Abstract
In Caenorhabditis elegans, cellular proteostasis is likely essential for longevity. Autophagy has been shown to be essential for lifespan extension of daf-2 insulin/IGF mutants. Therefore, it can be hypothesized that daf-2 mutants achieve this phenotype by increasing protein turnover. However, such a mechanism would exert a substantial energy cost. By using classical 35S pulse-chase labeling, we observed that protein synthesis and degradation rates are decreased in young adults of the daf-2 insulin/IGF mutants. Although reduction of protein turnover may be energetically favorable, it may lead to accumulation and aggregation of damaged proteins. As this has been shown not to be the case in daf-2 mutants, another mechanism must exist to maintain proteostasis in this strain. We observed that proteins isolated from daf-2 mutants are more soluble in acidic conditions due to increased levels of trehalose. This suggests that trehalose may decrease the potential for protein aggregation and increases proteostasis in the daf-2 mutants. We postulate that daf-2 mutants save energy by decreasing protein turnover rates and instead stabilize their proteome by trehalose.
Keywords: Caenorhabditis, Protein metabolism, Trehalose, Radiolabeling.
Progressively declining rates of protein synthesis and degradation with age have been observed in many species from yeast to mammals, including nematodes (1,2). These declining protein turnover rates are accompanied by increased accumulation and aggregation of biochemically altered and misfolded proteins (3), which are in turn linked to the development of age-related pathologies and suggested to contribute to the aging process (4). The autophagic–lysosomal and ubiquitin–proteasome system are primarily responsible for the proteolytic clearance of aberrant proteins and their age-associated impairment has been suggested to be the main cause of the progressive loss of proteostasis (5–7). Interventions that promote longevity such as dietary restriction and reduced insulin/IGF-1 signaling (IIS) are therefore thought to stimulate proteolytic turnover of proteins (and whole organelles), thereby delaying the accumulation of cellular damage and slowing-down aging (8,9). Consistent with this notion, dietary restriction was found to stimulate macroautophagy (10,11) and proteasome function (12,13), and to increase turnover of proteins in aging rats (14) and liver mitochondria in mice (15).
In Caenorhabditis elegans, dietary restriction and reduced IIS lead to increased activation of macroautophagy as observed by a GFP-tagged reporter protein that localizes to the autophagosomal membrane upon autophagic induction (16,17). Although autophagic activation is required for increased longevity in these animals, whether bulk protein degradation is also increased under these conditions has not been tested to date. On the other hand, inhibition of overall protein synthesis rates is now a well-established manner to increase longevity from yeast to mice, but it is difficult to reconcile with the turnover paradigm (18,19). Moreover, we have shown that protein synthesis rates are strongly decreased in young adults of the IIS receptor mutant daf-2 (20).
Using a classical pulse-chase approach, we assessed the effect of aging on overall protein synthesis and bulk degradation in long-lived insulin/IGF-1-like signaling (IIS) mutants of C elegans. As expected, we found a strong age-dependent decline in protein turnover rates in normal-lived C elegans. Counter to the turnover paradigm, long-lived IIS mutants display very low protein synthesis and degradation levels throughout life. Instead, we found that their proteins are much more soluble in trichloroacetic acid (TCA) and that this solubility depends on the presence of trehalose, suggesting that this carbohydrate may support the maintenance of proteostasis in these animals.
Our work thus implies that enhanced proteostasis in the long-lived IIS mutant is obtained by stabilizing the proteome with protectants such as trehalose, rather than by enhancing protein turnover rates to minimize damage accumulation.
Materials and Methods
Caenorhabditis elegans Strains and Culturing
The following strains were used: glp-4(bn2ts)I; daf-2(e1370)III, glp-4(bn2ts)I; daf-2(m577)III, and glp-4(bn2ts)I; daf-16(mgDf50)I; daf-2(e1370ts)III. The temperature-sensitive glp-4(bn2) background mutation disrupts normal postembryonic proliferation of the germ line resulting in sterile adults without eggs (21). This prevents the loss of 35S-labeled protein due to egg laying. Synchronized cohorts of worms were grown as described previously (22). Worms were grown on Escherichia coli K12-seeded nutrient agar plates until third larval stage (L3) at 16°C and then shifted to 24°C for the remainder of the experiment. As development of the daf-2 mutant is slightly slower than that of the control strain, L1 plates of the long-lived mutants were initiated approximately 8-hour upfront. Hence, both strains reached adulthood simultaneously and could be sampled together. At fourth larval stage, worms were transferred into Fernbach flasks containing 250-mL S-basal at densities not exceeding 1,500 worms/mL and shaken at 120 rounds per minute. Frozen E coli K12 cells were added twice daily to the culture medium to maintain the desired OD550 level of 1.8 (approximately 3×109 cells/mL).
