Abstract
Background
Reduced lung function in patients with chronic obstructive pulmonary disease (COPD) is likely due to both environmental and genetic factors. We report here a targeted high-throughput DNA sequencing approach to identify new and previously known genetic variants in a set of candidate genes for COPD.
Methods
Exons in 22 genes implicated in lung development as well as 61 genes and 10 genomic regions previously associated with COPD were sequenced using individual DNA samples from 68 cases with moderate or severe COPD and 66 controls matched for age, gender and smoking. Cases and controls were selected from the Obstructive Lung Disease in Northern Sweden (OLIN) studies.
Results
In total, 37 genetic variants showed association with COPD (p < 0.05, uncorrected). Several variants previously discovered to be associated with COPD from genetic genome-wide analysis studies were replicated using our sample. Two high-risk variants were followed-up for functional characterization in a large eQTL mapping study of 1,111 human lung specimens. The C allele of a synonymous variant, rs8040868, predicting a p.(S45=) in the gene for cholinergic receptor nicotinic alpha 3 (CHRNA3) was associated with COPD (p = 8.8 x 10−3). This association remained (p = 0.003 and OR = 1.4, 95 % CI 1.1-1.7) when analysing all available cases and controls in OLIN (n = 1,534). The rs8040868 variant is in linkage disequilibrium with rs16969968 previously associated with COPD and altered expression of the CHRNA5 gene. A follow-up analysis for detection of expression quantitative trait loci revealed that rs8040868-C was found to be significantly associated with a decreased expression of the nearby gene cholinergic receptor, nicotinic, alpha 5 (CHRNA5) in lung tissue.
Conclusion
Our data replicate previous result suggesting CHRNA5 as a candidate gene for COPD and rs8040868 as a risk variant for the development of COPD in the Swedish population.
Electronic supplementary material
The online version of this article (doi:10.1186/s12890-016-0309-y) contains supplementary material, which is available to authorized users.
Keywords: COPD, Sequencing, eQTL, Association, Lung development, CHRNA5
Background
Chronic obstructive pulmonary disease (COPD), characterised by a persistent airflow obstruction [1], is a life-threatening disease accounting for 6 % of all deaths globally in 2012 [2]. The development of the disease is influenced by environmental determinants, most commonly cigarette smoking, genetic risk factors and possible genetic protective factors [3]. Candidate gene association studies have suggested several potential COPD susceptibility genes, and genome-wide association studies (GWAS) have identified multiple COPD susceptibility loci [4]. However, genetic mapping in families with high penetrance for a disease gene variant can be helpful in pinpointing new susceptibility genes even for multifactorial traits. Recently, we reported mutations in the gene for fibroblast growth factor 10 (FGF10) involved in lung development, as a possible cause of COPD in families from Sweden [5]. Hence, a monogenic form of COPD could result from mutations in FGF10. To date, the only other known monogenic form of COPD is alpha 1-antitrypsin deficiency caused by disruption of the alpha-1-antiproteinase (SERPINA1) gene [6].
Typically in GWAS, common polymorphisms are tested for association. In this study, we provide an alternative approach with the aim to perform an in-depth analysis of exons of candidate genes for COPD by using high-throughput sequencing. This allowed us to detect the full spectrum of single nucleotide variation at any frequency in selected genomic regions and to also capture variants with a potential functional effect on gene expression levels. We show here that targeted high throughput sequencing using a well-defined population-based case–control sample can i) assess the impact of common variants in genes important for lung development, and ii) test genetic variants in a large set of candidate genes and genomic regions for association with COPD. To accomplish this we captured and sequenced 22 genes implicated in lung development as well as 61 genes and 10 genomic regions previously associated with COPD. The sample used here is comprised of cases and controls from The Obstructive Lung Disease in Northern Sweden (OLIN) studies. The population in northern Sweden, an admixture of three different ethnic groups (Swedes, Finns and Saami), showed a dramatic growth of population size since the 18th century from a relatively small founder population [7]. This resulted in founder effects that significantly reduced the heterogeneity of this population, making it suitable for genetic association studies of multifactorial phenotypes, such as COPD [8].
This study assessed Swedish COPD cases and controls and assessed detected variants in candidate genes for association with COPD. We replicated a previous described association signal in CHRNA3, which also associated with lower CHRNA5 gene expression. The DNA capture design and targeted sequencing used here show potential to detect known single nucleotide variants in association with COPD with the additional potential to also detect low-frequent variants. The result presented here using the relative limited sample size could be replicated using our targeted capture design in larger samples from different populations.
Methods
Patient material and ethics statement
The OLIN studies are an on-going research program focused on asthma, allergy and COPD. It started 30 years ago [9] and now involves more than 50,000 subjects from northern Sweden. Within OLIN, a COPD-cohort was identified at re-examination of several cohorts in 2002–2004 [10]. At recruitment, COPD (n = 993) was defined using the fixed ratio of FEV1 / FVC < 0.70 (forced expiratory volume in 1 s / forced vital capacity). When calculating the ratio FEV1 / FVC, the highest values of FEV1 and the highest value of forced vital capacity (FVC) or slow vital capacity (SVC) were used. This has support in the GOLD documents [1] and is acknowledged in the recent ERS task force guidelines for epidemiological studies on COPD [11]. An age and gender matched control population (n = 993) without obstructive lung function impairment was also recruited [10]. Since 2005 the OLIN COPD cohort with corresponding controls is followed up annually with a basic program including spirometry and interviews regarding symptoms and morbidity [12]. We initially selected 96 COPD cases (18 non-smokers, 43 former smokers and 35 smokers) from those who had an FEV1 < 80 % of predicted value in 2005 and either FEV1 / FVC < LLN (lower limit of normal) in 2010 or were rapid decliners with an annual FEV1 decline of ≥ 60 ml between 2005 and 2010. We also identified a set of 96 age- and gender-matched controls (33 smokers and 63 former smokers) with normal lung function. These 96 cases and 96 controls are henceforth termed the OLIN discovery sample (Table 1). Furthermore, we defined an OLIN replication sample consisting of individuals from the OLIN COPD study for which DNA was available (n = 1,534). From this group we classified individuals as cases when FEV1 / FVC was lower than LLN in 2010, or if they had a yearly FEV1 decline from 2005 to 2010 of at least 60 ml (n = 256). The remaining individuals were used as a reference group (n = 1,278). The physiological parameters for the OLIN replication sample included average age (cases 64 years, SD 11; controls 66 years, SD 11; P = 0.0012), gender (cases 123 females, 133 males; controls 569 females, 709 males; P = 0.30), smoking habits (cases 11 pack/year, SD 15; controls 23 pack/year, SD 17; P = 4.2 × 10−9), weight (cases 73 kg, SD 15; controls 77 kg, SD 14; P = 1.2 × 10−4) and height (cases 168 cm, SD 9; controls 168 cm, SD 10; P = 0.8). The phenotype description of this sample included measures of FVC (cases 3.29, SD 1.01; controls 3.50, SD 1. 03; P = 0.0031), FEV1 (cases 1.90, SD 0.67; controls 2.64, SD 0.81; P = 9.5 × 10−43) and FEV1% of predicted values (cases 67 %, SD 17 %; controls 95 % SD 16 %; P = 5.3 × 10−73), as well as the FEV1/FVC ratio (cases 0.57, SD 0.08; controls 0.75, SD 0.07; P = 2.0 × 10−108) when investigated the fifth year of the evaluations. For description of individual reference values, see Additional file 1.
