Abstract
Repair of DNA damage is critical for maintaining the genomic integrity of cells. DNA polymerase lambda (POLL/Pol λ) is suggested to function in base excision repair (BER) and non-homologous end-joining (NHEJ), and is likely to play a role in damage tolerance at the replication fork. Here, using next-generation sequencing, it was discovered that the POLL rs3730477 single nucleotide polymorphism (SNP) encoding R438W Pol λ was significantly enriched in the germlines of breast cancer patients. Expression of R438W Pol λ in human breast epithelial cells induces cellular transformation and chromosomal aberrations. The role of estrogen was assessed as it is commonly used in hormone replacement therapies and is a known breast cancer risk factor. Interestingly, the combination of estrogen treatment and the expression of the R438W Pol λ SNP drastically accelerated the rate of transformation. Estrogen exposure produces 8-oxoguanine lesions that persist in cells expressing R438W Pol λ compared to WT Pol λexpressing cells. Unlike WT Pol λ, which performs error-free bypass of 8-oxoguanine lesions, expression of R438W Pol λ leads to an increase in mutagenesis and replicative stress in cells treated with estrogen. Together, these data suggest that individuals who carry the rs3730477 POLL germline variant have an increased risk of estrogen-associated breast cancer.
Keywords: DNA repair, mutagenesis, genomic instability, cancer, 8-oxoG
Introduction
Faithful DNA repair is required to maintain genomic integrity. If left unrepaired, damaged DNA can lead to a mutator phenotype and cancer (1). In fact, germline mutations in key genes involved in DNA repair have been shown to induce genomic instability and cellular transformation (2–4). DNA is estimated to be endogenously damaged more than 20,000 times per cell per day (5,6). This damage arises from normal cellular processes such as metabolism, which produces reactive oxygen species (ROS) that are highly reactive with DNA. One of the most common mutagenic lesions produced by ROS is 8-oxoguanine (8-oxoG) (5) with more than 1000 lesions produced per cell per day (7). 8-oxoG possesses a two atom difference compared to guanine (8) and its ability to functionally resemble thymine allows replicative polymerases to insert adenines leading to G:T transversions (9–11).
DNA polymerase lambda (Pol λ) plays an important role in protecting the cells against 8-oxoG-induced damage, as it is the main polymerase capable of correctly bypassing the lesion and preventing mutagenesis (12,13). In addition, Pol λ is also involved in base excision repair (BER), the DNA repair pathway that repairs oxidative DNA lesions (14). Although Pol β is the primary polymerase in BER, Pol λ has a minor, compensatory role in the pathway (15,16). During BER, the specific DNA glycosylases MutY homology DNA glycosyslase (MUTYH) or 8-oxoguanine glycosylase (OGG1) recognize 8-oxoG:A or 8-oxoG:C mispairs, respectively, excise the damaged base, and initiate BER (17–19). In recent studies, it has been demonstrated that Pol λ is recruited, along with MUTYH, to 8-oxoG:A mispairs to facilitate repair (20). Coupled with its error-free bypass of 8-oxoG, it is clear that Pol λ plays an important role in the repair of oxidative damage in cells.
Pol λ is a member of X family of DNA polymerases, which also includes Pol β, Pol μ, and terminal transferase (Tdt). Although there is low sequence homology ranging from 23–44% (21), the tertiary structures of these polymerase are similar (22). All members of the X family possess four subdomains: 8 kD, thumb, palm, and fingers. The 8 kD subdomain is structurally similar between the proteins, but only Pols β and λ possess dRP lyase activity. The 8 kD and thumb subdomains bind the DNA, the palm subdomain contains three conserved aspartic acid residues required for catalysis, and the fingers subdomain binds nucleotide. Pol λ, along with Pol μ and Tdt, possess an additional subdomain, an N-terminal BRCT subdomain that facilitates interactions with proteins involved in non-homologous end joining (NHEJ), implicating a role for Pol λ in this pathway (21).
The rs3730477 single nucleotide polymorphism (SNP) encodes the R438W Pol λ variant and is present in 12% of the population. Residue 438 is located in the palm subdomain and is distant from the active site, but in close proximity to LoopI, which has been demonstrated to be important for fidelity of DNA synthesis, but not catalytic activity (23). LoopI stabilizes the template strand of DNA during the change from the inactive to active conformation (23) and its motion is affected by the change from R to W at residue 438.
There are two reported germline SNPS in the POLL gene that may be associated with cancer, R438W and T221P (24). Swett et al.(24) used a novel bioinformatic approach in which they sought to determine correlations between SNPs, provided from existing large databases, with deleterious phenotypes. Through this analysis, named hypothesis driven single nucleotide polymorphism search (HyDn-SNP-S), the Pol λ R438W variant was associated with a 3-fold increased incidence of breast cancer. Previous work by others showed that overexpression of the R438W Pol λ variant in Chinese Hamster Ovary (CHO) cells induces chromosomal aberrations and point mutations including an 8-fold increase in G:T transversions (25).
Long-term exposure to estrogen is a risk factor for breast cancer (26). Although the main effect of estrogen exposure is by promoting transcription of genes involved in proliferation (27), estrogen metabolism produces ROS as a by-product leading to DNA damage, and most commonly induces 8-oxoG in the DNA, as wells as estrogen-DNA adducts (28,29). Estrogen is non-carcinogenic on its own, but its ability to produce ROS, coupled with a defect in repair of ROS-dependent DNA damage may synergize to promote a malignant phenotype.
In a small-scale screen, we identified the rs3730477 SNP encoding R438W Pol λ as being enriched in the germlines of women with breast cancer. Given this and previous work suggesting that overexpression of this variant in CHO cells induces genomic instability, we hypothesized that expression of R438W in immortal but non-transformed human breast cells could drive cellular transformation. Our data show that R438W induces cellular transformation in human breast cells. The slow DNA polymerase activity of R438W results in replication fork stalling and collapse, which lead to genomic instability cellular transformation of human breast cells. Most importantly, our results demonstrate that the phenotype of R438W Pol λis linked to estrogen exposure. Specifically, we show that treatment with estrogen produces persistent oxidative damage in cells expressing R438W, significantly decreasing the latency of cellular transformation, consistent with a critical role for Pol λ in bypassing 8-oxoG in DNA. Our results suggest that the R438W Pol λ variant is linked to breast cancer risk involving estrogen exposure.