35S Protein Assays
35S-labeled bacteria were obtained by growing E coli K12 overnight at 37°C in low-sulfate medium (44mM Na2HPO4, 22mM KH2PO4, 85mM NaCl, 20mM NH4Cl, 1.25mg/L thiamine, 0.1% (w/v) glucose, 2mM MgCl2) (23) supplemented with lysogeny broth medium (1% final concentration) and 5 µCi/mL [35S]sulfate (PerkinElmer, Waltman, MA). These quantities were carefully chosen as they optimize the balance between bacterial growth and efficient label incorporation. Bacterial concentrations were determined by measuring optical density at 550nm. During pulse labeling, 35S bacteria (at 1.8 OD550) were fed to worms cultured in 10-mL S-basal in tissue culture flasks (approximately 1,000 worms/mL). The rate of protein synthesis was calculated as the upward slope of the 35S signal obtained from worm protein extracts from six samples taken over a 6-hour time period. For measuring protein degradation, worms were pulse labeled by feeding 35S bacteria overnight, cleansed from radioactive bacteria (cfr. sampling procedure below) and chased in liquid culture containing nonradioactive K12 (OD550 = 1.8). The protein degradation rate was calculated as the downward slope of log-transformed protein radioactivity from five samples taken over a 48-hour chase period. To prevent reincorporation of excreted 35S, the chase medium was refreshed twice daily. During the sampling procedure, worms were washed five times over a period of 15 minutes in S-buffer supplemented with nonradioactive E coli K12 to purge the intestine from undigested 35S-labeled bacteria. Negative controls were produced by incubating worms in 35S bacteria for less than 1min. To isolate proteins, worms were first boiled for 15 minutes in 50% Tris–sodium dodecyl sulfate buffer (25mM Tris, 250mM NaCl, 5% sodium dodecyl sulfate, pH 7.4), and debris was pelleted by centrifugation for 5 minutes at 20,000 rcf. To precipitate proteins in the supernatant, TCA (final concentration 9.3%) was added to the supernatant and allowed to incubate at room temperature for 1 hour. Precipitated proteins were centrifuged at 20,000 rcf for 5 minutes and washed once with 1mL of 10% TCA. The protein pellet (TCA insoluble fraction) was dissolved in 150 µL 350mM NaOH for at least 1 hour at room temperature. To quantify 35S, 100 µl of TCA supernantant (sTCA fraction) or dissolved protein pellet (pTCA fraction) was added to 5-mL Ultima Gold LSC-cocktail (PerkinElmer, Waltman, MA) for liquid scintillation counting in a Tri-Carb 2800TR Liquid Scintillation Counter (PerlinElmer). Counts per minute were normalized to total protein concentration as determined with a BCA Protein Assay Kit (Thermo Scientific, Rockford, IL).
Determination of Free and Bound Amino Acid Content
Determination of amino acid concentrations by high-performance liquid chromatography (HPLC) was performed as described before (24). Free amino acids were extracted by treating worm homogenates with 15% TCA and taking the supernatant. Bound amino acids were released by acid hydrolysis in 12M HCl containing 0.1% phenol and 0.1% Na2SO3 for 24 hours at 105°C followed by neutralization of the mixture. All amino acids were derivatized with o-phthaldialdehyde in the injector of the Agilent 1100 system HPLC (Agilent Technologies, Switzerland) and quantified by fluorometry (ex 340nm/em 450nm). Free and bound amino acid content was normalized to the total amount of protein (TCA-soluble and TCA-insoluble fraction) in each sample determined with a BCA Protein Assay Kit (Thermo Scientific).
Trehalose and Glutathione Quantification
Worms were grown to second day of adulthood on standard agar plates seeded with E coli K12. The glp-4(bn2ts) and daf-2(e1370ts) mutations required a temperature switch from 16°C to 24°C at the L3 stage. Young adult worms were washed in S-basal and were frozen immediately. Samples were thawed and homogenized by bead beating as described in ref. (22). Trehalose was measured in microplates using a trehalose assay kit according to the manufacturer’s instructions (Megazyme, Wicklow, Ireland) (25). Glutathione was measured in microplates using a standard glutathione assay kit (Oxford Biomedical Research) according to the manufacturer’s instructions.
Regression Analysis
Regression analysis was performed to assess whether rates of protein degradation and synthesis change with age using the mixed linear regression model (LMM) PROC MIXED in SAS 9.2 (SAS Institute Inc., Cary, NC, 2002–2003).
Trehalose and protein stability data were assessed for normal distribution using Shapiro and Levene tests. Significance was assessed by using analysis of variance and Tukey’s honest significance post hoc test.
Results
Reduced IIS Reduces Protein Synthesis and Degradation in Young Worms
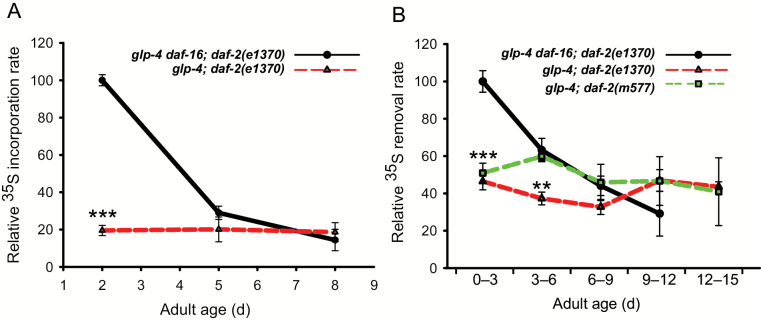
Increased protein turnover has been suggested to be beneficial for the animal because this process removes and replaces damaged proteins, thereby delaying progressive damage accumulation that causes aging (26). Protein synthesis and degradation rates were determined by pulse-chasing worms that were fed with 35S-radiolabeled bacteria. We compared the normal-lived reference strain glp-4 daf-16;daf-2 with the long-lived glp-4;daf-2 strain. The glp-4 mutant background, causing sterility, was used in both strains to avoid purging of radioactive signal by egg laying. Under the conditions used, the glp-4 mutation does not affect lifespan. As the DAF-16 protein is necessary and sufficient to cause the lifespan-extending effect of daf-2 mutants, the daf-2;daf-16 double mutation was used in the reference strain. Similar mutant backgrounds have been previously used by us and others (20,27–29). We found that the overall rate of protein synthesis declines rapidly with age in the reference strain (slope = −13.5±2.5; p < .001; LMM) (Figure 1A). Surprisingly, the rate of protein synthesis in young adult (day 2) IIS mutants is approximately five times lower compared to the age-matched control population (p < .0001, LMM), and this low level of 35S incorporation remains unchanged during the experiment (slope = −0.5±1.35; p = .72; LMM). We showed earlier that young daf-2 mutants that do not carry the glp-4 background mutation and mutants carrying the less pleiotropic daf-2(m577) allele also show reduced protein synthesis levels (20).