Table 1.
Pulmonary function in patients and controls
| Average | FVC (cm3) | FVC LLN (cm3) | FVC pred (cm3) | FEV1 (cm3) | FEV1 LLN (cm3) | FEV1 pred (cm3) | FEV1%pred (%) | FEV1 / FVC | FEV1 / FVC LLN | FEV / FVC pred |
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | 2.69 (0.95) | 2.87 (0.81) | 3.79 (0.81) | 1.50 (0.59) | 2.01 (0.55) | 2.78 (0.61) | 53 (14) | 0.56 (0.11) | 0.62 (0.03) | 0.73 (0.03) |
| Controls | 3.96 (0.90) | 2.77 (0.77) | 3.83 (0.77) | 3.11 (0.69) | 2.05 (0.55) | 2.82 (0.59) | 110 (9) | 0.79 (0.05) | 0.62 (0.03) | 0.74 (0.03) |
| P value | 6.3 × 10−18 | 0.42 | 0.75 | 4.0 × 10−41 | 0.62 | 0.58 | 2.5 × 10−81 | 2.5 × 10−45 | 0.24 | 0.23 |
Values in parenthesis denote standard deviations. FVC Forced vital capacity, LLN Lower limit of normal, FVC pred, predicted FVC based on age, gender and length in the population. FEV1 Forced expiratory volume in 1 s, FEV1 Predicted FEV1 based on age, gender and length in the population, FEV1%pred, FEV1 Divided by predicted FEV1 in the population, FEV1 / FVC ratio when investigated the fifth year of the evaluations. The lower limit of normal (LLN) values where calculated by subtracting 1.645 × RSD (residual standard deviation) from the predicted value (pred). FEV1%pred is the measured FEV1 value divided by the predicted value (i.e., FEV1 / FEV1pred). P values are calculated using two-sided Student’s t-test assuming equal variance. SD Standard deviation
P values for differences in parameters between cases and controls were calculated using two-sided Student’s t-tests, assuming equal variance. The ethics board of Umeå University (Dnr 04-045 M, supplement approved 2005-06-13) approved the use of individual phenotypic data and DNA samples for genetic research.
Sequencing and quality controls
In total 22 genes implicated in lung development, 61 genes and 10 genomic regions previously associated with COPD (Additional files 1 and 2), were investigated using targeted sequencing of captured genomic regions (HaloPlex Protocol Version A, Agilent Technologies, Santa Clara, CA). Regions of 1.5 kb of genomic sequence, including specific intergenic polymorphisms, was also included in the design. The regions of interest (ROI) were designed to target all known exons of major/known transcripts and at least 20 base pairs (bp) of intronic sequences flanking each exon. The sequence capture design included 953 target regions spanning 204.384 bp with 95.9 % (196.066 bp) coverage an average. Captured genomic regions were subjected to high throughput paired-end (100 bp read module) sequencing (HiSeq2000, Illumina, San Diego, CA) at the Science for Life Laboratory in Uppsala, Sweden. Sequence reads were aligned to the hg19 reference genome and single nucleotide variants (SNVs) were called using GATK Unified Genotyper (GATK bundle v.2.2) [13]. Next, we enriched for high quality SNVs by removing SNVs with low confidence (QD < 1.5), Phred scaled quality score (<50) and SNVs within SNV clusters. These high quality variants are henceforth referred to as ‘variants’. We also removed individual cases and controls with sequencing read depth consistently < 10 reads. The strategy for filtering and quality controls are illustrated in Fig. 1.
Fig. 1.

Flow chart of the strategy for variant calling, quality control and association tests. QC = quality control
Statistical analyses
Test for genetic association and genetic effect was performed for each predicted variant separately using the discovery sample. In addition, rs8040868 and rs11728716 were tested for association using the available OLIN sample (replication sample) as, according to RegulomeDB, these two markers present with potential functional effects on gene regulation (Table 2). Tests for allelic association of individual variants with COPD were performed using the Fisher’s exact test. Results were considered statistically significant when p < 0.05. No adjustment for multiple testing was performed in these analyses. Effect size was measured using odds-ratios (OR) with 95 % confidence intervals (CI).
Table 2.
