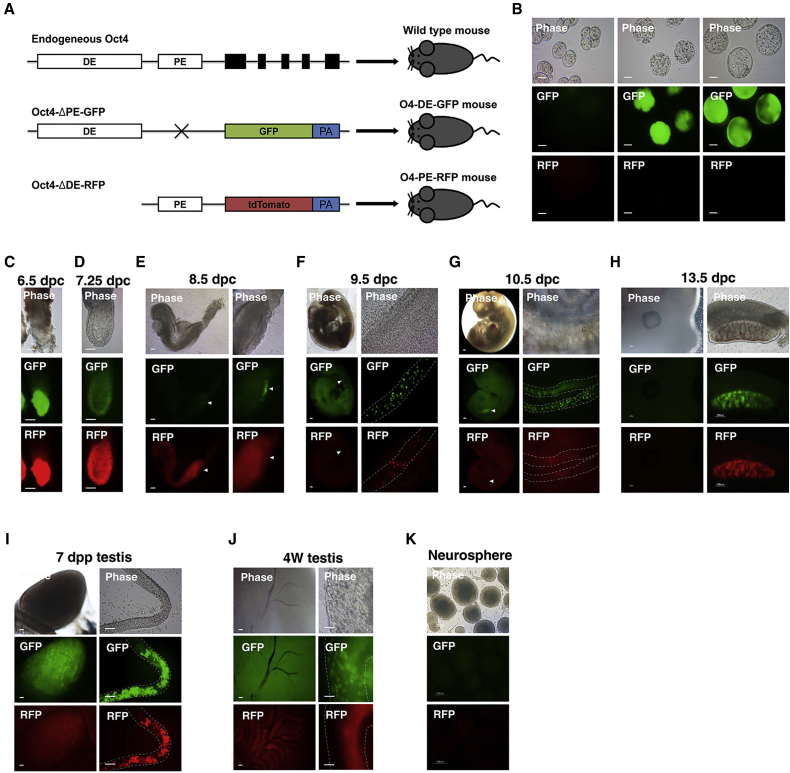
Oct4 is expressed in both naive and primed PSCs. However, Oct4 expression in naive and primed pluripotent cells is differentially controlled by two regulatory elements, DE and PE, respectively. We intended to understand how Oct4 is regulated by DE and PE during development (Figure 1). Therefore, we generated double transgenic mice expressing GFP and RFP under the control of either DE or PE of Oct4, respectively. O4-DE-GFP mice carried the Oct4-ΔPE-GFP transgene, originally termed OG2 (Szabo et al., 2002), and O4-PE-RFP mice carried the Oct4-ΔDE-RFP transgene (Figure 1A). Five O4-PE-RFP founder transgenic mice (two male and three female) were generated (Figure S1). O4-PE-RFP mice were crossed with homozygous O4-DE-GFP mice, and subsequently O4-DE-GFP+/−/O4-PE-RFP+/+ double transgenic mice were derived (Figure 1A). O4-DE-GFP+/−/O4-PE-RFP+/− embryos were obtained from wild-type female mice after crossing them with O4-DE-GFP+/−/O4-PE-RFP+/+ male mice. All transgenic animals that were studied for their expression had one allele of each transgene (O4-DE-GFP+/−/O4-PE-RFP+/−). In all transgenic animals both endogenous Oct4 alleles were present.
Figure 1.
Generation of Dual Transgenic Mice (O4-DE-GFP/O4-PE-RFP) and the Distinct Oct4 Regulatory Elements in the Totipotent Cycle
(A) Physical maps of wild-type endogenous Oct4, Oct4-ΔPE-GFP (O4-DE-GFP), and Oct4-ΔDE-RFP (O4-PE-RFP).
(B) The expression pattern of O4-DE-GFP and O4-PE-RFP in pre-implantation embryos (2C, blastocyst). The O4-DE-GFP/O4-PE-RFP pre-implantation embryos expressed only O4-DE-GFP. Scale bars, 50 μm.
(C–H) The expression pattern of O4-DE-GFP and O4-PE-RFP in post-implantation embryos. Phase and fluorescence images of (C) 6.5-dpc embryo (scale bar, 100 μm); (D) 7.25-dpc embryo (scale bar, 100 μm); (E) 8.5-dpc embryo (scale bar, 100 μm); (F) 9.5-dpc embryo and migrating PGCs (scale bar, 100 μm); (G) 10.5-dpc embryo and PGCs (scale bar, 100 μm); and (H) 13.5-dpc embryo and gonad (scale bar, 100 μm). Arrowheads indicate GFP-positive areas in the developing gonads.
(I and J) The expression pattern of O4-DE-GFP and O4-PE-RFP in testis. Phase and fluorescence image of (I) 7-dpp testis and seminiferous tubules and (J) 4-week-old adult mouse testis and seminiferous tubules. Scale bars, 100 μm.
(K) The neurosphere did not express either O4-DE-GFP or O4-PE-RFP. Scale bar, 100 μm.
O4-DE-GFP and O4-PE-RFP Recapitulate the Stage-Specific Expression of Oct4 during Mouse Embryo Development
Two-cell-stage embryos did not express either GFP nor RFP (Figure 1B), in agreement with the zygotic genome not being active at this stage. GFP was initially detected in eight-cell embryos and was strongly expressed at the ICM of the blastocyst stage, whereas RFP was not detected even at the blastocyst stage (Figure 1B), indicating that PE is dispensable for Oct4 expression in the pre-implantation embryo.
Next, we observed the expression of O4-DE-GFP and O4-PE-RFP during the post-implantation stages (6.5–13.5 days post coitum [dpc]). The 5.5- and 6.5-dpc epiblasts were positive both for GFP and RFP (Figures 1C and S2). At 7.25 dpc the intensity of the GFP signal decreased, but the RFP signal remained strong in epiblast cells (Figure 1D). Primordial germ cells (PGCs) were not distinguishable at this stage. However, at 8.5 dpc, GFP-positive cells were localized to the posterior regions of the embryos where the PGCs form a cluster and begin migrating into the genital ridge (Figure 1E). Although RFP-positive cells were detected extensively at the posterior regions of the embryos, these cells did not overlap with the GFP-positive cells, indicating that early PGCs do not require PE for Oct4 expression. At 9.5 dpc, GFP-positive cells were detected in the hindgut area (Figure 1F). RFP-positive cells disappeared from the soma; however, some cells in the hindgut expressed both RFP and GFP (approximately 34.7%), indicating that migratory PGCs at 9.5 dpc can be divided into two populations: GFP+ and GFP+/RFP+ cells. At the 10.5-dpc stage, when getting close to the genital ridge, most PGCs expressed both Oct4-GFP and -RFP (Figure 1G). This was also the case once the PGCs arrived at and proliferated in the gonads (13.5 dpc; Figure 1H). These results demonstrate that the two regulatory regions, DE and PE, dynamically control Oct4 expression during embryonic development and that founder PGCs use DE while migratory as well as post-migratory PGCs employ both DE and PE to drive Oct4 expression.
Oct4 has been shown to be expressed in mitotically arrested prospermatogonia and type A spermatogonia, but is downregulated in type B spermatogonia and spermatocytes in adult testis (Pesce et al., 1998). Expression of both GFP and RFP was detected 7 days postpartum (dpp) in the seminiferous tubules of male transgenic testis (Figure 1I). Interestingly, although both GFP+ and RFP+ cells were detected in 4-week-old adult male mouse testis, only GFP+ cells were localized to the periphery (near the basement membrane) of the seminiferous tubules while RFP+ cells were detected at the center of the seminiferous tubules (Figure 1J). Immunohistochemistry analysis confirmed that the GFP+ cells are present at the periphery (type A spermatogonia niche) of the seminiferous tubules and that the RFP+ cells are detected at the center (differentiated germ cell niche) of the seminiferous tubules (Figure S3). These results indicate that type A spermatogonia at the pre-meiotic division stage (7 dpp) can use both DE and PE to express Oct4 but that in adult testis only DE drives Oct4 expression in type A spermatogonia of 4-week-old mice. Committed germ cells located near the lumen also express Oct4, which is controlled by PE.
In this study, we focused on spermatogenic cells since RFP was not detected in the transgenic ovary. We did not detect transgenic expression in somatic cells. For example, we detected neither a GFP nor an RFP signal in neural stem cells (NSCs) (Figure 1K), demonstrating that our transgenic system is specific for the germline, i.e., PSCs and germ cells, but not for somatic stem cells. Taken together, our results indicate that DE and PE activities stage-specifically regulate expression of Oct4 in totipotent/pluripotent cells and germ cells in the developing mouse embryo.