Figure 1.
(A) Relative rate of 35S incorporation in worm proteins with age after feeding 35S-labeled bacteria to controls (glp-4 daf-16; daf-2(e1370)) and the insulin/IGF-1 receptor mutant glp-4; daf-2(e1370). (B) Relative rate of 35S removal in the trichloroacetic acid–precipitated protein fraction over age. *p < .05, **p < .005, ***p < .0005 (linear regression model). Averages ± SEM are from four independent replicate experiments.
Protein degradation rates were quantified by pulsing worms overnight with 35S radioactive K12 bacteria, followed by a chase in fresh culture medium supplemented with nonradioactive K12. During the chase, samples were taken at regular time intervals. The rate at which 35S label was lost from the TCA-precipitated (pTCA) protein was taken as a measure for protein degradation rate. Protein degradation rates in the reference strain mirrored the pattern found for protein synthesis: protein degradation strongly decreases with age (Figure 1B). Despite the genetic evidence that autophagy (16) and the ubiquitin–proteasome system (30) are indispensible for lifespan extension in IIS mutants, we found that both young daf-2(e1370) and daf-2(m577) mutants retain 35S much longer in the pTCA protein fraction compared to controls. This low level of protein degradation remains fairly constant over adult age (daf-2(e1370) slope = −0.97±2.74; p = .72; daf-2(m577) slope = −1.1±0.86; p = .16; LMM).
The daf-2 Proteome Shows Increased Resistance to TCA-Mediated Precipitation
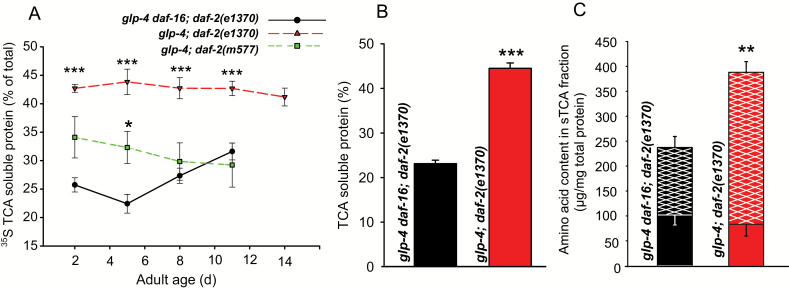
After TCA precipitation in the pulse-chase experiments, both TCA-precipitated protein (pTCA) and the TCA-soluble fraction (sTCA) were collected. We found that the 35S activity in the sTCA fraction was much higher in the long-lived daf-2(e1370) mutant compared to the reference strain, irrespective of age (Figure 2A). A similar trend was also observed for the daf-2(m577) mutant allele, albeit with only borderline statistical significance. The positive signal obtained for the sTCA fraction using a bicinchoninic acid (BCA) protein quantitation assay suggests that the increase in sTCA 35S activity in daf-2 nematodes was proteinaceous in nature (Figure 2B). In earlier studies we found that the thiol-containing tripeptide glutathione is maintained at higher levels in middle-aged and old, but not in young daf-2 mutants compared to wild type (31). Therefore, we verified whether the high 35S levels in the sTCA of the young daf-2 worms used in our experiment could be due to elevated glutathione levels. We did not find elevated glutathione levels in young daf-2 worms, confirming our earlier findings (Supplementary Figure 1).
Figure 2.
(A) Percentage of total 35S activity remaining soluble in the presence of trichloroacetic acid (TCA) (10% final concentration) measured after an 8-h chase period as a function of adult age. Mean values ± SEM of three to six independent experiments. (B) Percentage of total proteinaceous material remaining soluble in the presence of TCA (10% final concentration) as determined with a bicinchoninic acid assay. Mean values ± SEM of three independent replicates. (C) Total amino acid content determined by HPLC analysis of free- (solid stacked bars) and bound- (hatched stacked bars) amino acids in the sTCA fraction. Bound amino acids originate from complete acid hydrolysis of peptides and proteins that remained soluble in the sTCA fraction. Full bar height is indicative for total amino acid content (free amino acids, peptides, and proteins) in the TCA-soluble (sTCA) fraction. Worm samples of (B) and (C) were taken at the second day of adulthood. Mean values ± SEM of three independent replicates are shown. *p < .05, **p < .005, and ***p < .0005 for Student’s two-sided unpaired t-test.
Next, we reasoned that intracellular recycling of amino acids (eg, by autophagy or proteasomal activity) may be upregulated in daf-2 mutants, resulting in high protein turnover rates that may not be readily detected by classical pulse-chase experiments because of reuse of internal unlabeled amino acids. We therefore wondered if the rise in sTCA proteinaceous content was the result of increased standing levels of free amino acids in daf-2 worms. To test this hypothesis, we determined amino acid content in the sTCA fraction by HPLC separation of o-phthaldialdehyde-derivatized amino acids followed by fluorometric detection. However, we found no significant overall change in the standing levels of free amino acids between the control and long-lived worms (shaded bars in Figure 2C). Hydrochloric acid hydrolysis was used to break peptide bonds and release the amino acid constituents of proteins and peptides present in the sTCA fraction. The amount of bound amino acids (ie, amino acids released from peptides or proteins in the sTCA fraction) was significantly elevated in daf-2 mutants (hatched bars in Figure 2C), further supporting increased abundance of proteins in the sTCA fraction. Other precipitation methods, such as ammonium acetate and polyethylene glycol, gave similar results, further confirming precipitation resistance of glp-4;daf-2 proteins (Supplementary Figure 2A and B). We therefore conclude that the observed increase in sTCA 35S activity in the daf-2(e1370) mutant is the result from peptides and proteins that resist precipitation in the presence of TCA.