Materials and Methods
Next-generation Sequencing (NGS)
Genomic DNA was isolated from the lymphocytes of 32 patients who had been diagnosed with breast cancer (BRCA1,2 non-mutant) on HIC protocol #0805003789 as previously described (30). The Agilent SureSelect Target Enrichment Platform was used to design primers to capture all the exons and 5′ and 3′ UTRs of all DNA repair genes in addition to genes that function in the DNA damage response (i.e. ATM, ATR, 53BP1) (for a full list of genes, see http://www.cgal.icnet.uk/DNA_Repair_Genes.html). The primer design included a total of 3500 unique features representing approximately 1 Mb of exonic sequence representing the 168 genes. Paired end sequencing enabled at least 50x coverage using multiplexing on the Illumina Genome Analyzer on a fee-for-service basis using the Yale Center for Genome Analysis. Following mapping of regions with the alignment software bwa (31), variants (both SNPs and short insertions/deletions) were identified with the Genome Analysis Toolkit [GATK: http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit] (32). We identified 150 single nucleotide polymorphisms (SNPs) in these genes and analyzed them using the SIFT (33) and Polyphen (34) prediction algorithms. SNPs that were predicted to affect protein function in either or both programs were selected and Fisher’s exact tests were used to determine if the SNP was significantly enriched in these breast cancer patients after correcting for ancestry.
Plasmids and Cloning
Human Pol λ cDNA was cloned into the pET28a expression plasmid (Novagen) for expression with an N-terminal 6xHis tag. Wildtype Pol λ cDNA sequence was verified by sequencing. For cell culture experiments, human Pol λ cDNA with a C-terminal hemagglutinin (HA) tag was cloned into the pRVYTet retroviral vector as described (35). The R438W variant was introduced into the human WT Pol λ cDNA sequence using site-directed mutagenesis (Stratagene) following the manufacturer’s protocols. The primers used for these reactions are available upon request. Positive clones were sequenced at the W.M. Keck facility at Yale University School of Medicine.
Cell Lines and Cell Culture
The GP2-293 packaging cell line (Clontech) was maintained in high glucose DMEM (Invitrogen) supplemented with 10% FBS (Gemini Bio-Products), 1% penicillin-streptomycin, 1% L-glutamine, and 220 μg/ml hygromycin B (used for selection) (Invitrogen). The human mammary epithelial cell line, HME1 (ATCC), was maintained in DMEM/F12 medium (Invitrogen) supplemented with 5% horse serum (Invitrogen), 1% penicillin-streptomycin, epidermal growth factor (20 ng/ml), hydrocortisone (0.5 μg/ml) cholera toxin (100 ng/ml), insulin (10 μg/ml) (Sigma-Aldrich) and hygromycin B (50 μg/ml) (used for selection) (Invitrogen). For estrogen experiments, cells were grown continuously in medium containing 1 mM estrogen and/or 1 mM tamoxifen (Sigma-Aldrich) in the medium described above with the exception that charcoal-stripped horse serum was used (Valley Biomedical, Inc). All cells were grown at 37°C in a 5% CO2 humidified incubator.
Transfection, Infection, and Expression Analysis
Human Pol λ WT and R438W constructs were packaged into retrovirus using the GP2-293 packaging line as previously described (4). Expression of exogenous HA-tagged Pol λ was verified by Western blot using monoclonal mouse anti-HA antibody (Covance). β-actin (Sigma-Aldrich) was used as a loading control.
Anchorage independent growth assay
Anchorage independent growth was assessed as previously described (35). Briefly, 1 × 104 HME1 cells expressing either WT or R438W Pol λ were mixed with media containing 0.7% noble agar (USB) and poured onto a layer of media containing 1.0% noble agar. The number of colonies present in each of ten microscope fields per well from a total of 3 wells per experiment were counted after 4 weeks of growth.
Cell Proliferation
HME1 cells expressing either WT or R438W Pol λ were plated at a density of 2 × 104 cells in 60 mm dishes. Cells were counted every day for five consecutive days. Data were plotted as change in cell number per day.
DNA Fiber Assay
HME1 cells expressing WT or R438W Pol λ were grown in the presence or absence of estrogen to approximately 30–40% confluence. Cells were pulsed with 25 μM IdU for 30 minutes, washed three times with PBS, and pulsed with 250 μM CldU for 30 minutes. Cells were harvested and resuspended in PBS at a concentration of 1.7 × 106 cells/ml and 3 μl (5000 cells) of the cell suspensions were placed on glass slides and mixed with 7 μl of lysis buffer (200 mM Tris, pH 7.6, 50 mM EDTA, 0.5%SDS) for 2 min. Slides were tilted at 20° to allow gravity flow. The slides were fixed in a 3:1 solution of methanol-acetic acid for 20 min at −20°C and treated with 2.5 M HCL for 30 min followed by washes with PBS before blocking in 5% BSA for 30 min at 37°C. To detect incorporated IdU and CldU, DNA fibers were incubated with mouse anti-BrdU (Becton Dikinson; 1:25) and rat anti-BrdU monoclonal antibody (Abcam; 1:400), respectively, for 1 h at room temperature (RT), followed by 3 washes with PBS, and incubation with sheep anti-mouse Cy3 (Sigma; 1:500) and goat anti-rat Alex Flour 488 (Invitrogen 1:400) for 1 h at RT. The slides were mounted with Vectashield mounting medium and covered with coverslips. Images were acquired using a Zeiss microscope at 63x magnification and processed using Image J software (http://imagej.nih.gov/ij/). Further details are available in the Supplemental Experimental Procedures.
Immunofluorescence
HME1 cells expressing WT or R438W Pol λ were grown in the presence or absence of estrogen on glass coverslips coated with poly-L-lysine (Sigma). Cells were fixed in a 3:1 solution of methanol-acetic acid for 20 min at −20°C and permeabilized in 0.5% Triton buffer (20 mM HEPES, pH 7.4, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 0.5% Triton x-100) for 10 min at RT. Coverslips were blocked in 3% BSA and goat serum for 30 min at RT and incubated with the primary antibodies, anti-PCNA (Santa Cruz), anti-γH2AX (cell signaling), or anti-8-oxoG (Millipore) overnight at 4°C. Coverslips were washed and incubated with secondary FITC or rhodamine antibodies. Coverslips were mounted on slides using SlowFade® Gold Antifade Mountant containing DAPI to stain the nuclei (Invitrogen).
Genomic Instability Analysis
Chromosomal aberrations were assessed as previously described (4). Cells were harvested by mitotic shake-off and lysed in 0.75% KCl at 37°C for 30 minutes before fixing in Carnoy’s Fixative (75% methanol, 25% acetic acid). Images were taken using Spot Camera software (Diagnostic Instruments). Metaphase spreads were de-identified and scored by eye for chromosomal fusions, breaks, and fragments.