Derivation of ESCs from Dual-Color Fluorescence Transgenic Blastocysts
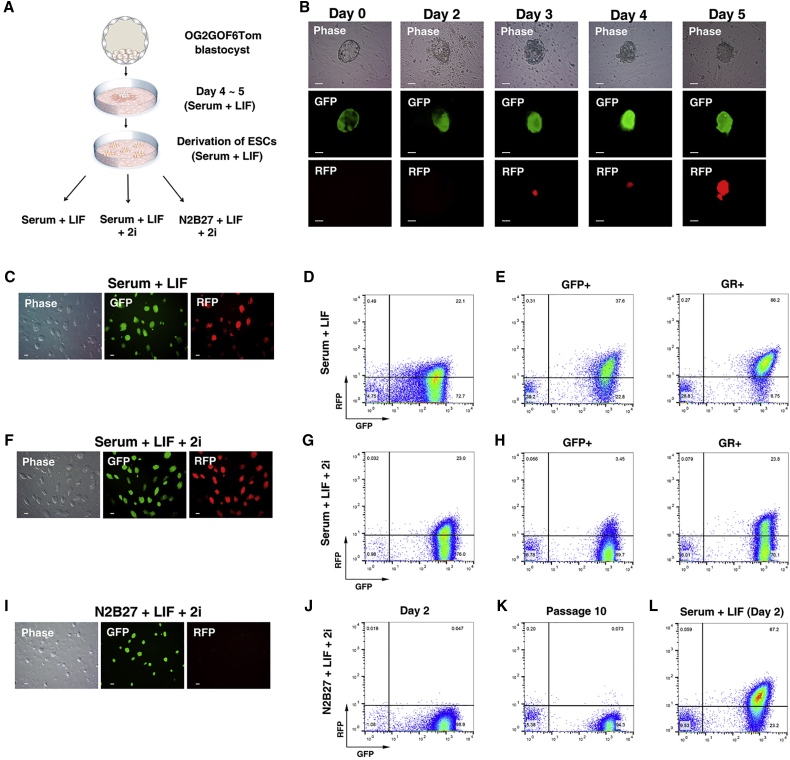
Previous studies have suggested that ESCs grown under conventional ESC culture conditions are a heterogeneous population (Hayashi et al., 2008, Martinez Arias and Brickman, 2011, Miyanari and Torres-Padilla, 2012). Thus, we attempted to verify the heterogeneity of ESCs and derive a pure population of naive pluripotent ESCs, using our double transgenic system. Blastocysts were plated on a feeder-layered dish in conventional mouse ESC medium (serum + leukemia inhibitory factor [LIF]) (Figures 2A and 2B). Initially the ICM of blastocysts expressed only GFP, whereas RFP was expressed after 3 days. As the ICM outgrowths expanded, the GFP+RFP− cells became GFP+RFP+ cells (Figure 2B). This result supports a previous study demonstrating that pluripotent ESC derivation requires epiblast specification (Boroviak et al., 2014). This in vitro culture system indirectly suggests that during the 4.5- to 5.5-dpc embryonic development, GFP+RFP− cells may be gradually changed into GFP+RFP+ cells in vivo. Trypsinized ICM outgrowth cells were transferred into a new dish with serum + LIF medium where two distinct ESC populations were cultured (GFP+RFP− and GFP+RFP+; Figure 2C). Fluorescence-activated cell sorting analysis showed that during the initial stage of ESC derivation in serum + LIF the GFP+RFP− cells, presumably naive PSCs, constituted only 72.7%, while GFP+RFP+ and GFP−RFP+ cells constituted 22.1% and 0.49%, respectively, of the population (Figure 2D). When GFP+RFP− cells were sorted and recultured in serum + LIF medium, GFP+RFP− cells reverted to GFP+RFP+ subsequent to ten passages (Figure 2E). In contrast, when GFP+RFP+ cells were sorted and cultured in serum + LIF medium, GFP+RFP+ cells reverted to GFP+RFP− cells subsequent to ten passages (Figure 2E). These results demonstrate that the ESCs in serum + LIF medium were heterogeneous; they contained two populations of cell types that were interconvertible. These GFP+RFP+ and GFP+RFP− cell populations could not have been distinguished using an Oct4-ΔPE-GFP mono-transgenic system.
Figure 2.
ESC Culture Conditions Influence the Activity of Oct4 Enhancers
(A and B) Derivation of ESCs from O4-DE-GFP/O4-PE-RFP blastocysts. O4-PE-RFP was initially expressed during the establishment of ESCs. Scale bars, 50 μm.
(C) Phase and fluorescence images of O4-DE-GFP/O4-PE-RFP ESCs in serum + LIF medium. Scale bars, 50 μm.
(D) Flow cytometry analysis of the proportion of O4-DE-GFP+ only cells or O4-DE-GFP+ and O4-PE-RFP+ cells in serum + LIF medium.
(E) Flow cytometry analysis of the proportion of O4-DE-GFP+ only cells or O4-DE-GFP+ and O4-PE-RFP+ cells from sorted O4-DE-GFP+ only cells or O4-DE-GFP+ and O4-PE-RFP+ cells cultured in serum + LIF medium for ten passages.
(F) Phase and fluorescence images of O4-DE-GFP/O4-PE-RFP ESCs in serum + LIF + 2i medium. Scale bars, 50 μm.
(G) Flow cytometry analysis of the proportion of O4-DE-GFP+ only cells or O4-DE-GFP+ and O4-PE-RFP+ cells in serum + LIF + 2i medium.
(H) Flow cytometry analysis of the proportion of O4-DE-GFP+ only cells or O4-DE-GFP+ and O4-PE-RFP+ cells from sorted O4-DE-GFP+only cells or O4-DE-GFP+ and O4-PE-RFP+ cells cultured in serum + LIF + 2i medium for ten passages.
(I) Phase and fluorescence images of O4-DE-GFP/O4-PE-RFP ESCs in N2B27 + LIF + 2i medium. Scale bars, 50 μm.
(J and K) Flow cytometry analysis of the proportion of O4-DE-GFP+ only cells or O4-DE-GFP+ and O4-PE-RFP+ cells cultured in N2B27 + LIF + 2i medium for 2 days (J) or ten passages (K).
(L) Flow cytometry analysis of the proportion of O4-DE-GFP+ only cells or O4-DE-GFP+ and O4-PE-RFP+ cells cultured in serum + LIF medium for 2 days from N2B27 + LIF + 2i medium.
Next, we modified the culture conditions by adding two inhibitors (2i), ERK1/2 (PD0325901) and GSK3 (CHIR99021), into the serum + LIF medium. Previous reports have demonstrated that these 2i contributed to the establishment of naive PSCs and shielded PSCs from differentiation triggers (Ying et al., 2008). Under serum + LIF + 2i conditions, only 76.0% of cells were GFP+RFP− and 23.0% were GFP+RFP+ (Figures 2F and 2G). When the GFP+RFP− cells were sorted and cultured for ten passages under serum + LIF + 2i conditions, 3.45% became GFP+RFP+ (Figure 2H, left panel). In contrast, the sorted GFP+RFP+ cells also converted into GFP+RFP− cells (70.1%) (Figure 2H, right panel). These findings indicate that serum + LIF + 2i culture conditions are insufficient for the maintenance of GFP+RFP− cells. Thus, ESCs are metastable and interconvertible between GFP+RFP− and GFP+RFP+ cells in serum-containing medium. Next, we used the new culture medium to obtain a pure population of naive PSCs. Serum was removed from the medium since mouse ESCs exhibited elevated levels of ERK1/2 phosphorylation, a key event in priming of ESCs for differentiation (Kunath et al., 2007, Wray et al., 2011, Yamaji et al., 2013), in serum-containing medium. Serum-free ESC culture medium (N2B27 medium) supplemented with 2i in combination with LIF (Marks et al., 2012, Silva et al., 2008, Ying et al., 2008) was used for ESC culturing. ESCs (from serum + LIF culture) were transferred into N2B27 + LIF + 2i medium. On day 2 after transfer to the N2B27 + LIF + 2i medium (Figure 2I), most GFP+RFP+ cells were converted into GFP+RFP− cells (98.9%; Figure 2J). Subsequent to further culturing, nearly all cells were maintained as GFP+RFP− cells (94.3%; Figure 2K). Next, the ESCs (from N2B27 + LIF + 2i medium) were transferred into serum + LIF medium. Subsequent to 2 days of culturing, nearly all of the GFP+RFP− cells were converted into GFP+RFP+ cells (67.2%; Figure 2L). Taken together, our double transgenic system unequivocally shows that the ESCs in serum-containing medium were heterogeneous. This heterogeneous ESC population became homogeneous subsequent to culturing in serum-free medium supplemented with 2i and LIF. Therefore, serum-free culture condition is an essential requirement for culturing a pure population of naive PSCs.
