Abstract
NAD(P)H:quinone oxidoreductase (NQO1) is essential for cell defense against reactive oxidative species, cancer, and metabolic stress. Recently, NQO1 was found in ribonucleoprotein (RNP) complexes, but NQO1-interacting mRNAs and the functional impact of such interactions are not known. Here, we used ribonucleoprotein immunoprecipitation (RIP) and microarray analysis to identify comprehensively the subset of NQO1 target mRNAs in human hepatoma HepG2 cells. One of its main targets, SERPINA1 mRNA, encodes the serine protease inhibitor α-1-antitrypsin, A1AT, which is associated with disorders including obesity-related metabolic inflammation, chronic obstructive pulmonary disease (COPD), liver cirrhosis and hepatocellular carcinoma. Biotin pulldown analysis indicated that NQO1 can bind the 3′ untranslated region (UTR) and the coding region (CR) of SERPINA1 mRNA. NQO1 did not affect SERPINA1 mRNA levels; instead, it enhanced the translation of SERPINA1 mRNA, as NQO1 silencing decreased the size of polysomes forming on SERPINA1 mRNA and lowered the abundance of A1AT. Luciferase reporter analysis further indicated that NQO1 regulates SERPINA1 mRNA translation through the SERPINA1 3′UTR. Accordingly, NQO1-KO mice had reduced hepatic and serum levels of A1AT and increased activity of neutrophil elastase (NE), one of the main targets of A1AT. We propose that this novel mechanism of action of NQO1 as RNA-binding protein may help to explain its pleiotropic biological effects.
Keywords: NQO1, RNA binding protein, SERPINA1, A1AT, neutrophil elastase, mRNA translation
Introduction
The enzyme NAD(P)H:quinone oxidoreductase 1(NQO1, EC 1.6.5.2) utilizes either NADH or NADPH as electron donor to catalyze the reduction of various quinones to hydroquinones [1]. NQO1 has been studied extensively as a component of the cellular defense program against the adverse effects of quinone xenobiotics, free radicals, and reactive oxidative species [2]. NQO1 protects against excessive oxidative stress by functioning as superoxide reductase and generating antioxidant forms of α-tocopherol and coenzyme Q [3–5]. In addition, induction of NQO1 by caloric restriction has been shown to attenuate age-related loss of antioxidative capacity in brain [6] and liver [7].
NQO1 is expressed ubiquitously in human tissues [8] and is transcriptionally regulated by the Kelch-like ECH-associated protein 1 (Keap1)-nuclear factor erythroid 2-related factor 2 (Nrf2)-antioxidant response elements (ARE) pathway [9–11]. NQO1 is expressed at high levels in human tumors including cancers of the liver, thyroid gland, adrenal gland, breast, ovary, colon and lung [8], prompting the hypothesis that cytotoxic quinones bioactivated by NQO1 could be used strategically to target NQO1-rich cancer cells [12–13].
Studies carried in NQO1-null mice have also revealed a role for NQO1 in the regulation of lipid metabolism and glucose homeostasis. NQO1-null mice display altered metabolic pathways resulting in significantly lower abdominal adipose tissue and increased NADPH concentrations compared to wild-type (WT) mice [14]. However, these animals have higher levels of triglycerides in liver and blood and develop insulin resistance [14]. Conversely, NQO1-dependent NADH oxidation by β-lapachone (β-lap, a naturally occurring quinone isolated from the bark of the Lapacho tree) has been shown to protect against obesity [15] and hypertension [16] in mice, suggesting that therapeutic stimulation of NQO1 activity may be beneficial in metabolic disorders. Moreover, a polymorphism in NQO1 (C609T, NQO1*2), which essentially causes a NQO1-null phenotype, has been shown to be associated with increased risk of complications related to metabolic syndrome [17,18].
Despite these studies, the full spectrum of metabolic functions controlled by NQO1 remains unknown. A recent study that aimed to define a comprehensive atlas of proteins interacting with mRNAs identified NQO1 as a potential RNA-binding protein (RBP) [19]. However, the RNA-binding properties of NQO1 are unknown. In the present study, we present evidence that NQO1 functions as a RBP that associates with several mRNAs. One of its targets, SERPINA1 mRNA, encodes alpha-1 antitrypsin (A1AT), an acute-phase protein with anti-protease, immunoregulatory and metabolic activities [20–22]. Our results indicate that NQO1 binding to SERPINA1 mRNA promotes A1AT translation. Short-term treatment with β-lap reduced the binding of NQO1 to SERPINA1 mRNA, causing a decline in A1AT translation and protein levels. NQO1 binding to SERPINA1 mRNA mapped primarily to the 3′ untranslated region (UTR) in liver and pancreatic cells. Reporter analysis revealed that NQO1 selectively enhances A1AT translation through the SERPINA1 3′UTR. Accordingly, the levels of A1AT were lower after silencing NQO1 in cultured cells and were constitutively lower in NQO1-null mice. We propose that the pleiotropic effects of NQO1 are linked, at least in part, to this novel RNA-binding function of NQO1.
Materials and Methods
Cell culture and reporter analysis
Human hepatoma (HepG2) were purchased from ATCC and upon receipt, cells were expanded for a few passages to enable the generation of new frozen stocks. Cells were resuscitated as needed and used for fewer than 6 months after resuscitation (no more than 10 passages). Cell line authentication was performed by ATCC utilizing Short Tandem Repeat (STR) profiling. The NQO1 overexpressing Panc-1 cells clone5 (Panc-1/C5) expressing NQO1 was previously generated form parental Panc-1 cells according to methods described by Siegel et al. [23].
Cells were cultured in DMEM supplemented with 2 mM Glutamax, 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen) under standard cell culture conditions. For reporter assays, 50 ng of control plasmid psiCHECK2 (Promega) or psiCHECK2 bearing the 3′UTR of SERPINA1 mRNA were transfected using lipofectamine 2000 (Invitrogen); 72 h later, renilla luciferase (RL) and firefly luciferase (FL) were measured using Dual-Glo Luciferase Assay System (Promega).
RNP immunoprecipitation (IP) and microarray analyses
Endogenous mRNA–protein complexes were immunoprecipitated as previously described [24]. Briefly, cytoplasmic cell lysates were prepared in polysome lysis buffer (PLB; 20 mM Tris-HCl at pH 7.5, 100 mM KCl, 5 mM MgCl2 and 0.5% NP-40) containing 100 U/ml RNase OUT (Invitrogen) and protease inhibitors (Complete Mini, Roche). For microarray analysis, 3 mg of lysate were incubated (2 h, 4°C) with 100 m μl of a 50% (v/v) suspension of protein-A Sepharose beads precoated with 20 mg each of mouse anti-NQO1 or mouse IgG (Santa Cruz Biotechnology). After washes with NT2 buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40), bound RNA was extracted from the beads using TRIzol (Thermo Fisher Scientific).