Associated genetic variants in the discovery sample set (n = 96 cases and n = 96 controls)
| Chr | Pos (bp) | SNP rs ID | Alt | Ref | F c (%) | F p (%) | OR (95 % CI) | P value | Gene | Coding | RG score | ExAc Eur frequencya | CADD_PHREDb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 110230569 | 72989301 | G | A | 10.0 | 22.1 | 2.5 (1.0-6.2) | 0.038 | GSTM1 | 4 | 0.24 | NA | |
| 1 | 110233057 | 111436983 | C | T | 11.6 | 26.7 | 2.8 (1.2-6.2) | 0.013 | GSTM1 | 7 | 0.24 | NA | |
| 2 | 216991935 | 12694384 | A | C | 29.4 | 17.9 | 0.5 (0.3-0.9) | 0.04 | XRCC5 | 5 | NA | NA | |
| 2 | 218669225 | 61741262 | C | T | 13.6 | 0 | NA | 0.027 | TNS1 | p.(Asp1722Ser) | 4 | 0.13 | 12.49 |
| 2 | 218746990 | 2303381 | T | A | 7.7 | 1.5 | 0.2 (0.0-0.9) | 0.019 | TNS1 | 6 | NA | NA | |
| 3 | 55520778 | 566926 | T | G | 39.2 | 25.0 | 0.5 (0.3-0.9) | 0.017 | WNT5A | 3a | NA | NA | |
| 4 | 106638697 | 72671840 | G | A | 2.3 | 11.4 | 5.3 (1.5-18.9) | 5.8 × 10−3 | GSTCD | 6 | NA | NA | |
| 4 | 106647679 | 72671858 | T | C | 3.0 | 10.5 | 3.7 (1.2-11.7) | 0.026 | GSTCD | 7 | NA | NA | |
| 4 | 106755996 | 11728716 | A | G | 3.4 | 21.0 | 7.4 (2.5-22.5) | 6.5 × 10−5 | GSTCD | 1f | NA | NA | |
| 5 | 58284208 | 3805557 | C | T | 22.0 | 11.9 | 0.5 (0.2-0.9) | 0.034 | PDE4D | 7 | NA | NA | |
| 5 | 58284283 | 3805556 | G | A | 22.0 | 11.8 | 0.5 (0.2-0.9) | 0.033 | PDE4D | 6 | 0.84 | NA | |
| 5 | 58286625 | 1553114 | C | T | 22.0 | 11.8 | 0.5 (0.2-0.9) | 0.033 | PDE4D | 7 | 0.84 | NA | |
| 5 | 141993867 | 17223611 | T | C | 10.6 | 3.0 | 0.3 (0.1-0.8) | 0.025 | FGF1 | 5 | NA | NA | |
| 6 | 142703137 | 2143390 | T | C | 2.9 | 19.2 | 7.9 (1.6-37.6) | 4.5 × 10−3 | GPR126 | p.(Asp373=) | 7 | 0.12 | NA |
| 6 | 151197501 | 9322290 | C | T | 17.4 | 9.0 | 0.5 (0.2-1.0) | 0.047 | MTHFD1L | 5 | NA | NA | |
| 6 | 151206894 | 147872265 | T | C | 7.6 | 2.2 | 0.3 (0.1-1.0) | 0.049 | MTHFD1L | 7 | 0.002 | NA | |
| 6 | 151263456 | 803451 | A | G | 41.8 | 55.7 | 1.8 (1.1-2.9) | 0.04 | MTHFD1L | 7 | NA | NA | |
| 6 | 151264132 | 803448 | T | C | 37.9 | 50.7 | 1.7 (1.0-2.7) | 0.037 | MTHFD1L | 6 | NA | NA | |
| 6 | 152183551 | 1643821 | A | G | 25.8 | 39.0 | 1.8 (1.1-3.1) | 0.026 | ESR1 | 6 | NA | NA | |
| 8 | 42552530 | 41272375 | C | G | 1.5 | 6.9 | 4.8 (1.0-22.8) | 0.034 | CHRNB3 | 5 | NA | NA | |
| 9 | 98239503 | 3780573 | A | G | 18.3 | 8.7 | 0.4 (0.2-0.9) | 0.041 | PTCH1 | 5 | NA | NA | |
| 10 | 81706281 | 6413520 | G | A | 0.8 | 5.9 | 8.2 (1.0-66.4) | 0.036 | SFTPD | p.(Ser45=) | 5 | 0.07 | NA |
| 10 | 123358096 | 41301039 | G | C | 25.0 | 2.8 | 0.1 (0.0-0.7) | 0.017 | FGFR2 | 4 | NA | NA | |
| 11 | 102738499 | 632009 | T | C | 29.6 | 50.8 | 2.5 (1.5-4.1) | 6.7 × 10−4 | MMP12 | 7 | NA | NA | |
| 12 | 23737566 | 11046992 | A | G | 21.2 | 33.1 | 1.8 (1.1-3.2) | 0.039 | SOX5 | 6 | NA | NA | |
| 12 | 110224916 | 60258652 | T | C | 2.4 | 10.5 | 4.7 (1.0-22.9) | 0.05 | TRPV4 | 5 | NA | NA | |
| 12 | 110224922 | 1861810 | A | C | 36.6 | 53.8 | 2.0 (1.1-3.8) | 0.04 | TRPV4 | 5 | NA | NA | |
| 12 | 110232032 | 59870578 | A | G | 7.8 | 1.6 | 0.2 (0.0-0.9) | 0.034 | TRPV4 | 4 | NA | NA | |
| 12 | 110232034 | 59940634 | T | G | 7.8 | 1.6 | 0.2 (0.0-0.9) | 0.034 | TRPV4 | 4 | NA | NA | |
| 15 | 71434029 | 2004101 | A | T | 0.8 | 5.6 | 7.5 (0.9-61.6) | 0.035 | THSD4 | 7 | NA | NA | |
| 15 | 78790189 | 2292115 | G | A | 4.8 | 0 | NA | 0.03 | IREB2 | 7 | NA | NA | |
| 15 | 78911181 | 8040868 | C | T | 31.8 | 47.8 | 2.0 (1.2-3.2) | 8.8 × 10−3 | CHRNA3 | p.(Val53=) | 1f | 0.41 | NA |
| 16 | 16130514 | 903880 | A | C | 15.9 | 27.2 | 2.0 (1.1-3.6) | 0.027 | ABCC1 | 4 | NA | NA | |
| 16 | 16205741 | 9673292 | C | G | 4.5 | 0 | NA | 0.013 | ABCC1 | 6 | NA | NA | |
| 16 | 16230290 | 212087 | A | G | 38.6 | 52.2 | 1.7 (1.1-2.8) | 0.028 | ABCC1 | 5 | 0.44 | NA | |
| 16 | 16235366 | 113328089 | A | G | 6.9 | 1.5 | 0.2 (0.0-1.0) | 0.034 | ABCC1 | 5 | NA | NA | |
| 20 | 15967390 | 41275442 | T | C | 9.1 | 17.7 | 2.1 (1.0-4.5) | 0.049 | MACROD2 | p.(Thr100Met) | 4 | 0.18 | 3.98 |
Alt, non-reference allele. Ref, reference allele, OR Odds ratio with 95 % confidence intervals, F c Frequency in controls, F p Frequency in cases (patients), RG Score, RegulomeDB score, NA Not available
aNon-Finnish European allele frequency extracted from ExAc v.0.3.1
bThe “PHRED-scaled” CADD score based on ranks of all SNV in the hg19 genome reference
Visualization SNPs associated with COPD located in the CHRNA3/5 region was made using LocusZoom v1.1 [14] available on http://locuszoom.sph.umich.edu/locuszoom/. RefSeq gene/transcript case–control tests for aggregation of genetic variants in the targeted genomic regions were performed using PLINK/SEQ v0.1 [15]. Tests were divided into an analysis of rare variants with minor allele frequency (MAF) < 5 % or common variants (MAF ≥ 5 %). The UNIQ test, which identifies unique risk alleles, was utilized using default parameters to count the total number of alleles found only in cases (risk variants). Similarly, the SKAT burden tests, which assesses excess of rare alleles in cases compared to controls, was also utilized. Since both UNIQ and SKAT burden are 1-sided tests, we also swapped the phenotype information and analyzed the effects in both directions (excess of alleles in cases or controls) separately to capture evidence of both risk and protective alleles. Due to the matched design of the case and control groups no covariate adjustments (age, sex, pack-years) were performed in analysis using the discovery sample.