Protein Expression and Purification
pET28a plasmids with human WT or R438W Pol λ cDNA were transformed into Rosetta 2(DE3) cells (Novagen). Luria broth cultures were incubated at 37°C until the OD600 reached approximately 0.6. Protein expression was induced by the addition of 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) (American Bioanalytical) and incubated at 26°C for four hours. Protein induction was verified by using 10% SDS-PAGE stained with Coommassie. WT and R438W Pol λ proteins were purified using fast protein liquid chromatography as described previously (36). Pellets were resuspended in buffer B (40 mM TrisCl, pH 8.0, 0.5 M NaCl, 5 mM imidazole) supplemented with 1 mM phenylmethanesulfonyl fluoride solution (PMSF) (Sigma-Aldrich) and complete EDTA-free protease inhibitor cocktail tablet (Roche) and lysed via sonication. The lysate was cleared by centrifugation and applied to a 5 ml HiTrap Chelating HP column (GE Healthcare) charged with NiSO4 using a linear imidazole gradient (from 5 to 500 mM) in buffer B. Pol λ was eluted in a broad peak at about 225 mM imidazole. The fractions were combined, concentrated to about 1 ml and diluted to 10 ml in buffer F, pH 7.0 (50 mM Tris-HCl pH 7.4, 1 mM EDTA, 10% glycerol, 50 mM NaCl). The diluted protein applied to a HiTrap SP HP column (GE Healthcare) using a linear NaCl gradient from 100 mM to 2000 mM. Purified protein fractions eluted at approximately 1000–1200 mM NaCl in a sharp peak. Pol λ containing fractions were combined and concentrated to less than 1 ml. Glycerol was added to a final concentration of 15% and aliquots were flash frozen in liquid nitrogen and stored at −80°C. All proteins were purified to >90% homogeneity based on Coomassie Blue staining of 10% SDS-PAGE gels. Final protein concentration was determine using the absorbance at 280 nm and the extinction coefficient for Pol λ (18700 M−1 cm&−1).
Ouabain Mutagenesis Assay
HME1 cells expressing WT or R438W Pol λ were grown in the presence of 1 mM estrogen to exponential growth. Cells were trypsinized and plated in 10 cm dishes at various concentrations. Cells were allowed to attach overnight and ouabain (Sigma) was added to a final concentration of 75 nM. After 3 weeks of growth, cells were stained with 0.25% crystal violet (Sigma). Colonies were counted and mutagenesis was calculated by dividing the total number of ouabain resistance colonies by the number of surviving colonies on plates grown in the absence of ouabain.
Preparation of DNA Substrates
Oligonucleotides were synthesized by the W.M. Keck facility and purified by polyacrylamide gel electrophoresis as described previously (37). Complete substrate annealing was confirmed using 12% native polyacrylamide gel electrophoresis and visualized using autoradiography.
Pre-steady State Kinetics
Radiolabeled DNA (300 nM) and Pol λ (100 nM) were combined with the correct dGTP and 10 mM MgCl2 in buffer L (50 mM TrisHCl, pH 8.4, 5 mM MgCl2, 100 mM NaCl, 0.1 mM EDTA, 5 mM DTT, 10% glycerol, 0.1 mg/ml BSA (38)) in a KinTek Chemical Quench-Flow apparatus at 37°C. The reactions were quenched by the addition of 0.5 M EDTA. The reaction products were separated on a 20% denaturing polyacrylamide gels, visualized, and quantified using a Storm 860 Phosphorimager with ImageQuant software. The concentration of extended product was plotted as a function of time and the data were fit to a single-exponential curve:
where A is the amplitude, kobs is the observed rate constant (37).
Gel Electrophoretic Mobility Shift Assay
The DNA binding constant was determined by gel electrophoretic mobility shift assay as described previously (4). The dissociation constant for DNA (KD) was determined by fitting the fraction bound protein (Y) versus protein concentration with the equation:
where Y is the amount of bound protein, m is a scaling factor, and b is the apparent minimum Y value.
Statistics
Unpaired two-tailed t-tests, one-way, or two-way analysis of variance (ANOVA) were used as appropriate to determine whether the mean of each cell line was different from the empty vector cells. Tukey’s or Sidak’s post hoc tests were used to determine significant differences between the means of each group. All statistics were performed using GraphPad Prism version 5 (GraphPad Software, San Diego, CA). Data are represented as mean ± SEM.
Results
Enrichment of the R438W SNP in breast cancer patients
To determine if there was a significant enrichment of SNPs in DNA repair genes in the germlines of individuals with breast cancer, we conducted a small-scale screen of 32 breast cancer patients using next-generation sequencing (NGS) to sequence the exons of all DNA repair and DNA damage response genes (Supplemental Fig S1). One DNA repair SNP that emerged from this analysis was the rs3730477 POLL germline SNP that encodes R438W Pol λ. The rs3730477 was found in 40% of the (13/32) cases from patients of predominantly European ancestry. Examination of dbSNP 147 indicates that the rs3730477 SNP is present at frequencies ranging from 22–29% in individuals with European ancestry (39). Therefore, this SNP in the POLL gene is present in our small cohort of breast cancer patients at frequencies that are slightly increased over the expected frequency for individuals with European ancestry. Given this and previous indications of a link to breast and ovarian cancer and genomic instability (24,25), we decided to characterize this SNP in human breast cells for its ability to exhibit a cancer-associated phenotype.