For Illumina microarray analysis, the RNA obtained after IP was assessed using an Agilent 2100 Bioanalyzer and RNA 6000 nanochips. The RNA was used to generate biotin- labeled cRNA using the Illumina TotalPrep RNA Amplification Kit (Ambion), which was then hybridized to Human HT-12 v4 Expression BeadChips (Illumina), containing 24,000 well-annotated RefSeq transcripts with ~30-fold redundancy. The arrays were scanned using an Illumina BeadStation 500X Genetic Analysis Systems scanner and the image data extracted using Illumina BeadStudio software, version 1.5, normalized by Z-score transformation and used to calculate differences in signal intensities. Significant values were calculated from three independent experiments, using a two-tailed Z-test and P < 0.01. The complete list of mRNAs enriched in NQO1 IP identified on the arrays is shown in https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81614.
RNA isolation and RT-qPCR analysis
RNA was isolated from cell extracts or from RIP samples using TRIzol following the manufacturer’s procedures. Reverse transcription was performed using iScript™ Advanced cDNA Synthesis Kit for RT-qPCR (Bio-Rad). For real-time quantitative (q)PCR analysis, iTaq™ Universal SYBR® Green Supermix (Bio-Rad) was used with 250 nM gene-specific primers (Table S1).
Renilla luciferase and Firefly luciferase mRNA levels were measured using primers TACAAGTACCTCACCGCTTGGT and TGATCTTGTCTTGGTGCTCGTA for RL mRNA, CTAAGAAGGGCCTGCAGAAGAT and AAGCCCTGGTAGTCGGTCTTAG for FL mRNA.
Western blot and biotin pull-down analyses
Whole-cell lysates were prepared using RIPA buffer and western blot analysis was performed with 5 μg of protein fractionated by electrophoresis through 4–15% Criterion™ TGX™ precast gels (Bio-Rad) and transferred to nitrocellulose membrane using Trans-Blot Turbo™ Transfer System (Bio-Rad). Primary antibodies recognizing NQO1, A1AT, β-actin (all from Abcam) and LC3B (Cell Signalling) were added over night at 4°C followed by incubation with the appropriate HRP-labeled secondary antibodies (Santa Cruz Biotechnology). Immunocomplexes were detected using enhanced chemiluminescence (Amersham). Cell media were concentrated in 30-kDa spin columns (Sigma) for detection of extracellular A1AT. Quantitation of the protein bands was performed by volume densitometry using ImageJ software (National Institutes of Health).
For biotin pull-down assays, PCR fragments generated using primers bearing the T7 RNA polymerase promoter sequence (CCAAGCTTCTAATACGACTCACTATAGGGAGA) were used as templates for in vitro transcription using MegaScript T7 kit (Ambion) followed by purification using Nuc-Away Spin Columns (Applied Biosystems). Biotinylated transcripts were incubated with cytoplasmic lysates from HepG2 or Panc-1 cells (150 μg lysate, 1 μg biotinylated RNA) for 30 min at room temperature, and complexes were isolated with streptavidin-coated magnetic Dynabeads (Dynal) and analyzed using western blot analysis to detect NQO1. The primers used to prepare biotinylated RNA fragments spanning SERPINA1 mRNA are listed in Table S2.
Polyribosome fractionation
Cells were incubated with cycloheximide (Calbiochem; 100 μg/ml, 15 min) and cytoplasmic lysates (500 μl) were fractionated by centrifugation through 10–50% linear sucrose gradients and divided into 12 fractions for RT-qPCR analysis, as described [25].
Experimental animals
Wild-type C57BL/6 control mice were initially purchased from The Jackson Laboratory. Nqo1-null mice were obtained from Dr. Frank Gonzalez (National Cancer Institute) and backcrossed into the C57BL/6 genomic background. The generation of the original Nqo1-null mice is described elsewhere [26]. Animals were housed four per cage in a room maintained at a constant temperature (20–22°C) in a light:dark 12:12-h schedule according to animal protocols and NIH guidelines. Mice were maintained on standard diet (Harlan rodent diet 2018).
Serum A1AT levels and elastase activity assay
Serum A1AT levels were measured with ELISA kits according to the instructions provided by Immunology Consultants Laboratory. Serum NE activity was measured using the highly specific synthetic NE substrate N-methoxysuccinyl-Ala-Ala-Pro-Val-p-nitroanilide (Sigma) according to the previously described method (24). Briefly, 40 μl of freshly isolated serum was incubated with 0.1 M Tris-HCl buffer (pH 8.0) containing 0.5 M NaCl and 1 mM N-methoxysuccinyl-Ala-Ala-Pro-Val-p-nitroanilide at 37°C for 24 h and liberated p-nitroanilide was measured spectrophotometrically at 405 nm.
Statistical analysis
Statistical differences were determined by unpaired Student’s t test and p values were generated using GraphPad Prism 6.0. Error bar represents SEM.
Results
Identification and validation of NQO1 target mRNAs
HepG2 cells were used to isolate NQO1-mRNA ribonucleoprotein (RNP) complexes using a specific antibody against NQO1; control IgG IP reactions were performed in parallel (Fig. 1A). To confirm that NQO1 was immunoprecipitated successfully, Western blot analysis was used to detect NQO1 (Fig. 1B). The immunopecipitation (IP) reactions were carried out under conditions that preserved protein–mRNA complexes as described (24), and the RNA in the NQO1 IP and control IgG IP samples was isolated and analyzed using Illumina microarrays (‘Materials and Methods’ section). The transcripts that were associated with NQO1 are potentially involved in many cellular processes (e.g. fatty acid metabolism, glucose utilization, cell cycle and apoptosis, insulin signaling, mitochondrial activity, protein homeostasis and translation) and included multiple members of the serine protease inhibitor (serpin) family.
Fig. 1.