Increased daf-2 TCA Resistance Is Mediated by tps-1 and tps-2
IIS mutants are characterized by elevated levels of the glucose disaccharide trehalose that result from the increased expression of the trehalose-6-phosphate synthase genes tps-1 and tps-2 (29,32–36). Besides its role in carbohydrate storage and transport, trehalose also acts as a cytoprotectant against cold, heat, dehydration, hypoxic, and oxidative insult in invertebrates, most likely by stabilizing the proteome and lipid membranes (25,37,38). High trehalose levels contribute to daf-2 longevity as RNAi knockdown of tps-1 and tps-2 shortens daf-2 lifespan significantly (36).
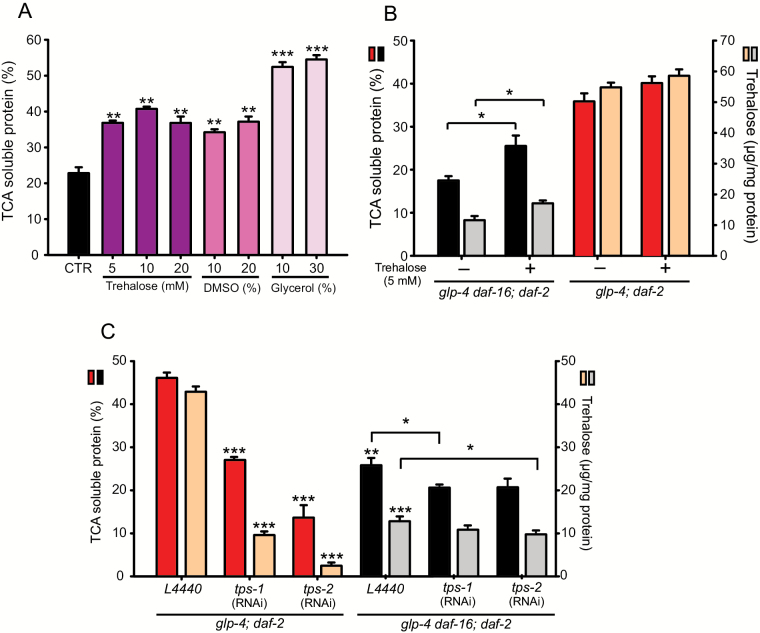
To test whether trehalose can protect proteins from TCA-mediated precipitation, we designed an in vitro approach in which different trehalose concentrations were added to worm homogenates. Addition of 5mM trehalose results in a significant decrease in protein precipitation in 10% TCA, but higher trehalose concentrations had no additional effect (Figure 3A). To confirm that the sTCA fraction is a valid measure for protein stabilization, we added glycerol, a common cosolvent for the storage of proteins, inhibiting their aggregation (39) and produced in C elegans upon osmotic stress (40,41). As predicted, this compound resulted in a strongly increased sTCA fraction (Figure 3A). Dimethyl sulfoxide can extend C elegans lifespan independently of IIS signaling, and it ameliorates paralysis induced by amyloid-β aggregation (42), suggesting that Dimethyl sulfoxide could improve protein homeostasis. Indeed, adding dimethyl sulfoxide to worm homogenates also significantly increased the sTCA fraction to levels comparable with trehalose treatment (Figure 3A).
Figure 3.
(A) The effect of trehalose, dimethyl sulfoxide (DMSO), and glycerol on protein solubility in the presence of 10% trichloroacetic acid (TCA). (B) The effect of adding 5mM trehalose to the culture medium on both protein solubility and worm trehalose concentration. (C) Protein solubility and trehalose content of worms fed either RNAi bacteria expressing tps-1 and tps-2 or empty vector (L4440; *p < .05, **p < .005, ***p < .0005). Asterisks on top of bars refer to comparison with the control (CTR in [A] and glp-4; daf-2 L4440 in [C]), whereas asterisks on brackets compare the connected bars.
Adding trehalose to the nutrient agar culture medium (5mM final concentration) also results in a modestly increased solubility of glp-4 daf-16;daf-2 worm proteins after homogenization and exposure to 10% TCA, which is likely the result of a small but significant trehalose uptake by the worms (Figure 3B). In the long-lived glp-4;daf-2, no additional effect in protein solubility is seen upon adding trehalose to the culture medium and, likewise, worm trehalose levels remained unchanged under these conditions (Figure 3B).
To test whether the high resistance to TCA-mediated protein precipitation in the glp-4;daf-2 mutant is dependent on its high intrinsic trehalose levels, we measured sTCA protein content upon treating the worms with tps-1 or tps-2 RNAi. Both tps-1 and tps-2 RNAi resulted in a strong decrease in worm trehalose levels, mirrored by decreased protein solubility in the presence of 10% TCA (Figure 3C). As tps RNAi in glp-4;daf-2 worms leads to comparable levels in both trehalose and protein solubility as in the reference strain, protein stability in daf-2 worms is mainly determined by trehalose levels.