Estrogen treatment decreases the latency of transformation in cells expressing R438W Pol λ
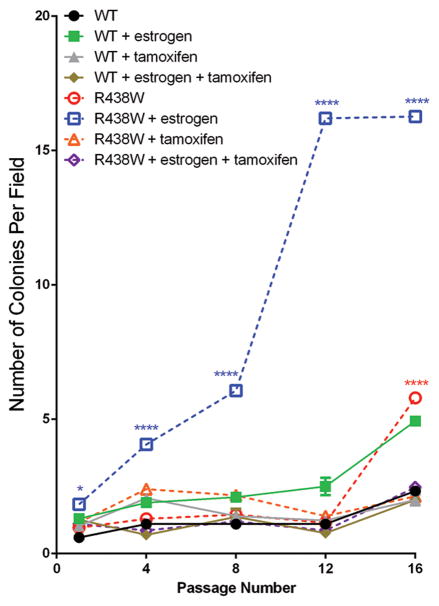
Previous biological studies of the R438W Pol λ variant were conducted predominantly in CHO cells and human fibroblasts (25). Although interesting phenotypes indicating that R438W induces genomic instability emerged from these studies, they were not conducted in human breast epithelial cells and did not assess the ability of R438W to induce cellular transformation. Importantly, the previous studies did not address the role of estrogen even though it induces ROS, which is processed by Pol λ. Therefore, given the link of the R438W Pol λ variant with breast cancer risk, we tested whether its expression in the normal, immortalized breast epithelial cell line, HME1, induced cell transformation. HME1 cells expressing either WT or R438W were serially passaged and tested for growth in soft agar as a marker of anchorage-independent growth and a transformed phenotype. At passage 16, cells expressing R438W had a significantly increased number of colonies that grew in an anchorage-independent manner compared to WT cells (Fig 1; open red circles compared to filled black circles). Since prolonged exposure to estrogen is a major risk factor for breast cancer (26) and has been shown to transform human breast epithelial cells (40,41), we sought to determine if exposure to estrogen, in combination with the expression of the R438W germline variant, would decrease the latency of cellular transformation compared to cells not treated with estrogen. Cells were treated continuously with either estrogen, the estrogen antagonist tamoxifen, or the combination of estrogen and tamoxifen, and tested for growth in soft agar. Remarkably, treatment with estrogen led to a significant increase in the ability of R438W-expressing cells to grow in soft agar with significant increases in growth observed as early as passage 1 (Fig 1; open blue squares compared to filled green squares). The numbers of colonies able to grow in an anchorage-independent manner continued to increase in R438W Pol λ-expressing cells until passage 16, when the experiment was discontinued. Moreover, the decreased latency of transformation in cells expressing R438W Pol λ was a direct effect exerted by estrogen as the anchorage-independent growth was effectively blocked in these cells by co-treatment with the estrogen antagonist, tamoxifen. The transformation was not due to different levels of expression since western blotting showed that total Pol λ protein expression was similar (Supplemental Fig S2). In addition, these results were recapitulated in three separate pools of HME1 cells with experiments performed in the presence (Fig 1) or absence of estrogen (Supplemental Fig S3). These data strongly suggest that R438W Pol λ induces cellular transformation in human breast cells and that estrogen plays a significant role in decreasing the latency of transformation in these cells compared to WT-expressing cells.
Fig. 1. Continuous exposure to estrogen decreases the latency of cellular transformation in cells expressing R438W Pol λ.
HME1 cells expressing WT or R438W were left untreated or were continuously treated with 1 μM estrogen, 1 μM tamoxifen, or both estrogen and tamoxifen. Cells were serially passaged and plated for growth in 0.7% soft agar at various passages. Data are presented as mean ± SEM (n=3). * or **** denote p<0.05 or 0.0001, respectively, comparing R438W to WT for each treatment group.
Expression of R438W induces replication fork collapse
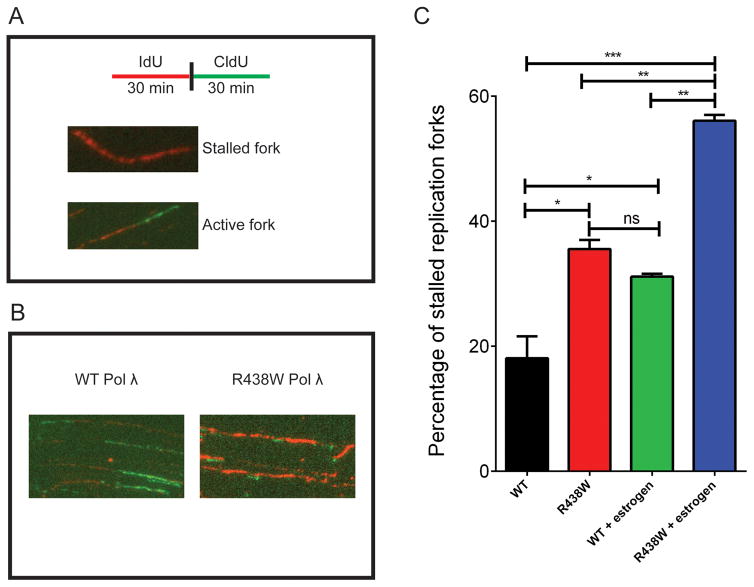
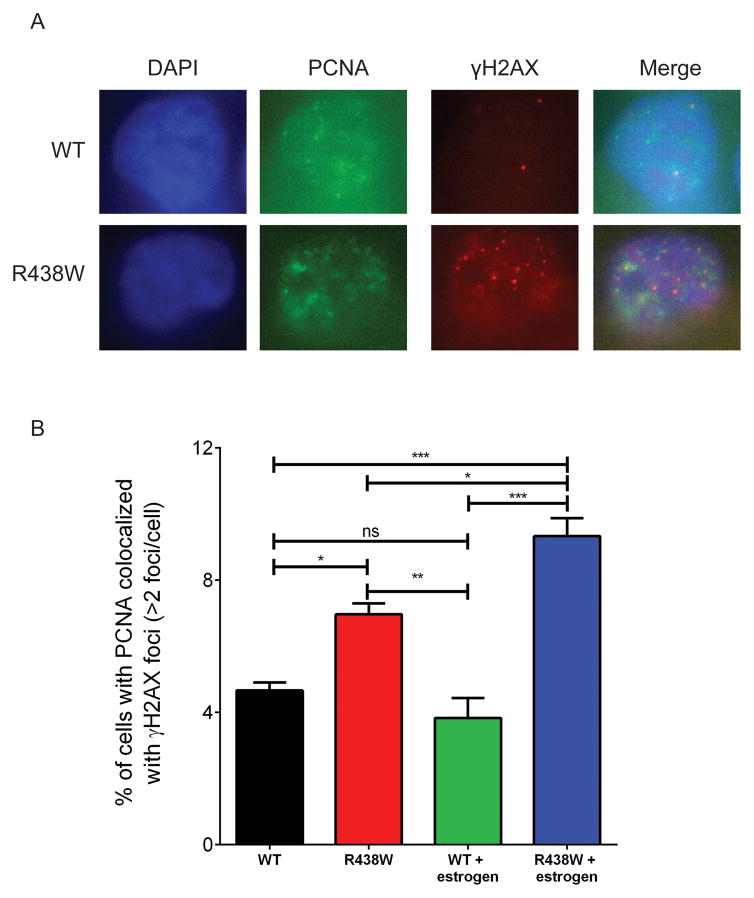
Pol λ is required at replication forks and its silencing leads to replicative stress (42). We hypothesized that replication fork stability was compromised in cells expressing R438W. To test this, we analyzed the DNA fibers to determine if stalled replication forks were present in cells expressing R438W. This approach labels the DNA by first pulsing with IdU (red) followed by pulsing with CldU (green) (Fig 2A). We labeled the actively replicating DNA of HME1 cells expressing either WT or R438W with the nucleotide analogs (Fig 2B) and found that cells expressing R438W had a significant increase in the percentage of stalled forks compared to WT (Fig 2C). Importantly, treatment with estrogen most significantly increased the percentage of stalled replication forks in cells expressing R438W. Next, we analyzed γH2AX colocalization with PCNA as another marker of replication fork stalling. We found that R438W-expressing cells had a significant increase in the percentage of cells with PCNA colocalized with γH2AX foci and this was increased further when the cells were treated with estrogen (Fig 3). Together, these data suggest that R438W Pol λ expression in cells leads to replicative stress in S phase, indicative of replication fork collapse, which is worsened in cells treated with estrogen.