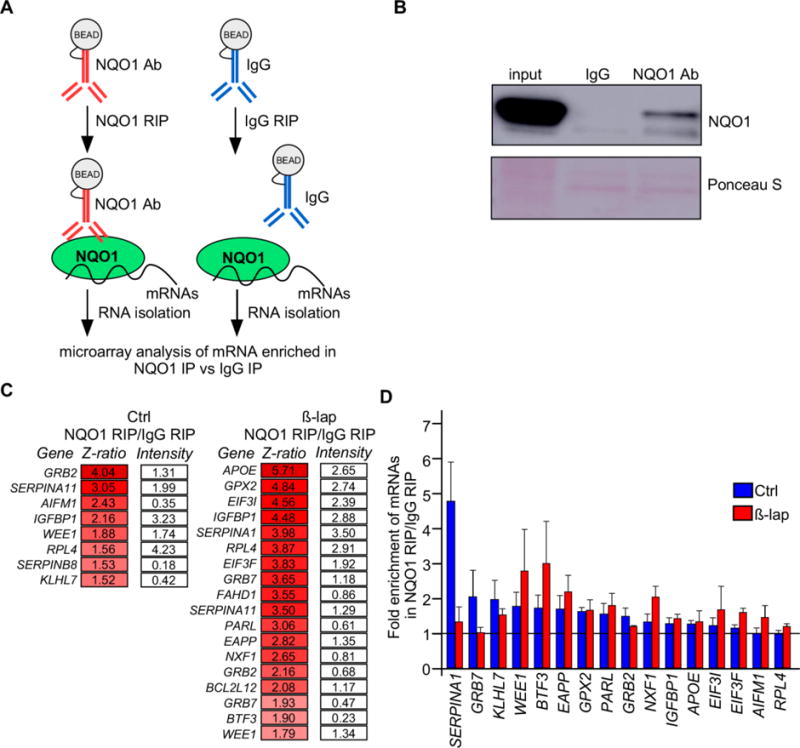
NQO1 binds SERPINA1 mRNA. (A) Schematic of the RIP assay using cytoplasmic HepG2 cell lysates under conditions that preserved mRNA-RBP associations. (B) Western blot analysis of NQO1 recovered in IP samples. (C) Data represent the Z-ratio and signal intensities of mRNAs in NQO1-IP relative to IgG-IP under CTRL and β-lap treatment. (D) RT-qPCR analysis revealed that the main NQO1 target among those tested was SERPINA1 mRNA. β-lap treatment caused a rapid loss in the amount of SERPINA1 mRNA bound to NQO1. Data represent the mean of three independent experiments ± SEM.
We chose to validate by RT-qPCR analysis the interaction between NQO1 and the mRNAs showing higher Z-scores in the two conditions and those that might be relevant to NQO1-related functions reported in the literature. A partial list of target transcripts that were highly enriched in NQO1 IP relative to IgG IP is shown (Fig. 1C, left). A similar analysis (NQO1 IP vs IgG IP) was carried out using cells that had been treated with 10 μM β-lap (a substrate of NQO1) and a partial list of enriched NQO1-bound transcripts is shown (Fig. 1C, right). A complete list of target mRNAs of NQO1 on arrays was deposited in the Gene Expression Omnibus (GEO) database (GSE81614). Although microarray analysis allows an unbiased transcriptome-wide approach for the identification of mRNAs isolated in RNP complexes, this interaction has to be validated by multiple approaches to avoid over-interpretation of the microarray data.
A subset of NQO1-bound target mRNAs was validated by reverse transcription (RT) followed by real-time, quantitative (q)PCR analysis employing gene-specific primer pairs (‘Materials and Methods’ section). In the validation step, we included RNA samples obtained from β-lap-treated cells (Fig. 1D). SERPINA1 [serpin peptidase inhibitor, clade A (α-1 antiproteinase, antitrypsin), member 1] mRNA was highly enriched in NQO1 IP samples compared to IgG IP samples, and binding was significantly reduced upon β-lap treatment, as assessed by RT-qPCR analysis (Fig. 1D and Fig. S1A). Enrichment of other transcripts like GRB7 (growth factor receptor-bound protein 7) and KLHL (Kelch-like family member 7) mRNAs was also measured by RT-qPCR analysis. Binding of NQO1 to GAPDH mRNA, which encodes a housekeeping protein, was used for normalization. Since GAPDH mRNA is highly abundant and binds IP components (e.g., beads, antibody, reaction tube) at background levels, assessment of GAPDH mRNA levels demonstrated that sample input was equivalent. Together, these findings indicate that NQO1 interacts specifically with various mRNAs.
NQO1 promotes SERPINA1 mRNA translation
Many RBPs associate with mRNAs to regulate their stability and/or translation. To investigate the functional influence of NQO1 on SERPINA1 mRNA, we specifically silenced NQO1 using small interfering (si)RNA directed at the NQO1 mRNA. Interestingly, NQO1 downregulation reduced A1AT levels in cell lysates and A1AT released in the cell culture medium, without influencing the steady-state levels of SERPINA1 mRNA (Fig. 2A and Fig. S1B). These data suggest that NQO1 did not influence SERPINA1 mRNA stability and may instead regulate its translation. To test if translation was altered by NQO1, we studied SERPINA1 mRNA distribution in the polysomes (‘Materials and Methods’ section) of cells with normal or reduced NQO1 levels. As shown in Fig. 2B, NQO1 downregulation did not alter appreciably the global profiles of polysomes compared with control cells (Fig. 2B). However, SERPINA1 mRNA was found in lighter (thus less actively translating) polysomes in cells with lower NQO1 levels than in control cells (Fig. 2C), supporting the hypothesis that NQO1 indeed enhanced SERPINA1 mRNA translation by promoting the formation of larger polysomes on SERPINA1 mRNA.
Fig. 2.
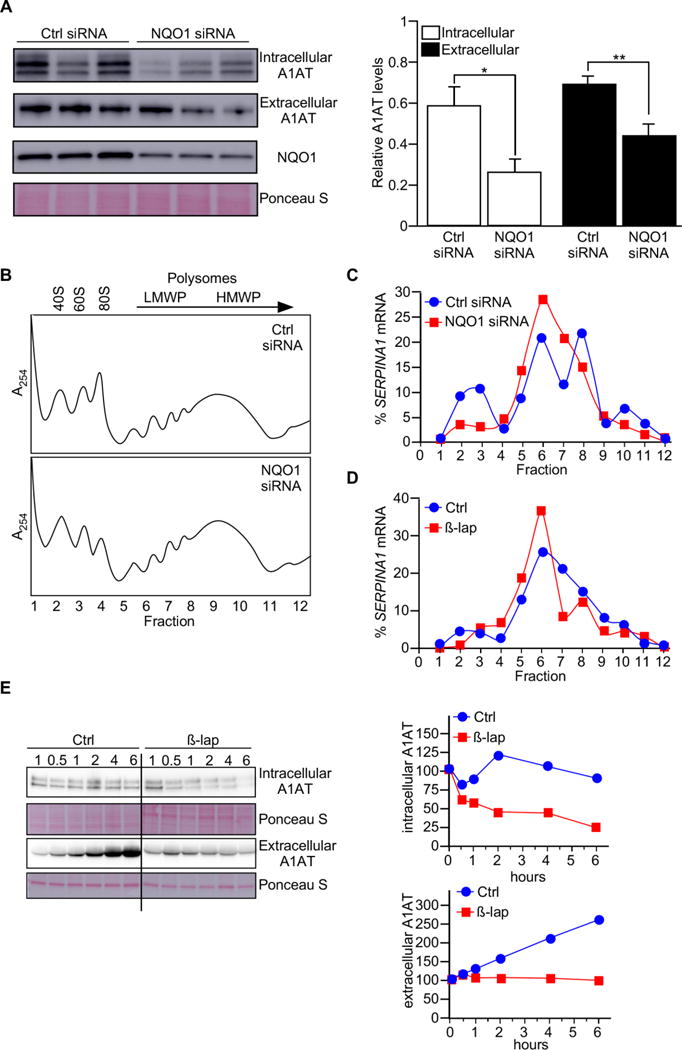
NQO1 promotes SERPINA1 mRNA translation. (A) Small interfering RNA (siRNA) was used to determine the effect of NQO1 knockdown on SERPINA1 mRNA and A1AT protein levels in HepG2 cells. A ~60% reduction in NQO1 protein levels was accompanied by significant reduction in A1AT levels, measured in cell lysate (intracellular) and in cell culture medium (extracellular). Data represent the mean of three independent experiments ± SEM. *P<0.05, **P<0.01. (B–C) Seventy-two hours after NQO1 silencing, cytoplasmic extracts were fractionated through sucrose gradients to obtain cytoplasmic components of progressively larger weight: ribosomal subunits (40S, 60S), monosomes (80S), low-molecular-weight (LMW) and high-molecular-weight (HMW) polysomes. (C) The relative distribution of SERPINA1 mRNA was measured by RT-qPCR analysis of RNA in each of the gradient fractions and represented as percentage of total RNA in the gradient. (D) β-lap caused a shift of SERPINA1 mRNA towards smaller polysomes. (E) Western blot analysis in HepG2 lysates and supernatants showing a substantial reduction in A1AT intracellularly and in cell culture media after β-lap treatment.