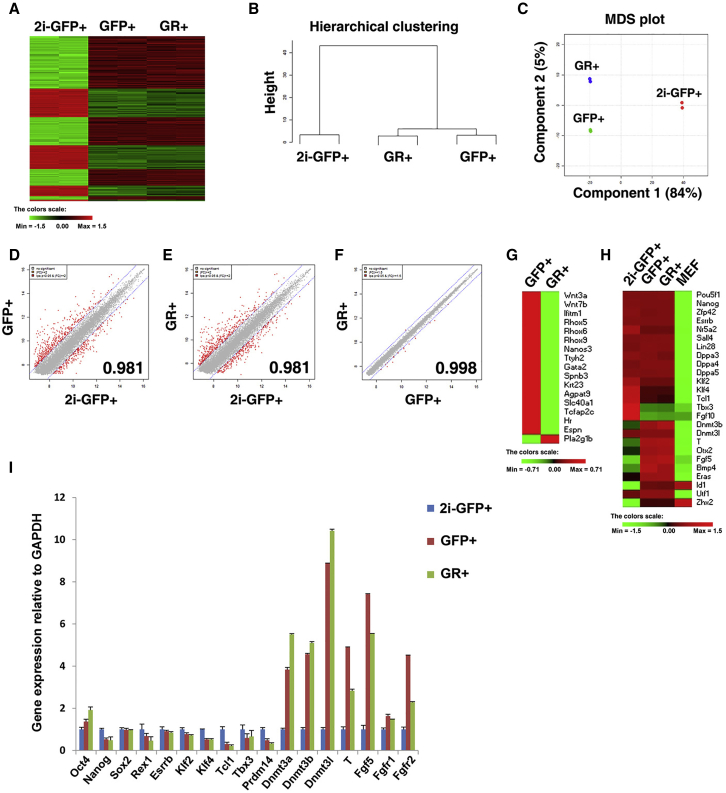
Distinct Gene Expression Patterns in 2i-GFP-, GFP-, and GFP/RFP-Positive ESCs
Oct4 expression is controlled by the DE in naive PSCs, while the PE regulates Oct4 expression in primed PSCs (Brons et al., 2007, Tesar et al., 2007, Yeom et al., 1996). Therefore, we investigated whether the expression of Oct4-ΔPE-GFP and Oct4-ΔDE-RFP accurately represents the naive and primed PSC states in an in vitro culture system. We compared the global gene expression patterns of GFP+RFP− (from N2B27 + LIF + 2i and serum + LIF medium) and GFP+RFP+ cells (from serum + LIF) using microarray analysis (Illumina MouseRef-8 v2 Expression BeadChip). Surprisingly, the gene expression profile of GFP+RFP− cells cultured in N2B27 + LIF + 2i (2i-GFP+RFP−) was distinct from that of GFP+RFP− cells cultured in serum + LIF (Figure 3A). The hierarchical clustering and multidimensional scaling (MDS) plot analyses showed that the gene expression pattern of GFP+RFP− cells was more similar to GFP+RFP+ cells than to 2i-GFP+RFP− cells (Figures 3B and 3C). Pairwise scatter-plot analyses also demonstrated that GFP+RFP+ cells were almost identical to GFP+RFP− cells (0.998), which were distinct from 2i-GFP+RFP− cells (Figures 3D–3F). Differentially expressed genes in GFP+RFP− and GFP+RFP+ cells were largely associated with germline development; GFP+RFP− cells highly expressed Wnt3a, Rhox5, Rhox6, Rhox9, Nanos3, and Tcfap2c (Figure 3G). We found that 249 genes were upregulated and 446 genes were downregulated in 2i-GFP+RFP− versus GFP+RFP− cells (Figure S4A). The expression levels of pluripotency factors such as Pou5f1 (Oct4), Nanog, Zfp42 (Rex1), Esrrb, Sall4, and Lin28 were almost identical in 2i-GFP+RFP−, GFP+RFP−, and GFP+RFP+ cells (Figure 3H). However, the naive pluripotency markers, Klf4, Tcl1, and Tbx3, were more highly expressed in 2i-GFP+RFP− than in GFP+RFP− and GFP+RFP+ cells. In contrast, differentiation-related genes such as Dnmt3b, Brachyury (T), Otx2, and Fgf5 were more highly upregulated in GFP+RFP− and GFP+RFP+ cells than in 2i-GFP+RFP− cells. These gene expression patterns were further confirmed by qRT-PCR analysis (Figure 3I). The expression levels of the pluripotency markers (Oct4, Nanog, Sox2, Rex1, Esrrb, and Klf2) in 2i-GFP+RFP− cells were similar to those in GFP+RFP− and GFP+RFP+ cells (Figure 3I). However, while naive pluripotency markers were upregulated in 2i-GFP+RFP− cells, the development-related genes were downregulated (Figure S4B). These findings suggest that although GFP+RFP− cells display more naive characteristics (such as higher expression of germ cell markers) under serum + LIF culture conditions, they are in a metastable state and are readily converted into GFP+RFP+ cells. Stable naive PSCs could be maintained only in serum-free N2B27 + LIF + 2i medium, since 2i-GFP+RFP− cells were found to be distinct from GFP+RFP− cells cultured in serum-containing medium and expressed a greater number of naive pluripotency markers.
Figure 3.
2i-GFP-Positive Cells Cultivated in N2B27 + LIF + 2i Medium and GFP- or GR-Positive Cells Cultivated in Serum + LIF Medium Exhibit Distinct Gene Expression Patterns
(A) Heatmap of global gene expression patterns in 2i-GFP+, GFP+RFP− (GFP), and GFP+RFP+ (GR) cells.
(B and C) Hierarchical clustering (B) and MDS plot (C) analysis showing that 2i-GFP-positive cells are distinct from GFP- or GR-positive cells.
(D–F) Scatter plots of global gene expression comparing (D) 2i-GFP- and GFP-positive cells, (E) 2i-GFP- and GR-positive cells, and (F) GFP- and GR-positive cells.
(G) Heatmap analysis shows that GFP-positive cells express germ cell-related genes including Wnt3a, Rhox5, Rhox6, Rhox9, Nanos3, and Tcfap2c at a higher level than do GR-positive cells.
(H) Heatmap analysis showing that 2i-GFP-positive cells highly express naive pluripotency-related genes and GFP- and GR-positive cells highly express differentiation markers.
(I) Quantitative gene expression analysis of pluripotency markers (Oct4, Nanog, Sox2, Esrrb), naive pluripotency markers (Rex1, Klf2, Klf4, Tcl1, Tbx3, Prdm14), and differentiation markers (Dnmt1, Dnmt3a, Dnmt3b, T, Fgf5). Data are presented as mean ± SEM for n = 3 independent experiments. GAPDH was used as a housekeeping gene.
Functional annotation clustering of differentially expressed genes using gene ontology (GO) analysis revealed that the upregulated genes (>2-fold) in 2i-GFP+RFP− cells were significantly enriched for GO terms linked to “sterol biosynthetic and metabolic process,” “cholesterol biosynthetic and metabolic process,” and “lipid biosynthetic process” (Marks et al., 2012) (Table S1). The upregulated genes in GFP+RFP− or GFP+RFP+ cells were enriched for GO terms linked to “gland, tube development, and developmental growth,” “collagen metabolic and catabolic process,” “membrane organization,” and “germ cell development” (Table S2).