Linkage disequilibrium information of associated variants was extracted from genotypes from the sequencing analysis using Haploview 4.2 [16].
Functional analysis of associated variants
To study the possible effect of associated variants on gene expression, we used information from RegulomeDB, a database that combines ENCODE data sets (chromatin immunoprecipitation sequencing (ChIP-seq) peaks, DNase I hypersensitivity peaks, DNase I footprints) with additional data sources (ChIP-seq data from the NCBI Sequence Read Archive, conserved motifs, expression quantitative trait loci (eQTL), and experimentally validated functional variants) [17]. A scoring system is based on the confidence of the functionality of variants, a lower score corresponding to stronger confidence. Subcategories are used to denote additional functional annotations. Combined Annotation Dependent Depletion (CADD) scores were used to assess potential structural and functional effect of associated nonsynonymous variants [18].
Lung expression quantitative trait loci analyses
The existence of expression quantitative trait loci (eQTLs) was investigated as previously described using genotyping and gene expression data from 1,111 patients who underwent lung surgery at one of three sites, Laval University (discovery sample), University of British Columbia, and University of Groningen (replication sample sets) (referred to as Laval, UBC, and Groningen) [19, 20]. The eQTL data is derived from non-tumour lung parenchymal samples and expression data were adjusted for age, gender, and smoking status. Estimated P-values for each region were Bonferroni-corrected for multiple testing based on the number of SNPs and probe sets (number of SNPs x number of probe sets) and were considered significant if corrected p < 0.05.
SNP genotyping for validation of rs11728716 and rs8040868
Individuals from the OLIN replication sample (n = 1,534) were genotyped for the rs11728716 and rs8040868 variants (99.2 % and 99.8 % success rate, respectively) at the Uppsala Genome Center (Uppsala, Sweden) using commercially available TaqMan assays (Life Technologies, Carlsbad, CA). Assay conditions were according to manufacturer’s recommendations. Effect size was estimated by comparing ORs with 95 % CI between cases and controls. Furthermore, to assess smoking dependence, we measured association and effect sizes also between the groups ‘non-smokers’ and ‘ever smokers’, and between ‘current smokers’ and ‘former smokers’.
Results
Selection of cases and controls for the discovery sample
The characteristics of each sample are listed in this section as value ± standard deviation. Cases and controls were matched for age (cases: 68 ± 10 years; controls: 66 ± 11 years; p = 0.15), gender (cases: 35 females, 61 males; controls: 31 females, 65 males; p = 0.65) and smoking habits (cases: 26 ± 19 pack/year; controls: 28 ± 12 pack/year; p = 0.53). Both groups were also closely matched for weight (cases: 76 ± 15 kg; controls: 77 ± 15 kg; p = 0.85) and height (cases: 169 ± 9 cm; controls: 169 ± 9 cm; p = 0.7). No non-smokers were included in the control group to avoid false negative results. The cases presented a significant reduction in lung function consistent with moderate or severe COPD. This is illustrated by a reduced FVC (cases: 2.69 ± 0.95 L; controls: 3.96 ± 0.90 L; p = 6.3 × 10−18), FEV1 (cases: 1.50 ± 0.59 L; controls: 3.11 ± 0.69 L; p = 4.0 × 10−41) and FEV1% of predicted values (cases: 53 ± 14 %; controls: 110 ± 9 %; p = 2.5 × 10−81), as well as the FEV1/FVC ratio (cases: 0.56 ± 0.11; controls: 0.79 ± 0.05; p = 2.5 × 10−45) when investigated the fifth year of the evaluations (Table 1).
Test for association between genetic variants and COPD
We identified 2,151 SNVs after analysis of the sequenced target regions. After variant and sample quality control procedures, 1588 SNVs and 68 cases and 66 controls were retained in the downstream analysis (Fig. 1). Out of the 1588 variants, we identified 37 variants with significantly different allele frequencies in cases and controls (henceforth referred to as ‘associated variants’) (Table 2). We initially detected two novel variants in the discovery sample: GRCh37.p13, 5:g.157002804C > G in the ADAM19 gene and GRCh37.p13, 7:g.73477874C > A in ELN. However, using Sanger sequencing of the same sample we excluded both variants, as they were monomorphic.
Three of the associated variants were shown to be unique to controls including missense variant rs61741262 (p.Asn1722Ser) in TNS1. The most significantly associated variants were all intronic (GSTCD, rs11728716, p = 6.5 × 10−5, OR = 7.4 (2.5-22.5) and MMP12, rs632009, p = 6.7 × 10−4, OR = 2.5 (1.5-4.1).
Although the majority of the associated variants were intronic (or intergenic), five were protein-coding (Table 2). Of these, two variants predicted amino-acid substitutions (missense variants): p.(Thr100Met) in MACROD2 and p.(Asn1722Ser) in TNS1 respectively. The p.(Asn1722Ser) variant could be potential damaging based on a relative high CADD score or 12.49. Of the coding variants, we found that rs2143390, predicting a p.(D373=) in GPR126 (p = 0.005, OR = 7.9 (1.6-37.6)), rs6413520 in SFTPD (p.(Ser45=), p = 0.036, OR = 8.2 (1.0-66.4)), rs8040868 in CHRNA3 (p.(Val53=), p = 8.8 × 10−3, OR = 2.0 (1.2-3.2)) and rs41275442 in MACROD2 (p.(Thr100Met), p = 0.049, OR = 2.1 (1.0-4.5)) conferred moderate to high risk for COPD (Table 2).