Since β-lap treatment induced the dissociation of the NQO1-SERPINA1 mRNA complex (Fig. 1D and S1A), we investigated the impact of β-lap treatment on SERPINA1 mRNA translation and A1AT levels. As shown in Fig. 2D, β-lap treatment decreased the levels of SERPINA1 mRNA in actively translating fractions (where heavy polysomes reside) compared to untreated cells, although SERPINA1 mRNA steady-state levels were not affected (Fig. S1C). Consequently, there was a substantial reduction in both cellular A1AT levels and in A1AT released into the cell culture medium (Fig. 2E). The rapid decline in A1AT protein after β-lap treatment may suggest an involvement of NQO1 in the regulation of A1AT stability. Surprisingly, however, stimulation of HepG2 cells with the proteasome inhibitor MG-132 increased protein degradation, as denoted by the reduction in the A1AT- immunoreactive band and the appearance of lower proteolitic bands (~37 kDa). Interestingly, this effect was paralleled by an increase in the autophagy marker LC3B-II (Fig. S1D). However, the combination of MG-132 and β-lap treatments had no additive effect on A1AT reduction (Fig. S1D).
Taken together, these results support the hypothesis that NQO1 binding to SERPINA1 mRNA enhances the translation of SERPINA1 mRNA and the accumulation and secretion of A1AT.
NQO1 regulates SERPINA1 mRNA translation through binding to the SERPINA1 3′UTR
To gain molecular details of how NQO1 binds to and promotes SERPINA1 mRNA translation, we sought to map the regions of NQO1 interaction with SERPINA1 mRNA. We generated biotinylated partial transcripts which spanned the 5′UTR, coding region (CR) and 3′UTR of SERPINA1 mRNA (Figure 3A, top). The biotinylated RNAs were incubated with lysates from HepG2 cells or from the human pancreatic carcinoma cell line Panc-1/C5 (34); the resulting biotinylated RNA-protein (RNP) complexes were pulled down using streptavidin-coated beads, and the presence of NQO1 in the RNPs was examined by Western blot analysis (‘Materials and Methods’ section). Binding assays using biotinylated transcripts indicated that NQO1 showed robust interaction with the 3′UTR (fragments 6 and 7) of SERPINA1 mRNA both in HepG2 and Panc-1 lysates, although NQO1 also showed some affinity for the CR fragments, particularly in HepG2 samples. Biotinylated GAPDH 3′UTR, included as a negative control, showed background binding of NQO1 and indicated that several SERPINA1 RNA segments bound to NQO1 specifically (Fig. 3A, bottom).
Fig. 3.

NQO1 binding to the 3′UTR of SERPINA1 mRNA promotes its translation. (A) Biotin-RNA pulldown indicated that NQO1 was capable of binding the 3′ untranslated region (UTR) and the coding region (CR) of SERPINA1 mRNA in two different human cell lines. (B) 3′UTR of SERPINA1 mRNA was cloned downstream of the renilla luciferase (RL) coding sequence, and firefly luciferase (FL) expressed from the same construct served as internal normalization control. After NQO1 silencing, HepG2 cells were transfected with the vector control (psiCHECK2) or with a construct containing SERPINA1 3′UTR. Twenty h later, the ratio of RL/FL activity in siNQO1 cells vs control siRNA cells was calculated using empty vector as reference. (C) NQO1 silencing inhibited RL/FL reporter activity in HepG2 cells. (D) RT-qPCR analysis of RL mRNA levels normalized to FL mRNA levels in the vector control (psi) and in the construct containing SERPINA1 3′UTR (psi-3′UTR). Data represent the mean of three independent experiments ± SEM. ****, P < 0.001.
To test if NQO1 binding to the SERPINA1 3′UTR affects SERPINA1 mRNA translation, we cloned the SERPINA1 3′UTR into a luciferase reporter construct (psiCHECK2) bearing the renilla luciferase (RL) coding region and measured luciferase production from the resulting chimeric RNA (Fig. 3B); this vector also expressed firefly luciferase (FL), which served as an internal control for transfection. Transfection of psiCHECK2-SERPINA1(3′UTR) and the parent vector psiCHECK2 into HepG2 cells that expressed either normal or reduced levels of NQO1 was followed by measurements of luciferase activities. As shown in Fig. 3C, NQO1 silencing significantly reduced luciferase expression without significant changes in the levels of the corresponding reporter mRNAs (Figure 3D), indicating that the 3′UTR was responsible for promoting translation of a heterologous reporter mRNA in the presence of NQO1. Together, these findings indicate that binding of NQO1 to the 3′UTR of SERPINA1 mRNA enhances translation of the encoded protein, A1AT.
NQO1 KO mice have reduced A1AT levels and increased neutrophil elastase activity
In order to study the impact of NQO1 in vivo, we assessed the levels of A1AT in WT mice and in mice in which both Nqo1 alleles have been knocked out (NQO1-KO). As shown in Fig. 4A, NQO1 was robustly expressed in liver from WT mice, but not from NQO1-KO mice; A1AT levels were significantly reduced in the livers of KO compared to WT mice. Similar results were observed both in males and females. In contrast, similar levels of Serpina1 mRNA levels were observed between KO and WT animals (Fig. S1E). These changes were consistent with low levels of A1AT after NQO1 silencing and supported the positive regulatory effect of NQO1 on SERPINA1 mRNA translation.
Fig. 4.