GFP−RFP+ Cells Represent Primed PSCs
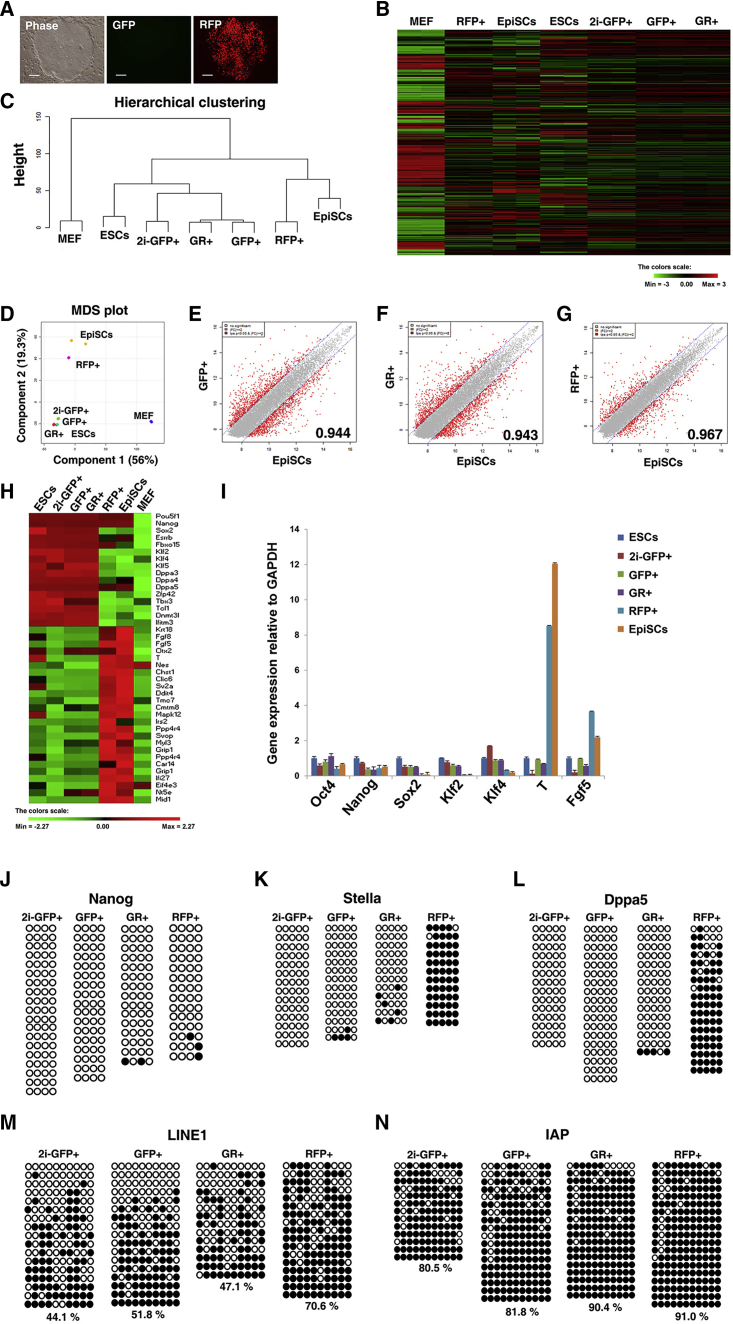
The results of our study show that GFP+RFP− ESCs stably maintain a naive pluripotent state when cultured in serum-free N2B27 + LIF + 2i medium. Next, we sought to identify primed PSCs using the double transgenic system. Since the PE regulates Oct4 expression in EpiSCs (Brons et al., 2007, Tesar et al., 2007), primed PSCs must be GFP−RFP+ cells and naive PSCs must be GFP+RFP− cells. During EpiSC derivation from O4-DE-GFP+/−/O4-PE-RFP+/− epiblasts, the initial expression of GFP decreased and only RFP+ cells were expanded (Figures S5A and S5B). Next, we tried to differentiate GFP+RFP+ ESCs into GFP−RFP+ EpiSC-like cells (EpiLCs). The GFP+RFP+ ESCs changed morphologically to form flat colonies that became GFP−RFP+ cells (Figure 4A). Heatmap and hierarchical clustering analyses showed that the gene expression patterns of GFP−RFP+ cells were more similar to EpiSCs than to ESCs (Figures 4B and 4C). The GFP−RFP+ cells were more similar to EpiSCs than to 2i-GFP+, GFP+RFP−, or GFP+RFP+ cells by MDS plot and scatter-plot analyses (Figures 4D–4G). These results demonstrate that transition of enhancer activity from DE to PE parallels the conversion from the naive to primed pluripotent state. Next, we examined the expression of core pluripotency-, naive pluripotency-, and differentiation-related genes in GFP−RFP+ EpiLCs (Figure 4H). Oct4 and Nanog were highly expressed in EpiLCs, EpiSCs, and all ESC lines (Figure 4H). However, GFP−RFP+ EpiLCs expressed very low levels of the naive pluripotency-related genes Klf2, Klf4, Klf5, Dppa3, Dppa4, Zfp42, Tbx3, and Tcl1, as shown for the EpiSCs. However, the differentiation-related genes Krt18, Fgf5, Fgf8, Otx2, T (Brachyury), and Nestin were highly expressed in GFP−RFP+ EpiLCs and EpiSCs. qRT-PCR analysis confirmed the high expression of the core pluripotency genes (Oct4 and Nanog) and EpiSC markers (T and Fgf5), and low expression of Sox2, Klf2, and Klf4 in GFP−RFP+ EpiLCs and EpiSCs (Figure 4G). These data show that GFP−RFP+ EpiLCs are similar to EpiSCs in terms of morphology and gene expression profiles.
Figure 4.
Oct4-ΔDE-RFP-Positive Cells Constitute Primed Pluripotent Stem Cells, as Determined by Gene Expression Patterns and Epigenetic Status
(A) Phase and fluorescence images of EpiSC-like cells (EpiLCs) from O4-DE-GFP/O4-PE-RFP ESCs. These EpiLCs express Oct4-ΔDE-RFP but not Oct4-ΔPE-GFP. Scale bars, 100 μm.
(B) Heatmap of global gene expression patterns in 2i-GFP+, GFP+RFP− (GFP+), GFP+RFP+ (GR), GFP−RFP+ (RFP) cells, ESCs, EpiSCs, and mouse embryonic fibroblasts (MEF).
(C and D) Hierarchical clustering and MDS plot analyses showing that RFP-positive cells are more similar to EpiSCs than to ESCs, 2i-GFP+, GFP+, or GR+ cells.
(E–G) Scatter plots of global gene expression comparing (E) EpiSCs and GFP-positive cells, (F) EpiSCs and GR-positive cells, and (G) EpiSCs and RFP-positive cells.
(H and I) Heatmap (H) and quantitative gene expression (I) analysis show that RFP-positive cells do not express naive pluripotency-related genes but highly express primed pluripotency-related genes. Data are presented as mean ± SEM for n = 3 independent experiments. GAPDH was used as a housekeeping gene.
(J–N) Bisulfite genomic sequencing of the Nanog (J), Stella (K), Dppa5 (L), LINE (M), and IAP (N) regions in 2i-GFP+, GFP+, GR+, and RFP+ cells. Black and white circles represent methylated and unmethylated CpGs, respectively.
Next, we investigated the DNA methylation status at the promoter regions of Nanog, Stella (Dppa3), and Dppa5 in GFP+RFP−, GFP+RFP+, 2i-GFP+RFP−, and GFP−RFP+ EpiLCs (Figures 4J–4L). The Nanog promoter regions were hypomethylated in all samples (Figure 4J). However, the promoter regions of Stella and Dppa5 were hypermethylated in GFP−RFP+ EpiLCs (Figures 4K and 4L). Long interspersed nuclear element 1 (LINE1) and Intracisternal A particle (IAP) have been shown to be hypermethylated in primed PSCs (Yamaji et al., 2013). Thus, we examined the DNA methylation status of LINE1 and IAP in GFP−RFP+ EpiLCs compared with GFP+RFP−, GFP+RFP+, and 2i-GFP+RFP− cells (Figures 4M and 4N). The LINE1 regions were more hypermethylated in GFP−RFP+ EpiLCs than in GFP+ (GFP+RFP−, GFP+RFP+, and 2i-GFP+RFP−) cells (Figure 4M). The IAP regions were more methylated in RFP+ cells (GFP+RFP+ and GFP−RFP+ EpiLCs) than in GFP+RFP− and 2i-GFP+RFP− cells (Figure 4N). Taken together, these results indicate that the Oct4-ΔPE-RFP reporter represents an applicable primed PSC marker. Thus, the conversion of ESCs into EpiLCs entails a shift from GFP+ to RFP+ (i.e., from DE to PE regulation), which also coincides with a change in transcriptome and epigenetic status specific for the primed pluripotent state.