Fig. 2. Expression of R438W induces replicative stress.
A. A schematic representation of the DNA combing assay. B. Representative images of DNA fibers from HME1 cells expressing either WT or R438W. C. The percentage of stalled forks in HME1 cells expressing either WT or R438W in the presence or absence of estrogen. Data are presented as mean ± SEM (n=3). *, **, *** denote p<0.05, 0.01, or 0.001, respectively.
Fig. 3. γH2AX colocalizes with PCNA in cells expressing R438W.
A. Representative images of WT and R438W-expressing cells immunostained with anti-PCNA (green) and anti-γH2AX (red). B. The percentage of cells with colocalization of PCNA and γH2AX. Data are presented as mean ± SEM (n=3). *, **, *** denote p<0.05, 0.01, or 0.001, respectively.
R438W Pol λ induces genomic instability in cells
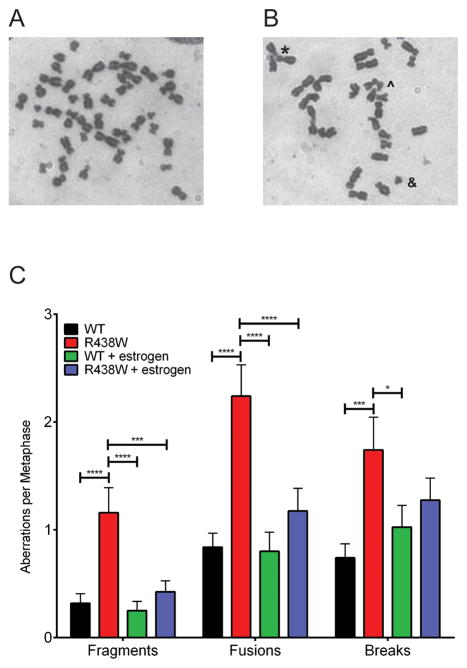
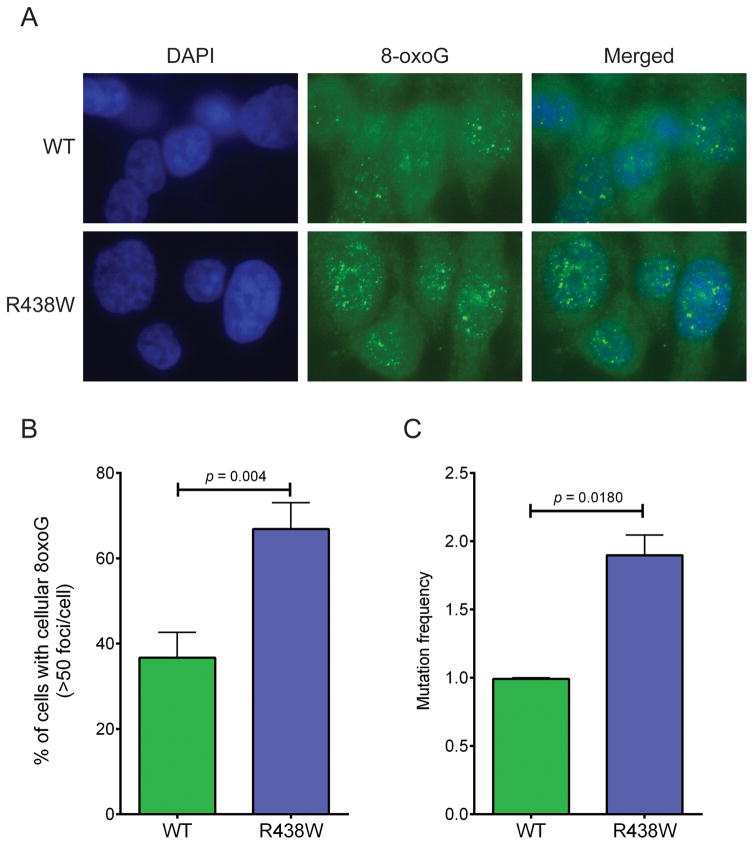
To determine if expression of R438W in HME1 cells induces genomic instability that may drive cellular transformation, we first characterized chromosomal aberrations in human breast epithelial cells expressing either WT or R438W at passages 2–5. In the absence of estrogen, expression of R438W significantly increased the levels of fragments, fusions, and breaks (Fig 4). Surprisingly, when treated with estrogen, R438W-expressing cells did not exhibit a significant increase in chromosomal aberrations compared to WT-expressing cells (Fig 4). To determine the mechanism by which estrogen induces cellular transformation in R438W-expressing cells, we first tested if treatment with estrogen affected the proliferation of cells, but found no difference in growth rates between the WT and R438W cells lines in the presence of estrogen (Supplemental Fig S4). Because estrogen is known to induce oxidative damage and 8-oxoG lesions and estrogen-DNA adducts, we examined whether cells expressing R438W had higher levels of 8-oxoG using an antibody that recognizes 8-oxoG and immunofluorescence in human breast cells continuously exposed to estrogen. Indeed, there was a significant increase in the percentage of cells expressing R438W with cellular 8-oxoG (Fig 5A–B) compared to WT-expressing cells. We next characterized the mutation frequency in estrogen-treated cells by assaying ouabain resistance. In concordance with the increase in 8-oxoG levels, R438W-expressing cells also exhibited a two-fold increase (p=0.0180) in mutation frequency to ouabain resistance in the presence of estrogen (Fig 5C). Together, these data suggest that treatment with estrogen increases 8-oxoG-mediated mutagenesis in cells expressing R438W.