We tested the presence of variants uniquely found in cases or controls as well as gene burden tests in cases against controls as specified in PLINK/SEQ v0.1 using standard settings. We noted that the ADAM19, WNT2, CHRNA5, NOS3 and PTCH1 genes all harbor rare variants (MAF < 5 %) uniquely found in cases (Additional file 3). Conversely, the FGF8, CTNNB1 and HHIP genes contain rare variants uniquely found in the control sample (Additional file 3). Neither gene burden analysis (SKAT) or analysis of rare alleles (MAF < 5 %) yielded significant results. However, by performing a joint analysis with only common alleles (MAF ≥ 5 %) in target regions using SKAT, we showed a significant gene burden for the genes GSTCD, FGF1, ELN and ESR1 (Additional file 4).
Haplotypes and linkage disequilibrium
We identified five regions with associated variants in pairwise LD (r2 > 0.7, D´ = 1.0). The regions were located at the GSTM1 gene locus on chromosome 1 (rs72989301-rs111436983), GSTCD on chromosome 4 (rs72671840-rs72671858), PDE4D on chromosome 5 (rs3805557- rs3805556- rs1553114), MTHFDIL on chromosome 6 (rs803451- rs803448) and TRPV4 on chromosome 12 (rs59870578-rs59940634) (Additional file 5). Furthermore, the variant rs8040868 on chromosome 15q21.1 is in pairwise LD (r2 = 0.76, D’ = 1.0) with rs16969968, a nonsynonymous variant previously associated with expression of the CHRNA5 gene [21]. The rs16969968 variant was included in our capture design but it did not reach significant association in the OLIN discovery sample (OR 1.6; p = 0.07) (Additional file 6).
In silico analysis of predicted functions of associated variants
According to RegulomeDB, all 37 associated variants were located within known and predicted regulatory elements in intergenic regions (Table 2). We noted that a variant in CHRNA3 (rs8040868) and a variant in GSTCD (rs11728716) each showed a RegulomeDB score of “1f”, denoting the presence of transcription binding site or DNAse peak.
Lung eQTL results
According to RegulomeDB, the rs8040868 (CHRNA3) and rs11728716 (GSTCD) variants could present with potential functional effects on gene regulation. To determine if these variant could represent eQTL, we analysed the genotypes and gene expression data in the discovery sample (Laval) as well as replication samples (UBC and Groningen). One of these variants, rs8040868:C > T, was confirmed to be significantly associated with gene expression of the nearby gene CHRNA5 in all three data sets, with the C allele (minor allele) associated with lower CHRNA5 expression (Fig. 2 and Additional file 7). Interestingly, we could also see a high correlation between rs8040868 and expression of an anti-sense transcript (AF147302) of unknown function from the adjacent IREB2 gene region (data not shown). AF147302 is likely a result of strong bi-directional promoter activity in this region [22].
Fig. 2.
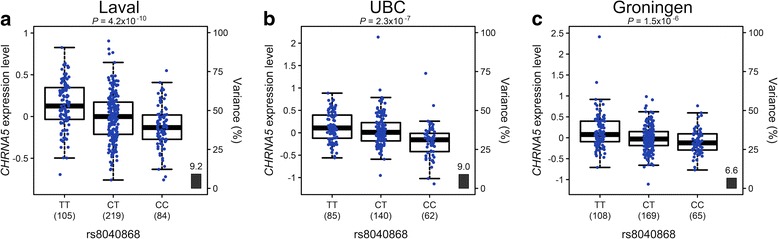
CHRNA5 gene expression levels in the lungs according to genotype groups for rs8040868. The left y-axis represents standardised gene expression levels in the lung with heterozygous genotype group set to zero. The x-axis represents the three genotyping groups, TT, CT and CC (risk allele C), for the variant in (a) the discovery set (Laval) n = 408; P = 4.2 × 10−10, and the replication sets (b) UBC, n = 287; P = 2.31 × 10−7, and (c) Groningen, n = 342; P = 1.5 × 10−6. The number of subjects per genotype group is indicated in parenthesis. The right y-axis shows the proportion of the gene expression variance explained by the variant (black bar)
The rs8040868 variant is associated with COPD in the OLIN replication sample
We also investigated pulmonary data from the replication sample (n = 1,534; cases = 256, controls = 1278). Analysis using RegulomeDB predicted both rs11728716 and rs8040868 variants as being functional (score 1f for both variants). We therefore selected these two variants for genotyping in all available OLIN samples (n = 1,534). The frequency of the rs8040868-C allele was 35 % in the reference group (n = 1,278) and 42 % in the cases (n = 256) resulting in a significant association (p = 0.003) and an OR of 1.4 (95 % CI 1.1-1.7) for COPD. SweGen variant frequency database reports a 39 % frequency of rs8040868 in 1000 whole genomes representing a cross-section of the Swedish population (https://swefreq.nbis.se). The frequency of the homozygous rs8040868-CC genotype was 12 % in the reference group and significantly higher (18 %) in the cases (p = 0.018). When comparing smoking status using all genotyped individuals, no significant difference in allele frequency between neither the groups ‘non-smokers’ (n = 589) and ‘ever smokers’ (n = 943) (OR 1.1 95 % CI 1.0-1.7; p = 0.09) nor between the groups ‘current smokers’ (n = 312) and ‘former smokers’ (n = 631) (OR 1.2 95 % CI 1.0-1.4; p = 0.11) was seen. The latter test was used to assess nicotine dependency and aptitude for smoking cessation under the assumption that a genetic variant associated with these traits would be underrepresented in a former smoking group as compared to a group of current smokers, i.e., harder to quit smoking. The tests for association with smoking must however be taken with caution as the confidence intervals are wide and a larger sample size would be needed for replication.
Analysis of rs11728716 using the OLIN replication sample (n = 1,534) revealed no association with COPD (p = 0.07; OR = 2.2). In order to test if rs11728716 is associated with severe COPD, we stratified the available COPD cases based on severity and selected cases with FEV1%pred < 40 % and FEV1/FVC < LLN. Our results show a significant association (p = 0.017) between rs11728716 and the group of severe COPD (n = 14). The allele frequency of rs11728716-A was 10 % among cases with severe COPD and 4 % in the controls.