Developmental Potential of GFP−RFP+ Cells
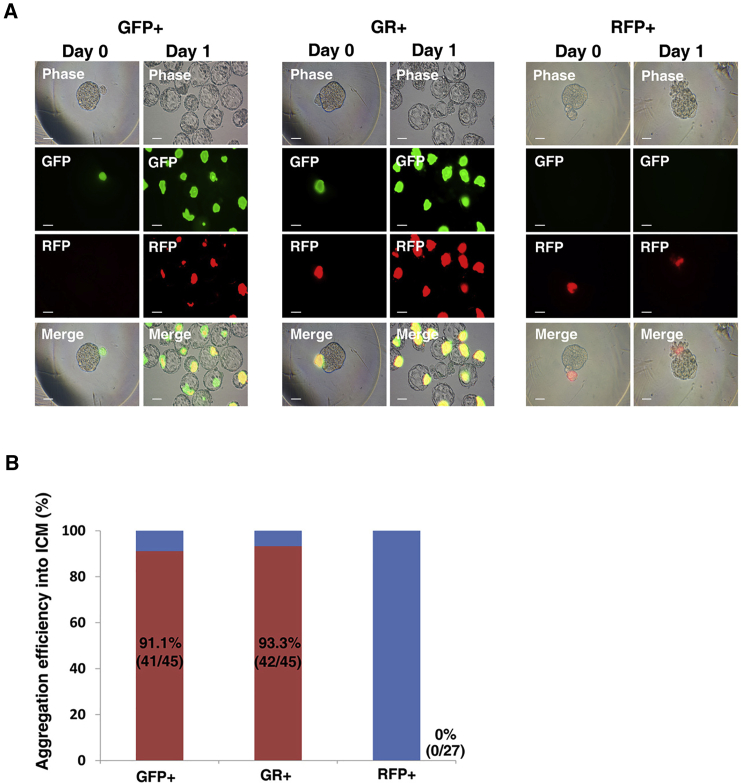
The ability of chimera formation is a strict criterion that distinguishes naive PSCs from primed PSCs (Chenoweth et al., 2010, Nichols and Smith, 2009, Nichols and Smith, 2011). Naive pluripotent ESCs contribute to chimeras; however, primed pluripotent EpiSCs barely contribute to chimeric embryos subsequent to blastocyst injection followed by transfer to a surrogate mother (Brons et al., 2007). GFP+RFP− and GFP+RFP+ cells as well as GFP−RFP+ EpiLCs were aggregated with wild-type eight-cell embryos and further cultured until the blastocyst stage. The GFP+RFP− and GFP+RFP+ cells were efficiently incorporated into the ICM of blastocysts (Figures 5A and 5B). Interestingly, at day 1 post aggregation, incorporated GFP+RFP− cells co-expressed RFP, similar to GFP+RFP+ cells (Figure 5A). The aggregation efficiency of GFP+RFP− and GFP+RFP+ cells was indistinguishable at 91.1% (41/45) and 93.3% (42/45), respectively (Figure 5B). However, basically all GFP−RFP+ EpiLCs failed to incorporate into the ICM of blastocysts, the aggregation efficiency being 0% (0/27) (Figures 5A and 5B). These results indicate that GFP+ cells exhibit characteristics of naive pluripotency and GFP−RFP+ EpiLCs demonstrate those of primed pluripotency-like EpiSCs.
Figure 5.
In Vivo Development Potential of Oct4-ΔDE-RFP-Positive Cells
(A) Aggregation of GFP-, GR-, and RFP-positive cells with normal embryo. RFP-positive cells did not incorporate into the embryos. Scale bars, 50 μm.
(B) Aggregation efficiency of GFP-, GR-, and RFP-positive cells.
Control of the DE and PE Elements in Naive and Primed PSCs
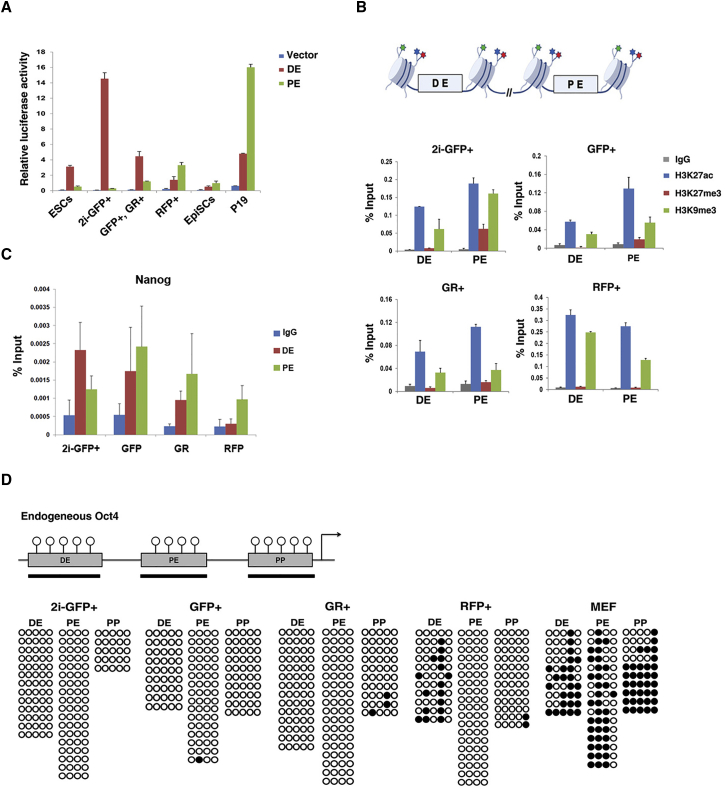
The activity of the Oct4 enhancers in naive and primed PSCs was evaluated in luciferase assays showing that 2i-GFP+ cells mainly use DE for Oct4 expression while PE was completely inactive; PE is slightly active in the control of ESCs cultured in serum + LIF medium (Figure 6A). GFP+RFP− and GFP+RFP+ cells in serum + LIF medium also mainly used DE, but the activity of PE was higher than in 2i-GFP+ cells. In contrast, the GFP−RFP+ cells mainly utilized PE for Oct4 expression, similar to EpiSCs and P19 embryonic carcinoma cells (Figure 6A). These results confirm that the expression of Oct4-ΔPE-GFP and Oct4-ΔDE-RFP accurately represents the naive and primed PSC state; thus, the double transgenic reporter system can be used as a tool for purifying populations of naive (2i-GFP+ cells) and primed (GFP−RFP+ cells) PSCs, since the regulatory mechanisms underlying the two different pluripotent states can be precisely defined.
Figure 6.
Epigenetic Status of Oct4 Regulatory Elements in Naive and Primed Pluripotent Stem Cells
(A) Analysis of Oct4 enhancer activity in ESCs, 2i-GFP+ cells, GFP+ GR+ cells, RFP+ cells, EpiSCs, and P19 embryonic carcinoma cells. Relative luciferase activity was normalized to the activity of the empty vector. Data are presented as mean ± SEM for n = 3 independent experiments.
(B) ChIP-qPCR analysis to determine immunoglobulin G (IgG), H3K27ac, H3K27me3, and H3K9me3 enrichment on the Oct4 distal and proximal enhancers. Data are presented as mean ± SEM for n = 3 independent experiments. Coloured stars indicate histone tails, such as H3K9 and H3K27.
(C) ChIP-qPCR analysis to determine IgG and Nanog enrichment on the Oct4 distal and proximal enhancers. Data are presented as mean ± SEM for n = 3 independent experiments.
(D) Bisulfite genomic sequencing of the regions of the Oct4 DE, PE, and PP in 2i-GFP+, GFP+, GR+, and RFP+ cells, and MEF. Black and white circles represent methylated and unmethylated CpGs, respectively.