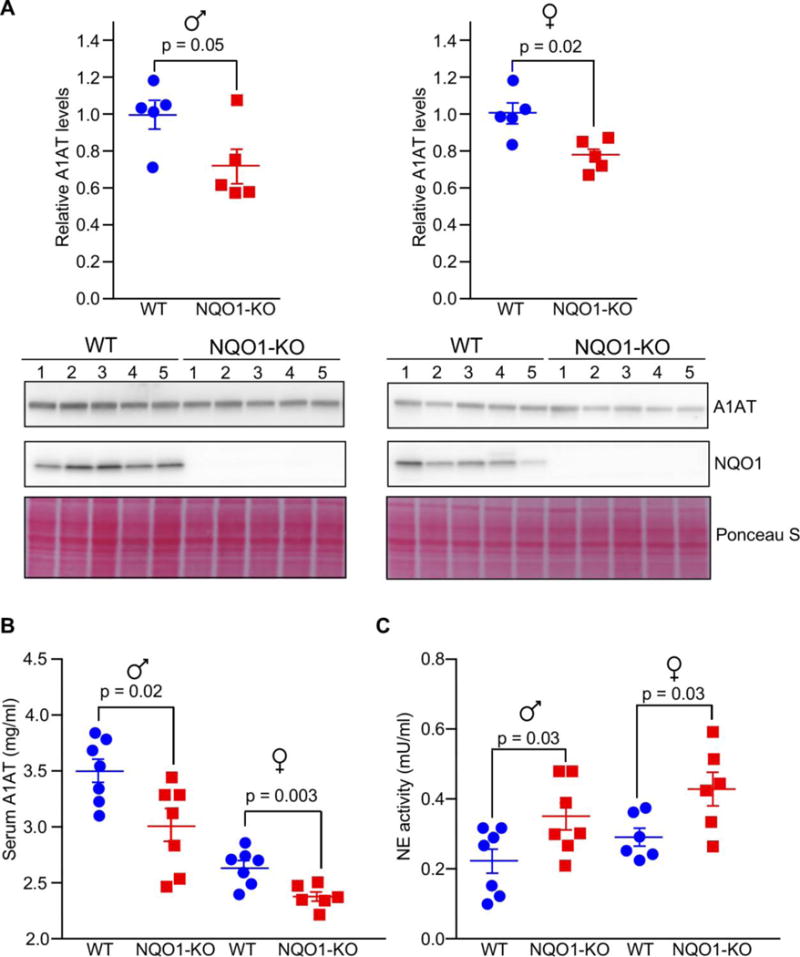
NQO1-null mice have reduced A1AT levels and increased NE activity. (A) Effect of NQO1 deletion on A1AT protein expression in livers of male (left) and female (right) animals. (B–C) Serum samples were collected from WT and NQO1 KO animals. Serum A1AT levels were measured by ELISA, and serum NE activity was detected by a colorimetric assay. In all cases, statistical differences were determined in diet-paired groups of mice of the same sex using a student’s t-test. Data are represented as the mean ± SEM.
Finally, since A1AT is the predominant physiologic inhibitor of neutrophil elastase (NE), we measured A1AT levels and NE activity in serum. Serum A1AT levels were lower in NQO1-KO than WT animals (Fig. 4B), consistent with the levels of A1AT observed in liver tissues. Importantly, we observed that NE activity was elevated in NQO1-KO compared to WT animals (Fig. 4C). Together, our findings indicate that NQO1-KO mice showed reduced A1AT levels and enhanced NE activity, and that NQO1 promoted A1AT expression via translation.
Discussion
This study provides evidence that NQO1 binds a subset of mRNAs in human HepG2 cells and that one major target was identified as SERPINA1 mRNA, which encodes A1AT, a circulating 52-kDa glycoprotein primarily known for its activity as a serine protease inhibitor [27,28]. NQO1 knockdown did not alter the stability of SERPINA1 mRNA, but reduced its translation. Biotin-RNA pulldown and reporter assays indicated that the regulatory effects on translation were elicited via the SERPINA1 3′UTR. We also found that the NQO1-SERPINA1 mRNA complex was dissociated by β-lap treatment, perhaps because, as a substrate of NQO1, β-lap may prevent binding of NQO1 to SERPINA1 mRNA. The impact of NQO1 on A1AT levels led to subsequent modulation of NE activity in vivo.
NQO1 has been proposed to mediate the 20S proteasomal degradation of specific proteins including p53, p63, p73 and ornithine decarboxylase [29]. Degradation of A1AT follows two major pathways: the proteasome is responsible for degrading the soluble forms of A1AT [30], while autophagy is specialized in the disposal of the insoluble polymers and aggregates [31] by the proteasome. The rapid decline in A1AT protein after β-lap treatment observed in this study suggests that NQO1 participates in A1AT degradation by the proteasome. However, this may not be the case, since co-incubation with the proteasome inhibitor MG-132 was not able to reverse the reduction in A1AT. On the contrary MG-132 increased A1AT degradation, probably by activation of a compensatory autophagy pathway [32], as indicated by the increased LC3B-II-to-LC3B-I ratio.
A1AT is the predominant physiologic inhibitor of NE, and a reduction in circulating A1AT results in deterioration of the lung tissue and promotes liver disease, a common observation in emphysema and COPD patients [27,33]. In light of our findings, this interaction between NQO1 and SERPINA1 mRNA may help explain previous observations on the biological roles of NQO1. Reduced Nrf2/NQO1 activity is associated with emphysema; in particular, Nrf2-deficient mice have exacerbated elastase-induced emphysema compared with control mice [34] and mice lacking NQO1 spontaneously develop emphysema [35]. Moreover, lung macrophages from patients with emphysema have reduced levels of NQO1 [36]. In these conditions, reduced A1AT levels may help explain the elevated NE activity and hence the development of emphysema.
Several reports have suggested additional roles for A1AT. In one set of studies, A1AT has been shown to reduce IL-1β–mediated pancreatic islet toxicity in vitro, and contribute to islet allograft survival by promoting antigen-specific immune tolerance in mice [37,38]. Accordingly, A1AT therapy also delays the development of diabetes in nonobese diabetic mice [39,40]. Recent studies have linked A1AT and NE to metabolic syndrome and insulin resistance in humans [22], as obese humans have significantly lower circulating levels of A1AT, but higher serum NE activity, an imbalance that contributes to the development of obesity, inflammation, liver steatosis, and insulin resistance [22,41]. Mice overexpressing human A1AT are resistant to diet-induced obesity, increased adiposity, and high fasting blood glucose, and are more insulin sensitive than WT littermates [22]. In keeping with these findings, our results showed that genetic deletion of NQO1 was accompanied by a significant reduction in A1AT levels and increased NE activity, thus providing a rationale for the metabolic abnormalities previously observed in these mice [14]. It remains to be evaluated whether interventions to elevate A1AT levels can alleviate the inflammation, insulin resistance and liver steatosis observed in NQO1-null mice.