Discussion and conclusion
Genetic variants influencing lung function in children and adults may ultimately lead to the development of COPD [23]. Since limited disease-specific therapy for COPD is available, an improved knowledge of genetic variants modulating the pathogenic mechanisms underlying COPD is greatly needed. We aimed here to identify genetic variants within, or close to, the coding regions of genes and loci previously associated with COPD, or in genes involved in lung development. We opted for a qualitative rather than a quantitative approach with the selection of cases with moderate or severe COPD and progressive decline in lung function. Furthermore, controls were all smokers without COPD that, in our study design, can aid the identification of potential protective genetic variants and aid detection of genetic variants associated with severe COPD. When applying a Bonferroni correction for the total number of variants detected, no variants showed statistically significant association. We did, however, identify several variants with a likely biological significance, as indicated by high effect sizes (odds ratio), that we believe warrants further investigation in a larger sample. Furthermore, potential functional effects of variants were investigated using data from a large number of lung samples and we describe here a COPD lung eQTL.
When comparing our association data with the lung eQTL data (discovery data set from Laval University), we could identify a variant associated with COPD that was also associated with level of gene expression (Fig. 2). This variant, synonymous variant (rs8040868) in CHRNA3 on chromosome 15, confers a risk for the development of COPD in both our OLIN discovery sample with moderate or severe COPD and our OLIN replication sample including all available COPD cases and controls in OLIN (OR 1.4, p = 0.003). In the lung eQTL data, we could see a correlation of the C allele of rs8040868 with lower expression levels of CHRNA5 (Fig. 2), and, to a lesser extent, also CHRNA3 and PSMA4, which are located in close proximity to CHRNA5. The α-nicotinic receptor (CHRNA3/5) gene locus on chromosome 15q25.1 is associated with COPD, lung cancer and peripheral arterial disease, as well as other smoking related conditions [24, 25] and nicotine addiction [26, 27]. Recently, the CHRNA3/5 locus was implicated in all-cause mortality among smokers in a Finnish cohort [2]. The rs8040868-C allele associates with both reduced pulmonary function and lung cancer [24, 25, 28, 29] and affects DNA-methylation and transcription of CHRNA5 [30]. Furthermore, rs8040868 is also in LD with a nearby variant (rs16969968) previously reported to be associated with expression levels of CHRNA5 in the lung [21]. The direction of effect is the same for both SNPs, with the minor alleles associated with reduced expression of CHRNA5. Also recently, rs16969968 was found to be the most significantly associated variant in an exome array analysis in a study including more than 6,100 COPD cases and 6,000 control subjects across five cohorts [31].
Several genetic variants showed association with COPD in our population, but did not correlate with gene expression levels in the lung, including previously identified variants in the genes glutathione S-transferase, c-terminal domain containing (GSTCD), surfactant protein D (SFTPD) and matrix metalloproteinase-12 (MMP12) [32–36]. We identified a haplotype consisting of three risk-conferring variants, rs72671840, rs72671858 and rs11728716 (G-T-A haplotype), at the GSTCD gene locus on chromosome 4q24. The variant rs11728716 has previously been associated with lung function [32–34] and is likely to affect the transcription of GSTCD. We show here that rs11728716 was associated with severe COPD using the OLIN replication sample. Although intriguing, due to the limited number of severe COPD cases used in this study, this result needs further verification in a larger sample. The other two variants (rs72671840 and rs72671858) are of unknown function [37]. GSTCD encodes a glutathione S-transferase C-terminal domain protein involved in detoxification by catalysing conjugation of glutathione to products of oxidative stress. We found association between COPD and rs6413520, a synonymous variant, p.(Ser45=), within SFTPD on chromosome 10q22.3. This variant conferred a high risk (OR = 8.2) for COPD in our study and has previously been reported to be associated with COPD susceptibility [36]. SFTPD encodes surfactant protein D, of importance for the regulation of oxidant production, inflammatory responses, and apoptotic cell clearance in the lung [38]. We also identified rs632009, in the MMP12 gene on chromosome 11q22.3, to confer moderate risk. Matrix metalloproteinases (MMPs) are involved in both tissue remodelling and repair and several members of the MMP family have been implicated in COPD pathology [35, 39, 40].
In this study, we also found association (uncorrected) with novel susceptibility variants. Several variants in the G-protein-coupled receptor 126 (GPR126) gene on chromosome 6q24.1 have previously been associated with FEV1 / FVC ratio [32]. GPR126 belongs to a superfamily of G protein-coupled receptors and is involved in cell signalling and adhesion. Studies in mice show an induction of Gpr126 expression between embryonic day 7 and 11 with expression in the developing heart and face as well as a high expression in the adult lung [41]. We found significant association between a synonymous variant in the GPR126 gene (rs2143390, p.(Asp373=)) and COPD. The alternative T allele is highly overrepresented in cases compared to controls (p = 4.5 × 10−3, OR = 7.9).
We also focused our attention to the chromosome 4q31 locus upstream of HHIP, previously shown to be associated with expression of the gene [20, 42]. The HHIP upstream region belongs to one of the so far strongest COPD association signals [43], but no association could be seen in our case–control groups for any upstream variants.
The sequencing approach allowed us to detect rare alleles in both cases and controls. We therefore performed gene burden tests to find evidence of overrepresented rare or common variants in individual genes or transcripts in the cases or controls, respectively. Interestingly, we found that the genes ADAM19, WNT2, CHRNA5, NOS3 and PTCH1 all contain rare variants (MAF < 5 %) uniquely found in cases of the OLIN discovery sample. These variants, and especially the coding variants with predicted functional effect, could be followed up in a larger case–control sample for verification and further genetic and functional analysis.
We assessed 83 genes and 10 genomic regions of 1.5 kb size for variants associated with COPD in a sample from Northern Sweden. Still, one limitation of our study is that the targeted capture design may exclude yet unknown genomic regions that can harbour genetic variation influencing COPD. Also, the two novel variants detected after sequencing were monomorphic and an assessment of the false discovery rate using HaloPlex with subsequent Illumina sequencing would be helpful in order to evaluate our set of candidate genes as a gene panel for COPD. Furthermore, we cannot rule out that some findings are influenced by population substructure and replication of our result in different populations is essential. It is also possible that some risk variants were not identified due to the limited number of cases and controls used for sequencing. Using a conservative Bonferroni correction based on the 1588 variants detected resulted in no variants reached significant association with COPD. However, we believe there is no definite consensus regarding the type of multiple testing procedures to use in targeted sequencing based approaches. Furthermore, many parameters such as variant quality checks, genotyping success rate and sequencing depth limit will influence the number of variants found, and consequently, multiple testing adjustments. Also, in addition to include genes including variants previously associated with COPD or asthma, we explored if a set of genes involved in lung development would harbour variants in association with COPD in the Swedish discovery sample. Therefore, as the study is exploratory with a mixed hypothesis the p values for association testing in this study are not corrected for multiple testing.