Fig. 4. Expression of R438W induces chromosomal aberrations in the absence of estrogen.
Metaphase spreads were prepared from HME1 cells expressing either (A) WT or (B) R438W. *, ^, or & denote fusion, break, or fragment, respectively. C. Quantification of the number of aberrations per metaphase spread. A total of at least 50 metaphase spreads were scored for each cell line. Data are presented as mean ± SEM. *, **, **** denote p<0.05, 0.01, or 0.0001, respectively.
Fig. 5. Continuous treatment with estrogen induces 8-oxoG damage in R438W-expressing cells.
A. Representative images of WT and R438W-expressing cells immunostained with anti-8-oxoG. B. The percentage of cells with cellular 8-oxoG DNA. C. Quantification of mutation frequency using the ouabain resistance mutation assay. Data are presented as mean ± SEM (n=3).
R438W Pol λ has reduced polymerase activity
To test whether R438W had a slower catalytic rate than WT, we performed pre-steady state kinetic analysis of the enzyme using 1 bp gapped DNA, a preferred substrate in BER. This type of experiment reveals both the initial reaction rate along with the steady-state rate of the enzyme, providing important information that is not observed under steady-state, and likely rate-limiting conditions (25). Under presteady-state burst conditions, R438W exhibited a 50% reduction in polymerization rate compared to WT (Fig 6A) suggesting that the genomic instability may be due to the lower catalytic efficiency of R438W, perhaps at the replication fork. Also, Pol λ is known to bypass 8-oxoG at the replication fork. Therefore, we wanted to determine if R438W is able to bypass 8-oxoG. First, we measured the ability of R438W Pol λ to bind to recessed DNA with 8-oxoG as the templating base. Gel shift assays demonstrated that although R438W protein binds undamaged DNA with similar affinity as WT Pol λ (KD= 70±10 nM or 130±20 nM for WT or R438W, respectively), R438W binds DNA with 8-oxoG poorly (KD= 70±10 nM or 1200±200 nM for WT or R438W, respectively) (Fig 6). Given this extremely low affinity for DNA, we were unable to measure the rate of bypass of 8-oxoG by R438W. Together, these data suggest that R438W Pol λ is a slow polymerase. In addition, 8-oxoG lesions are unlikely to be recognized and bypassed by R438W, resulting in mutagenesis in cells expressing R438W, and this likely leads to cellular transformation.
Fig. 6. R438W Pol λ has a defect in DNA synthesis and binding.
A. Pre-steady state burst experiments were performed by incubating 1 bp gap DNA and the correct dGTP with purified WT or R438W Pol λ protein. The time course of the reactions ranged from 0–60 s. Products were resolved on 20% denaturing gels and imaged using a phosphorimager and quantified. Data were plotted as product formed versus time and fit to a single exponential equation to obtain the kobs (WT=0.22±0.02 s−1; R438W=0.10±0.01 s−1). B. Various concentrations of purified WT or R438W protein were incubated with 0.1 nM undamaged recessed DNA (filled symbols) or recessed DNA with an 8-oxoG lesion on the template strand (open symbols) for 15 min at room temperature. Data were plotted as fraction product bound versus Pol λ concentration to obtain the KD(DNA) (undamaged DNA: WT, 70±10 nM; R438W, 130±20 nM; damaged DNA: WT, 70±10 nM; R438W, 1200±200 nM).
Discussion
In the current study, we provide evidence that the rs3730477 SNP encoding R438W Pol λ induces cellular transformation in human breast epithelial cells, and that the latency of cellular transformation is decreased in the presence of estrogen. R438W has a slower catalytic rate than WT on 1 bp gapped DNA substrate and poorly binds to DNA harboring 8-oxoG damage that persists and/or accumulates in cells expressing R438W Pol λ treated with estrogen. Our results suggest that the impaired DNA repair capacity of R438W Pol λ leads to genomic instability and mutagenesis in cells, likely resulting in cellular transformation. These findings suggest that individuals harboring R438W may be at an increased risk for breast cancer and that treatment with estrogen could enhance the risk.
Pol λ is involved in several distinct DNA repair pathways. It functions as a backup to Pol β in BER (16,43), in NHEJ (44), and has a specialized role in translesion synthesis (12,13). In BER, after a lesion-specific glycosylase excises the damaged base, APEI remodels the DNA ends, allowing for the single nucleotide gap to be filled by a polymerase. In vitro kinetics on 1 bp gapped DNA substrate showed that R438W has a 50% reduction in catalytic activity. This finding was in contrast to what was previously reported (25). However, those kinetic experiments were performed at one ten minute time point and under steady-state conditions, whereas our experiments were performed on a much shorter time scale (0–60 s) and under presteady-state conditions. Under these conditions and at shorter time points, we were able to reveal more subtle changes in the catalytic rates between the WT and variant Pol λ proteins.
The slow DNA polymerase activity of R438W likely leads to an accumulation of unrepaired breaks in cells expressing this variant polymerase which can then proceed to replication. If not repaired, this can lead to replication fork collapse and genomic instability. Indeed, we observe an increase in stalled forks and chromosomal aberrations in cells expressing R438W compared to WT cells. Therefore, we suggest that the inefficient gap-filling of Pol λ during BER results in replicative stress and genomic instability. Although Pol λ is not the major polymerase activity in BER, Pol λ is preferred to Pol β in repairing 8-oxoG:A mispairs (20,45). MUTYH glycosylase recognizes the damage and recruits Pol λ, as well as other repair factors including PCNA, FEN1 and DNA ligases to repair the DNA (20). Since endogenous oxidative lesions occur frequently, the decreased repair capacity of R438W is likely to lead to genomic instability. Alternatively, the increase in chromosomal aberrations may be due to the role of impaired NHEJ as others have suggested (25). However, the precise mechanism by which this would occur is not clear since residue 438 is located in the palm subdomain containing the active site, and is not proximal to the BRCT subdomain required for NHEJ.
Prolonged lifetime exposure to estrogen is a major risk factor for hormone-regulated cancers such as breast cancer (for review see (26)). Estrogen exposure comes from a variety of sources including hormone replacement therapies in post-menopausal women. There are both estrogen receptor (ER) dependent and independent mechanisms by which estrogen is proposed to drive cancer. The canonical ER-dependent mechanism involves driving the expression of genes involved in cellular proliferation through the binding ER α and β, (27). Rapidly replicating cells can elude DNA repair and cell cycle checkpoints leading to an increase in mutations. However, cells expressing WT and R438W Pol λ grew at comparable rate in the presence of estrogen.