The specific mechanism by which NQO1 promotes SERPINA1 mRNA translation was not elucidated in this study. NQO1 may compete with an RBP that suppresses translation or cooperates with an RBP that enhances translation; alternatively, NQO1 may compete with the binding of a microRNA that suppresses SERPINA1 mRNA translation. These possibilities await systematic analysis. Furthermore, the fact that NQO1 can bind a subset of mRNAs opens new avenues for studying NQO1 as an RBP that might influence the stability and/or translation of other target mRNAs.
NQO1 binding to target mRNAs was altered by the redox cycling quinone β-lap indicating that the redox state of the cell could alter the affinity of NQO1 for target mRNAs, or the binding sequences recognized by NQO1 affecting mRNA translation or turnover. While we only investigated SERPINA1 mRNA there may be other NQO1 target mRNAs whose association with NQO1 may be influenced by β-lap.
In addition, NQO1 can undergo post-translational modifications such as phosphorylation [42] and ubiquitination [43]. It has been shown previously that phosphorylation and other post-translational modifications of RBPs including HuR, BRF1, TTP, KSRP, TIAR/TIA-1 have been linked to changes in their subcellular localization and their association with target mRNAs [44]. Reduction of β-lap by NQO1 results in an increased ratio of NAD+/NADH and this in turn may activate the deacetylase SIRT1 [45]; of interest, NQO1 is itself target of acetylation [46] and this PTM may also contribute to alter its binding to target mRNAs. Further studies are needed to understand in detail if any of the known post-translational modifications might affect the RBP properties of NQO1.
In closing, these data demonstrate that NQO1 targets several mRNAs, including the SERPINA1 mRNA, which encodes A1AT. NQO1 promotes SERPINA1 mRNA translation by interacting with the 3′UTR. Whether the influence of NQO1 upon SERPINA1 mRNA translation is modulated by its interactions with other RBPs or microRNAs awaits systematic study. It also remains to be tested whether NQO1 is capable of binding other RNA types (e.g., long noncoding RNAs, microRNAs, or circular RNAs), if posttranslational modification alters NQO1 binding to target mRNAs, and whether NQO1 inhibitors or activators affect its binding to target mRNAs. Our in vitro data demonstrates that the NQO1-SERPINA1 mRNA interaction weakens as the NAD+/NADH ratio increased due to β-lap redox cycling. It follows then that NQO1 may function as a molecular switch responding to changes in pyridine nucleotide ratios resulting in the capture or release of mRNA targets. This hypothesis is also supported by major changes in the number of transcripts observed in the microarray analysis in response to β-lap treatment. Addressing these questions in depth will provide critical information about the role of NQO1 as an RBP, its cellular functions, and its potential usefulness in therapy.
Supplementary Material
Fig. S1. (A) RIP assays followed by RT-qPCR showed that β-lap treatment caused a rapid loss in the amount of SERPINA1 mRNA bound to NQO1. (B) RT-qPCR analysis showed no effect of NQO1 silencing on SERPINA1 mRNA levels. (C) RT-qPCR analysis showed no effect of β-lap on SERPINA1 mRNA levels. Data represent the means of 3 or 4 independent experiments ± SEM. (D) Western blot analysis in HepG2 lysates showing the effects of the proteasome inhibitor MG-132 on A1AT and LC3B-I/LC3B-II levels. (E) RT-qPCR analysis showed no difference in Serpina1 levels between NQO1 KO and WT mice. Data are represented as the mean ± SEM of n = 10 per group.
Highlights.
NQO1 is a novel RNA-binding protein
Binding of NQO1 to SERPINA1 mRNA enhances translation of the encoded protein, A1AT
NQO1-KO mice have reduced A1AT levels and increased neutrophil elastase activity
Acknowledgments
We thank Dawn Phillips, Dawn Nines and Justine Lucas for animal care, Lynn Wu for technical assistance, Drs. Kevin Becker and Yongquing Zhang for microarray analysis. We also thank Drs. Frank Gonzalez and Anil K. Jaiswal for providing the NQO1 null mice. This work was supported by the Intramural Research Program of the National Institute on Aging, NIH (grant number AG000362-02).
List of Abbreviations
- ARE
antioxidant response elements
- A1AT
α-1-antitrypsin
- COPD
chronic obstructive pulmonary disease
- CR
coding region
- FL
firefly luciferase
- GRB
growth factor receptor-bound protein
- IP
immunoprecipitation
- Keap1
Kelch-like ECH-associated protein 1
- KLHL
Kelch-like family member 7
- Nrf2
nuclear factor erythroid 2-related factor
- NQO1
NAD(P)H:quinone oxidoreductase 1
- (q)PCR
quantitative real-time polymerase chain reaction
- RBP
RNA-binding protein
- RIP
ribonucleoprotein immunoprecipitation
- RL
renilla luciferase
- RNP
ribonucleoprotein
- RT
reverse transcription
- SERPINA1
serpin peptidase inhibitor, Clade A (α-1 Antiproteinase, Antitrypsin), member 1
- (si)RNA
small interfering RNA
- β-lap
β-lapachone
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Winski SL, Koutalos Y, Bentley DL, Ross D. Subcellular localization of NAD(P)H:quinone oxidoreductase 1 in human cancer cells. Cancer Res. 2002;62:1420–1424. [PubMed] [Google Scholar]
- 2.Dinkova-Kostova AT, Talalay P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys. 2010;501:116–123. doi: 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel D, Bolton EM, Burr JA, Liebler DC, Ross D. The reduction of alpha-tocopherolquinone by human NAD(P)H:quinone oxidoreductase: the role of alpha-tocopherolhydroquinone as a cellular antioxidant. Mol Pharmacol. 1997;52:300–305. doi: 10.1124/mol.52.2.300. [DOI] [PubMed] [Google Scholar]
- 4.Siegel D, Gustafson DL, Dehn DL, Han JY, Boonchoong P, Berliner LJ, Ross D. NAD(P)H:quinone oxidoreductase 1: Role as a superoxide scavenger. Mol Pharmacol. 2004;65:1–10. doi: 10.1124/mol.65.5.1238. [DOI] [PubMed] [Google Scholar]