Recent studies have shown that histone modifications are closely associated with the activity of enhancers (Creyghton et al., 2010, Favaedi et al., 2012). Acetylation of histone H3 lysine 27 (H3K27) is an indicator of active enhancers (Bonn et al., 2012), and deacetylation of H3K27 is associated with decreased gene expression or poised enhancers. H3K27me3 and H3K9me3 have also been suggested to be poised enhancer marks (Creyghton et al., 2010, Favaedi et al., 2012). H3K9me3 can distinguish poised from active enhancers independently of H3K27me3 (Zentner et al., 2011). Thus, we investigated the chromatin enrichment state of H3K27ac, H3K27me3, and H3K9me3 in 2i-GFP+, GFP+RFP−, GFP+RFP+, and GFP−RFP+ cells (Figure 6B) using chromatin immunoprecipitation (ChIP)-qPCR analysis. The results of the ChIP-qPCR analysis showed that H3K27ac was highly enriched on the DE and PE of Oct4 in all Oct4-expressing cells, naive (2i-GFP+), metastable (GFP+RFP− and GFP+RFP+), and primed (GFP−RFP+) PSCs (Figure 6B). The poised enhancer marks H3K27me3 and H3K9me3 were enriched on PE rather than on DE in 2i-GFP+ cells. The level of H3K27me3 on the PE was slightly higher than that on DE in metastable cells (GFP+RFP− and GFP+RFP+), but the level of H3K9me3 on the DE and PE did not differ significantly. Therefore, in the metastable state, epigenetic marks seem to fluctuate and the activity of PE and DE cannot be distinguished by histone marks. In primed pluripotent GFP−RFP+ cells, H3K9me3 was enriched on DE rather than on PE while the level of H3K27me3 was not different. These results suggest that enrichment of H3K9me3 and H3K27me3 on PE indicates the naive pluripotent state and that enrichment of H3K9me3 on DE marks the primed pluripotent state.
Transcription factors can regulate Oct4 expression by binding to DE and PE (Wu and Scholer, 2014). Galonska et al. (2015) showed that 2i condition alters the binding enrichment of Nanog on PE in ESCs. Thus we next checked the binding profiles of Nanog on PE and DE of Oct4 in different cell populations. ChIP-qPCR analysis showed that Nanog was highly enriched on the DE rather than on PE in 2i-GFP+ cells, naive PSCs (Figure 6C). On the other hand, Nanog was more enriched on the PE in GFP+RFP− and GFP+RFP+ cells. These results suggest that the differential activity of DE and PE is closely related to binding affinity of transcription factors.
Since both DNA methylation and histone modification regulate gene expression and affect each other (Cedar and Bergman, 2009), we next examined the DNA methylation pattern of the Oct4 regulatory elements. The DNA methylation status of DE, PE, and PP of Oct4 were analyzed by bisulfite DNA sequencing analysis (Figure 6D). The DE, PE, and PP of Oct4 in naive PSCs (2i-GFP+), GFP+RFP−, and GFP+RFP+ cells were completely unmethylated. However, primed PSCs (GFP−RFP+) showed relatively hypermethylated patterns in the DE of Oct4, indicating that the DE of Oct4 in primed PSCs is regulated by DNA methylation as well as H3K9me3 (Figures 6B–6D).
Discussion
In this study, we show that DE and PE coordinately regulate Oct4 expression during embryonic and germ cell development in vivo and naive and primed pluripotency in vitro. The cell-specific and temporal control of DE and PE could be precisely determined using the dual reporter system. Approximately two decades ago, we first determined the activity of the genomic fragment containing Oct4 using a β-galactosidase reporter and found that DE activity was ESC- and germ cell-specific and that PE was epiblast- and embryonic carcinoma cell (P19)-specific (Yeom et al., 1996). Oct4-GFP transgenic mice have been generated and used for the detection of PGCs and PSCs and for monitoring reprogramming through nuclear transfer, cell fusion, and the transduction of reprogramming factors (Boiani et al., 2002, Choi et al., 2014, Do and Schöler, 2004). However, these existing transgenic systems cannot concurrently discriminate between the activity of DE and PE. In this study, a developed dual reporter system allowed us to monitor the DE, PE, and DE + PE activity simultaneously.
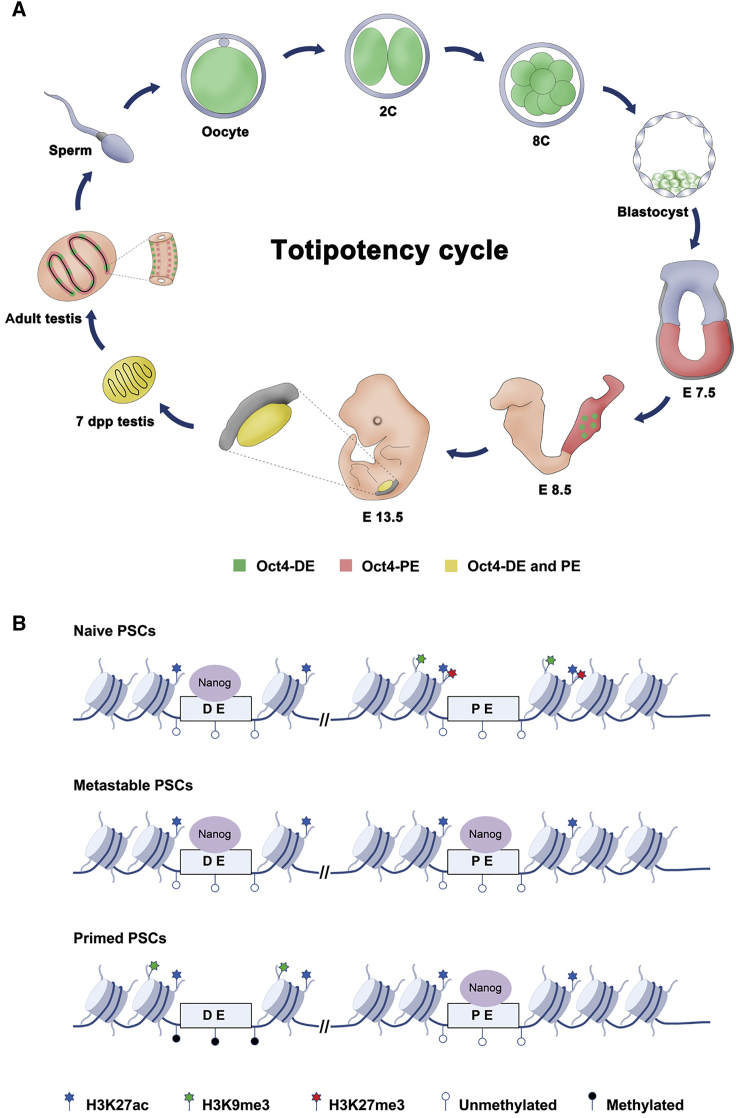
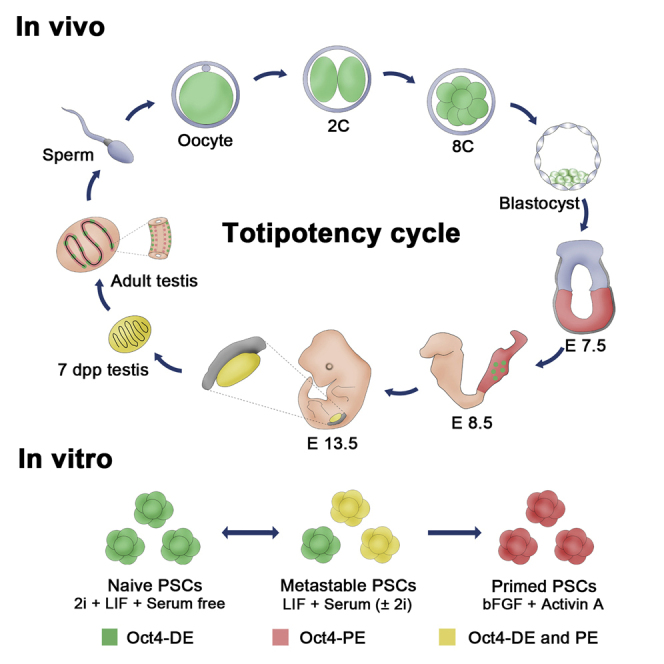
Enhancers play a central role in cell-type-specific and stage-specific regulation of gene expression and are capable of acting over a distance ranging from several to hundreds, and in rare cases even thousands of kilobases from their target genes (Bulger and Groudine, 2011, Ong and Corces, 2011, Ong and Corces, 2012). In developing mouse embryos, Oct4 gene expression is regulated spatially and temporally by at least two enhancers, DE and PE (Yeom et al., 1996). Here we have elucidated the control of Oct4 expression by its DE and PE elements during the totipotent cycle (Figure 7A). The Oct4 gene is expressed under the control of the DE during the pre-implantation stage, but its expression is controlled by PE in the epiblast subsequent to implantation (Figure 7A; Scholer, 1991). Expression of the Oct4 gene is downregulated in epiblasts and expressed exclusively in the developing germline. In founder PGCs, Oct4 is initially controlled by DE but subsequently by both DE and PE (Figures 1E–1J and 7A). Interestingly, we found that Oct4 was expressed not only in spermatogonia but also in differentiating germ cells; in adult mice, Oct4 expression was regulated by DE in spermatogonia but by PE in differentiating germ cells. It is well known that PGCs first emerge at the proximal region of the epiblast adjacent to the extraembryonic ectoderm and then migrate to form the PGC cluster at 7.25 dpc (Ginsburg et al., 1990, Saitou et al., 2002). However, while our dual reporter system could not distinguish the PGC population at 7.25 dpc, Oct4-ΔPE-GFP PGCs were apparent in 8.5-dpc embryos. Intriguingly, migrating PGCs in 9.5- and 10.5-dpc embryos contained two subpopulations, GFP+RFP− and GFP+RFP+ cells, and the GFP+RFP+ cell population increased during PGC migration into the genital ridge. Since genome-wide epigenetic modification, including imprinting erasure, occurs in migrating PGCs (Sasaki and Matsui, 2008), further experiments are required to examine the differences between these two migrating PGC populations and whether they can be distinguished using genome-wide epigenetic markers.