Discussion
Protein Turnover Decreases Over Age
Classical pulse-chase labeling showed that protein synthesis as well as degradation rates drastically decrease over age in the C elegans reference strain used in this study. Age-related decreases in protein synthesis rates have been known for long and were described for nematodes (43,44) as well as many other species (1). The reason for this decline is still unclear but a decrease in translation efficiency or ribosome abundance may be involved (26,45). We observed that the decrease in protein synthesis rates was paralleled by a similar decrease in protein degradation rates. This may point to age-related deterioration of the lysosomal and/or proteasomal system. Indeed, three lysosomal protease activities decline 2.5- to 10-fold with age in C elegans (46). Also, impairment of the ubiquitin–proteasome system was reported in the dorsorectal neurons of aged C elegans worms although ageing did not affect the ubiquitin–proteasome system in the body wall muscle cells (47). It is often assumed that the age-related reduction in protein turnover is causal to increased protein damage and aggregation that is observed in old individuals (1,2,26,48).
daf-2 Mutants Show Very Low Protein Turnover at Young Adult Age
If protein damage and aggregation, both hallmarks of aging, can be cleared by increased protein turnover, it is expected that long-lived daf-2 mutants display increased turnover rates. However, our results do not support this prediction. On the contrary, young adult daf-2 mutants show severely reduced protein synthesis and degradation rates compared to the reference strain. Food intake in daf-2(e1370) worms is lower than in the control strain, especially after three days of adulthood (29), which should result in decreased label uptake. This could be interpreted as a confounding variable, but it is important to note that this is not the case. On the contrary, low food intake (hence, low intake of aminoacids) may be a prime mechanism to reduce protein synthesis rates (eg, via target of rapamycin nutrient sensing). Hypothetical forced feeding of daf-2(e1370) mutants would equalize label intake but it would likely change daf-2 physiology and protein metabolism. Alternatively, feeding daf-2(e1370) with more intensely labeled E coli to compensate for the difference in label uptake would make any quantitative comparison between both strains illegitimate. Our pulse-chase experiments confirm the results of earlier proteomic studies describing global downregulation of protein synthesis machinery including ribosomal subunits, tRNA synthetases, s-adenosyl methionine synthase-I, vigilin, and RACK-1 in IIS mutants (20,49). Conversely, when protein synthesis is downregulated by genetic intervention, lifespan of C elegans is also increased (50–54), which seems to be in accordance with our results. Thus, daf-2 mutants do not maintain overall increased protein turnover rates to support their longevity. On the contrary, recent evidence suggests that translation in daf-2 is actively repressed by the long noncoding RNA tts-1 (55). Also, it should be noted that DAF-2 is part of an insulin/IGF-like pathway, known for its growth-promoting properties. Hence, it would not be surprising if mutation in this pathway causes a downregulation of the protein synthesis machinery.
daf-2 Proteins Are More Resistant to TCA Precipitation
The daf-2 mutants showed a higher retention of 35S radiolabel in the TCA-soluble protein fraction (sTCA) compared to the control. We reasoned that this signal may represent increased levels of free amino acids that may point at efficient internal recycling of proteins in daf-2 mutants. Label reutilization is a blind spot in radioisotope pulse-chase studies, and there are no straightforward solutions to this artifact (15). Furthermore, the possibility of high internal recycling is supported by the observation that autophagic activity in C elegans daf-2 mutants is a prerequisite for their longevity (16,17,56). Also, increased proteasomal activity may underlie an elevated free aminoacid pool in daf-2 animals (57). However, we found that RNAi inhibition of proteasomal subunits did not lower the elevated 35S sTCA signal in the daf-2 mutants (data not shown). Moreover, HPLC showed that sTCA fraction of daf-2 mutants contained higher protein levels, not free amino acids, compared to the control. The resistance of the daf-2 proteome to precipitation with TCA, ammonium acetate, or polyethylene glycol, suggests increased protein stability. Hence, protein stability rather than protein turnover may be key to daf-2 longevity. This view is in line with the finding that the daf-2 proteome is less prone to aggregation (58). Protein stabilization or protection is governed by chemical or molecular chaperones (eg, trehalose, glycerol, sucrose, proline, heat shock proteins). Small heat shock proteins may sequester surplus proteins and help to maintain protein balance in daf-2 (59). Because it was found earlier that trehalose (34,35) and trehalose synthase (29) levels are increased in IIS mutants and that their lifespan extension is partially dependent on the trehalose synthase genes tps-1 and tps-2 (36), we extended our functional analysis to this protective disaccharide.
Trehalose Stabilizes the Worm Proteome
It was shown earlier that addition of trehalose extends lifespan of wild-type but not daf-2 mutant worms and that lifespan extension in daf-2 worms is partially dependent on trehalose synthesis (36). We found that trehalose, added directly to worm homogenates, can act as an in vitro stabilizing agent for the worm proteome, thus making it more resistant to TCA precipitation. Similar effects were obtained with other stabilizers such as dimethyl sulfoxide, an organosulfur compound, and glycerol, a polyol of which the concentration is not increased in daf-2 (35). Control worms, cultured on trehalose-enriched medium, showed significant uptake of this disaccharide resulting in a concurrent increase in sTCA protein content. Additional trehalose uptake and proteome stabilization was not observed in daf-2 mutants, suggesting that these animals reach maximal proteome stability via endogenous trehalose synthesis. This stability can be decreased by knocking down the trehalose synthase genes tps-1 and tps-2, confirming that the stabilization is trehalose specific.
Finally, the hypertrehalosemic response to IIS disruption may be evolutionary conserved in ecdysosoa as increased trehalose levels have been found in the hemolymph of Drosophila insulin-like peptide mutant lines (60). Also, trehalose levels and protein degradation rates may be interlinked although the molecular basis of this connection is still unclear. Blocking the proteasome specifically causes a buildup of trehalose in Saccharomyces cerevisiae (61). Whether this response also occurs in C elegans is currently unknown. On the other hand, in mammalian cell cultures, addition of trehalose induces autophagy (62). However, as we have shown that the protein degradation rate is very low in daf-2, it seems unlikely that C elegans would show a similar response.