Alternatively, in an ER-independent mechanism, estrogen exposure induces ROS and estrogen-DNA adducts during its metabolism (28,29,46). After treatment with estrogen, we observed elevated levels of 8-oxoG DNA in cells expressing R438W compared to WT cells indicating that there is less or delayed repair. Cells expressing R438W also have a significant increase in mutagenesis compared to WT cells when grown in estrogen. These mutations are likely enriched for G:T transversions since R438W Pol λ was previously shown to have an 8-fold increase in this type of mutation (25). Also, this specific mutation would result from error-prone bypass of 8-oxoG DNA. However, we cannot exclude that some of the effects we observe are not due in part to depurinating estrogen-DNA adducts that would be repaired by BER.
In addition to replicative polymerases, translesion polymerases can bypass 8-oxoG lesions during the S or G2 phases of the cell cycle. Pol κ, Pol β, and Pol μ bypass 8-oxoG in an error-prone manner (12,47,48). Pol λ and pol η are the main translesion polymerases that bypass 8-oxoG lesions and preferentially insert the correct nucleotide. Pol λ has a much higher efficiency in bypass than Pol η and PCNA and RPA increase the fidelity of both Pol λ and η 1200- and 68-fold, respectively (12). However, in vitro gel shift studies revealed that R438W binds 8-oxoG-containing DNA substrates with 17-fold lower affinity than WT protein. This was specifically due to the 8-oxoG moiety, and not the DNA template since there was less than 2-fold difference in DNA affinities between the WT and R438W Pol λ proteins with undamaged DNA substrate. The increase in stalled forks in R438W-expressing cells is likely due to the low affinity of R438W for the 8-oxoG-modified DNA resulting in defective error-free bypass by this enzyme variant. Since we do not observe estrogen-dependent DNA breaks in the metaphase spreads, it suggests that the stalled forks are restarted after an error-prone polymerase, such as Pol η, binds to the DNA and bypasses 8-oxoG. However, this occurs in an error-prone manner resulting in an increase in mutagenesis, which we observe in R438W Pol λexpressing cells treated with estrogen. Alternatively, R438W Pol λ could preferentially insert the incorrect A. This is unlikely due to its overall poor affinity for the 8-oxoG-containing DNA. The question remains as to why WT Pol λ, which is present in the cells expressing R438W Pol λ, is unable to compensate. The cell may be overwhelmed with damage that WT Pol λ cannot repair, or R438W may be sequestering other required auxiliary proteins, preventing WT Pol λ from being able to properly function.
The faithful repair of DNA is important to maintaining genomic integrity and therefore it is critical to understand the mechanism of how functional germline SNPs in DNA repair genes can influence cancer susceptibility. Using NGS, we identified R438W Pol λ as a potential driver of breast cancer. We have further detailed how the R438W SNP can induce genomic instability and cellular transformation, and that increased oxidative stress by prolonged exposure to estrogen exacerbates the cancer-associated phenotypes induced by R438W. Therefore, our findings provide novel insight into how individuals carrying this variant may have increased cancer susceptibility, especially resulting from estrogen exposure.
Supplementary Material
Statement of Implication.
The Pol λ R438W mutation can serve as a biomarker to predict cancer risk and implicates that treatment with estrogen in individuals with this mutation may further increase their risk of breast cancer.
Acknowledgments
Financial Support: This work was supported by pilot funding from the Yale Comprehensive Cancer Center and RO1 ES019172 (JBS).
Footnotes
The authors have no conflicts to disclose.
References
- 1.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34(9):2311–21. [PubMed] [Google Scholar]
- 2.Galick HA, Kathe S, Liu M, Robey-Bond S, Kidane D, Wallace SS, et al. Germ-line variant of human NTH1 DNA glycosylase induces genomic instability and cellular transformation. Proc Natl Acad Sci U S A. 2013;110(35):14314–9. doi: 10.1073/pnas.1306752110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sjolund A, Nemec AA, Paquet N, Prakash A, Sung P, Doublie S, et al. A germline polymorphism of thymine DNA glycosylase induces genomic instability and cellular transformation. PLoS Genet. 2014;10(11):e1004753. doi: 10.1371/journal.pgen.1004753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamtich J, Nemec AA, Keh A, Sweasy JB. A germline polymorphism of DNA polymerase Beta induces genomic instability and cellular transformation. PLoS Genet. 2012;8(11):e1003052. doi: 10.1371/journal.pgen.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–76. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 7.Collins AR. Oxidative DNA damage, antioxidants, and cancer. Bioessays. 1999;21(3):238–46. doi: 10.1002/(SICI)1521-1878(199903)21:3<238::AID-BIES8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434(7033):612–8. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 9.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29(30):7024–32. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 10.Moriya M, Grollman AP. Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G:C-->T:a transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Molecular & general genetics: MGG. 1993;239(1–2):72–6. doi: 10.1007/BF00281603. [DOI] [PubMed] [Google Scholar]
- 11.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349(6308):431–4. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 12.Maga G, Crespan E, Wimmer U, van Loon B, Amoroso A, Mondello C, et al. Replication protein A and proliferating cell nuclear antigen coordinate DNA polymerase selection in 8-oxo-guanine repair. Proc Natl Acad Sci U S A. 2008;105(52):20689–94. doi: 10.1073/pnas.0811241106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, et al. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447(7144):606–8. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 14.Horton JK, Baker A, Berg BJ, Sobol RW, Wilson SH. Involvement of DNA polymerase beta in protection against the cytotoxicity of oxidative DNA damage. DNA repair. 2002;1(4):317–33. doi: 10.1016/s1568-7864(02)00008-3. [DOI] [PubMed] [Google Scholar]
- 15.Braithwaite EK, Kedar PS, Lan L, Polosina YY, Asagoshi K, Poltoratsky VP, et al. DNA polymerase lambda protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J Biol Chem. 2005;280(36):31641–7. doi: 10.1074/jbc.C500256200. [DOI] [PubMed] [Google Scholar]