Figure 7.
Schematic Representation of the Oct4 Regulatory Elements
(A) The regulatory element of Oct4 during the totipotent cycle.
(B) Epigenetic status of the Oct4 enhancer in naive and primed pluripotency.
This study showed that ICM cells initially express only GFP but express both GFP and RFP during the derivation of ESCs (Figure 2B). This result supports the findings of recent studies indicating that the ability of ICM cells to self-renew as ESCs is acquired during epiblast specification (Boroviak et al., 2014, Tang et al., 2010). Boroviak et al. (2014) also suggested that early ICM cells are distinct from ESCs and that ESCs exhibit the greatest degree of identity to the embryonic day 4.5 (E4.5) epiblast. Our results confirm that the switch in enhancer activity occurs at approximately the E4.5 stage (Yeom et al., 1996), and enhancer activity of DE and PE overlapped in early post-implantation epiblast (5.5–6.5 dpc).
Self-renewing ESCs cultivated in conventional ESC culture medium (supplemented with LIF and serum) constitute a cell population at various stages of pluripotency (Hayashi et al., 2008, Martinez Arias and Brickman, 2011, Miyanari and Torres-Padilla, 2012). A defined ESC culture system, containing 2i (MEK inhibitor PD0325901 and glycogen synthase kinase 3 inhibitor CHIR99021) (Ying et al., 2008) together with LIF (2i + LIF) has been used to select naive pluripotent cells (Silva et al., 2008). Here we show that ESCs from 2i + LIF medium still contain two cell populations when cultured in serum-containing medium and that the mixed population could be converted to homogeneous naive PSCs by using serum-free N2B27 medium. This result indicates that serum contains a factor(s) that induce differentiation or inhibit maintenance of naive pluripotency. Previous reports have suggested that serum contains ERK phosphorylation-inducing factors (Yamaji et al., 2013). Silva et al. (2008) also showed that the presence of MEK inhibitor in serum-containing medium accelerated the activation of Nanog and Rex1 during reprogramming. This is also supported by the findings that GFP+RFP− ESCs cultured in serum-containing medium were molecularly distinguishable from GFP+RFP− ESCs cultured in serum-free medium. Collectively, the GFP+RFP− state is a necessary but not sufficient condition for naive PSCs, since special medium is also required for maintaining naive PSCs. Several reporter systems suggested that undifferentiated ESCs heterogeneously expressed Nanog (Chambers et al., 2007), Rex1 (Marks et al., 2012, Toyooka et al., 2008), Stella (Hayashi et al., 2008), Esrrb (van den Berg et al., 2008), and Tbx3 (Niwa et al., 2009). The heterogeneity of transcription factors in ESCs regulated self-renewal capacity, expression of developmental genes, and differentiation potential (Torres-Padilla and Chambers, 2014). However, the GFP+RFP− and GFP+RFP+ ESCs in serum-containing medium are similar to each other at the transcriptional, epigenetic, and functional levels. Our dual transgenic system allows monitoring of enhancer activity of DE and PE, but not Oct4 expression levels. Although enhancer activity fluctuates between GFP+RFP− and GFP+RFP+ in serum-containing medium, these cells highly expressed Oct4 gene.
Mixed populations of PSCs were also converted to naive PSCs in 2i + LIF medium without serum or to primed EpiLCs in bFGF + activin medium. EpiLCs induced from ESCs expressed only RFP, and the RFP+ EpiLCs were indistinguishable from EpiSCs in terms of morphology, gene expression patterns, epigenetic status, and defective incorporation into ICM of blastocysts. Therefore, GFP−RFP+ state constitute a definitive marker of primed PSCs; GFP−RFP+ cells were not observed in any of the three different ESC culture media.
The conversion from GFP+RFP+ to GFP+RFP− cells and vice versa takes place within 2 days of changing the culture medium (Figure 2), indicating that the conversion of enhancer activity occurs within 2 days. It has been suggested that genome-wide demethylation and transcriptional changes occur in mouse ESCs within the first 24 hr of 2i addition (Ficz et al., 2013). Therefore, the regulatory machinery of Oct4 enhancers seems to change rapidly in response to external cues accompanying epigenetic modification.
GFP−RFP+ EpiLCs derived from transgenic ESCs showed the molecular signature of EpiSCs, including expression of EpiSC markers and hypermethylation of germ cell markers (Stella and Dppa5 region), LINE1, and IAP. Primed PSCs are derived from post-implantation stage epiblasts and express Oct4 for self-renewal under the control of the PE (Brons et al., 2007, Tesar et al., 2007). RFP+ cells were not incorporated into the ICM by aggregation with normal embryos. Although a single aggregated embryo with RFP+ cells was observed, the GFP−RFP+ cells did not contribute to the ICM (Figure S6). This result could support the observation that GFP−RFP+ cells formed a pure population of primed PSCs.
Finally, we showed that the activity of the two Oct4 cis-regulatory elements was controlled by DNA methylation and histone modification concurring with the naive or primed pluripotency state. Consequently, the differential activity of DE and PE was closely related to binding affinity of Nanog on the two enhancers; Nanog was highly enriched on the DE in naive PSCs, but highly enriched on the PE in primed PSCs (Figures 6C and 7B). We also found that the activity of Oct4 DE and PE is affected by the repressive histone marks, H3K9me3 and H3K27me3, and DNA methylation in a cell-type-specific manner. The enhancers of Oct4 in primed PSCs is regulated by DNA methylation as well as H3K9me3, but those in naive PSCs is regulated by H3K9me3 and H3K27me3 but not by DNA methylation; DNA methylation was only correlated with inactive DE in primed PSCs and not with inactive PE in naive PSCs (Figure 7B). Thus, demethylation of the PE region seems to be the default state for PSCs and methylation of DE could constitute an epigenetic marker for primed PSCs. In contrast, H3K27ac was enriched in both the DE and PE of naive and primed PSCs, indicating that H3K27ac cannot be a histone mark for the active state of the Oct4 DE and PE. Another explanation is that although one of the enhancers is not active based on fluorescent reporter expression, epigenetically these enhancer elements of Oct4 are basically not completely silenced as H3K27ac remains enriched in both naive and primed PSCs. Similarly the PE is not methylated in 2i/LIF conditions, although the fluorescent mark is not on. These results suggest that overall both enhancers are active and never fully silenced in any of the states, although their level of activity is different under the tested conditions. Thus we should be more careful against using of terms “off” and “on” when describing enhancer activity of Oct4, the more accurate term rather being “dominant activity.” This fact could also be the reason for the need of a dual reporter system for accurate separation of naive and primed PSCs.
In this study, we showed that enhancer-specific regulation of Oct4 could constitute a determinant for distinguishing between naive and primed pluripotency. This is based on the ability to accurately separate naive and primed PSCs using the dual reporter system, thereby obtaining pure populations of naive and primed pluripotent cells, which could be used for accurate analysis of distinct cell states. The dual reporter system could also be a useful tool for monitoring cellular reprogramming to naive or primed states as well as embryonic development under live-cell conditions.