In conclusion, we found that long-lived daf-2 mutants do not spend much of their energy resources on protein turnover but rather invest in the protection of their proteome by trehalose (and possibly other compounds). With this strategy, they seem to phenocopy the physiological characteristics of the dauer diapause stage (63), which is not entirely surprising as the IIS pathway is involved in dauer formation. It is still enigmatic why the lifespan extension of daf-2, a mutant showing low overall protein turnover, is strongly dependent on the autophagic pathway. Possibly, very low or local autophagic activity is crucial to extend lifespan in daf-2. Alternatively, other functions of bec-1, such as endosome-to-Golgi retrograde transport, may be required to support long life, rather than autophagy. It may also be considered that long-lived daf-2 worms or dietary-restricted worms are more susceptible to the deleterious consequences of inactivation of autophagy, leading to a shortened lifespan. Adding to the confusing relationship between autophagy and lifespan is the finding that RNAi knockdown of several autophagy genes in adult wild-type worms and daf-2 mutants extends lifespan in a daf-16 and sir-2.1 independent way (64).
Supplementary Material
Supplementary material can be found at: http://biomedgerontology.oxfordjournals.org/
Funding
This work was supported by the Fund for Scientific Research-Flanders (grant number G.04371.0N, 11E6415N to M.R., and 1187014N to I.D.) and the Special Research Fund of Ghent University (BOF01J04208 to G.D.).
Supplementary Material
Acknowledgements
The strains glp-4(bn2ts)I; daf-2(e1370)III, glp-4(bn2ts)I; daf-2(m577)III, and glp-4(bn2ts)I, daf-16(mgDf50)I, daf-2(e1370)III were kindly provided by David Gems. All other strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
REFERENCES
- 1. Van Remmen H, Ward W, Sabia R, Richardson A. Gene expression and protein degradation. In: Masoro EJ, eds. Handbook of Physiology: Section 11: Aging. Oxford, UK: Oxford University Press; 1995:71–234. [Google Scholar]
- 2. Rattan SI. Synthesis, modifications, and turnover of proteins during aging. Exp Gerontol. 1996;31:33–47. doi:10.1016/0531-5565(95)02022-5 [DOI] [PubMed] [Google Scholar]
- 3. Hipkiss AR. Accumulation of altered proteins and ageing: causes and effects. Exp Gerontol. 2006;41:464–473. doi:10.1016/j.exger.2006.03.004 [DOI] [PubMed] [Google Scholar]
- 4. Lindner AB, Demarez A. Protein aggregation as a paradigm of aging. Biochim Biophys Acta. 2009;1790:980–996. doi:10.1016/j.bbagen.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 5. Carrard G, Bulteau AL, Petropoulos I, Friguet B. Impairment of proteasome structure and function in aging. Int J Biochem Cell Biol. 2002;34:1461–1474. doi:10.1016/S1357-2725(02)00085-7 [DOI] [PubMed] [Google Scholar]
- 6. Martinez-Vicente M, Sovak G, Cuervo AM. Protein degradation and aging. Exp Gerontol. 2005;40:622–633. doi:10.1016/j.exger.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 7. Terman A, Gustafsson B, Brunk UT. Autophagy, organelles and ageing. J Pathol. 2007;211:134–143. doi:10.1002/path.2094 [DOI] [PubMed] [Google Scholar]
- 8. Bergamini E, Cavallini G, Donati A, Gori Z. The anti-ageing effects of caloric restriction may involve stimulation of macroautophagy and lysosomal degradation, and can be intensified pharmacologically. Biomed Pharmacother. 2003;57:203–208. doi:10.1016/S0753-3322(03)00048-9 [DOI] [PubMed] [Google Scholar]
- 9. Terman A, Kurz T, Navratil M, Arriaga EA, Brunk UT. Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxid Redox Signal. 2010;12:503–535. doi:10.1089/ars.2009.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cavallini G, Donati A, Gori Z, Pollera M, Bergamini E. The protection of rat liver autophagic proteolysis from the age-related decline co-varies with the duration of anti-ageing food restriction. Exp Gerontol. 2001;36:497–506. doi:10.1016/S0531-5565(00)00224-2 [DOI] [PubMed] [Google Scholar]
- 11. Donati A, Cavallini G, Paradiso C, et al. Age-related changes in the autophagic proteolysis of rat isolated liver cells: effects of antiaging dietary restrictions. J Gerontol A Biol Sci Med Sci. 2001;56:B375–B383. doi:10.1093/gerona/56.9.B375 [DOI] [PubMed] [Google Scholar]
- 12. Hayashi T, Goto S. Age-related changes in the 20S and 26S proteasome activities in the liver of male F344 rats. Mech Ageing Dev. 1998;102:55–66. doi:10.1016/S0047-6374(98)00011-6 [DOI] [PubMed] [Google Scholar]
- 13. Radák Z, Takahashi R, Kumiyama A, et al. Effect of aging and late onset dietary restriction on antioxidant enzymes and proteasome activities, and protein carbonylation of rat skeletal muscle and tendon. Exp Gerontol. 2002;37:1423–1430. doi:10.1016/S0531-5565(02)00116-X [DOI] [PubMed] [Google Scholar]
- 14. Goto S, Takahashi R, Kumiyama AA, et al. Implications of protein degradation in aging. Ann N Y Acad Sci. 2001;928:54–64. doi:10.1111/j.1749-6632.2001.tb05635.x [DOI] [PubMed] [Google Scholar]