- 16.Tano K, Nakamura J, Asagoshi K, Arakawa H, Sonoda E, Braithwaite EK, et al. Interplay between DNA polymerases beta and lambda in repair of oxidation DNA damage in chicken DT40 cells. DNA Repair (Amst) 2007;6(6):869–75. doi: 10.1016/j.dnarep.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girard PM, Guibourt N, Boiteux S. The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic acids research. 1997;25(16):3204–11. doi: 10.1093/nar/25.16.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjoras M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, et al. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997;16(20):6314–22. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, et al. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic acids research. 2000;28(6):1355–64. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Loon B, Hubscher U. An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase lambda. Proc Natl Acad Sci U S A. 2009;106(43):18201–6. doi: 10.1073/pnas.0907280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamtich J, Sweasy JB. DNA polymerase family X: function, structure, and cellular roles. Biochim Biophys Acta. 2010;1804(5):1136–50. doi: 10.1016/j.bbapap.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moon AF, Garcia-Diaz M, Batra VK, Beard WA, Bebenek K, Kunkel TA, et al. The X family portrait: structural insights into biological functions of X family polymerases. DNA repair. 2007;6(12):1709–25. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bebenek K, Garcia-Diaz M, Zhou RZ, Povirk LF, Kunkel TA. Loop 1 modulates the fidelity of DNA polymerase lambda. Nucleic acids research. 2010;38(16):5419–31. doi: 10.1093/nar/gkq261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swett RJ, Elias A, Miller JA, Dyson GE, Andres Cisneros G. Hypothesis driven single nucleotide polymorphism search (HyDn-SNP-S) DNA repair. 2013;12(9):733–40. doi: 10.1016/j.dnarep.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terrados G, Capp JP, Canitrot Y, Garcia-Diaz M, Bebenek K, Kirchhoff T, et al. Characterization of a natural mutator variant of human DNA polymerase lambda which promotes chromosomal instability by compromising NHEJ. PLoS One. 2009;4(10):e7290. doi: 10.1371/journal.pone.0007290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clemons M, Goss P. Estrogen and the risk of breast cancer. The New England journal of medicine. 2001;344(4):276–85. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- 27.Pike MC, Spicer DV, Dahmoush L, Press MF. Estrogens, progestogens, normal breast cell proliferation, and breast cancer risk. Epidemiol Rev. 1993;15(1):17–35. doi: 10.1093/oxfordjournals.epirev.a036102. [DOI] [PubMed] [Google Scholar]
- 28.Felty Q. Estrogen-induced DNA synthesis in vascular endothelial cells is mediated by ROS signaling. BMC Cardiovasc Disord. 2006;6:16. doi: 10.1186/1471-2261-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mense SM, Remotti F, Bhan A, Singh B, El-Tamer M, Hei TK, et al. Estrogen-induced breast cancer: alterations in breast morphology and oxidative stress as a function of estrogen exposure. Toxicol Appl Pharmacol. 2008;232(1):78–85. doi: 10.1016/j.taap.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelletier C, Speed WC, Paranjape T, Keane K, Blitzblau R, Hollestelle A, et al. Rare BRCA1 haplotypes including 3′UTR SNPs associated with breast cancer risk. Cell Cycle. 2011;10(1):90–9. doi: 10.4161/cc.10.1.14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26(5):589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 34.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sweasy JB, Lang T, Starcevic D, Sun KW, Lai CC, Dimaio D, et al. Expression of DNA polymerase {beta} cancer-associated variants in mouse cells results in cellular transformation. Proc Natl Acad Sci U S A. 2005;102(40):14350–5. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy DL, Jaeger J, Sweasy JB. A triad interaction in the fingers subdomain of DNA polymerase beta controls polymerase activity. J Am Chem Soc. 2011;133(16):6279–87. doi: 10.1021/ja111099b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamtich J, Starcevic D, Lauper J, Smith E, Shi I, Rangarajan S, et al. Hinge residue I174 is critical for proper dNTP selection by DNA polymerase beta. Biochemistry. 2010;49(11):2326–34. doi: 10.1021/bi901735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiala KA, Abdel-Gawad W, Suo Z. Pre-steady-state kinetic studies of the fidelity and mechanism of polymerization catalyzed by truncated human DNA polymerase lambda. Biochemistry. 2004;43(21):6751–62. doi: 10.1021/bi049975c. [DOI] [PubMed] [Google Scholar]
- 39.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kastrati I, Edirisinghe PD, Hemachandra LP, Chandrasena ER, Choi J, Wang YT, et al. Raloxifene and desmethylarzoxifene block estrogen-induced malignant transformation of human breast epithelial cells. PloS one. 2011;6(11):e27876. doi: 10.1371/journal.pone.0027876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russo J, Fernandez SV, Russo PA, Fernbaugh R, Sheriff FS, Lareef HM, et al. 17-Beta-estradiol induces transformation and tumorigenesis in human breast epithelial cells. FASEB J. 2006;20(10):1622–34. doi: 10.1096/fj.05-5399com. [DOI] [PubMed] [Google Scholar]
- 42.Zucca E, Bertoletti F, Wimmer U, Ferrari E, Mazzini G, Khoronenkova S, et al. Silencing of human DNA polymerase lambda causes replication stress and is synthetically lethal with an impaired S phase checkpoint. Nucleic acids research. 2013;41(1):229–41. doi: 10.1093/nar/gks1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Braithwaite EK, Prasad R, Shock DD, Hou EW, Beard WA, Wilson SH. DNA polymerase lambda mediates a back-up base excision repair activity in extracts of mouse embryonic fibroblasts. J Biol Chem. 2005;280(18):18469–75. doi: 10.1074/jbc.M411864200. [DOI] [PubMed] [Google Scholar]
- 44.Lee JW, Blanco L, Zhou T, Garcia-Diaz M, Bebenek K, Kunkel TA, et al. Implication of DNA polymerase lambda in alignment-based gap filling for nonhomologous DNA end joining in human nuclear extracts. J Biol Chem. 2004;279(1):805–11. doi: 10.1074/jbc.M307913200. [DOI] [PubMed] [Google Scholar]
- 45.Markkanen E, van Loon B, Ferrari E, Parsons JL, Dianov GL, Hubscher U. Regulation of oxidative DNA damage repair by DNA polymerase lambda and MutYH by cross-talk of phosphorylation and ubiquitination. Proc Natl Acad Sci U S A. 2012;109(2):437–42. doi: 10.1073/pnas.1110449109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sastre-Serra J, Valle A, Company MM, Garau I, Oliver J, Roca P. Estrogen down-regulates uncoupling proteins and increases oxidative stress in breast cancer. Free Radic Biol Med. 2010;48(4):506–12. doi: 10.1016/j.freeradbiomed.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Wu X, Guo D, Rechkoblit O, Taylor JS, Geacintov NE, et al. Lesion bypass activities of human DNA polymerase mu. J Biol Chem. 2002;277(46):44582–7. doi: 10.1074/jbc.M207297200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z. Response of human DNA polymerase iota to DNA lesions. Nucleic acids research. 2001;29(4):928–35. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.