Despite the limited size of the discovery sample used here, we identified several high-risk genetic variants for COPD and we replicated several previous GWAS results. In particular, our results support the CHRNA5 gene as a likely candidate gene for COPD where the rs8040868-C allele confers a risk for the disease in the Swedish population. Furthermore, we indicate the advantage of using less heterogeneous populations in the studies of complex disorders.
Acknowledgments
The authors would like to especially thank the participants and staff in the OLIN studies. The authors would further like to thank the staff at the Respiratory Health Network Tissue Bank of the FRQS for their valuable assistance with the lung eQTL dataset at Laval University.
Funding
J. Klar is funded by Svenska Sällskapet för Medicinsk Forskning (SSMF) and Magnus Bergvalls Stiftelse (014–00163). The lung eQTL study at Laval University was supported by the Chaire de pneumologie de la Fondation JD Bégin de l’Université Laval, the Fondation de l’Institut universitaire de cardiologie et de pneumologie de Québec, the Respiratory Health Network of the FRQS, the Canadian Institutes of Health Research (MOP - 123369), and the Cancer Research Society and Read for the Cure. M. Lamontagne is the recipient of a doctoral studentship from the Fonds de recherche Québec - Santé (FRQS). Y. Bossé holds a Canada Research Chair in Genomics of Heart and Lung Diseases.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the European Nucleotide Archive repository as study accession number PRJEB13652, http://www.ebi.ac.uk/ena/data/view/PRJEB13652.
Author contributions
HM, JK and BL designed the study. HM, CS and JK drafted the manuscript. HM and JK performed DNA capture experiments and COPD association analysis. EE performed gene burden analysis. SG performed Sanger sequencing validation. BL, HB, AL and ER contributed to sample selection and phenotype characterisation. ML, YB and DS conducted eQTL analyses. All authors revised the manuscript and approved the final version to be published.
Competing interests
The authors declare that they have no competing of interests.
Ethics approval and consent to participate
Informed consent of research use of spirometry data and DNA samples were obtained from the participants in the OLIN studies that include all participants in our study presented here. The ethics board of Umeå University (Dnr 04-045 M, supplement 2005-06-13) approved the use of phenotypic and genetic data for research purposes. Results in this study are presented as groups without personal identifiers.
Abbreviations
- ChIP-seq
Chromatin immunoprecipitation sequencing
- COPD
Chronic Obstructive Pulmonary Disease
- eQTLs
Expression quantitative trait loci
- FEV1
Expiratory volume in 1 s FVC: forced vital capacity
- GWAS
Genome-wide association studies
- LLN
Lower limit of normal
- OLIN
The Obstructive Lung Disease in Northern Sweden studies
- SNVs
Single nucleotide variants forced
- SVC
Slow vital capacity
Additional files
Supplementary methods. Detailed methods description regarding classification of COPD, sequencing protocol and linkage disequilibrium analysis. (DOCX 123 kb)
Targeted genes and variants for targeted high throughput sequencing. The table lists the selected genes and variants included in the study. (DOCX 79 kb)
Number of rare variants found uniquely in cases and controls. The table present data of the UNIQ test of rare variants in cases versus controls. (DOCX 78 kb)
Gene burden analysis of common variants. The table present the SKAT analysis of gene burden of common variants. (DOCX 86 kb)
Pairwise linkage disequilibrium (LD) of associated variants. A list of detected genetic variants found to be in LD. (DOCX 46 kb)
A plot containing genomic positions and p-values of variants in the CHRNA3/CHRNA5 gene locus with rs8040869 and rs16969968 highlighted. (PDF 144 kb)
Probe sets replicated in both replication sets (UBC and Groningen) in the lung eQTL analyses. A table of replicated probe sets in the lung eQTL analysis. (DOCX 38 kb)
Contributor Information
Hans Matsson, Phone: +46 733925213, Email: hans.matsson@ki.se.
Cilla Söderhäll, Email: cilla.soderhall@ki.se.
Elisabet Einarsdottir, Email: elisabet.einarsdottir@ki.se.
Maxime Lamontagne, Email: maxime.lamontagne@criucpq.ulaval.ca.
Sanna Gudmundsson, Email: sanna.gudmundsson@igp.uu.se.
Helena Backman, Email: helena.backman@nll.se.
Anne Lindberg, Email: anne.lindberg@algmed.se.
Eva Rönmark, Email: eva.ronmark@nll.se.
Juha Kere, Email: juha.kere@ki.se.
Don Sin, Email: dsin@mrl.ubc.ca.
Dirkje S. Postma, Email: d.s.postma@umcg.nl
Yohan Bossé, Email: Yohan.Bosse@criucpq.ulaval.ca.
Bo Lundbäck, Email: bo.lundback@gu.se.
Joakim Klar, Email: joakim.klar@igp.uu.se.
References
- 1.GOLD. From the Global Strategy for the Diagnosis, Management and Prevention of COPD, Global Initiative for Chronic Obstructive Lung Disease (GOLD). 2015. http://www.goldcopd.org/. Accessed date 26 Apr 2016.
- 2.WHO. Chronic obstructive pulmonary disease (COPD). 2015. Fact sheet N°315. http://www.who.int/en/. Accessed 27 Apr 2016.