- 15. Miwa S, Lawless C, von Zglinicki T. Mitochondrial turnover in liver is fast in vivo and is accelerated by dietary restriction: application of a simple dynamic model. Aging Cell. 2008;7:920–923. doi:10.1111/j.1474-9726.2008.00426.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans . Science. 2003;301:1387–1391. doi:10.1126/science.1087782 [DOI] [PubMed] [Google Scholar]
- 17. Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans . PLoS Genet. 2008;4:e24. doi:10.1371/journal.pgen.0040024.eor [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hipkiss AR. On why decreasing protein synthesis can increase lifespan. Mech Ageing Dev. 2007;128:412–414. doi:10.1016/j.mad.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 19. Kaeberlein M, Kennedy BK. Hot topics in aging research: protein translation and TOR signaling, 2010. Aging Cell. 2011;10:185–190. doi:10.1111/j.1474-9726.2010.00665.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Depuydt G, Xie F, Petyuk VA, et al. Reduced insulin/insulin-like growth factor-1 signaling and dietary restriction inhibit translation but preserve muscle mass in Caenorhabditis elegans . Mol Cell Proteomics. 2013;12:3624–3639. doi:10.1074/mcp.M113.027383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans . Development. 1992;116:755–766. [DOI] [PubMed] [Google Scholar]
- 22. Braeckman BP, Houthoofd K, De Vreese A, Vanfleteren JR. Assaying metabolic activity in ageing Caenorhabditis elegans . Mech Ageing Dev. 2002;123:105–119. doi:10.1016/S0047-6374(01)00331-1 [DOI] [PubMed] [Google Scholar]
- 23. Lewis JA, Fleming JT. Methods in Cell Biology. London, UK: Academic Press Limited; 1995. [Google Scholar]
- 24. Kerkaert B, Mestdagh F, Cucu T, Shrestha K, Van Camp J, De Meulenaer B. The impact of photo-induced molecular changes of dairy proteins on their ACE-inhibitory peptides and activity. Amino Acids. 2012;43:951–962. doi:10.1007/s00726-011-1157-y [DOI] [PubMed] [Google Scholar]
- 25. Erkut C, Penkov S, Khesbak H, et al. Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr Biol. 2011;21:1331–1336. doi:10.1016/j.cub.2011.06.064 [DOI] [PubMed] [Google Scholar]
- 26. Ryazanov AG, Nefsky BS. Protein turnover plays a key role in aging. Mech Ageing Dev. 2002;123:207–213. doi:10.1016/S0047-6374(01)00337-2 [DOI] [PubMed] [Google Scholar]
- 27. McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi:10.1074/jbc.M406207200October [DOI] [PubMed] [Google Scholar]
- 28. Patel DS, Garza-Garcia A, Nanji M, et al. Clustering of genetically defined allele classes in the Caenorhabditis elegans DAF-2 insulin/IGF-1 receptor. Genetics. 2008;178:931–946. doi:110.1534/genetics.107.070813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Depuydt G, Xie F, Petyuk VA, et al. LC-MS proteomics analysis of the insulin/IGF-1-deficient Caenorhabditis elegans daf-2(e1370) mutant reveals extensive restructuring of intermediary metabolism. J Proteome Res. 2014;13:1938–1956. doi:10.1021/pr401081b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ghazi A, Henis-Korenblit S, Kenyon C. Regulation of Caenorhabditis elegans lifespan by a proteasomal E3 ligase complex. Proc Natl Acad Sci USA. 2007;104:5947–5952. doi:10.1073/pnas.0700638104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brys K, Vanfleteren JR, Braeckman BP. Testing the rate-of-living/oxidative damage theory of aging in the nematode model Caenorhabditis elegans . Exp Gerontol. 2007;42:845–851. doi:10.1016/j.exger.2007.02.004 [DOI] [PubMed] [Google Scholar]
- 32. McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi:10.1046/j.1474-9728.2003.00043.x [DOI] [PubMed] [Google Scholar]
- 33. Murphy CT, McCarroll SA, Bargmann CI, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans . Nature. 2003;424:277–283. doi:10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- 34. Lamitina ST, Strange K. Transcriptional targets of DAF-16 insulin signaling pathway protect C. elegans from extreme hypertonic stress. Am J Physiol Cell Physiol. 2005;288:C467–C474. doi:10.1152/ajpcell.00451.2004 [DOI] [PubMed] [Google Scholar]
- 35. Fuchs S, Bundy JG, Davies SK, Viney JM, Swire JS, Leroi AM. A metabolic signature of long life in Caenorhabditis elegans . BMC Biol. 2010;8:14. doi:10.1186/1741-7007-8-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Honda Y, Tanaka M, Honda S. Trehalose extends longevity in the nematode Caenorhabditis elegans . Aging Cell. 2010;9:558–569. doi:10.1111/j.1474-9726.2010.00582.x [DOI] [PubMed] [Google Scholar]
- 37. Hottiger T, De Virgilio C, Hall MN, Boller T, Wiemken A. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vitro. Eur J Biochem. 1994;219:187–193. doi:10.1111/j.1432-1033.1994.tb19929.x [DOI] [PubMed] [Google Scholar]
- 38. Jain NK, Roy I. Effect of trehalose on protein structure. Protein Sci. 2009;18:24–36. doi:10.1002/pro.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vagenende V, Yap MG, Trout BL. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry. 2009;48:11084–11096. doi:10.1021/bi900649t [DOI] [PubMed] [Google Scholar]