- 5.Chan TS, Teng S, Wilson JX, Galati G, Khan S, O’Brien PJ. Coenzyme Q cytoprotective mechanisms for mitochondrial complex I cytopathies involves NAD(P)H:quinone oxidoreductase 1 (NQO1) Free Radic Res. 2002;36:421–427. doi: 10.1080/10715760290021270. [DOI] [PubMed] [Google Scholar]
- 6.Hyun DH, Hernandez JO, Mattson MP, de Cabo R. The plasma membrane redox system in aging. Ageing Res Rev. 2006;5:209–220. doi: 10.1016/j.arr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Kim JD, McCarter RJ, Yu BP. Influence of age, exercise, and dietary restriction on oxidative stress in rats. Aging (Milan) 1996;8:123–129. doi: 10.1007/BF03339566. [DOI] [PubMed] [Google Scholar]
- 8.Siegel D, Ross D. Immunodetection of NAD(P)H:quinone oxidoreductase 1 (NQO1) in human tissues. Free Radic Biol Med. 2000;29:246–253. doi: 10.1016/s0891-5849(00)00310-5. [DOI] [PubMed] [Google Scholar]
- 9.Ishii T, Itoh K, Yamamoto M. Roles of Nrf2 in activation of antioxidant enzyme genes via antioxidant responsive elements. Methods Enzymol. 2002;348:182–190. doi: 10.1016/s0076-6879(02)48637-5. [DOI] [PubMed] [Google Scholar]
- 10.Tanigawa S, Fujii M, Hou DX. Action of Nrf2 and Keap1 in ARE-mediated NQO1 expression by quercetin. Free Radic Biol Med. 2007;42:1690–1703. doi: 10.1016/j.freeradbiomed.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Pearson KJ, Lewis KN, Price NL, Chang JW, Perez E, Cascajo MV, Tamashiro KL, Poosala S, Csiszar A, Ungvari Z, Kensler TW, Yamamoto M, Egan JM, Longo DL, Ingram DK, Navas P, de Cabo R. Nrf2 mediates cancer protection but not prolongevity induced by caloric restriction. Proc Natl Acad Sci USA. 2008;105:2325–2330. doi: 10.1073/pnas.0712162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siegel D, Yan C, Ross D. NAD(P)H:quinone oxidoreductase 1 (NQO1) in the sensitivity and resistance to antitumor quinones. Biochem Pharmacol. 2012;83:1033–1040. doi: 10.1016/j.bcp.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bey EA, Bentle MS, Reinicke KE, Dong Y, Yang CR, Girard L, Minna JD, Bornmann WG, Gao J, Boothman DA. An NQO1- and PARP-1-mediated cell death pathway induced in non-small-cell lung cancer cells by beta-lapachone. Proc Natl Acad Sci USA. 2007;104:11832–11837. doi: 10.1073/pnas.0702176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaikwad A, Long DJ, 2nd, Stringer JL, Jaiswal AK. In vivo role of NAD(P)H:quinone oxidoreductase 1 (NQO1) in the regulation of intracellular redox state and accumulation of abdominal adipose tissue. J Biol Chem. 2001;276:22559–22564. doi: 10.1074/jbc.M101053200. [DOI] [PubMed] [Google Scholar]
- 15.Hwang JH, Kim DW, Jo EJ, Kim YK, Jo YS, Park JH, Yoo SK, Park MK, Kwak TH, Kho YL, Han J, Choi HS, Lee SH, Kim JM, Lee I, Kyung T, Jang C, Chung J, Kweon GR, Shong M. Pharmacological stimulation of NADH oxidation ameliorates obesity and related phenotypes in mice. Diabetes. 2009;58:965–974. doi: 10.2337/db08-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YH, Hwang JH, Kim KS, Noh JR, Gang GT, Oh WK, Jeong KH, Kwak TH, Choi HS, Lee IK, Lee CH. Enhanced activation of NAD(P)H:quinone oxidoreductase 1 attenuates spontaneous hypertension by improvement of endothelial nitric oxide synthase coupling via tumor suppressor kinase liver kinase B1/adenosine 5′-monophosphate-activated protein kinase-mediated guanosine 5′-triphosphate cyclohydrolase 1 preservation. J Hypertens. 2014;32:306–317. doi: 10.1097/HJH.0000000000000018. [DOI] [PubMed] [Google Scholar]
- 17.Ramprasath T, Murugan PS, Kalaiarasan E, Gomathi P, Rathinavel A. Selvam,G.S. Genetic association of Glutathione Peroxidase-1 (GPx-1) and NAD(P)H:Quinone Oxidoreductase 1 (NQO1) variants and their association of CAD in patients with type-2 diabetes. Mol Cell Biochem. 2012;361:143–150. doi: 10.1007/s11010-011-1098-5. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Hernandez A, Córdova EJ, Rosillo-Salazar O, García-Ortíz H, Contreras-Cubas C, Islas-Andrade S, Revilla-Monsalve C, Salas-Labadía C, Orozco L. Association of HMOX1 and NQO1 Polymorphisms with Metabolic Syndrome Components. PLoS One. 2015;10:e0123313. doi: 10.1371/journal.pone.0123313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 20.Jonigk D, Al-Omari M, Maegel L, Müller M, Izykowski N, Hong J, Hong K, Kim SH, Dorsch M, Mahadeva R, Laenger F, Kreipe H, Braun A, Shahaf G, Lewis EC, Welte T, Dinarello CA, Janciauskiene S. Anti-inflammatory and immunomodulatory properties of α1-antitrypsin without inhibition of elastase. Proc Natl Acad Sci USA. 2013;110:15007–15012. doi: 10.1073/pnas.1309648110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tawara I, Sun Y, Lewis EC, Toubai T, Evers R, Nieves E, Azam T, Dinarello CA, Reddy P. Alpha-1-antitrypsin monotherapy reduces graft-versus-host disease after experimental allogeneic bone marrow transplantation. Proc Natl Acad Sci USA. 2012;109:564–569. doi: 10.1073/pnas.1117665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansuy-Aubert V, Zhou QL, Xie X, Gong Z, Huang JY, Khan AR, Aubert G, Candelaria K, Thomas S, Shin DJ, Booth S, Baig SM, Bilal A, Hwang D, Zhang H, Lovell-Badge R, Smith SR, Awan FR, Jiang ZY. Imbalance between neutrophil elastase and its inhibitor α1-antitrypsin in obesity alters insulin sensitivity, inflammation, and energy expenditure. Cell Metab. 2013;17:534–548. doi: 10.1016/j.cmet.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siegel D, Shieh B, Yan C, Kepa JK, Ross D. Role for NAD(P)H:quinone oxidoreductase 1 and manganese-dependent superoxide dismutase in 17-(allylamino)-17-demethoxygeldanamycin-induced heat shock protein 90 inhibition in pancreatic cancer cells. J Pharmacol Exp Ther. 2011;336:874–880. doi: 10.1124/jpet.110.176438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panda AC, Abdelmohsen K, Martindale JL, Di Germanio C, Yang X, Grammatikakis I, Noh JH, Zhang Y, Lehrmann E, Dudekula DB, De S, Becker KG, White EJ, Wilson GM, de Cabo R, Gorospe M. Novel RNA-binding activity of MYF5 enhances Ccnd1/Cyclin D1 mRNA translation during myogenesis. Nucleic Acids Res. 2016;44:393–2408. doi: 10.1093/nar/gkw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdelmohsen K, Tominaga K, Lee EK, Srikantan S, Kang MJ, Kim MM, Selimyan R, Martindale JL, Yang X, Carrier F, Zhan M, Becker KG, Gorospe M. Enhanced translation by Nucleolin via G-rich elements in coding and non-coding regions of target mRNAs. Nucleic Acids Res. 2011;39:8513–8530. doi: 10.1093/nar/gkr488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radjendirane V, Joseph P, Lee YH, Kimura S, Klein-Szanto AJ, Gonzalez FJ, Jaiswal AK. Disruption of the DT diaphorase (NQO1) gene in mice leads to increased menadione toxicity. J Biol Chem. 1998;273:7382–7389. doi: 10.1074/jbc.273.13.7382. [DOI] [PubMed] [Google Scholar]