- 3.Eisner MD, Anthonisen N, Coultas D, Kuenzli N, Perez-Padilla R, Postma D, et al. An official American Thoracic Society public policy statement: Novel risk factors and the global burden of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182:693–718. doi: 10.1164/rccm.200811-1757ST. [DOI] [PubMed] [Google Scholar]
- 4.Bosse Y. Updates on the COPD gene list. Int J Chron Obstruct Pulmon Dis. 2012;7:607–31. doi: 10.2147/COPD.S35294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klar J, Blomstrand P, Brunmark C, Badhai J, Hakansson HF, Brange CS, et al. Fibroblast growth factor 10 haploinsufficiency causes chronic obstructive pulmonary disease. J Med Genet. 2011;48:705–9. doi: 10.1136/jmedgenet-2011-100166. [DOI] [PubMed] [Google Scholar]
- 6.Silverman EK, Speizer FE. Risk factors for the development of chronic obstructive pulmonary disease. Med Clin North Am. 1996;80:501–22. doi: 10.1016/S0025-7125(05)70451-X. [DOI] [PubMed] [Google Scholar]
- 7.Einarsdottir E, Egerbladh I, Beckman L, Holmberg D, Escher SA. The genetic population structure of northern Sweden and its implications for mapping genetic diseases. Hereditas. 2007;144:171–80. doi: 10.1111/j.2007.0018-0661.02007.x. [DOI] [PubMed] [Google Scholar]
- 8.Kristiansson K, Naukkarinen J, Peltonen L. Isolated populations and complex disease gene identification. Genome Biol. 2008;9:109. doi: 10.1186/gb-2008-9-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lundback B, Nystrom L, Rosenhall L, Stjernberg N. Obstructive lung disease in northern Sweden: respiratory symptoms assessed in a postal survey. Eur Respir J. 1991;4:257–66. [PubMed] [Google Scholar]
- 10.Lindberg A, Lundback B. The Obstructive Lung Disease in Northern Sweden Chronic Obstructive Pulmonary Disease Study: design, the first year participation and mortality. Clin Respir J. 2008;2(Suppl 1):64–71. doi: 10.1111/j.1752-699X.2008.00086.x. [DOI] [PubMed] [Google Scholar]
- 11.Bakke PS, Ronmark E, Eagan T, Pistelli F, Annesi-Maesano I, Maly M, et al. Recommendations for epidemiological studies on COPD. Eur Respir J. 2011;38:1261–77. doi: 10.1183/09031936.00193809. [DOI] [PubMed] [Google Scholar]
- 12.Stridsman C, Mullerova H, Skar L, Lindberg A. Fatigue in COPD and the impact of respiratory symptoms and heart disease--a population-based study. COPD. 2013;10:125–32. doi: 10.3109/15412555.2012.728642. [DOI] [PubMed] [Google Scholar]
- 13.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.PLINK/SEQ. O. source. 2014. https://atgu.mgh.harvard.edu/plinkseq/start-pseq.shtml. Accessed 20 Oct 2015.
- 16.Haploview 4.2. B. Institute. 2009. http://www.broadinstitute.org/haploview/haploview. Accessed 23 Oct 2015.
- 17.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao K, Bosse Y, Nickle DC, Pare PD, Postma DS, Laviolette M, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamontagne M, Couture C, Postma DS, Timens W, Sin DD, Pare PD, et al. Refining susceptibility loci of chronic obstructive pulmonary disease with lung eqtls. PLoS One. 2013;8:e70220. doi: 10.1371/journal.pone.0070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen JD, Lamontagne M, Couture C, Conti M, Pare PD, Sin DD, et al. Susceptibility loci for lung cancer are associated with mRNA levels of nearby genes in the lung. Carcinogenesis. 2014;35:2653–9. doi: 10.1093/carcin/bgu184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uesaka M, Nishimura O, Go Y, Nakashima K, Agata K, Imamura T. Bidirectional promoters are the major source of gene activation-associated non-coding RNAs in mammals. BMC Genomics. 2014;15:35. doi: 10.1186/1471-2164-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai TJ, Cardoso WV. Growth factors in lung development and disease: friends or foe? Respir Res. 2002;3:2. doi: 10.1186/rr169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–90. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–2. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–25. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock DB, Artigas MS, Gharib SA, Henry A, Manichaikul A, Ramasamy A, et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 2012;8:e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scherf DB, Sarkisyan N, Jacobsson H, Claus R, Bermejo JL, Peil B, et al. Epigenetic screen identifies genotype-specific promoter DNA methylation and oncogenic potential of CHRNB4. Oncogene. 2013;32:3329–38. doi: 10.1038/onc.2012.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hobbs BD, Parker MM, Chen H, Lao T, Hardin M, Qiao D, et al. Exome Array Analysis Identifies a Common Variant in IL27 Associated with Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2016;194:48–57. doi: 10.1164/rccm.201510-2053OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soler Artigas M, Wain LV, Repapi E, Obeidat M, Sayers I, Burton PR, et al. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. Am J Respir Crit Care Med. 2011;184:786–95. doi: 10.1164/rccm.201102-0192OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haq I, Chappell S, Johnson SR, Lotya J, Daly L, Morgan K, et al. Association of MMP - 12 polymorphisms with severe and very severe COPD: A case control study of MMPs - 1, 9 and 12 in a European population. BMC Med Genet. 2010;11:7. doi: 10.1186/1471-2350-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foreman MG, Kong X, DeMeo DL, Pillai SG, Hersh CP, Bakke P, et al. Polymorphisms in surfactant protein-D are associated with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2011;44:316–22. doi: 10.1165/rcmb.2009-0360OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–9. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 38.Pastva AM, Wright JR, Williams KL. Immunomodulatory roles of surfactant proteins A and D: implications in lung disease. Proc Am Thorac Soc. 2007;4:252–7. doi: 10.1513/pats.200701-018AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunninghake GM, Cho MH, Tesfaigzi Y, Soto-Quiros ME, Avila L, Lasky-Su J, et al. MMP12, lung function, and COPD in high-risk populations. N Engl J Med. 2009;361:2599–608. doi: 10.1056/NEJMoa0904006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallace AM, Sandford AJ. Genetic polymorphisms of matrix metalloproteinases: functional importance in the development of chronic obstructive pulmonary disease? Am J Pharmacogenomics. 2002;2:167–75. doi: 10.2165/00129785-200202030-00002. [DOI] [PubMed] [Google Scholar]
- 41.Moriguchi T, Haraguchi K, Ueda N, Okada M, Furuya T, Akiyama T. DREG, a developmentally regulated G protein-coupled receptor containing two conserved proteolytic cleavage sites. Genes Cells. 2004;9:549–60. doi: 10.1111/j.1356-9597.2004.00743.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhou X, Baron RM, Hardin M, Cho MH, Zielinski J, Hawrylkiewicz I, et al. Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Hum Mol Genet. 2012;21:1325–35. doi: 10.1093/hmg/ddr569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the European Nucleotide Archive repository as study accession number PRJEB13652, http://www.ebi.ac.uk/ena/data/view/PRJEB13652.