Experimental Procedures
Generation of O4-DE-GFP/O4-PE-RFP Mouse
To generate the Oct4-ΔDE-RFP transgene, we inserted the tdTomato (RFP) gene into genomic Oct4 fragments of 6 kb in length (GOF6), which lack DE (Yeom et al., 1996). Oct4-ΔDE-RFP transgene expresses tdTomato transgene under the control of Oct4 PE and PP. O4-PE-RFP embryos were generated by injecting Oct4-ΔDE-RFP (O4-PE-RFP) plasmid into normal zygote, which were developed to blastocysts. O4-PE-RFP blastocysts were transferred into the uterus of a pseudo-pregnant female mouse. We first generated five O4-PE-RFP founder transgenic mice, two male and three female (Figure S1). OG2 mice were purchased from the Jackson Laboratory and maintained in the mouse facility, and used for O4-DE-GFP founder mice (Szabo et al., 2002). The O4-PE-RFP mouse was crossed with homozygous O4-DE-GFP mouse. Finally, we generated O4-DE-GFP+/− O4-PE-RFP+/− mice carrying both Oct4-ΔPE-GFP and Oct4-ΔDE-RFP.
ESC Derivation and Culture
ESC lines from blastocyst were derived essentially as described previously (Nichols et al., 2009). Embryos at the eight-cell stage were flushed from oviducts of C57/BL6 mice mated with male O4-DE-GFP/O4-PE-RFP mice at 2.5 dpc, cultured in G2 medium for 2 days, and transferred to conventional ESC medium with 15% fetal bovine serum (FBS), 1× penicillin/streptomycin/glutamine, 0.1 mM nonessential amino acids, 1 mM β-mercaptoethanol, and 103 units/mL LIF on feeder layers. The established ESCs in conventional ESC medium were transferred into conventional ESC medium supplemented with LIF and 2i inhibitors, CHIR99021(3 μM) and PD0325901 (1 μM), or serum-free N2B27 medium supplemented with LIF and 2i inhibitors. The serum-free N2B27 was prepared as described by Ying and Smith (2003).
EpiSCs-like Cell Differentiation from ESCs
EpiLCs from ESCs were derived essentially as described previously (Hayashi et al., 2011). The EpiLCs were induced by plating 1.0 × 105 ESCs on a well of a 12-well plate on a feeder layer or feeder free in N2B27 medium containing activin A (20 ng/mL), bFGF (12 ng/mL), and knockout serum replacement (1%). The medium was changed every day. The differentiated EpiLCs (Oct4-ΔDE-RFP-positive cells) were passaged every 2–3 days by dissociation with 1 mg/mL collagenase type IV (Invitrogen) or by pipetting with a glass pipette.
Author Contributions
The study was conceived and designed by H.W.C and J.T.D. H.W.C. carried out the majority of experiments, performed data analysis, and created the figures. J.Y.J., Y.J.H., G.W., H.S., and J.S.K. performed some data analysis. Oct4-ΔDE-RFP vector was constructed by J.W.L. H.R.S. performed data interpretation. The study was supervised by J.T.D. The manuscript was written by H.W.C. and J.T.D.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (grant nos. 2015R1A2A2A01003604 and 2015R1A5A1009701) and the Next-Generation BioGreen 21 Program (grant no. PJ01133802) funded by the Rural Development Administration, Republic of Korea.
Published: October 27, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.09.012.
Supplemental Information
References
- Bao S., Tang F., Li X., Hayashi K., Gillich A., Lao K., Surani M.A. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M., Eckardt S., Schöler H.R., McLaughlin K.J. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 2002;16:1209–1219. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S., Zinzen R.P., Girardot C., Gustafson E.H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczyński B., Riddell A., Furlong E.E. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Boroviak T., Loos R., Bertone P., Smith A., Nichols J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Bulger M., Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H., Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chenoweth J.G., McKay R.D., Tesar P.J. Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev. Growth Differ. 2010;52:293–301. doi: 10.1111/j.1440-169X.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- Choi H.W., Kim J.S., Choi S., Hong Y.J., Kim M.J., Seo H.G., Do J.T. Neural stem cells differentiated from iPS cells spontaneously regain pluripotency. Stem Cells. 2014;32:2596–2604. doi: 10.1002/stem.1757. [DOI] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do J.T., Schöler H.R. Nuclei of embryonic stem cells reprogram somatic cells. Stem Cells. 2004;22:941–949. doi: 10.1634/stemcells.22-6-941. [DOI] [PubMed] [Google Scholar]
- Favaedi R., Shahhoseini M., Akhoond M.R. Comparative epigenetic analysis of Oct4 regulatory region in RA-induced differentiated NT2 cells under adherent and non-adherent culture conditions. Mol. Cell. Biochem. 2012;363:129–134. doi: 10.1007/s11010-011-1165-y. [DOI] [PubMed] [Google Scholar]
- Ficz G., Hore T.A., Santos F., Lee H.J., Dean W., Arand J., Krueger F., Oxley D., Paul Y.L., Walter J. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351–359. doi: 10.1016/j.stem.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Galonska C., Ziller M.J., Karnik R., Meissner A. Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell. 2015;17:462–470. doi: 10.1016/j.stem.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg M., Snow M.H., McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- Greber B., Wu G., Bernemann C., Joo J.Y., Han D.W., Ko K., Tapia N., Sabour D., Sterneckert J., Tesar P. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J.S., Eyres I., Mansfield W., Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Markoulaki S., Mitalipova M., Cheng A.W., Cassady J.P., Staerk J., Carey B.W., Lengner C.J., Foreman R., Love J. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J.H., Saha K., Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Lopes S.M., Tang F., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Kim J.B., Sebastiano V., Wu G., Arauzo-Bravo M.J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kunath T., Saba-El-Leil M.K., Almousailleakh M., Wray J., Meloche S., Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A., Brickman J.M. Gene expression heterogeneities in embryonic stem cell populations: origin and function. Curr. Opin. Cell Biol. 2011;23:650–656. doi: 10.1016/j.ceb.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Miyanari Y., Torres-Padilla M.E. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nichols J., Smith A. The origin and identity of embryonic stem cells. Development. 2011;138:3–8. doi: 10.1242/dev.050831. [DOI] [PubMed] [Google Scholar]
- Nichols J., Silva J., Roode M., Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Ogawa K., Shimosato D., Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Ong C.T., Corces V.G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C.T., Corces V.G. Enhancers: emerging roles in cell fate specification. EMBO Rep. 2012;13:423–430. doi: 10.1038/embor.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce M., Wang X., Wolgemuth D.J., Scholer H. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech. Dev. 1998;71:89–98. doi: 10.1016/s0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Saitou M., Barton S.C., Surani M.A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 2008;9:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- Scholer H.R. Octamania: the POU factors in murine development. Trends Genet. 1991;7:323–329. doi: 10.1016/0168-9525(91)90422-m. [DOI] [PubMed] [Google Scholar]
- Scholer H.R., Dressler G.R., Balling R., Rohdewohld H., Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.E., Hubner K., Scholer H., Mann J.R. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 2002;115:157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., Surani M.A. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen T.W., Powell B.E., Wang H., Mitalipova M., Faddah D.A., Reddy J., Fan Z.P., Maetzel D., Ganz K., Shi L. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla M.E., Chambers I. Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Vallier L., Touboul T., Chng Z., Brimpari M., Hannan N., Millan E., Smithers L.E., Trotter M., Rugg-Gunn P., Weber A. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS One. 2009;4:e6082. doi: 10.1371/journal.pone.0006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg D.L., Zhang W., Yates A., Engelen E., Takacs K., Bezstarosti K., Demmers J., Chambers I., Poot R.A. Estrogen-related receptor beta interacts with Oct4 to positively regulate Nanog gene expression. Mol. Cell. Biol. 2008;28:5986–5995. doi: 10.1128/MCB.00301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R., Smith A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 2011;13:838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Scholer H.R. Role of Oct4 in the early embryo development. Cell Regen. 2014;3:7. doi: 10.1186/2045-9769-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M., Ueda J., Hayashi K., Ohta H., Yabuta Y., Kurimoto K., Nakato R., Yamada Y., Shirahige K., Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Yeom Y.I., Fuhrmann G., Ovitt C.E., Brehm A., Ohbo K., Gross M., Hubner K., Scholer H.R. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Ying Q.-L., Smith A.G. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner G.E., Tesar P.J., Scacheri P.C. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.