- 27.Silverman EK, Sandhaus RA. Clinical practice. Alpha1-antitrypsin deficiency. N. Engl J Med. 2009;360:2749–2757. doi: 10.1056/NEJMcp0900449. [DOI] [PubMed] [Google Scholar]
- 28.Hunt JM, Tuder R. Alpha 1 anti-trypsin: one protein, many functions. Curr Mol Med. 2012;12:827–835. doi: 10.2174/156652412801318755. [DOI] [PubMed] [Google Scholar]
- 29.Sollner S, Macheroux P. New roles of flavoproteins in molecular cell biology: an unexpected role for quinone reductases as regulators of proteasomal degradation. FEBS J. 2009;276:4313–4324. doi: 10.1111/j.1742-4658.2009.07143.x. [DOI] [PubMed] [Google Scholar]
- 30.Teckman JH, Burrows J, Hidvegi T, Schmidt B, Hale PD, Perlmutter DH. The proteasome participates in degradation of mutant alpha 1-antitrypsin Z in the endoplasmic reticulum of hepatoma-derived hepatocytes. J Biol Chem. 2001;276:44865–44872. doi: 10.1074/jbc.M103703200. [DOI] [PubMed] [Google Scholar]
- 31.Kamimoto T, Shoji S, Hidvegi T, Mizushima N, Umebayashi K, Perlmutter DH, Yoshimori T. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J Biol Chem. 2006;281:4467–4476. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- 32.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007;171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janciauskiene SM, Bals R, Koczulla R, Vogelmeier C, Köhnlein T, Welte T. The discovery of α1-antitrypsin and its role in health and disease. Respir Med. 2011;105:1129–1139. doi: 10.1016/j.rmed.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Ishii Y, Itoh K, Morishima Y, Kimura T, Kiwamoto T, Iizuka T, Hegab AE, Hosoya T, Nomura A, Sakamoto T, Yamamoto M, Sekizawa K. Transcription factor Nrf2 plays a pivotal role in protection against elastase-induced pulmonary inflammation and emphysema. J Immunol. 2005;175:6968–6975. doi: 10.4049/jimmunol.175.10.6968. [DOI] [PubMed] [Google Scholar]
- 35.Potts-Kant EN, Li Z, Tighe RM, Lindsey JY, Frush BW, Foster WM, Hollingsworth JW. NAD(P)H:quinone oxidoreductase 1 protects lungs from oxidant-induced emphysema in mice. Free Radic Biol Med. 2012;52:705–715. doi: 10.1016/j.freeradbiomed.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Goven D, Boutten A, Leçon-Malas V, Marchal-Sommé J, Amara N, Crestani B, Fournier M, Lesèche G, Soler P, Boczkowski J, Bonay M. Altered Nrf2/Keap1-Bach1 equilibrium in pulmonary emphysema. Thorax. 2008;63:916–924. doi: 10.1136/thx.2007.091181. [DOI] [PubMed] [Google Scholar]
- 37.Lewis EC, Shapiro L, Bowers OJ, Dinarello CA. Alpha1-antitrypsin monotherapy prolongs islet allograft survival in mice. Proc Natl Acad Sci USA. 2005;102:12153–12158. doi: 10.1073/pnas.0505579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis EC, Mizrahi M, Toledano M, Defelice N, Wright JL, Churg A, Shapiro L, Dinarello CA. Alpha1-antitrypsin monotherapy induces immune tolerance during islet allograft transplantation in mice. Proc Natl Acad Sci USA. 2008;105:16236–16241. doi: 10.1073/pnas.0807627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koulmanda M, Bhasin M, Hoffman L, Fan Z, Qipo A, Shi H, Bonner-Weir S, Putheti P, Degauque N, Libermann TA, Auchincloss H, Jr, Flier JS, Strom TB. Curative and beta cell regenerative effects of alpha1-antitrypsin treatment in autoimmune diabetic NOD mice. Proc Natl Acad Sci USA. 2008;105:16242–16247. doi: 10.1073/pnas.0808031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, Lu Y, Campbell-Thompson M, Spencer T, Wasserfall C, Atkinson M, Song S. Alpha1-antitrypsin protects beta-cells from apoptosis. Diabetes. 2007;56:1316–1323. doi: 10.2337/db06-1273. [DOI] [PubMed] [Google Scholar]
- 41.Talukdar S, Oh da Y, Bandyopadhyay G, Li D, Xu J, McNelis J, Lu M, Li P, Yan Q, Zhu Y, Ofrecio J, Lin M, Brenner MB, Olefsky JM. Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nat Med. 2012;18:1407–1412. doi: 10.1038/nm.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai CF, Wang YT, Yen HY, Tsou CC, Ku WC, Lin PY, Chen HY, Nesvizhskii AI, Ishihama Y, Chen YJ. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat Commun. 2015;6:6622. doi: 10.1038/ncomms7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee EK. Post-translational modifications of RNA-binding proteins and their roles in RNA granules. Curr Protein Pept Sci. 2012;13:331–336. doi: 10.2174/138920312801619411. [DOI] [PubMed] [Google Scholar]
- 45.Kim HJ, Oh GS, Shen A, Lee SB, Choe SK, Kwon KB, Lee S, Seo KS, Kwak TH, Park R, So HS. Augmentation of NAD(+) by NQO1 attenuates cisplatin-mediated hearing impairment. Cell Death Dis. 2014;5:e1292. doi: 10.1038/cddis.2014.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (A) RIP assays followed by RT-qPCR showed that β-lap treatment caused a rapid loss in the amount of SERPINA1 mRNA bound to NQO1. (B) RT-qPCR analysis showed no effect of NQO1 silencing on SERPINA1 mRNA levels. (C) RT-qPCR analysis showed no effect of β-lap on SERPINA1 mRNA levels. Data represent the means of 3 or 4 independent experiments ± SEM. (D) Western blot analysis in HepG2 lysates showing the effects of the proteasome inhibitor MG-132 on A1AT and LC3B-I/LC3B-II levels. (E) RT-qPCR analysis showed no difference in Serpina1 levels between NQO1 KO and WT mice. Data are represented as the mean ± SEM of n = 10 per group.