- 40. Lamitina ST, Morrison R, Moeckel GW, Strange K. Adaptation of the nematode Caenorhabditis elegans to extreme osmotic stress. Am J Physiol Cell Physiol. 2004;286:C785–C791. doi:10.1152/ajpcell.00381.2003 [DOI] [PubMed] [Google Scholar]
- 41. Burkewitz K, Choe KP, Lee EC, Deonarine A, Strange K. Characterization of the proteostasis roles of glycerol accumulation, protein degradation and protein synthesis during osmotic stress in C. elegans . PLoS One. 2012;7:e34153. doi:10.1371/journal.pone.0034153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pellerone FI, Archer SK, Behm CA, Grant WN, Lacey MJ, Somerville AC. Trehalose metabolism genes in Caenorhabditis elegans and filarial nematodes. Int J Parasitol. 2003;33:1195–1206. doi:10.1016/S0020-7519(03)00173-5 [DOI] [PubMed] [Google Scholar]
- 43. Prasanna HR, Lane RS. Protein degradation in aged nematodes (Turbatrix aceti). Biochem Biophys Res Commun. 1979;86:552–559. doi:10.1016/0006-291X(79)91749-2 [DOI] [PubMed] [Google Scholar]
- 44. Johnson TE, McCaffrey G. Programmed aging or error catastrophe? An examination by two-dimensional polyacrylamide gel electrophoresis. Mech Ageing Dev. 1985;30:285–297. [DOI] [PubMed] [Google Scholar]
- 45. Payao SLM, Smith MDC, Winter LPF, Bertolucci PHF. Ribosomal RNA in Alzheimer’s disease and ageing. Mech Ageing Dev. 1998;105:265–272. doi:10.1016/S0047-6374(98)00095-5 [DOI] [PubMed] [Google Scholar]
- 46. Sarkis GJ, Ashcom JD, Hawdon JM, Jacobson LA. Decline in protease activities with age in the nematode Caenorhabditis elegans . Mech Ageing Dev. 1988;45:191–201. [DOI] [PubMed] [Google Scholar]
- 47. Hamer G, Matilainen O, Holmberg CI. A photoconvertible reporter of the ubiquitin-proteasome system in vivo. Nat Methods. 2010;7:473–478. doi:10.1038/nmeth.1460 [DOI] [PubMed] [Google Scholar]
- 48. Shringarpure R, Davies KJ. Protein turnover by the proteasome in aging and disease. Free Radic Biol Med. 2002;32:1084–1089. doi:10.1016/S0891-5849(02)00824-9 [DOI] [PubMed] [Google Scholar]
- 49. Stout GJ, Stigter EC, Essers PB, et al. Insulin/IGF-1-mediated longevity is marked by reduced protein metabolism. Mol Syst Biol. 2013;9:679. doi:10.1038/msb.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans . Aging Cell. 2007;6:95–110. doi:10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- 51. Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans . Nature. 2007;445:922–926. doi:10.1038/nature05603 [DOI] [PubMed] [Google Scholar]
- 52. Pan KZ, Palter JE, Rogers AN, et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans . Aging Cell. 2007;6:111–119. doi:10.1111/j.1474-9726.2006.00266.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tohyama D, Yamaguchi A, Yamashita T. Inhibition of a eukaryotic initiation factor (eIF2Bdelta/F11A3.2) during adulthood extends lifespan in Caenorhabditis elegans . FASEB J. 2008;22:4327–4337. doi:10.1096/fj.08-112953 [DOI] [PubMed] [Google Scholar]
- 54. Wang J, Robida-Stubbs S, Tullet JM, Rual JF, Vidal M, Blackwell TK. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 2010;6:8. doi:10.1371/journal.pgen.1001048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Essers PB, Nonnekens J, Goos YJ, et al. A long noncoding RNA on the ribosome is required for lifespan extension. Cell Rep. 2015;10:1–7. doi:10.1016/j.celrep.2014.12.029 [DOI] [PubMed] [Google Scholar]
- 56. Hars ES, Qi H, Ryazanov AG, et al. Autophagy regulates ageing in C. elegans . Autophagy. 2007;3:93–95. doi:10.4161/auto.3636 [DOI] [PubMed] [Google Scholar]
- 57. Matilainen O, Arpalahti L, Rantanen V, Hautaniemi S, Holmberg CI. Insulin/IGF-1 signaling regulates proteasome activity through the deubiquitinating enzyme UBH-4. Cell Rep. 2013;3:1980–1995. doi:10.1016/j.celrep.2013.05.012 [DOI] [PubMed] [Google Scholar]
- 58. David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans . PLoS Biol. 2010;8:e1000450. doi:10.1111/j.1474-9726.2006.00267.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Walther DM, Kasturi P, Zheng M, et al. Widespread proteome remodeling and aggregation in aging C. elegans . Cell. 2015;161:919–932. doi:10.1016/j.cell.2015.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Belgacem YH, Martin JR. Disruption of insulin pathways alters trehalose level and abolishes sexual dimorphism in locomotor activity in Drosophila . J Neurobiol. 2006;66:19–32. doi:10.1002/neu.20193 [DOI] [PubMed] [Google Scholar]
- 61. Lee DH, Goldberg AL. Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae . Mol Cell Biol. 1998;18:30–38. doi:10.1128/MCB.18.1.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi:10.1074/jbc.M609532200 [DOI] [PubMed] [Google Scholar]
- 63. Burnell AM, Houthoofd K, O’Hanlon K, Vanfleteren JR. Alternate metabolism during the dauer stage of the nematode Caenorhabditis elegans . Exp Gerontol. 2005;40:850–856. doi:10.1016/j.exger.2005. 09.006 [DOI] [PubMed] [Google Scholar]
- 64. Hashimoto Y, Ookuma S, Nishida E. Lifespan extension by suppression of autophagy genes in Caenorhabditis elegans . Genes Cells. 2009;14:717–726. doi:10.1111/j.1365-2443.2009.01306.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.